Abstract
Metallothioneins are small, cysteine-rich proteins that avidly bind heavy metals such as zinc, copper, and cadmium to reduce their concentration to a physiological or nontoxic level. Metallothionein gene transcription is induced by several stimuli, notably heavy metal load and oxidative stress. Transcriptional induction of metallothionein genes is mediated by the metal-responsive transcription factor 1 (MTF-1), an essential zinc finger protein that binds to specific DNA motifs termed metal-response elements. In cell-free DNA binding reactions with nuclear extracts, MTF-1 requires elevated zinc concentrations for efficient DNA binding but paradoxically is inactivated by other in vivo inducers such as cadmium, copper, and hydrogen peroxide. Here we have developed a cell-free, MTF-1-dependent transcription system which accurately reproduces the activation of metallothionein gene promoters not only by zinc but also by these other inducers. We found that while transcriptional induction by zinc can be achieved by elevated zinc concentration alone, induction by cadmium, copper, or H2O2 additionally requires the presence of zinc-saturated metallothionein. This is explained by the preferential binding of cadmium or copper to metallothionein or its oxidation by H2O2; the concomitant release of zinc in turn leads to the activation of transcription factor MTF-1. Conversely, thionein, the metal-free form of metallothionein, inhibits activation of MTF-1. The release of zinc from cellular components, including metallothioneins, and the sequestration of zinc by newly produced apometallothionein might be a basic mechanism to regulate MTF-1 activity upon cellular stress.
All living organisms are able to cope with a variety of stress situations by immediately adapting their gene expression program to the stress stimulus. For example, metallothioneins, small cysteine-rich proteins, are strongly upregulated upon heavy metal load (2, 27, 29, 49, 63, 70, 71). They have the ability to bind and hence neutralize toxic (such as Cd and Hg) and excess nontoxic (such as Zn and Cu) heavy metal ions and also act as radical scavengers. Metal response element binding transcription factor 1, also called metal-responsive transcription factor 1 (MTF-1), plays an important role in the cellular response to heavy metal stress (2, 21, 38, 55, 75) and is essential for embryonic liver development in the mouse (24). MTF-1 contains six zinc fingers of the C2H2 type. C-terminal to the zinc fingers are three distinct activation domains, an acidic, a proline-rich, and a serine/threonine-rich domain. Via the zinc fingers, it binds to DNA sequence motifs with the consensus binding site TGCRCNC, known as metal response elements (MREs). MREs are present in the promoters of metallothionein genes (41, 61, 64) and other target genes, including zinc transporter ZnT1 and γ-glutamylcysteine synthetase heavy chain (24, 34, 37). MTF-1 is also involved in the responses to oxidative stress (16, 24), hypoxia (23, 46), and amino acid starvation (1). In quiescent cells, MTF-1 preferentially resides in the cytoplasm but translocates to the nucleus upon several stress conditions, notably heavy metal load (59, 62, 67). Furthermore, changes in phosphorylation state have been shown to contribute to MTF-1 activity (35, 58).
At least three domains of MTF-1 are involved in sensing heavy metal load. Among these, the zinc fingers appear most important for metal response, as indicated by several findings: (i) they are the most conserved region of MTF-1 from insects to mammals, (ii) chimeric MTF-1 that is essentially reduced to the zinc fingers and fused to a heterologous activation domain retains partial responsiveness (43), (iii) finger deletions in conjunction with DNA binding and metal binding studies have implicated zinc fingers 5 and 6 as well as 1 in metal responsiveness (11, 30). A second region was found by systematic domain swapping between mouse and human MTF-1. The latter shows a lower basal but higher metal-inducible activity than mouse MTF-1, and this species difference could be narrowed to the acidic activation domain (43). Most recently, a third domain was found to contribute to metal inducibility, a cysteine-rich segment C-terminal to the serine/threonine-rich activation domain. Cysteine substitutions or deletions did not affect nuclear translocation or DNA binding of MTF-1 but impaired metal induction in vivo (D. Giedroc, personal communication). These two regions involved in metal sensing and metal response probably act in concert with the zinc finger domain, since a chimeric MTF-1 in which the zinc fingers were replaced with a POU DNA binding domain displayed metal-independent, constitutive activity (E. Brugnera and W. Schaffner, unpublished data).
MTF-1 is likely to act as an intracellular zinc sensor, since in a competitive situation, it requires a higher zinc concentration for DNA binding/function than other zinc-binding factors (7, 26, 41; this study). This suggests a molecular mechanism for why MTF-1-dependent genes are transcriptionally activated by zinc load. However, these metal binding studies also revealed an apparent paradox: in vivo, other heavy metals, notably cadmium and copper, induce metallothionein gene transcription via MTF-1, yet these metals are unable to functionally substitute for zinc in cell-free DNA binding assays of MTF-1; rather, they have a deleterious effect in vitro by inhibiting MTF-1's ability to bind DNA (7, 22, 26, 45, 50, 80). Furthermore, it was reported that the activity of mammalian MTF-1 expressed in Saccharomyces cerevisiae was inducible by zinc but not by cadmium or H2O2 (17). It was thus speculated that all other stress conditions (except zinc load) that activate MTF-1 in vivo exert their effect mostly indirectly, that is, by liberating zinc from intracellular stores (38, 50). However, the nature of such an intracellular supply remains to be identified.
Metallothioneins, which are mostly saturated with zinc under steady-state conditions (9, 48), are an obvious candidate zinc source for MTF-1 in the response to nonzinc stimuli (2, 38). Metallothioneins have been shown in cell-free reactions to remove or supply zinc to the zinc finger transcription factors Sp1, TFIIIA, estrogen receptor, and tramtrack (10, 56, 78, 79), but unlike MTF-1, these are apparently not activated by heavy metal stress. Sp1 might even counteract the effect of MTF-1 (47), and certain zinc finger proteins function best at very low zinc concentrations (6).
Biochemical studies have shown that the particularly high affinity of metallothioneins for cadmium and copper and the oxidation of their sulfhydryl groups by reaction products of hydrogen peroxide can liberate zinc (31, 54, 63, 71). Moreover, metallothionein genes are well-defined targets of MTF-1, i.e., their transcription is induced by MTF-1 in response to heavy metals. We have previously shown that MTF-1 is essential not only for zinc-induced transcription of metallothionein genes and for the induction by other heavy metals like cadmium and copper, but also for their basal-level expression under nonstress conditions (26). Here we provide evidence that metallothionein itself can contribute to the regulation of MTF-1 activity. We show that addition of cadmium, copper, or hydrogen peroxide together with zinc-saturated metallothionein results in specific activation of MTF-1, while addition of thionein (the metal-free form) results in preferential inhibition of MTF-1. Based on the data reported here, we consider it very likely that an elevated intracellular zinc concentration can be used for a direct activation of MTF-1.
MATERIALS AND METHODS
Construction of reporter, reference, and effector genes.
The reporter genes (4xMREd and MT-I promoter, −728 to −8) and reference genes are based on the rabbit beta-globin OVEC (oligonucleotide vector) system (73). TATA-only OVEC refers to the original vector used for the cloning of the upstream promoter elements of interest. 4xSp1-OVEC and 4xOct-OVEC (p4HO) were constructed according to Radtke et al. (55), Matsuo et al. (40), and Müller et al. (44).
The expression vector for human MTF-1 for transfection into HEK293 cells is driven by the cytomegalovirus promoter (26), as is the expression vector for human Sp1.
Cell culture.
HeLa (human cervix carcinoma) S3 cells, BJA-B (human Burkitt's lymphoma) B-lymphocyte cells, and human embryonic kidney (HEK) 293 cells were cultured as described before (5, 80).
Preparation of basal nuclear extracts from HeLa and BJA-B cells for in vitro transcription reaction.
The basal nuclear extract provided RNA polymerase II basal transcription components (5, 73). Initial experiments were done with HeLa cell nuclear extract; similar results were obtained with BJA-B cell nuclear extract. Nuclear extracts of HeLa and especially BJA-B cells had been used before in our laboratory for transcription studies, and we could rely on high-quality batches (42, 73).
Transient transfection of 293 cells and preparation of nuclear miniextract for complementation of human MTF-1 or Sp1 to in vitro transcription.
Transient transfections were done with the calcium phosphate coprecipitation technique. Nuclear protein miniextract (the complementing nuclear extract) was prepared according to Schreiber et al. (60). Transiently overexpressed MTF-1 was largely found in the nucleus, which permitted the use of nuclear extracts as a source of MTF-1 protein, while in serum-starved, resting cells, most of the MTF-1 localizes to the cytoplasm and only translocates to the nucleus upon heavy metal load and other stress (59, 62, 67). HEK293 cells were used because of their highly efficient uptake and expression of transfected DNA.
In vitro transcription and nuclease S1 mapping.
