Abstract
It is widely accepted that the redox status of protein thiols is of central importance to protein structure and folding and that glutathione is an important low-molecular-mass redox regulator. However, the total cellular pools of thiols and disulfides and their relative abundance have never been determined. In this study, we have assembled a global picture of the cellular thiol–disulfide status in cultured mammalian cells. We have quantified the absolute levels of protein thiols, protein disulfides, and glutathionylated protein (PSSG) in all cellular protein, including membrane proteins. These data were combined with quantification of reduced and oxidized glutathione in the same cells. Of the total protein cysteines, 6% and 9.6% are engaged in disulfide bond formation in HEK and HeLa cells, respectively. Furthermore, the steady-state level of PSSG is <0.1% of the total protein cysteines in both cell types. However, when cells are exposed to a sublethal dose of the thiol-specific oxidant diamide, PSSG levels increase to >15% of all protein cysteine. Glutathione is typically characterized as the “cellular redox buffer”; nevertheless, our data show that protein thiols represent a larger active redox pool than glutathione. Accordingly, protein thiols are likely to be directly involved in the cellular defense against oxidative stress.
Keywords: cysteine, glutathione, protein
Because of the thiol group (SH), cysteine is the most chemically reactive natural amino acid found in cells. SH is mainly found in proteins (PSH) and in low-molecular-mass metabolites such as the highly abundant glutathione (GSH). Two SH groups can be oxidized to form a disulfide bond, and, in the cell, disulfides are mainly found in proteins (PSSP), in glutathione disulfide (GSSG), or as mixed disulfides between protein and glutathione (PSSG). In addition, SH groups can be reversibly oxidized by reactive oxygen species to nitrosothiols or sulfenic acids. Higher oxidation states, such as sulfinic and sulfonic acids, are generally considered to be irreversible.
Disulfides are important for protein structure and folding, and formation of disulfide bonds can regulate protein function (1). Furthermore, glutathionylation, here used to denote the formation of PSSG, has been implicated as a mechanism for protection against irreversible protein oxidation during oxidative stress (2).
The cytosolic glutathione pool is highly reducing (3) and consequently, disulfide bond formation is infrequent and typically only takes place transiently during catalysis of redox processes. In contrast, disulfide bond formation is favored in other compartments of the cell, such as the endoplasmic reticulum (ER) and the intermembrane space of mitochondria (4). The overall distribution of thiols and disulfides in different cellular pools defines the global thiol–disulfide redox status of the cell. Maintenance of thiol–disulfide redox homeostasis in different subcellular compartments is important, and failure to do so can be detrimental to the structure, stability and activity of various proteins, including enzymes, chaperones, and transcription factors.
Over recent years several methods have been developed for studying the protein thiol status of the cell. These methods typically involve differential labeling of protein thiols and disulfides, followed by chromatographic or electrophoretic separation of cellular proteins. Finally, cysteine containing proteins are identified by mass spectrometry and in some cases information of the thiol–disulfide status of peptides are also determined (5, 6). Some studies have thus quantified changes in the ratio of reduced to oxidized thiols in specific proteins (6–10). Although these studies have supplied pieces to the puzzle, the global picture of the distribution between reduced and oxidized thiols in proteins have not been determined. Furthermore, the relative abundance of GSH and PSH, and the total amount of PSSG, under normal conditions is unknown. To fill these gaps, we have developed a new methodology for the quantitative determination of the absolute thiol and disulfide levels in all proteins, including membrane proteins, in cultured mammalian cells. These data are combined with quantifications of GSH and GSSG in the same cells. Furthermore, by exposing cells to the thiol-specific oxidant diamide and quantifying the redistribution of the thiol and disulfide pools we show that PSH constitutes a pool of active reducing equivalents that is larger than that of GSH.
Results
Experimental Approach.
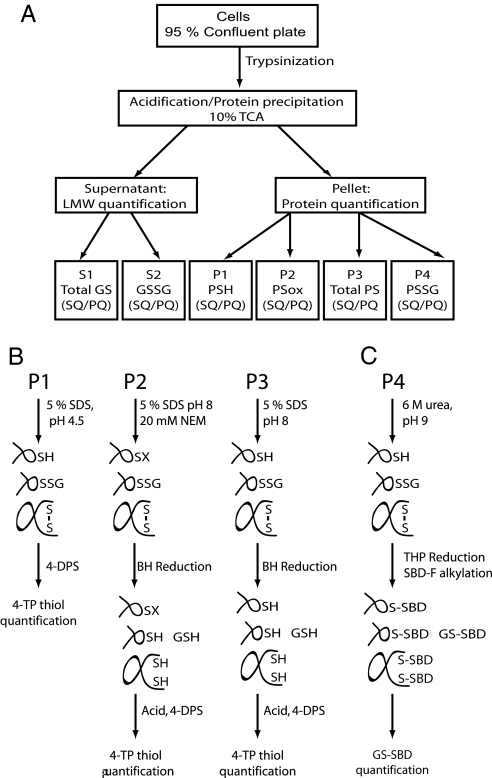
The experimental strategy for quantification of cellular redox species is illustrated in Fig. 1A. A crucial point, when working with redox biology, is to avoid oxidation and thiol–disulfide exchange during sample preparation. The combined acidifying and denaturing effects of trichloroacetic acid (TCA) makes it an ideal choice for quenching redox reactions in cell extracts. Furthermore, addition of TCA to 10% results in protein precipitation, whereas low-molecular-mass metabolites stay soluble.
Fig. 1.