In vitro transcription was performed according to Mueller-Storm (42) with 10 to 40 μg of basal nuclear extract (1 to 2 μl) from HeLa or BJA-B cells, 5 to 40 ng of reporter constructs, and 20 to 40 ng of reference plasmids. The reference gene was driven by the simian virus 40 enhancer (OVEC-REF) except for Fig. 1B, in which CMV-REF driven by the cytomegalovirus promoter was used. From 10 to 50 μg of complementing nuclear extract from 293 cells was included when indicated. The transcription reaction (15 μl) was carried out at 30°C for 60 min in a buffer containing 10 mM HEPES, pH 7.9, 8.5% glycerol, 20 mM KCl, 7 mM NaCl, 5 mM MgCl2, 0.15 mM dithiothreitol, 0.1 mM EDTA, 1.5 units of RNasin, 5 mM creatine phosphate, and 0.5 mM each of the four nucleoside triphosphates. RNA was quantified by nuclease S1 mapping as described previously (5, 72, 73).
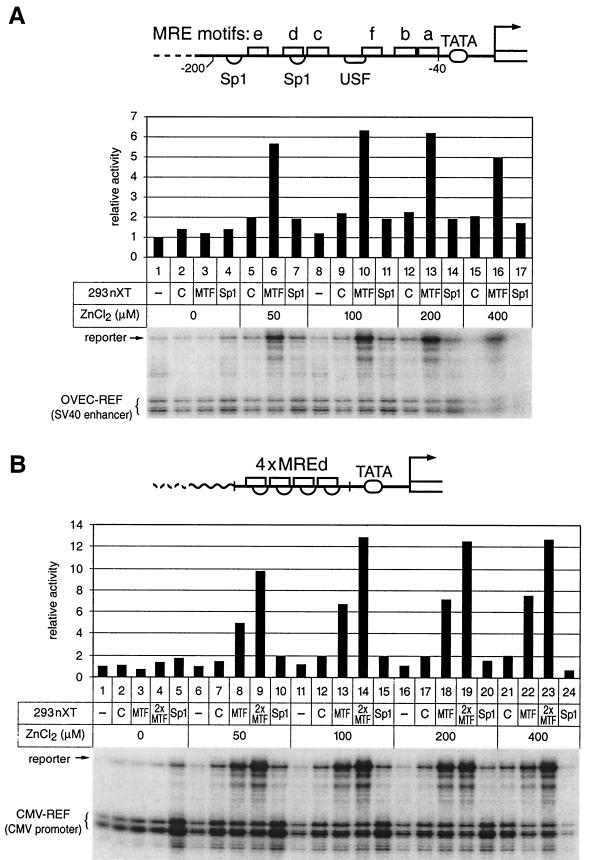
FIG.1.
Cell-free transcription system that responds to transcription factor MTF-1 and zinc. (A) Response of the mouse metallothionein-I (MT-I) promoter. The promoter construct with six metal response elements (MRE a to f) and binding sites for Sp1 and USF (3) is shown on top. The basic transcription reaction contains 37 μg of protein from nuclear extracts of BJA-B cells (−) (basal nuclear extract) or is complemented with 10 μg of protein from nuclear extracts of HEK293 cells that had not been transfected (C, control) or had been transfected with a human MTF-1 (MTF) or Sp1 (Sp1) expression vector. ZnCl2 was included at the concentrations indicated. The lower group of bands are from the internal reference gene OVEC-REF driven by the simian virus 40 enhancer. 293 nXT, complementing nuclear extract from HEK293 cells. The bar diagram depicts reporter gene activity normalized for reference gene expression, and the relative activity of the reporter gene in each lane was further normalized with that of lane 1, which reflects the basal activity of the reporter. All quantification of expression values shown was done the same way. (B) Response of the synthetic 4xMREd promoter. A schematic view of the promoter construct is shown on top. A promoter construct with four tandem copies of MREd is highly responsive to MTF-1 and zinc. (−), basic transcription reaction with 19 μg of basal nuclear extract from BJA-B cells; C, supplemented with 10 μg of nuclear extract from untransfected HEK293 cells; MTF, supplemented with 10 μg of nuclear extract from HEK293 cells transfected with the human MTF-1 expression vector; 2xMTF, supplemented with 20 μg of the same extract; Sp1, supplemented with 10 μg of nuclear extract from HEK293 cells transfected with the human Sp1 expression vector. Note that in this experiment the reference gene (CMV-REF) is driven by the cytomegalovirus (CMV) promoter containing strong proximal binding sites for Sp1. As a consequence, the reference gene but not the metal-responsive reporter is consistently activated by supplementation with extra Sp1 factor (lanes 5, 10, 15, and 20, lower group of bands). In these four lanes, other reference values were used for calibration (see Materials and Methods for details). Note that the 4xMREd promoter was slightly more active than the MT-I promoter; twice the amount of BJA-B basal extract was needed for the latter to obtain the same signal. 293 nXT, HEK293 cell complementing nuclear extract.
The gels were developed with a PhosphorImager (Molecular Dynamics), and bands were quantified with ImageQuant software. For the calculation of relative activity in bar diagrams, the band intensity corresponding to reporter gene expression in each lane was normalized to the group of bands from the reference gene (OVEC-REF, except for Fig. 1B, in which CMV-REF was used) expression. The values obtained were further normalized to the first lane, which shows the basal transcriptional activity and therefore was taken as 1.0. However, in Fig. 1B the cotransfection of an Sp1 transactivator boosted reference gene (CMV-REF) activity. Therefore, in lanes 5, 10, 15, and 20, the reference gene value was discounted, and the average reference gene activity from the preceding three lanes was taken instead.
Electrophoretic mobility shift assay.
Mini nuclear extract from untransfected HEK293 cells was prepared as for the transfected cells (see before). The electrophoretic mobility shift assay was done according to Radtke et al. (55). MRE-s is a consensus MRE sequence without overlapping Sp1 binding sites (55). The Sp1 oligonucleotide was derived from herpes simplex virus as described before (8). The Oct factor-binding site is based on the murine 104-2 immunoglobulin heavy-chain gene promoter (44).
The typical electrophoretic mobility shift assay pattern of Sp1 and MTF-1 bands was observed (24).
Purification of metallothionein and apo-MT from rabbit liver (MT-1).
Metallothionein from rabbit liver (MT-1), fully (Zn7-MT) or incompletely (Zn4-MT and apo-MT) loaded with zinc was prepared as described (68, 69). Note that copper is bound to metallothionein as copper(I) ion; in our studies, the reducing conditions of our incubation mixture (containing dithiothreitol) generated and stabilized this oxidation state.
Preparation of antisera.
Polyclonal rabbit antibodies against human and mouse MTF-1 were obtained by using purified, bacterially expressed protein representing either the N-terminal or the C-terminal region of MTF-1, always excluding amino acids 140 to 320 (i.e., the zinc finger region) to avoid possible cross-reactivity with other C2H2 zinc finger proteins.
RESULTS
Metal-inducible promoters are activated in vitro by MTF-1 and zinc.
We intended to develop an in vitro transcription system that would depend on MTF-1 and on an activating stimulus, notably heavy metals. Therefore, HeLa cell or human B lymphocyte (BJA-B) nuclear extract provided basal transcription components (basal nuclear extract) and was complemented with nuclear extract from human embryonic kidney (HEK293) cells transfected with a human MTF-1 expression vector (complementing extract). As a control, nuclear extract from untransfected HEK293 cells was used. Reporter and reference gene constructs were based on the OVEC (oligonucleotide vector) beta-globin gene, and transcriptional activity was quantified by nuclease S1 mapping (72, 73). The reporter genes tested were driven either by a synthetic promoter containing four copies of the strong MREd element (4xMREd) derived from the mouse metallothionein-I (MT-I) promoter or the natural MT-I promoter containing all six known MREs (Fig. 1).
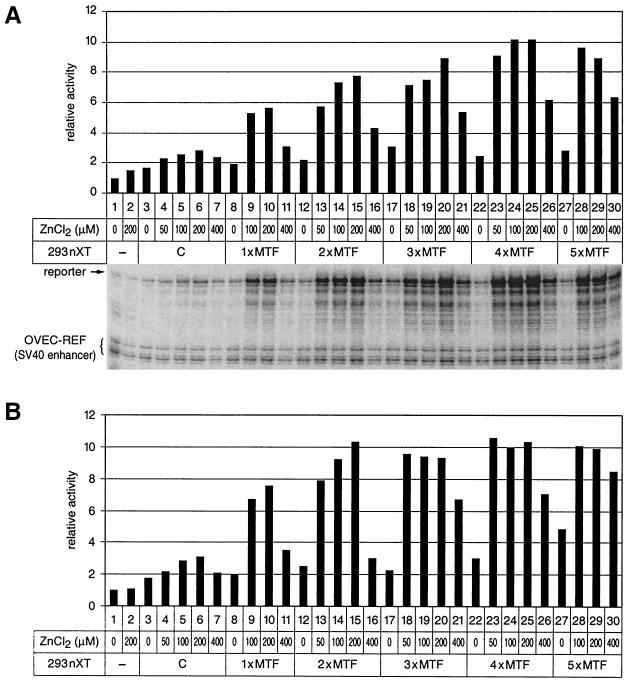
Our in vitro transcription system showed only basal transcription activity unless it was supplemented with extra zinc and human MTF-1. The response was both zinc concentration dependent and MTF-1 dose dependent (Fig. 1 and Fig. 2). Zinc at 50 to 100 μM yielded optimal induction for both promoters; higher concentrations did not increase transcript levels any further. The zinc concentration for optimal DNA binding of MTF-1 was previously reported to be at least 15 μM (15, 28). The fact that higher zinc concentrations were needed in our experiments may reflect the presence of the chelator EDTA in the transcription reactions. As is also evident from Fig. 1 and 2, both the synthetic 4×MREd promoter and the natural MT-I promoter behaved similarly in response to zinc and MTF-1. However, the response of the synthetic promoter was generally somewhat stronger than that of the full-length MT-I promoter. The small increase of the reporter signal in the presence of untransfected 293 complementing nuclear extract and zinc is most likely due to the endogenous human MTF-1 from HEK293 cells (Fig. 1 and Fig. 2; see lanes 4 to 7 in Fig. 2).
FIG. 2.
Optimizing the conditions for in vitro transcription. The reporter constructs and basic transcription reactions are as in Fig. 1. The transcription reactions contain increasing amounts of complementing nuclear extract from HEK293 cells that had been transfected with human MTF-1 expression vector (1xMTF, 2xMTF, 3xMTF, 4xMTF, and 5xMTF correspond to 10, 20, 30, 40, and 50 μg of transfected 293 cell nuclear extract, respectively). C, control nuclear extract from untransfected HEK293 cells. 293 nXT, complementing nuclear extract from HEK293 cells. (A) In vitro transcription with the mouse MT-I promoter. The transcription reaction is driven by the mouse MT-I promoter in 37 μg of basal nuclear extract from BJA-B cells. OVEC-REF driven by the simian virus 40 enhancer was used as the reference gene. (B) In vitro transcription with the 4xMREd promoter. The transcription reactions are the same as in panel A except that the reporter was driven by the synthetic 4xMREd promoter and 19 μg of basal nuclear extract from BJA-B cells was used.
The MREd sequence, which is the strongest among the six known MREs of the mouse MT-I promoter (12, 64), also contains an overlapping binding site for the general transcription factor Sp1 (47) (see also Fig. 1 and Fig. 5). To test whether Sp1, another transcription factor containing three C2H2-type zinc fingers, contributes to the zinc response of our reporter constructs, complementing nuclear extract derived from HEK293 cells transfected with a human Sp1 expression vector was compared side by side with MTF-1-transfected complementing nuclear extract in our in vitro transcription. In contrast to MTF-1, Sp1 did not induce transcription of either reporter gene in response to elevated zinc concentrations (Fig. 1), even though Sp1 is also a zinc finger protein that depends on zinc for its DNA binding (74, 78). Rather, transcription of the reference gene driven by the CMV promoter (which contains Sp1 binding sites) was boosted by cotransfection of Sp1, confirming the activity of this factor on an appropriate promoter (Fig. 1B).
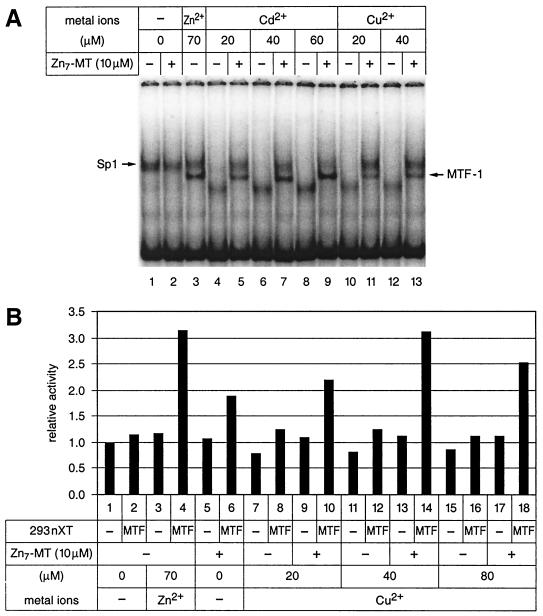
FIG. 5.
Zinc-saturated metallothionein (Zn7-MT) alleviates the inhibitory effects of heavy metals on MTF-1 DNA binding and transcriptional activity. (A) MTF-1 DNA binding activity facilitated by the combination of cadmium or copper and zinc-loaded metallothionein. The mobility shift assay was done with 11 μg of nuclear extract from untransfected HEK293 cells and the MREd sequence as the probe. Lane 1, extract not supplemented with extra metal shows a band shift with Sp1 factor (Sp1 has a higher affinity for zinc than MTF-1 (26) but no band characteristic for MTF-1.) Lane 2, addition of Zn7-MT alone had no influence on this situation. Addition of zinc (lane 3) yielded a strong band for MTF-1. Conversely, the addition of cadmium (lanes 4, 6, and 8) or copper (lanes 10 and 12) prevented the appearance of both the Sp1 and MTF-1 bands. Addition of cadmium or copper to the reaction with zinc-saturated metallothionein (Zn7-MT) restored both MTF-1 and Sp1 binding. (The lower band seen with cadmium and copper in the absence of metallothionein is not MTF-1, since it did not react with the antibody, and was not further characterized). (B) Transcription induction in vitro of the 4xMREd promoter by copper and metallothionein. MTF, complementing nuclear extract from human MTF-1-transfected HEK293 cells.
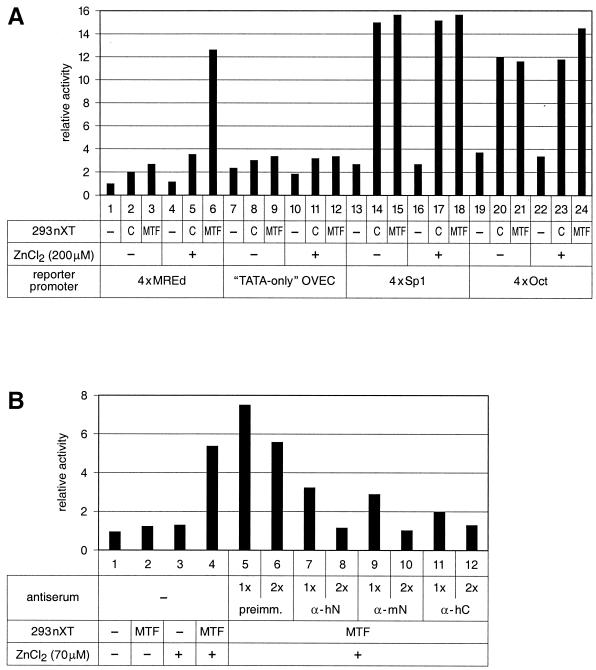
As further controls, the following constructs were tested: (i) the TATA-only OVEC reporter gene which contains only the core promoter sequence (beta-globin TATA box and initiation site), (ii) 4xSp1-OVEC (with four tandem sites for the three-zinc finger transcription factor Sp1), and (iii) 4xOct-OVEC, which contains the “octamer” consensus sequence ATGCAAAT and is driven by the zinc-independent POU domain transcription factors Oct1 and 2 (Fig. 3A). As expected, none of the control promoters responded to elevated concentrations of either zinc or MTF-1. Note that both 4xSp1 and 4xOct reporter signals increased upon addition of complementing nuclear extract from HEK293 cells, which is in good agreement with previous findings that Sp1 and Oct factors are quite abundant in these cells. Taken together, the transcriptional response to zinc and MTF-1 is specific for MRE-containing promoters.
FIG. 3.
Stimulation of transcription by zinc and MTF-1 is specific for MRE-containing promoters and can be inhibited by anti-MTF-1 antibodies. (A) In vitro transcription with different control promoters in 12 μg of BJA-B cell basal nuclear extract. The typical induction of the 4xMREd promoter (lanes 5 and 6) by a combination of zinc and complementing nuclear extract from human MTF-1-transfected HEK293 cells was not observed with three control promoters. C, control nuclear extract from untransfected HEK293 cells; MTF, complementing nuclear extract from human MTF-1-transfected HEK293 cells. (B) Zinc-responsive transcription is inhibited by anti-MTF-1 antibodies. The transcription reactions were performed in HeLa cell nuclear extract with the 4xMREd promoter as reporter. The typical activation by a combination of zinc and MTF-1 (lane 4) was not affected by addition of control serum (preimm., preimmune serum, lanes 5 and 6) but strongly affected by various antibody preparations reactive to mammalian MTF-1 (lanes 7 and 8, α-hN, anti-human MTF-1 N terminus; lanes 9 and 10, α-mN, anti-mouse MTF-1 N terminus; lanes 11 and 12, α-hC, anti-human MTF-1 C terminus). Anti-mouse and anti-human MTF-1 antibodies show cross-reactivity due to strong conservation of MTF-1 between the two species. 1x, 1 μl of anti-MTF-1 serum; 2x, 2 μl of anti-MTF-1 serum. MTF, complementing nuclear extract from HEK293 cells transfected with the human MTF-1 expression vector.