Experimental approach for quantifying the cellular thiol/disulfide environment. (A) Flow chart defining individual fractions quantified. To avoid contribution of protein disulfides from serum in the growth medium, cells were trypsinized and washed in PBS before addition of TCA to 10%. After centrifugation, the supernatant contains low-molecular-weight thiol species, and proteins are pelleted. The supernatant is divided in two for the quantification of total (S1) and oxidized (S2) glutathione. The TCA pellet is divided into 4 samples, used for quantification of the following redox species: PSH (P1); PSox = PSSP + PSSG (P2), Total PS (P3), and PSSG (P4). In addition to redox species, the total protein content in each pellet is quantified and used as a common denominator for the individual samples. SQ, sulfhydryl quantification; PQ, protein quantification. (B) Quantification of protein thiols and disulfides. Pellets were solubilized by sonication in 5% SDS buffers appropriate for either thiol or disulfide quantification. PSH was quantified from P1 by use of 4-DPS, essentially as described in ref. 12. PSox was quantified from P2 by first alkylating PSH with NEM followed by disulfide reduction with BH. Before thiol quantification with 4-DPS, BH was destroyed by addition of acid. As a control, Total PS was experimentally measured by solubilizing P3 in SDS without NEM and subjecting the sample to BH reduction and 4-DPS quantification, as described above. (C) Quantification of PSSG. Glutathione bound to protein (P4) was modified by reaction with the fluorogenic thiol label SBD-F under reducing conditions. The SBD-GS derivative was subsequently detected by HPLC as described in SI Text. This method quantitatively detected GSSG exogenously added to P4 samples, confirming the reliability of the method.
To selectively quantify the different protein sulfhydryl species TCA pellets are divided into fractions and solubilized by sonicating in buffers appropriate for thiol and disulfide quantifications. Importantly, this procedure renders even thiols that are buried in the native protein structures accessible to the thiol detection agent.
Thiol detection reactions are generally dependent on the deprotonated thiolate anion, and because pKa values of SH groups are typically ≈8–9, a pH of at least 7 is usually required for these reactions to occur. At this pH, however, unwanted spontaneous redox reactions, such as oxidation, take place on the microsecond time scale (11). To circumvent this potential problem we have quantified protein sulfhydryl equivalents by use of the thiol specific reagent 4-dithiodipyridine (4-DPS), which reacts with thiols to form stoichiometric amounts of the chromogenic compound 4-thiopyridone (4-TP), absorbing at 324 nm (Fig. 1B). In this context, the primary virtue of 4-DPS is its reactivity at low pH where thiol–disulfide reactions do not normally occur (12).
Although detection of thiols is fairly straightforward, accurate quantification of disulfides is more complicated as it requires alkylation of free thiols followed by reduction of disulfides and their subsequent quantification. However, reagents for disulfide reduction, thiol alkylation, and thiol quantification are frequently found to cross-react. Disulfide levels can, for example, be underestimated if left-over alkylating agent cross-reacts with newly reduced thiols. In contrast, overestimation of disulfide amounts can occur, if the reducing agent cross-reacts with the quantification reagent.
To selectively quantify disulfides involving proteins (PSox), we alkylated with N-ethylmaleimide (NEM) followed by reduction with sodium borohydride (BH) (Fig. 1). The main benefits of this strategy are that BH inactivates NEM (13) so alkylation of the newly reduced thiol groups is avoided. In addition, excess BH is easily removed by the addition of acid and cross-reactivity with 4-DPS is eliminated. By use of this strategy thiol alkylation, disulfide reduction and quantification can be performed in the same test tube.
Finally, PSSG was quantified by reduction of disulfide bonds with tris(hydroxypropyl)phosphine (THP) and fluorescent labeling of sulfhydryl compounds with 7-fluorobenzo-2-oxa-1,3-diazole-4-sulfonate (SBD-F; (14)) by incubating at pH 8.5 at 60 °C for 1 h (Fig. 1C). By applying HPLC with fluorescence detection the SBD-GS derivative can be selectively determined. The method exploits that phosphines do not cross-react with SBD-F, and reduction and derivatization can take place in the same test tube (15). This means that the rather harsh reaction conditions do not cause artificial thiol oxidation, because reducing agent is present during the derivatization.
In addition to sulfhydryl quantification, the total protein content in the pellet fractions were determined (Fig. 1A) and used as a common denominator to compare the individual samples. This is a crucial step as it eliminates any bias from a possible uneven division of the TCA pellet into fractions. In addition, it enables us to compare samples obtained under different growth conditions or from different cell types.
We found that no standard method was reliable for quantitative determination of protein content of crude TCA pellets containing total protein (soluble and integral membrane). The problem was circumvented by hydrolysis of proteins in hydrochloric acid, followed by amino acid determination using ninhydrin. This constitutes a highly reproducible and sensitive method (Fig. S1), and, with a proper standard, it yields numbers that can be calibrated to “amino acids in protein.” The potential interference of primary amines released from polynucleotides in the TCA pellet is likely to be insignificant compared with the amino acids in protein, thus, all following data will be shown as sulfhydryl per amino acid (SH/aa).
Total Cellular Protein Thiols and Disulfides.
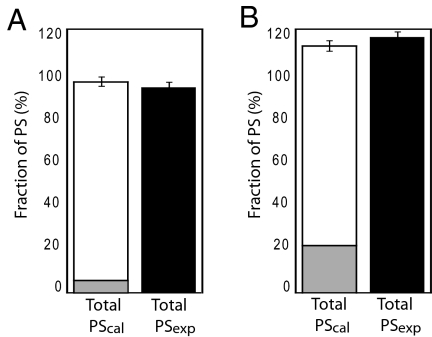
By summation of the SH/aa values of PSH and PSox the value of total PS/aa was calculated, and to verify the method, this value was also determined experimentally (P3) (Fig. 1B). As shown in Fig. 2A, for HEK293 cells, there is excellent agreement between the experimentally determined value (Total PSexp) and the calculated value. As an additional control, PSox and Total PSexp samples were spiked with similar levels of purified lysozyme, and a full recovery of protein sulfhydryls was obtained (Fig. 2B).