The specificity of the metal-responsive transcription reaction was confirmed with two further experiments. Upon addition of rabbit anti-MTF-1 antibodies, the transcriptional induction on both 4xMREd and the natural mouse MT-I promoter was strongly inhibited or completely abrogated (Fig. 3B and not shown). The same anti-MTF-1 antibodies had no effect on transcription from 4xOct-OVEC; moreover, an excess of binding site oligonucleotides for MTF-1 but not for Sp1 was also inhibitory (not shown).
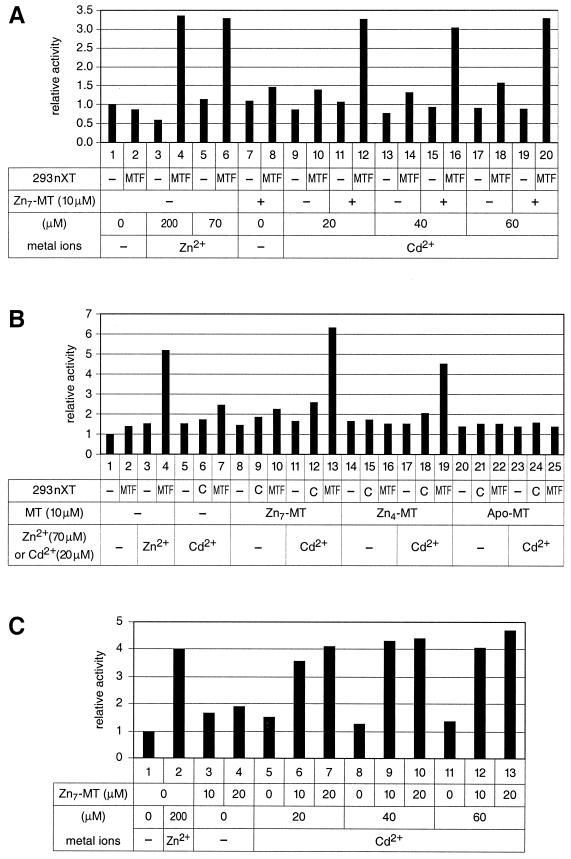
Promoter induction by cadmium and copper depends on the presence of zinc-saturated metallothionein (Zn7-MT).
Metallothionein gene transcription in vivo is readily activated not only by zinc but also by a number of other conditions, notably cadmium load. MTF-1 plays a key role in this process (26), although other transcription factors may contribute to this activation as well (3, 18, 36). By contrast, binding of MTF-1 to DNA in a cell-free system responds to zinc but not to any other heavy metal added (7, 80). This suggests that zinc can activate MTF-1 directly, whereas other heavy metals like cadmium and copper might exert their effect indirectly via zinc release from cellular components. It has also long been known that cadmium can displace zinc from metallothioneins due to its higher affinity to the latter (71).
We reasoned that complementation of our transcription reaction with purified metallothioneins might result in binding of cadmium to metallothionein with a concomitant release of zinc, which in turn would induce MTF-1 DNA binding and activation of transcription. This was indeed found to be the case. As seen with the synthetic 4xMREd promoter (Fig. 4A and 4B), addition of cadmium alone yielded poor, if any, stimulation, while cadmium in combination with zinc-saturated metallothionein (Zn7-MT) activated transcription to the same extent as if the system had been activated by zinc (Fig. 4A, lanes 12, 16, and 20; Fig. 4B, lane 13). Partially zinc-loaded metallothionein (Zn4-MT) and metal-deprived metallothionein (referred to as thionein or apo-MT) yielded reduced and no transcriptional activation, respectively (Fig. 4B). Metallothionein loaded with cadmium (Cd7-MT) failed to give any activation upon addition of cadmium (not shown). As another control, Zn7-MT did not have any influence on the response of the reporter gene to zinc (not shown).
FIG.4.
Transcriptional response to the toxic heavy metal cadmium depends on zinc-saturated metallothionein (Zn7-MT). (A) Transcription from the 4xMREd promoter in the presence of cadmium and Zn7-MT in HeLa cell basal nuclear extract. MTF, complementing nuclear extract from human MTF-1-transfected HEK293 cells. (B) Response of 4xMREd promoter to Zn4-MT and apo-MT (thionein). In vitro transcription was done in HeLa cell basal nuclear extract. C, control nuclear extract from untransfected HEK293 cells. MTF, complementing nuclear extract from human MTF-1-transfected HEK293 cells. (C) In vitro transcription with the mouse MT-I promoter in the presence of cadmium and Zn7-MT. The reactions were done in 32 μg of HeLa cell basal nuclear extract. All samples were supplemented with 20 μg of nuclear extract from HEK293 cells transfected with the human MTF-1 expression vector.
Similar to the synthetic 4xMREd, the genuine MT-I promoter was also inducible by cadmium in our cell-free system, again only in combination with Zn7-MT (Fig. 4C). To see whether the combination of Zn7-MT with cadmium or copper can also activate the DNA binding activity of MTF-1, a band shift was performed with nuclear miniextract from untransfected HEK293 cells with the MREd sequence as the probe. As seen before, MTF-1 but not Sp1 required elevated zinc concentrations for efficient DNA binding. Zn7-MT alone did not change this situation, while the combination with either cadmium or copper resulted in activation of MTF-1 in DNA binding (Fig. 5A). To test whether either metallothionein or cadmium exerted an unspecific stimulatory effect, the experiment was also performed with a reporter gene driven by an “octamer” promoter (4xOct-OVEC). As expected, this promoter did not respond to any of the inducers (not shown).
Copper is an essential component of many cellular proteins such as Cu/Zn-superoxide dismutase and tyrosinase, but its concentration has to be particularly well controlled due to its severe toxicity. Copper is also an inducer of metallothionein gene transcription (26, 45, 77, 80) and, similar to cadmium, has a particularly high affinity for metallothioneins (63, 71). We tested copper in our cell-free system and found that it behaved like cadmium; on its own it was deleterious for MTF-1 in both DNA binding and transcription assays, while in the presence of zinc-saturated metallothionein it strongly stimulated both DNA binding and transcription (Fig. 5).
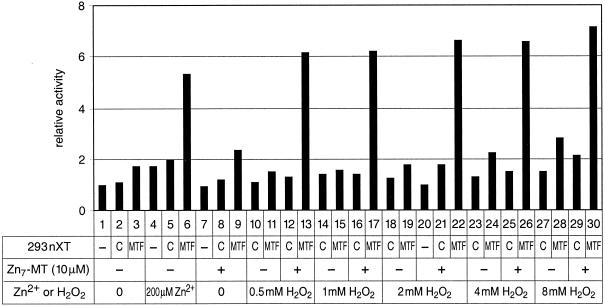
Promoter induction by hydrogen peroxide also depends on zinc-metallothionein.
Metallothionein gene transcription was shown to be induced in hepatoma cells by hydrogen peroxide (H2O2) (14). Also, primary mouse embryo fibroblasts with targeted disruption of MTF-1 are more sensitive to H2O2 than wild-type cells (24). Thus, we were interested to see whether H2O2 could also, in combination with zinc-loaded metallothionein, activate MTF-1-driven transcription in vitro. We found that H2O2 by itself did not produce a band shift with MTF-1, but in the presence of Zn7-MT the DNA binding activity of MTF-1 was restored (not shown). Similarly, H2O2 on its own also failed to induce transcription by MTF-1. Upon complementation of the reaction with zinc-saturated metallothionein (Zn7-MT), transcription from the MT-I promoter and the synthetic 4xMREd promoter was fully induced (Fig. 6 and not shown). As expected, transcription of the reference gene driven by the simian virus 40 enhancer (OVEC-REF) was not affected by either H2O2 or metallothionein.
FIG. 6.
In vitro transcription of mouse MT-I promoter also responds to hydrogen peroxide (H2O2). As a positive control, MTF-1 and zinc induced transcription (lane 6), as shown before. Transcription was also strongly induced by the combination of MTF-1, zinc-saturated metallothionein (Zn7-MT), and various concentrations of hydrogen peroxide (0.5 mM, 1 mM, 2 mM, 4 mM, and 8 mM, in lanes 13, 17, 22, 26, and 30, respectively). (−), basal reaction in BJA-B cell nuclear extract without complementing HEK293 extract; C, supplemented with nuclear extract from untransfected HEK293 cells; MTF, supplemented with nuclear extract from HEK293 cells previously transfected with the human MTF-1 expression vector.
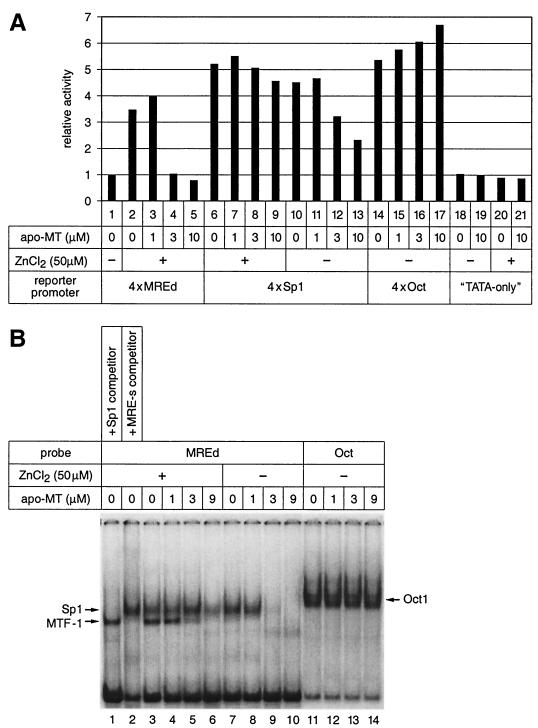
Apo-MT preferentially abrogates activation of MTF-1 by zinc.
Under the assumption that metallothionein can act as a zinc donor for MTF-1, and since metallothioneins are known target genes of MTF-1, we reasoned that de novo synthesis of thionein (apo-MT) would result in a deprivation of zinc and loss of MTF-1 binding and thus would counteract zinc-mediated activation of MTF-1 (50). This was tested with increasing amounts of apo-MT added to the transcription reaction. Indeed, MTF-1 turned out to be exquisitely sensitive to the presence of apo-MT (Fig. 7A): while 1 μM apo-MT did not significantly reduce transcription, 3 or 10 μM strongly reduced MTF-1-driven transcription down to the basal level even in the presence of 50 μM zinc, which is nominally in excess over the binding capacity of 3 μM apo-MT.
FIG. 7.
Apo-MT counteracts the activation of MTF-1 by zinc. (A) Apo-MT abrogates reporter activation by MTF-1 and zinc on the 4xMREd promoter. Four different promoter constructs as used in Fig. 3A were tested in our in vitro transcription system with 12 μg of BJA-B cell basal nuclear extract in the presence of increasing amounts of apo-MT. Complementing nuclear extract from human MTF-1-transfected HEK293 cells was included in all the reactions; 50 μM ZnCl2 was used where indicated. All reporter signals were quantified against the reference. (B) Apo-MT abolishes MTF-1 DNA binding activity in the presence of zinc. Band shifting was performed with nuclear extract from untransfected HEK293 cells. The MREd sequence was used as the probe to visualize the binding of Sp1 and MTF-1 proteins, and an octamer binding site (Oct) was used as the probe to show the binding of octamer factors. Where indicated, a 300-fold (5 pmol) excess of cold MRE-s or Sp1 binding site oligonucleotide was included as a competitor in the reaction.
Considering the results presented in Fig. 4, the inhibitory effect of apo-MT on MTF-1-driven transcription appears disproportionately high: 3 μM apo-MT, which is expected to bind merely 21 μM zinc, inhibited transcription in the presence of 50 μM zinc. On the other hand, 20 μM cadmium, which would be expected to release 20 μM zinc from Zn7-MT, could restore transcription. A possible explanation, which deserves further investigation, is that apo-MT is particularly efficient in removing zinc ions that are required for MTF-1's activity.
A promoter driven by the ubiquitous zinc finger transcription factor Sp1 was far less sensitive to apo-MT addition, and a promoter driven by the zinc-independent “octamer” transcription factor Oct1/2 (4xOct-OVEC) was completely refractory; if anything, the presence of apo-MT resulted in a slightly increased activity. Moreover, the TATA-only OVEC reporter activity was also not influenced by apo-MT, even though zinc is generally required for RNA polymerase II function. These effects are specific for apo-MT because addition of Zn7-MT instead of apo-MT did not change the zinc response at all (not shown). DNA binding of the three factors reflected their transcriptional activity: at increasing apo-MT concentrations, MTF-1 was the first to lose its DNA binding ability, Sp1 was more resistant, and DNA binding of Oct1 was not affected at all by apo-MT (Fig. 7B).
DISCUSSION
We have established a cell-free transcription system based on human cell nuclear extract that activates reporter gene transcription in response to the heavy metals zinc, cadmium, and copper and to hydrogen peroxide, similar to MTF-1-dependent induction of metallothionein gene transcription in vivo. Reporter constructs containing either a synthetic promoter with four tandem metal response elements (4xMREd) or the natural metallothionein-I promoter (MT-I) are poorly transcribed unless the transcription reactions are complemented with MTF-1-enriched nuclear extract and with elevated concentrations of zinc or, in case of cadmium, copper, and H2O2, with zinc-saturated metallothionein. The general zinc finger transcription factor Sp1 does not substitute for MTF-1 in this reaction (Fig. 1), nor can reporters with other promoters (including an Sp1-dependent or octamer factor-dependent promoter) respond to elevated zinc concentrations and MTF-1 (Fig. 3A). Interference with anti-MTF-1 antibodies (Fig. 3B) or competitor MRE oligonucleotides confirms the specificity of the observed transcriptional induction.
Our finding that induction by cadmium and other nonzinc inducers depends on the presence of zinc-loaded metallothionein may well be of physiological relevance because any cell under nonstress conditions contains a basal amount of metallothioneins and other zinc-binding components. In spite of a particularly high affinity for cadmium, copper, and some other heavy metals, most intracellular metallothioneins are normally saturated with the highly abundant zinc (19, 66, 70). We reasoned that cadmium would bind with high avidity to metallothioneins, thereby liberating zinc and making it available for the transcription factor MTF-1. Indeed, such an effect was seen both with the synthetic 4xMREd promoter and with a metallothionein-I promoter but not with an octamer factor-dependent control promoter (Fig. 4).
Electrophoretic mobility shift assay confirmed the situation observed with transcription. In extracts of cell nuclei, MTF-1 but not Sp1 binding depended on elevated zinc concentration. Cadmium and copper eliminated the band shifts of both Sp1 and MTF-1, while in the presence of zinc-saturated metallothionein, the same metals in fact induced DNA binding (Fig. 5A). MTF-1 and metallothioneins have been implicated before in the handling of oxidative stress, notably in response to hydrogen peroxide (14, 16, 24). It was therefore comforting to see that treatment with H2O2, similar to treatment with toxic heavy metals, induces transcription only in the presence of zinc-loaded metallothionein. These data suggest that metallothioneins are not merely passive targets of MTF-1 but may rather contribute to the regulation of its activity.
Another aspect concerns the downregulation of MTF-1 following its induction by zinc or other stimuli, including cadmium, copper, and H2O2. Metallothioneins are likely to play a role in this process (50), since thioneins, the metal-free form of metallothioneins, bind zinc with high affinity. Furthermore, it has been shown that, presumably by removing zinc, apo-MT is able to inhibit DNA binding of the C2H2-type zinc finger proteins Sp1, TFIIIA, and Tramtrack (56, 78, 79) and other zinc finger transcription factors such as estrogen receptor (10).
To see whether apo-MT can also inhibit MTF-1, we compared its activity with that of Sp1, another zinc finger transcription factor harboring the same type of metal-binding motifs (C2H2-type zinc fingers), in the presence of apo-MT. Since RNA polymerase II also needs zinc as a cofactor, we chose the zinc-independent octamer promoter as well as the TATA-only OVEC, which contains only the TATA box and initiation site, as controls for the influence of apo-MT on the basal transcription machinery. Consistent with previous findings (78), RNA polymerase II maintained its activity even at high concentrations of apo-MT, while Sp1 was sensitive only to relatively high apo-MT concentrations. Among these three zinc-binding proteins, MTF-1 is clearly the most sensitive to zinc deprivation, while the zinc of RNA polymerase II is apparently not available to apo-MT (Fig. 7). The preferential inhibition of MTF-1 activity by apo-MT shown here is the first experimental evidence for a negative-feedback loop involving newly synthesized thionein.
Our studies imply that metallothioneins are crucial components in the modulation of MTF-1 activity, which has independently been suggested before (2, 32, 38, 50). The question arises whether the mechanism of MTF-1 activation by zinc released from metallothioneins is also relevant in vivo, e.g., whether the steady-state concentration of zinc-saturated metallothionein is sufficiently high for such a mechanism. Conservative estimates arrive at approximately 100,000 metallothioneins per cell, each of which can bind seven zinc (or cadmium) ions, a large excess over the approximately 10,000 molecules of MTF-1, in which only one or two of the six zinc fingers are critically involved in conditional zinc binding (4, 11). Besides, cellular zinc-binding proteins other than metallothioneins may well contribute to the effect, as long as they preferentially release zinc upon exposure to cadmium, copper, or hydrogen peroxide.