Fig. 2.
Validation of the method for quantification of cellular protein thiols and disulfides. Percentage of measured SH/aa relative to the calculated value of total SH/aa (Total PScal) is shown. (A) % PSox (gray bar) is stacked with % PSH (white bar). This bar shows Total PScal. The black bar shows the value of the experimentally determined total PS (Total PSexp). (B) P2 and P3 samples (defined in Fig. 1A) were spiked with equal amounts of purified lysozyme, which contains 4 disulfide bonds. Exogenously added lysozyme PS equivalents (PSlys) in P2 and P3 were recovered by (100 ± 5)% and (106 ± 3)%, respectively. The percentage of PSH (white bar) is stacked with the percentage of (PSox + PSlys) shown in the gray bar. The black bar shows percentage of (Total PSexp + PSlys).
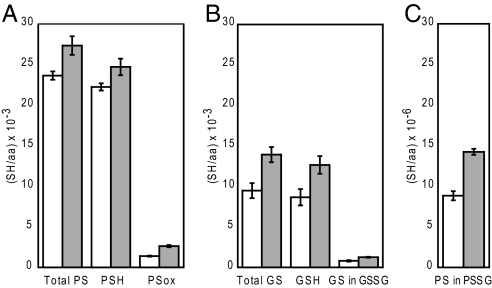
The assay was performed on HEK and HeLa cells as shown in Fig. 3A. These numbers are the absolute SH/aa values and include all protein sulfhydryls within the cells, including membrane proteins. From these numbers we could calculate the fraction of PSox relative to total PS in HEK and HeLa to (5.9 ± 0.3)% and (9.6 ± 0.6)%, respectively. However, the method cannot distinguish intra- and intermolecular disulfide bonds and glutathione bound to protein is included in these numbers. Consequently the next step was to selectively quantify mixed disulfides between protein and low-molecular-weight compounds, such as glutathione.
Fig. 3.
Distribution of sulfhydryl equivalents in HEK and HeLa cells. SH/aa values for HEK cells (white bars) and HeLa cells (gray bars) are shown. (A) Protein sulfhydryl equivalents were determined as described in Fig. 1B. (B) Total soluble glutathione equivalents (Total GS = GSH + GS in GSSG) and oxidized glutathione equivalents (GS in GSSG) were quantified from the TCA supernatant by use of the NPM-HPLC assay, as described experimental procedures, whereas GSH was quantified by subtracting GS in GSSG from total GS. Error bars for GSH are calculated by using the rules of error propagation (38). (C) Glutathionylated protein equivalents (Note: in units of SH/aa × 10−6) were quantified with SBD-F as illustrated in Fig. 1C.
Analysis of Low-Molecular-Weight Thiols in HEK and HeLa Cells.
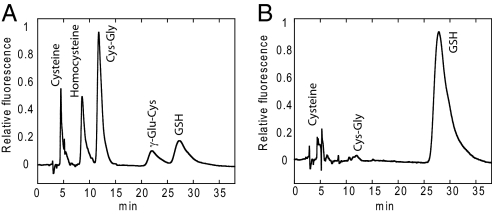
Before quantification of soluble or protein-bound low-molecular-weight sulfhydryls we decided to determine the relative amount of the different thiol species inside the cells. This was done by fluorescent labeling of sulfhydryl compounds with SBD-F followed by HPLC separation. Fig. 4A shows a chromatogram of SBD-derivatives of the typical thiol compounds, likely to be found in cells. However, as shown in Fig. 4B, glutathione was by far the most abundant species in the TCA supernatants. Only very small peaks from cysteine and cysteinylglycine (Cys-Gly) were observed in HEK cells, and similar results were obtained for HeLa cells (data not shown).
Fig. 4.
HPLC chromatograms of SBD-labeled thiol compounds. Sulfhydryl compounds were labeled with SBD-F as described in experimental procedures. Samples were injected onto a Phenomenex Luna C18 (2) column and eluted isocractically in 75 mM sodium citrate, pH 2.9, 2% methanol. The SBD-derivatized thiols were detected by fluorescence with excitation and emission at 386 and 516 nm, respectively. Cys-Gly, cysteinylglycine; γ-Glu-Cys = γ-glutamylcysteine. (A) Standard thiol compounds in 10% TCA. (B) TCA supernatant from HEK cells.
Global PSSG Levels Are Extremely Low.
As glutathione constitutes the majority of the soluble sulfhydryl pool, we anticipated that it would also be the most common low-molecular-weight thiol bound to protein. Indeed, the only peak observed in the HPLC chromatogram from HEK and HeLa, after protein disulfide reduction and SBD-F thiol labeling (Fig. 1C), was the SBD-GS derivative. By relating this peak area to a glutathione standard curve (Fig. S2) the level of PSSG could be quantified.
The absolute values of protein cysteine equivalents involved in glutathionylation in HEK and HeLa cells are shown in Fig. 3C. By comparing with the numbers given in Fig. 3A, we could conclude that the level of PSSG is very low in both cell types, with values of a factor of 1,000 below the value of total PS. Furthermore, PSSG contributed only with <1% to the PSox value.
It was now possible to calculate the absolute value of PSSP by subtracting the PSSG value from the PSox value. The relative distributions of the different protein sulfhydryl equivalents are given in Table 1, and the absolute SH/aa-values are shown in Table S1.
Table 1.