Some in vivo experiments pertinent to this model of MTF-1 activation were performed by Palmiter (50), who found that induction with copper and cadmium was compromised in zinc-starved cells. We have also attempted to test the model of MTF-1 activation in vivo, with fibroblast-type embryonal cells with a targeted disruption of both stress-inducible metallothionein genes (MT-I and -II) derived from the appropriate knockout mouse strain (39). We found weak induction of an MTF-1-dependent reporter in response to both zinc and cadmium. Full inducibility of the reporter gene by cadmium and zinc was restored by transfection of a human MTF-1 expression plasmid, which was unexpected at first glance because these metallothionein knockout cells already expressed MTF-1 of their own. However, further analysis by semiquantitative RT-PCR revealed a low but significant mRNA expression of the two remaining metallothionein mRNAs (MT-III and MT-IV). These metallothioneins also contain MRE-like motifs in their promoter regions but are usually referred to as noninducible and tissue specific, with preferential expression in the central nervous system and squamous epithelia, respectively (33, 51, 53). The expression of these extra metallothioneins precluded a clear analysis, in particular because the expression of MT-IV was even boosted by transfection of MTF-1 (not shown). Taken together, the in vivo experiments do not prove the model but are compatible with it.
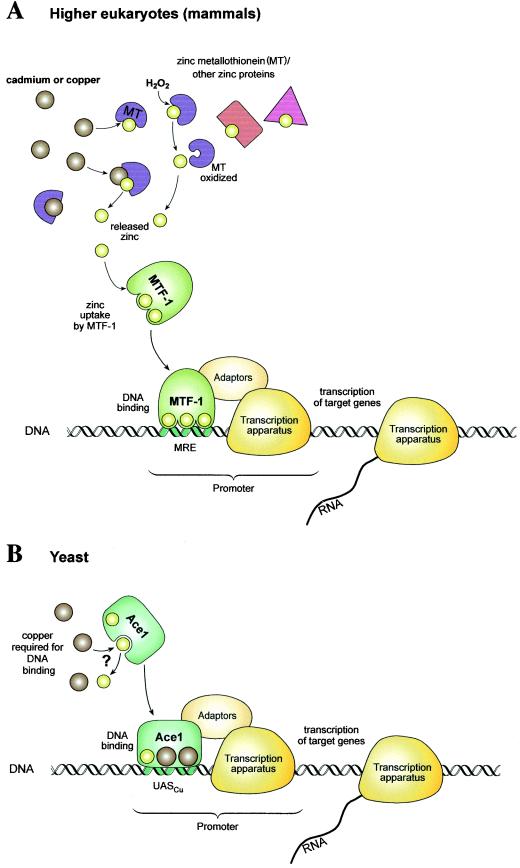
In this context, it is worth mentioning that Saccharomyces cerevisiae has evolved its own metal transcription factor/metallothionein system which is similar yet unrelated to the one relying on MTF-1, which is predominant from insects to humans. The yeast transcriptional activator AceI acts as a copper sensor which is unable to bind DNA efficiently in normal growth medium. Upon copper load, a tetracopper cluster forms in the DNA binding domain of AceI, which is a prerequisite for promoter recognition at the copper metallothionein gene CUP1 (7a, 13, 20, 52, 76). AceI also binds zinc, some of which may be displaced by copper. The system is thus conceptually similar to the one presented here for MTF-1, with the important difference that in S. cerevisiae, copper binding occurs directly to the transcription factor itself rather than to a separate protein (metallothionein). A possible drawback of this seemingly simpler system may be an increased risk of oxidative DNA damage due to the close proximity of redox-active copper (Fig. 8B). We note that also in the case of iron, mammalian cells avoid the presence of a redox-active metal in a DNA binding transcription factor; transferrin receptor and ferritin expression is regulated by posttranscriptional mechanisms (25, 57).
FIG.8.
Heavy metal-responsive transcription in mammals and S. cerevisiae. (A) Model for MTF-1 activation by cadmium, copper, and hydrogen peroxide (H2O2) via zinc-loaded metallothionein (Zn7-MT). Cadmium and copper bind to metallothionein (MT) with a much higher affinity than zinc, but due to the great abundance of the latter, the majority of metallothionein under physiological conditions is present as Zn7-MT. Upon cadmium or copper loading, zinc is released from metallothionein and presumably from other cellular proteins and allows zinc saturation of MTF-1, which requires a higher zinc concentration for DNA binding than other typical zinc finger proteins such as Sp1. H2O2 also induces zinc release from metallothionein via oxidation of sulfhydryl groups. In contrast to cadmium and other agents, which are postulated to activate MTF-1 indirectly, elevated zinc concentrations activate MTF-1 directly, i.e., without the need for zinc release from other proteins. The scheme is not meant to be drawn to scale or numerically accurate (e.g., in reality, seven to eight zinc ions bind to one metallothionein molecule, and MTF-1 has six zinc fingers). Furthermore, aspects of MTF-1 regulation such as nucleocytoplasmic shuttling and phosphorylation are not addressed in this model, which is proposed to depict the primordial mechanism of heavy metal-induced gene activation in higher eukaryotes. (B) Model of the regulation of metallothionein transcription in S. cerevisiae. In S. cerevisiae, copper metallothionein (CUP1) gene transcription is controlled by the copper/zinc transcription factor AceI (52, 65, 76). In the absence of elevated copper concentrations, some or all metal binding sites of AceI are filled with zinc. Upon copper loading, four copper ions form a tetracopper cluster, thereby possibly replacing some zinc. Of the four copper(I) ions, only two are shown schematically. This induces an allosteric conformation change which permits DNA binding at control sequences (UASCu) and activation of metallothionein transcription.
Also in other aspects, the mammalian MTF-1 system appears more complex than the yeast AceI /CUP1 system: Recently, nuclear-cytoplasmic shuttling of MTF-1 was found to be regulated by heavy metal and other stress conditions (59, 62, 67), and phosphorylation, presumably by more than one kinase, contributes to the activity of MTF-1 (35, 58). MTF-1 has also been implicated in the hypoxic response via activation of the angiogenic placenta growth factor gene (23, 46). Finally, MTF-1 was recently reported to bind to the mRNA for ribosomal protein S25 under conditions of amino acid starvation, implying a role in stress response and apoptotic cell death upon prolonged starvation (1). MTF-1 no doubt has multiple roles in different stress-related pathways, and we are only beginning to understand the many facets of this essential transcription factor. Even though the regulation of MTF-1 is known to be rather complex, we suggest that activation of MTF-1 by zinc liberated from metallothioneins and other zinc-binding components and subsequent inactivation of MTF-1 by newly synthesized apo-MT are evolutionarily ancient mechanisms for regulating MTF-1 activity upon heavy metal and oxidative stress (Fig. 8A). Phosphorylation and nucleocytoplasmic shuttling, which are also known to affect MTF-1 function, might be more recent control mechanisms.
Acknowledgments
We are indebted to David N. Arnosti for preparing HeLa and BJA-B nuclear extracts. We also thank Bernd Roschitzki for providing apo-MT and for verifying zinc and cadmium concentrations, Fritz Ochsenbein for graphic artwork, and E. Randal Hofmann, David N. Arnosti, and David P. Giedroc for critical reading of the manuscript.
This work was supported by the Schweizerischer Nationalfonds and by the Kanton Zürich.
REFERENCES
- 1.Adilakshmi, T., and R. O. Laine. 2002. Ribosomal protein S25 mRNA partners with MTF-1 and La to provide a p53-mediated mechanism for survival or death. J. Biol. Chem. 277:4147-4151. [DOI] [PubMed]
- 2.Andrews, G. K. 2000. Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem. Pharmacol. 59:95-104. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, G. K., D. K. Lee, R. Ravindra, P. Lichtlen, M. Sirito, M. Sawadogo, and W. Schaffner. 2001. The transcription factors MTF-1 and USF1 cooperate to regulate mouse metallothionein-I expression in response to the essential metal zinc in visceral endoderm cells during early development. EMBO J. 20:1114-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apuy, J. L., X. Chen, D. H. Russell, T. O. Baldwin, and D. P. Giedroc. 2001. Ratiometric pulsed alkylation/mass spectrometry of the cysteine pairs in individual zinc fingers of MRE-binding transcription factor-1 (MTF-1) as a probe of zinc chelate stability. Biochemistry 40:15164-15175. [DOI] [PubMed] [Google Scholar]
- 5.Arnosti, D. N., A. Merino, D. Reinberg, and W. Schaffner. 1993. Oct-2 facilitates functional preinitiation complex assembly and is continuously required at the promoter for multiple rounds of transcription. EMBO J. 12:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird, A. J., H. Zhao, H. Luo, L. T. Jensen, C. Srinivasan, M. Evans-Galea, D. R. Winge, and D. J. Eide. 2000. A dual role for zinc fingers in both DNA binding and zinc sensing by the Zap1 transcriptional activator. EMBO J. 19:3704-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bittel, D., T. Dalton, S. L. Samson, L. Gedamu, and G. K. Andrews. 1998. The DNA binding activity of metal response element-binding transcription factor-1 is activated in vivo and in vitro by zinc, but not by other transition metals. J. Biol. Chem. 273:7127-7133. [DOI] [PubMed] [Google Scholar]
- 7a.Brown, K. R., G. l. Keller, I. J. Pickering, H. H. Harris, G. N. George, and D. R. Woinge. 2002. Structures of cuprous-thiolate clusters of the Mac1 and Ace1 transcriptional activators. Biochemistry 41:6469-6476. [DOI] [PubMed] [Google Scholar]
- 8.Brugnera, E., O. Georgiev, F. Radtke, R. Heuchel, E. Baker, G. R. Sutherland, and W. Schaffner. 1994. Cloning, chromosomal mapping and characterization of the human metal-regulatory transcription factor MTF-1. Nucleic Acids Res. 22:3167-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buhler, R. H., and J. H. Kagi. 1974. Human hepatic metallothioneins. FEBS Lett. 39:229-234. [DOI] [PubMed] [Google Scholar]
- 10.Cano-Gauci, D. F., and B. Sarkar. 1996. Reversible zinc exchange between metallothionein and the estrogen receptor zinc finger. FEBS Lett. 386:1-4. [DOI] [PubMed] [Google Scholar]
- 11.Chen, X., M. Chu, and D. P. Giedroc. 1999. MRE-Binding transcription factor-1: weak zinc-binding finger domains 5 and 6 modulate the structure, affinity, and specificity of the metal-response element complex. Biochemistry 38:12915-12925. [DOI] [PubMed] [Google Scholar]
- 12.Culotta, V. C., and D. H. Hamer. 1989. Fine mapping of a mouse metallothionein gene metal response element. Mol. Cell. Biol. 9:1376-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Culotta, V. C., T. Hsu, S. Hu, P. Furst, and D. Hamer. 1989. Copper and the ACE1 regulatory protein reversibly induce yeast metallothionein gene transcription in a mouse extract. Proc. Natl. Acad. Sci. 86:8377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalton, T., R. D. Palmiter, and G. K. Andrews. 1994. Transcriptional induction of the mouse metallothionein-I gene in hydrogen peroxide-treated Hepa cells involves a composite major late transcription factor/antioxidant response element and metal response promoter elements. Nucleic Acids Res. 22:5016-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalton, T. P., D. Bittel, and G. K. Andrews. 1997. Reversible activation of mouse metal response element-binding transcription factor 1 DNA binding involves zinc interaction with the zinc finger domain. Mol. Cell. Biol. 17:2781-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalton, T. P., Q. Li, D. Bittel, L. Liang, and G. K. Andrews. 1996. Oxidative stress activates metal-responsive transcription factor-1 binding activity. Occupancy in vivo of metal response elements in the metallothionein-I gene promoter. J. Biol. Chem. 271:26233-26241. [DOI] [PubMed] [Google Scholar]
- 17.Daniels, P. J., D. Bittel, I. V. Smirnova, D. R. Winge, and G. K. Andrews. 2002. Mammalian metal response element-binding transcription factor-1 functions as a zinc sensor in yeast, but not as a sensor of cadmium or oxidative stress. Nucleic Acids Res. 30:3130-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datta, P. K., and S. T. Jacob. 1997. Activation of the metallothionein-I gene promoter in response to cadmium and USF in vitro. Biochem. Biophys. Res. Commun. 230:159-163. [DOI] [PubMed] [Google Scholar]
- 19.Davis, S. R., and R. J. Cousins. 2000. Metallothionein expression in animals: a physiological perspective on function. J. Nutr. 130:1085-1088. [DOI] [PubMed] [Google Scholar]
- 20.Furst, P., S. Hu, R. Hackett, and D. Hamer. 1988. Copper activates metallothionein gene transcription by altering the conformation of a specific DNA binding protein. Cell 55:705-717. [DOI] [PubMed] [Google Scholar]
- 21.Giedroc, D. P., X. Chen, and J. L. Apuy. 2001. Metal response element (MRE)-binding transcription factor-1 (MTF-1): structure, function, and regulation. Antioxid. Redox Signal. 3:577-596. [DOI] [PubMed] [Google Scholar]
- 22.Giedroc, D. P., X. Chen, M. A. Pennella, and A. C. LiWang. 2001. Conformational heterogeneity in the C-terminal zinc fingers of human MTF-1: an NMR and zinc-binding study. J. Biol. Chem. 276:42322-42332. [DOI] [PubMed] [Google Scholar]
- 23.Green, C. J., P. Lichtlen, N. T. Huynh, M. Yanovsky, K. R. Laderoute, W. Schaffner, and B. J. Murphy. 2001. Placenta growth factor gene expression is induced by hypoxia in fibroblasts: a central role for metal transcription factor-1. Cancer Res. 61:2696-2703. [PubMed] [Google Scholar]
- 24.Gunes, C., R. Heuchel, O. Georgiev, K. H. Muller, P. Lichtlen, H. Bluthmann, S. Marino, A. Aguzzi, and W. Schaffner. 1998. Embryonic lethality and liver degeneration in mice lacking the metal-responsive transcriptional activator MTF-1. EMBO J. 17:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hentze, M. W., and L. C. Kuhn. 1996. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. 93:8175-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heuchel, R., F. Radtke, O. Georgiev, G. Stark, M. Aguet, and W. Schaffner. 1994. The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression. EMBO J. 13:2870-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hidalgo, J., M. Aschner, P. Zatta, and M. Vasak. 2001. Roles of the metallothionein family of proteins in the central nervous system. Brain Res. Bull. 55:133-145. [DOI] [PubMed] [Google Scholar]
- 28.Hider, R. C., D. Bittel, and G. K. Andrews. 1999. Competition between iron(III)-selective chelators and zinc finger domains for zinc(II). Biochem. Pharmacol. 57:1031-1035. [DOI] [PubMed] [Google Scholar]
- 29.Jacob, S. T., K. Ghoshal, and J. F. Sheridan. 1999. Induction of metallothionein by stress and its molecular mechanisms. Gene Expr. 7:301-310. [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang, H., P. J. Daniels, and G. K. Andrews. 2003. Putative zinc-sensing zinc fingers of metal response element-binding transcription factor-1 stabilize a metal-dependent chromatin complex on the endogenous metallothionein-I promoter. J. Biol. Chem. 278:30394-30402. [DOI] [PubMed] [Google Scholar]
- 31.Jimenez, I., M. Gotteland, A. Zarzuelo, R. Uauy, and H. Speisky. 1997. Loss of the metal binding properties of metallothionein induced by hydrogen peroxide and free radicals. Toxicology 120:37-46. [DOI] [PubMed] [Google Scholar]
- 32.Kimura, T., N. Itoh, M. Takehara, I. Oguro, J. Ishizaki, T. Nakanishi, and K. Tanaka. 2002. MRE-binding transcription factor-1 is activated during endotoxemia: a central role for metallothionein. Toxicol. Lett. 129:77-84. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi, H., Y. Uchida, Y. Ihara, K. Nakajima, S. Kohsaka, T. Miyatake, and S. Tsuji. 1993. Molecular cloning of rat growth inhibitory factor cDNA and the expression in the central nervous system. Brain Res. Mol. Brain Res. 19:188-194. [DOI] [PubMed] [Google Scholar]
- 34.Langmade, S. J., R. Ravindra, P. J. Daniels, and G. K. Andrews. 2000. The transcription factor MTF-1 mediates metal regulation of the mouse ZnT1 gene. J. Biol. Chem. 275:34803-34809. [DOI] [PubMed] [Google Scholar]
- 35.LaRochelle, O., V. Gagne, J. Charron, J. W. Soh, and C. Seguin. 2001. Phosphorylation is involved in the activation of metal-regulatory transcription factor 1 in response to metal ions. J. Biol. Chem. 276:41879-41888. [DOI] [PubMed] [Google Scholar]
- 36.Li, Q., N. Hu, M. A. Daggett, W. A. Chu, D. Bittel, J. A. Johnson, and G. K. Andrews. 1998. Participation of upstream stimulator factor (USF) in cadmium-induction of the mouse metallothionein-I gene. Nucleic Acids Res. 26:5182-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lichtlen, P., Y. Wang, T. Belser, O. Georgiev, U. Certa, R. Sack, and W. Schaffner. 2001. Target gene search for the metal-responsive transcription factor MTF-1. Nucleic Acids Res. 29:1514-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lichtlen, P., and W. Schaffner. 2001. Putting its fingers on stressful situations: the heavy metal-regulatory transcription factor MTF-1. Bioassays 23:1010-1017. [DOI] [PubMed] [Google Scholar]