Relative distribution of protein sulfhydryl equivalents in HEK and HeLa cells
| Thiol or disulfide | Percentage relative to total PScal |
|||
|---|---|---|---|---|
| HEK |
HeLa |
|||
| No treatment | With diamide | No treatment | With diamide | |
| PSH | 94 ± 3 | 61 ± 5 | 90.4 ± 5.3 | 56 ± 10 |
| PS in PSSP* | 5.8 ± 0.3 | 24 ± 3 | 9.5 ± 0.6 | 25 ± 11 |
| PS in PSSG | 0.037 ± 0.003 | 15 ± 1 | 0.052 ± 0.003 | 19 ± 4 |
Values are given as the means ± SD.
*PS in PSSP was calculated by subtracting (2 × PS in PSSG) from PSox.
Quantification of Glutathione.
To fully describe the cellular thiol/disulfide state, we quantified the low-molecular-weight thiol pool in the same cell samples as the protein quantifications were performed with. Because glutathione was by far the most abundant low-molecular-weight sulfhydryl species in these cells, we decided to limit the analysis to the quantification of GSH and GSSG. The SBD-F assay was not reliable for determination of GSH/GSSG ratios, thus we applied an assay based on reduction of disulfides with tris(2-carboxyethyl)phosphine (TCEP), followed by fluorescent labeling of thiols with N-(1-pyrenyl)maleimide (NPM).
The majority of soluble glutathione equivalents were found as GSH (Fig. 3B). Furthermore, comparison of HEK and HeLa cells revealed that HeLa contain ≈30% more total glutathione than HEK. Because we had total protein amino acids as a common denominator we could map the global distribution of GS equivalents in the cells. The relative distribution is shown in Table 2 and the SH/aa values are given in Table S2. Although HeLa cells in general contained more glutathione, the relative amount of GSSG was very similar in HEK and HeLa cells. In addition, the fraction of the glutathione in PSSG was ≈0.1% for both cell types.
Table 2.
Relative distribution of glutathione equivalents in HEK and HeLa cells
| Thiol or disulfide | Percentage relative to total GS* |
|||
|---|---|---|---|---|
| HEK |
HeLa |
|||
| No treatment | With diamide† | No treatment | With diamide† | |
| GS in PSSG | 0.09 ± 0.01 | 36 ± 2 | 0.10 ± 0.01 | 23 ± 4 |
| GS in GSSG | 8.5 ± 1.2 | 50 ± 12 | 9.1 ± 0.7 | 41 ± 10 |
| GSH‡ | 91 ± 14 | 14 ± 15 | 91 ± 10 | 37 ± 14 |
Values are given as the means ± SD.
*Total GS is calculated by addition of [total soluble GS] and [GS in PSSP].
†Quantified GSSG levels were corrected for postlysis diamide oxidation, as described in Experimental Procedures.
‡GSH is calculated by subtracting [GS in GSSG] from [total soluble GS].
Is the Protein Thiol Pool an Active Redox Buffer in the Cell?
By comparing the PSH and GSH numbers in Fig. 3 it was observed that the PSH pool constituted the majority (up to 70%) of the reduced thiol equivalents within cells (Table 3). This observation implied that protein thiols could be important players in cellular redox homeostasis. However, it would require that a considerable fraction of PSH was accessible on the surface of protein. To investigate this we exposed cells to a sublethal dose of oxidant and quantified the redistribution of thiol and disulfide pools. We selected diamide (16) as oxidant, because it, in contrast to H2O2, reacts specifically with thiols and is not readily metabolized in the cell. Thus, trypsinized HEK and HeLa cells were exposed to 5 mM diamide for 5 min before quenching with 10% TCA. This concentration was chosen to produce a noticeable response in the cellular redox status without killing the cells (Table S3).
Table 3.
Relative distribution of GSH and PSH in the total pool of reduced thiol equivalents
| Thiol | Percentage |
|
|---|---|---|
| HEK | HeLa | |
| GSH | 28 ± 4 | 34 ± 4 |
| PSH | 72 ± 4 | 66 ± 5 |
The total pool of reduced thiol equivalents is calculated by the addition of GSH and PSH values. Values are given as the means ± SD.
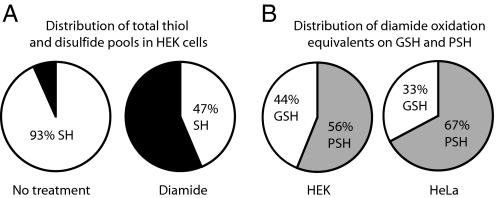
The diamide exposure resulted in dramatic changes in the global distribution of reduced and oxidized sulfhydryl pools, where more than half of the total reduced thiol pool (PSH + GSH) became oxidized in HEK and HeLa cells (Fig. 5A and Fig. S3). The most dramatic effects were seen in the levels of PSSG that had increased by a factor of ≈300 in both cell types (Tables 1 and 2). For HEK cells this meant that ≈1/3 of the total glutathione pool was bound to protein, corresponding to glutathionylation of 15% of the total protein sulfhydryl pool. It should be noted that there was no significant difference between the total amount of glutathione equivalents before and after diamide treatment (Fig. S4).
Fig. 5.
Diamide oxidation. Cells were exposed to 5 mM diamide for 5 min before TCA quenching. (A) Redistribution of total thiol and disulfide pools upon diamide exposure of HEK cells. Total thiols pools (GSH + PSH) are shown in white, and total disulfide pools (PSSP + GSSG + PSSG) are shown in black. Detailed calculations are shown in SI Text. Similar results were obtained in HEK cells as shown in Fig. S3 (B) Consumption of diamide equivalents by the GSH (white) and PSH (gray) pools. The difference in the PSH pool (ΔPSH) and GSH pool (ΔGSH) before and after diamide treatment was used to calculate the relative distribution of diamide equivalents between GSH and PSH according to the following equations: (i) Total diamide oxidation = ΔPSH + ΔGSH; (ii) % diamide consumed by PSH = ΔPSH/(ΔPSH + ΔGSH); (iii) % diamide consumed by GSH = ΔGSH/(ΔPSH + ΔGSH). Detailed calculations can be found in the SI Text.
Compared with the increase in glutathionylation, the increases of GSSG and PSSP pools in HEK cells, by factors of 6 and 3.5, respectively, were more modest. Similar results were obtained in HeLa cells (Tables 1 and 2).
The major increase in PSSG implied that the cellular PSH pool was active as a redox buffer during diamide oxidation. The diamide exposure resulted in oxidation of 43% and 56% of the PSH equivalents in HEK and HeLa cells, respectively (calculations are shown in SI Text). From this result we conclude that approximately half of all PSH equivalents are accessible to diamide oxidation and are not buried by the protein structure. We found no other low-molecular-weight thiols, like cysteine, to sequester a significant number of oxidizing equivalents under diamide stress (data not shown).
To address the specific contribution of GSH vs. PSH pools in consuming diamide oxidation we calculated how the oxidizing equivalents were distributed among GSH and PSH (Fig. 5B). Indeed, both in HEK and HeLa cells the PSH pool consumed >50% of the diamide-induced oxidation, indicating directly that the protein thiol pool is an active player in the antioxidant defense system of the cell.
Recovery from Diamide Stress Is Fast.
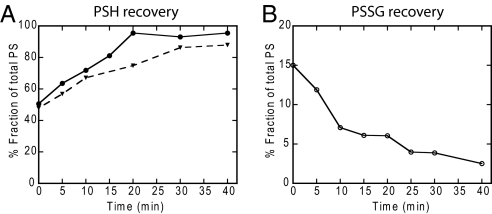
As the exposure to diamide was not lethal (cells were alive and dividing 48 h after exposure, Table S3) we wished to gauge the recovery from the diamide oxidation directly on the thiol–disulfide level. As shown in Fig. 6, the kinetics for the decrease in PSSG and the increase in PSH were very similar. Furthermore, the PSH recovery was nearly completed 20 min after removal of diamide. The PSH recovery in HeLa was monitored as well, and here a full recovery of PSH was observed after 30 min. To make sure the PBS incubation, as such, did not affect the protein redox state, the experiment was also performed without diamide. Here, PSH and PSSG levels remained stable throughout the experiment over a 1-h period (data not shown).
Fig. 6.
Protein sulfhydryl recovery from diamide oxidation. Trypsinized cells were treated with 5 mM diamide for 5 min and rapidly washed twice in 1 mL of PBS. Cells were resuspended in 1 mL of PBS and incubated at 37 °C with gentle shaking. At indicated time points samples were taken and directly quenched by addition of TCA to 10% followed by quantification of PSH and PSSG as described in Fig. 1. The numbers are given relative to total PS equivalents in each cell type. (A) PSH recovery in HEK (filled circles) and HeLa (inverted filled triangles). (B) PSSG recovery in HEK.
Discussion
Global Cellular Inventory of Thiols and Disulfides.
The fact that cysteine residues in proteins are found either as thiols or parts of disulfide bonds has long been text-book knowledge. Similarly, it is generally accepted that glutathione to a large degree functions as “the cellular thiol–disulfide redox buffer.” It is therefore surprising that the relative amounts of these species have remained obscure and that the potential involvement of protein thiols in redox homeostasis has never been addressed. In this study, we have accurately mapped the distribution of oxidized and reduced thiol pools in glutathione and in proteins. We have focused on obtaining a complete picture of the cellular content so that all protein thiols/disulfides were detected, both in soluble and membrane proteins.
We have determined absolute numbers of total PS equivalents per amino acid (Fig. 3A) and conclude that the frequency of the cysteine residue in proteins is 2.4% and 2.7% for HEK and HeLa, respectively. This is in excellent agreement with studies made on single-residue compositions of proteomes where an average cysteine frequency of ≈2% has been calculated for eukaryotes (17).
It has been shown that the majority of the cellular protein sulfhydryl equivalents are found in the reduced form (6, 7, 9). In this study, we have established that PSSP constitute 6% and 9.6% of the total protein cysteine equivalents in HEK and HeLa cells, respectively (Table 1). The higher levels of PSSP in HeLa cells could imply higher expression levels of disulfide containing proteins compared with HEK cells.
In terms of distribution of glutathione equivalents there were no significant differences between HEK and HeLa cells with GSSG levels constituting ≈4.5% of the total GS equivalents (Table 2). However, the absolute levels of glutathione were ≈30% higher in HeLa cells than in HEK cells (Fig. 3B). A higher oxidative load on HeLa cells could lead to an up-regulation of glutathione synthesis, because glutathione have been suggested to function as an antioxidant in the ER, consuming excess oxidizing equivalents followed by export of GSSG (18).
By using a genetically encoded glutathione-specific redox probe the concentrations of GSH and GSSG in the yeast cytosol have been estimated to be 13 mM and 4 μM, respectively (3). The cytosolic glutathione environment has been suggested to be even more reducing in HeLa cells (R.E.H. and J.R.W., unpublished data, and ref. 19). If the GSH concentrations are assumed to be approximately the same as in yeast, a cytosolic GSSG concentration of 0.2 μM in HeLa cells, corresponding to a GSH/GSSG ratio of 65,000, can be calculated. This suggests that the vast majority of the total GSSG is found in secretory compartments and possibly in the mitochondrial intermembrane space.
Protein Glutathionylation in Nonstressed Cultured Cells.
The cellular levels and physiological role of protein glutathionylation is a matter of some controversy. Although the suggested mechanisms of PSSG formation under physiological conditions are many (20), thiol–disulfide exchange could seem as a likely mechanism (Eqs. 1 and 2).