- 39.Masters, B. A., E. J. Kelly, C. J. Quaife, R. L. Brinster, and R. D. Palmiter. 1994. Targeted disruption of metallothionein I and II genes increases sensitivity to cadmium. Proc. Natl. Acad. Sci. 91:584-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuo, K., J. Silke, O. Georgiev, P. Marti, N. Giovannini, and D. Rungger. 1998. An embryonic demethylation mechanism involving binding of transcription factors to replicating DNA. EMBO J. 17:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller, P. R., S. J. Salser, and B. Wold. 1988. Constitutive and metal-inducible protein:DNA interactions at the mouse metallothionein I promoter examined by in vivo and in vitro footprinting. Genes Dev. 2:412-427. [DOI] [PubMed] [Google Scholar]
- 42.Mueller-Storm, H. P., J. M. Sogo, and W. Schaffner. 1989. An enhancer stimulates transcription in trans when attached to the promoter via a protein bridge. Cell 58:767-777. [DOI] [PubMed] [Google Scholar]
- 43.Muller, H. P., Brugnera, E., Georgiev, O., Badzong, M., Müller, K. H., and Schaffner, W. 1995. Analysis of the heavy metal-responsive transcription factor MTF-1 from human and mouse. Somat. Cell Mol. Genet. 21:289-297. [DOI] [PubMed] [Google Scholar]
- 44.Muller, M. M., S. Ruppert, W. Schaffner, and P. Matthias. 1988. A cloned octamer transcription factor stimulates transcription from lymphoid-specific promoters in non-B cells. Nature 336:544-551. [DOI] [PubMed] [Google Scholar]
- 45.Murata, M., P. Gong, K. Suzuki, and S. Koizumi. 1999. Differential metal response and regulation of human heavy metal-inducible genes. J. Cell Physiol. 180:105-113. [DOI] [PubMed] [Google Scholar]
- 46.Murphy, B. J., G. K. Andrews, D. Bittel, D. J. Discher, J. McCue, C. J. Green, M. Yanovsky, A. Giaccia, R. M. Sutherland, K. R. Laderoute, and K. A. Webster. 1999. Activation of metallothionein gene expression by hypoxia involves metal response elements and metal transcription factor-1. Cancer Res. 59:1315-1322. [PubMed] [Google Scholar]
- 47.Ogra, Y., K. Suzuki, P. Gong, F. Otsuka, and S. Koizumi. 2001. Negative regulatory role of Sp1 in metal responsive element-mediated transcriptional activation. J. Biol. Chem. 276:16534-16539. [DOI] [PubMed] [Google Scholar]
- 48.Onosaka, S., K. S. Min, C. Fukuhara, K. Tanaka, S. Tashiro, I. Shimizu, M. Furuta, T. Yasutomi, K. Kobashi, and K. Yamamoto. 1986. Concentrations of metallothionein and metals in malignant and non-malignant tissues in human liver. Toxicology 38:261-268. [DOI] [PubMed] [Google Scholar]
- 49.Palmiter, R. D. 1998. The elusive function of metallothioneins. Proc. Natl. Acad. Sci. 95:8428-8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmiter, R. D. 1994. Regulation of metallothionein genes by heavy metals appears to be mediated by a zinc-sensitive inhibitor that interacts with a constitutively active transcription factor, MTF-1. Proc. Natl. Acad. Sci. 91:1219-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmiter, R. D., S. D. Findley, T. E. Whitmore, and D. M. Durnam. 1992. MT-III, a brain-specific member of the metallothionein gene family. Proc. Natl. Acad. Sci. 89:6333-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pena, M. M., K. A. Koch, and D. J. Thiele. 1998. Dynamic regulation of copper uptake and detoxification genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:2514-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quaife, C. J., S. D. Findley, J. C. Erickson, G. J. Froelick, E. J. Kelly, B. P. Zambrowicz, and R. D. Palmiter. 1994. Induction of a new metallothionein isoform (MT-IV) occurs during differentiation of stratified squamous epithelia. Biochemistry 33:7250-7259. [DOI] [PubMed] [Google Scholar]
- 54.Quesada, A. R., R. W. Byrnes, S. O. Krezoski, and D. H. Petering. 1996. Direct reaction of H2O2 with sulfhydryl groups in HL-60 cells: zinc-metallothionein and other sites. Arch. Biochem. Biophys. 334:241-250. [DOI] [PubMed] [Google Scholar]
- 55.Radtke, F., R. Heuchel, O. Georgiev, M. Hergersberg, M. Gariglio, Z. Dembic, and W. Schaffner. 1993. Cloned transcription factor MTF-1 activates the mouse metallothionein I promoter. EMBO J. 12:1355-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roesijadi, G., R. Bogumil, M. Vasak, and J. H. Kagi. 1998. Modulation of DNA binding of a tramtrack zinc finger peptide by the metallothionein-thionein conjugate pair. J. Biol. Chem. 273:17425-17432. [DOI] [PubMed] [Google Scholar]
- 57.Rouault, T., and R. Klausner. 1997. Regulation of iron metabolism in eukaryotes. Curr. Top. Cell Regul. 35:1-19. [DOI] [PubMed] [Google Scholar]
- 58.Saydam, N., T. K. Adams, F. Steiner, W. Schaffner, and J. H. Freedman. 2002. Regulation of metallothionein transcription by the metal-responsive transcription factor MTF-1: identification of signal transduction cascades that control metal-inducible transcription. J. Biol. Chem. 277:20438-20445. [DOI] [PubMed] [Google Scholar]
- 59.Saydam, N., O. Georgiev, M. Y. Nakano, U. F. Greber, and W. Schaffner. 2001. Nucleo-cytoplasmic trafficking of metal-regulatory transcription factor 1 is regulated by diverse stress signals. J. Biol. Chem. 276:25487-25495. [DOI] [PubMed] [Google Scholar]
- 60.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with mini-extracts prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Searle, P. F. 1990. Zinc dependent binding of a liver nuclear factor to metal response element MRE-a of the mouse metallothionein-I gene and variant sequences. Nucleic Acids Res. 18:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smirnova, I. V., D. C. Bittel, R. Ravindra, H. Jiang, G. K. Andrews. 2000. Zinc and cadmium can promote rapid nuclear translocation of metal response element-binding transcription factor-1. J. Biol. Chem. 275:9377-9384. [DOI] [PubMed] [Google Scholar]
- 63.Stillman, M. J. 1995. Metallothioneins. Coord. Chem. Rev. 144:461-511. [Google Scholar]
- 64.Stuart, G. W., P. F. Searle, and R. D. Palmiter. 1985. Identification of multiple metal regulatory elements in mouse metallothionein-I promoter by assaying synthetic sequences. Nature 317:828-831. [DOI] [PubMed] [Google Scholar]
- 65.Thiele, D. J. 1988. ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene. Mol. Cell. Biol. 8:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vallee, B. L., and K. H. Falchuk. 1993. The biochemical basis of zinc physiology. Physiol. Rev. 73:79-118. [DOI] [PubMed] [Google Scholar]
- 67.Vanacore, R. M., J. D. Eskew, P. J. Morales, L. Sung, and A. Smith. 2000. Role for copper in transient oxidation and nuclear translocation of MTF-1, but not of NF-kappa B, by the heme-hemopexin transport system. Antioxid. Redox Signal. 2:739-752. [DOI] [PubMed] [Google Scholar]
- 68.Vasak, M. 1991. Metal removal and substitution in vertebrate and invertebrate metallothioneins. Methods Enzymol. 205:452-458. [DOI] [PubMed] [Google Scholar]
- 69.Vasak, M. 1991. Standard isolation procedure for metallothionein. Methods Enzymol. 205:41-44. [DOI] [PubMed] [Google Scholar]
- 70.Vasak, M., and D. W. Hasler. 2000. Metallothioneins: new functional and structural insights. Curr. Opi. Chem. Biol. 4:177-183. [DOI] [PubMed] [Google Scholar]
- 71.Vasak, M., and J. H. Kagi. 1994. Metallothioneins, vol. 4. Wiley and Sons, Chichester, England.
- 72.Weaver, R. F., and C. Weissmann. 1979. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5′ termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 7:1175-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Westin, G., T. Gerster, M. M. Müller, G. Schaffner, and W. Schaffner. 1987. OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 15:6787-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Westin, G., and W. Schaffner. 1988. Heavy metal ions in transcription factors from HeLa cells: Sp1, but not octamer transcription factor requires zinc for DNA binding and for activator function. Nucleic Acids Res. 16:5771-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Westin, G., and W. Schaffner. 1988. A zinc-responsive factor interacts with a metal-regulated enhancer element (MRE) of the mouse metallothionein-I gene. EMBO J. 7:3763-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Winge, D. R. 1998. Copper-regulatory domain involved in gene expression. Prog. Nucleic Acid Res. Mol. Biol. 58:165-195. [DOI] [PubMed] [Google Scholar]
- 77.Yagle, M. K., and R. D. Palmiter. 1985. Coordinate regulation of mouse metallothionein I and II genes by heavy metals and glucocorticoids. Mol. Cell. Biol. 5:291-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zeng, J., R. Heuchel, W. Schaffner, and J. H. Kagi. 1991. Thionein (apometallothionein) can modulate DNA binding and transcription activation by zinc finger containing factor Sp1. FEBS Lett. 279:310-312. [DOI] [PubMed] [Google Scholar]
- 79.Zeng, J., B. L. Vallee, and J. H. Kagi. 1991. Zinc transfer from transcription factor IIIA fingers to thionein clusters. Proc. Natl. Acad. Sci. 88:9984-9988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang, B., D. Egli, O. Georgiev, and W. Schaffner. 2001. The Drosophila homolog of mammalian zinc finger factor MTF-1 activates transcription in response to heavy metals. Mol. Cell. Biol. 21:4505-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]