The equilibrium constant, K, of this reaction is typically close to 1 (21, 22) and in combination with the reducing environment of the cytosol, steady-state levels of PSSG is expected to be extremely low with a ratio of PSSG/PSH of 1.5 × 10−5. This implies that the vast majority of PSSG should be found in secretory compartments.
Previous methods for detection of PSSG require manipulation of the cell system by either preventing protein synthesis (23–26) or by loading cells with biotin-labeled glutathione (27, 28), which could affect the steady-state levels of PSSG. We have quantified the absolute basal levels of glutathionylation directly in acid quenched cells. This direct method has shown the global level of PSSG to be extremely low in both HEK and HeLa cells, consistent with results where basal levels of PSSG in platelets were undetectable by immunoblotting (29). However, in liver microsomes >50% of the glutathione equivalents in ER were found to be bound to protein (30). Although we cannot directly determine subcellular distributions of PSSG equivalents, we can estimate the maximal fraction of PSSG in the ER, assuming that all PSSG equivalents are found in the ER, and that the concentration of GS equivalents is the same in all compartments. If we further conservatively assume that the ER constitutes <5% of the total cell volume (31, 32) we can calculate that maximum 2% (0.1% PSSG in 5% of the cell volume) of the GS equivalents in ER are found as PSSG. Thus, we cannot confirm the previously estimated numbers of PSSG in ER. In addition, our results are consistent with the recent suggestion that extensive oxidation of glutathione takes place during isolation of microsomes (33).
Protein Thiols Constitute a Redox-Active Pool That Is Similar in Size to the Glutathione Pool.
We observe that protein thiols constitute a redox active pool in living cells that is just as important as glutathione. A sublethal diamide exposure resulted in 50% oxidation of the total cellular SH groups and we could calculate that PSH consumed more than half of the oxidizing equivalents (Fig. 5C). It should be noted that the cells are treated under completely native conditions, so little or no protein unfolding is expected to take place. This allows us to conclude that close to half of the PSH equivalents, rather than being sequestered in the cores of proteins, is accessible to either glutathionylation or formation of intra- or interprotein disulfide formation. Surprisingly, this tendency is most pronounced in HeLa cells, which has a larger pool of GSH relative to PSH than HEK cells (Table 3). This suggests that the diamide-accessible PSH pool is larger in HeLa cells. Furthermore, the distribution of diamide equivalents in HeLa cells is very similar to the distribution of GSH and PSH in the total pool of reduced thiol equivalents in unstressed cells (Table 3). Thus, contrary to previous suggestions (16) diamide oxidation in the cell appears not to have any special preference of the GSH pool over the redox-active PSH pool.
Protein thiols have been proposed to serve as cellular antioxidants (34) because of the presence of abundant cellular proteins whose activity is not affected by glutathionylation. Likewise, hemoglobin has been shown to be a cellular antioxidant in erythrocytes, where the highly reactive thiol groups of this extremely abundant protein have been shown to intercept oxidative species more effectively than glutathione (35). Because the cellular concentration of different proteins may vary quite dramatically this could reflect properties of a subset highly abundant proteins such as cytoskeletal proteins, which have been shown to be glutathionylated during diamide stress (36, 37).
Previous studies have assayed recovery from oxidant exposure by measuring the decrease in PSSG (23, 24). However, a decrease can be caused by either a reduction of the PSSG disulfide or a degradation of glutathionylated proteins. Furthermore, such a strategy cannot provide information about recovery of thiols oxidized to PSSP. Consequently, we decided to monitor diamide recovery by quantifying the increase in PSH, and the decrease in PSSG.
The kinetics for protein deglutathionylation upon removal of diamide correlates very well with previous observations (23, 24). That the kinetics for the increase in PSH and the decrease in PSSG are identical (Fig. 6) suggests that oxidation most likely does not lead to massive protein degradation and further supports the notion that the protein thiols are redox active in their native state.
In HEK cells, the majority of GSH was oxidized by diamide. We did attempt to measure GSH recovery by following the decrease in GSSG but basal levels were reached within 5 min, which was the first possible obtainable data point (data not shown). This suggests that the soluble glutathione pool is reduced extremely rapidly.
Together, these data illustrates that protein thiols are directly involved in the cellular defense mechanism against oxidants, which seems to contribute to a remarkable ability to recover from thiol–disulfide stress.
Experimental Procedures
For a detailed description of the materials and methods, see SI Text.
Cell Culture, Diamide Exposure, and Sample Preparation.
Cell culturing and diamide exposure is described in detail in SI Text. Cells from a 95% confluent 10-cm dish were harvested by trypsinization, washed in PBS, and directly quenched by addition of 10% TCA. GS equivalents were quantified from the supernatant and PS equivalents were quantified from the pellet. To eliminate traces of soluble thiols the TCA pellets were washed in 10% TCA by 4 rounds of sonication followed by centrifugation.
Quantification of Glutathione.
Total GS were quantified by reducing disulfides with TCEP followed by thiol derivatization with NPM. For quantification of GSSG, free thiols were first blocked by NEM followed by TCEP reduction and NPM derivatization. GSH equivalents were calculated by subtracting GS in GSSG from total GS. Materials and methods are described in detail in ref. 3 and SI Text. For diamide experiments the TCA precipitation left excess diamide in the supernatant. Because diamide is stable in TCA it can react with GSH when pH was increased to 7. Thus, to avoid postlysis diamide oxidation, NEM is required to alkylate thiols faster than diamide can oxidize them. We tested this by comparing the GSSG content in samples containing equivalent amounts of GSH and GSSG in 10% TCA with or without diamide. In samples containing diamide, the GSSG content was overestimated by a factor of 1.2 ± 0.3 (data not shown). Consequently, the cellular GSSG measurements were corrected for this.
Protein Sulfhydryl Quantifications.
Quantification of PSH, PSox, and Total PS was performed with 4-DPS and BH using a modification of the HPLC method described by Hansen, et al. (12), optimized for cell extracts. PSH was quantified by directly solubilizing P1 (Fig. 1A) in 0.4 M sodium citrate, 1 mM EDTA, 5% SDS, pH 4.5 and addition of 4-DPS to a final concentration of 0.5 mM followed by HPLC analysis. For quantification of PSox, P2 was solubilized in 0.5 M Tris·HCl, 1 mM EDTA, 5% SDS, 20 mM NEM, pH 8.3, and thiol alkylation was allowed to proceed for 15 min at 50 °C. Protein disulfides were reduced by the addition of BH to a final concentration of 3.3% (wt/vol) and incubated at 50 °C for 30 min. Before quantification with 4-DPS, BH was destroyed by the addition of HCl.
Quantification of PSSG.
Quantification of PSSG was performed by solubilizing P4 in 200 mM Bicine, 8 mM EDTA, 6 M urea, pH 9.16. Protein thiols were reduced with 2.1 mM THP and thiols were derivatized with 6.4 mM SBD-F by incubating 1 h at 60 °C. Selective quantification of GS-SBD was performed with HPLC analysis as illustrated in Fig. S2 and described in detail in SI Text. Urea was chosen as protein denaturant as it, in contrast to SDS, did not interfere with thiol peaks in the HPLC chromatogram (Fig. S2). In addition, low-molecular-weight thiols in the TCA supernatant were identified with SBD-F as described in SI Text.
Quantification of Total Protein.
The quantification of total protein in TCA pellets was performed by HCl hydrolysis of proteins followed by quantification of amino acids with ninhydrin, which reacts with primary amino groups to form a blue color absorbing at 570 nm. Details of assay conditions and preparation of ninhydrin reagent are described in SI Text.
Data Presentation.
All data shown are the mean ± SD (standard deviation of the mean) of 4–6 independent experiments, each performed in triplicate. When combining of uncertainties was necessary the rules of error propagation were used according to ref. 38.
Supplementary Material
Acknowledgments.
We thank Andrea Vala and Kristine Steen Jensen for critically reading the manuscript. This work was supported by the Danish Natural Science Research Council (J.R.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812149106/DCSupplemental.
References
- 1.Linke K, Jakob U. Not every disulfide lasts forever: Disulfide bond formation as a redox switch. Antioxid Redox Signal. 2003;5:425–434. doi: 10.1089/152308603768295168. [DOI] [PubMed] [Google Scholar]
- 2.Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Ostergaard H, Tachibana C, Winther JR. Monitoring disulfide bond formation in the eukaryotic cytosol. J Cell Biol. 2004;166:337–345. doi: 10.1083/jcb.200402120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrmann JM, Kohl R. Catch me if you can! Oxidative protein trapping in the intermembrane space of mitochondria. J Cell Biol. 2007;176:559–563. doi: 10.1083/jcb.200611060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eaton P. Protein thiol oxidation in health and disease: Techniques for measuring disulfides and related modifications in complex protein mixtures. Free Radic Biol Med. 2006;40:1889–1899. doi: 10.1016/j.freeradbiomed.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 6.Leichert LI, Jakob U. Global methods to monitor the thiol-disulfide state of proteins in vivo. Antioxid Redox Signal. 2006;8:763–772. doi: 10.1089/ars.2006.8.763. [DOI] [PubMed] [Google Scholar]
- 7.Hochgrafe F, Mostertz J, Albrecht D, Hecker M. Fluorescence thiol modification assay: Oxidatively modified proteins in Bacillus subtilis. Mol Microbiol. 2005;58:409–425. doi: 10.1111/j.1365-2958.2005.04845.x. [DOI] [PubMed] [Google Scholar]
- 8.Le MN, Clement G, Le MS, Tacnet F, Toledano MB. The Saccharomyces cerevisiae proteome of oxidized protein thiols: Contrasted functions for the thioredoxin and glutathione pathways. J Biol Chem. 2006;281:10420–10430. doi: 10.1074/jbc.M513346200. [DOI] [PubMed] [Google Scholar]
- 9.Leichert LI, et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci USA. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sethuraman M, McComb ME, Heibeck T, Costello CE, Cohen RA. Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol Cell Proteomics. 2004;3:273–278. doi: 10.1074/mcp.T300011-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Creighton TE. Disulfide bond formation in proteins. Methods Enzymol. 1984;107:305–329. doi: 10.1016/0076-6879(84)07021-x. [DOI] [PubMed] [Google Scholar]
- 12.Hansen RE, Ostergaard H, Norgaard P, Winther JR. Quantification of protein thiols and dithiols in the picomolar range using sodium borohydride and 4,4′-dithiodipyridine. Anal Biochem. 2007;363:77–82. doi: 10.1016/j.ab.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Svardal AM, Mansoor MA, Ueland PM. Determination of reduced, oxidized, and protein-bound glutathione in human plasma with precolumn derivatization with monobromobimane and liquid-chromatography. Anal Biochem. 1990;184:338–346. doi: 10.1016/0003-2697(90)90691-2. [DOI] [PubMed] [Google Scholar]
- 14.Abukhalaf IK, et al. High performance liquid chromatographic assay for the quantitation of total glutathione in plasma. J Pharm Biomed Anal. 2002;28:637–643. doi: 10.1016/s0731-7085(01)00658-6. [DOI] [PubMed] [Google Scholar]
- 15.Kirley TL. Reduction and fluorescent labeling of cyst(e)ine-containing proteins for subsequent structural-analyses. Anal Biochem. 1989;180:231–236. doi: 10.1016/0003-2697(89)90422-3. [DOI] [PubMed] [Google Scholar]
- 16.Kosower NS, Kosower EM. Diamide—an oxidant probe for thiols. Methods Enzymol. 1995;251:123–133. doi: 10.1016/0076-6879(95)51116-4. [DOI] [PubMed] [Google Scholar]
- 17.Pe'er I, et al. Proteomic signatures: Amino acid and oligopeptide compositions differentiate among phyla. Proteins. 2004;54:20–40. doi: 10.1002/prot.10559. [DOI] [PubMed] [Google Scholar]
- 18.Cuozzo JW, Kaiser CA. Competition between glutathione and protein thiols for disulphide-bond formation. Nat Cell Biol. 1999;1:130–135. doi: 10.1038/11047. [DOI] [PubMed] [Google Scholar]
- 19.Dooley CT, et al. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem. 2004;279:22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 20.Ghezzi P, Di Simplicio P. Glutathionylation pathways in drug response. Curr Opin Pharmacol. 2007;7:398–403. doi: 10.1016/j.coph.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert HF. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol. 1995;251:8–28. doi: 10.1016/0076-6879(95)51107-5. [DOI] [PubMed] [Google Scholar]
- 22.Hansen RE, Ostergaard H, Winther JR. Increasing the reactivity of an artificial dithiol-disulfide pair through modification of the electrostatic milieu. Biochemistry. 2005;44:5899–5906. doi: 10.1021/bi0500372. [DOI] [PubMed] [Google Scholar]
- 23.Schuppe-Koistinen I, Gerdes R, Moldeus P, Cotgreave IA. Studies on the reversibility of protein S-thiolation in human endothelial-cells. Arch Biochem Biophys. 1994;315:226–234. doi: 10.1006/abbi.1994.1494. [DOI] [PubMed] [Google Scholar]
- 24.Seres T, et al. Protein S-thiolation and dethiolation during the respiratory burst in human monocytes—A reversible post-translational modification with potential for buffering the effects of oxidant stress. J Immunol. 1996;156:1973–1980. [PubMed] [Google Scholar]
- 25.Fratelli M, et al. Identification by redox proteomics of glutathionylated proteins in oxidatively stressed human T lymphocytes. Proc Natl Acad Sci USA. 2002;99:3505–3510. doi: 10.1073/pnas.052592699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lind C, et al. Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch Biochem Biophys. 2002;406:229–240. doi: 10.1016/s0003-9861(02)00468-x. [DOI] [PubMed] [Google Scholar]
- 27.Brennan JP, et al. The utility of N,N-biotinyl glutathione disulfide in the study of protein S-glutathiolation. Mol Cell Proteomics. 2006;5:215–225. doi: 10.1074/mcp.M500212-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Sullivan DM, Wehr NB, Fergusson MM, Levine RL, Finkel T. Identification of oxidant-sensitive proteins: TNF-alpha induces protein glutathiolation. Biochemistry. 2000;39:11121–11128. doi: 10.1021/bi0007674. [DOI] [PubMed] [Google Scholar]
- 29.Dalle-Donne I, Giustarini D, Colombo R, Milzani A, Rossi R. S-glutathionylation in human platelets by a thiol-disulfide exchange-independent mechanism. Free Radic Biol Med. 2005;38:1501–1510. doi: 10.1016/j.freeradbiomed.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 30.Bass R, Ruddock LW, Klappa P, Freedman RB. A major fraction of endoplasmic reticulum-located glutathione is present as mixed disulfides with protein. J Biol Chem. 2004;279:5257–5262. doi: 10.1074/jbc.M304951200. [DOI] [PubMed] [Google Scholar]
- 31.Dean PM. Ultrastructural morphometry of pancreatic beta-cell. Diabetologia. 1973;9:115–119. doi: 10.1007/BF01230690. [DOI] [PubMed] [Google Scholar]
- 32.Weibel ER, Staubli W, Gnagi HR, Hess FA. Correlated morphometric and biochemical studies on liver cell. I. Morphometric model, stereologic methods, and normal morphometric data for rat liver. J Cell Biol. 1969;42:68–91. doi: 10.1083/jcb.42.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon BM, Heath SHD, Kim R, Suh JH, Hagen TM. Assessment of endoplasmic reticulum glutathione redox status is confounded by extensive ex vivo oxidation. Antioxid Redox Signal. 2008;10:963–972. doi: 10.1089/ars.2007.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas JA, Poland B, Honzatko R. Protein sulfhydryls and their role in the antioxidant function of protein S-thiolation. Arch Biochem Biophys. 1995;319:1–9. doi: 10.1006/abbi.1995.1261. [DOI] [PubMed] [Google Scholar]
- 35.Rossi R, et al. Fast-reacting thiols in rat hemoglobins can intercept damaging species in erythrocytes more efficiently than glutathione. J Biol Chem. 1998;273:19198–19206. doi: 10.1074/jbc.273.30.19198. [DOI] [PubMed] [Google Scholar]
- 36.Brennan JP, et al. Detection and mapping of widespread intermolecular protein disulfide formation during cardiac oxidative stress using proteomics with diagonal electrophoresis. J Biol Chem. 2004;279:41352–41360. doi: 10.1074/jbc.M403827200. [DOI] [PubMed] [Google Scholar]
- 37.Cumming RC, et al. Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem. 2004;279:21749–21758. doi: 10.1074/jbc.M312267200. [DOI] [PubMed] [Google Scholar]
- 38.Taylor JR. In: An Introduction to Error Analysis. McGuire A, editor. Mill Valley, CA: University Science Books; 1997. pp. 45–79. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.