Abstract
Microtubules are polarized polymers that exhibit dynamic instability, with alternating phases of elongation and shortening, particularly at the more dynamic plus-end. Microtubule plus-end tracking proteins (+TIPs) localize to and track with growing microtubule plus-ends in the cell. +TIPs regulate microtubule dynamics and mediate interactions with other cellular components. The molecular mechanisms responsible for the +TIP tracking activity are not well understood, however. We reconstituted the +TIP tracking of mammalian proteins EB1 and CLIP-170 in vitro at single-molecule resolution using time-lapse total internal reflection fluorescence microscopy. We found that EB1 is capable of dynamically tracking growing microtubule plus-ends. Our single-molecule studies demonstrate that EB1 exchanges rapidly at microtubule plus-ends with a dwell time of <1 s, indicating that single EB1 molecules go through multiple rounds of binding and dissociation during microtubule polymerization. CLIP-170 exhibits lattice diffusion and fails to selectively track microtubule ends in the absence of EB1; the addition of EB1 is both necessary and sufficient to mediate plus-end tracking by CLIP-170. Single-molecule analysis of the CLIP-170–EB1 complex also indicates a short dwell time at growing plus-ends, an observation inconsistent with the copolymerization of this complex with tubulin for plus-end-specific localization. GTP hydrolysis is required for +TIP tracking, because end-specificity is lost when tubulin is polymerized in the presence of guanosine 5′-[α,β-methylene]triphosphate (GMPCPP). Together, our data provide insight into the mechanisms driving plus-end tracking by mammalian +TIPs and suggest that EB1 specifically recognizes the distinct lattice structure at the growing microtubule end.
Keywords: +TIP, single molecule, total internal reflection fluorescence (TIRF) microscopy, dynamic instability
Microtubules are highly dynamic polymers of tubulin that are essential for various fundamental processes within eukaryotic cells, including vesicle transport, cell division, and cell motility. In most mammalian cells, microtubules are nucleated at the microtubule organizing center (MTOC), where their minus-ends may remain stably anchored (1). Microtubule plus-ends growing out from the MTOC are highly dynamic, undergoing stochastic switching between growing and shortening phases (2). These dynamics allow microtubule plus-ends to explore the cellular environment. A “search-and-capture” model has been proposed in which the randomly directed outgrowth of a microtubule from the MTOC may lead to a chance encounter with a binding target, such as an organelle or the cell cortex (3). Interaction of the microtubule plus-end with a target protein may lead to preferential stabilization of end dynamics; the transiently stabilized microtubule may then serve as a marker for the establishment of cell polarity or act to stabilize a cellular protrusion or extension (4). Stabilized microtubules also may serve as preferential tracks for intracellular transport, allowing localized delivery within the cell (5). Locally captured microtubules may play a more active role, transmitting a force between the cell cortex and the MTOC. This generation of force may further facilitate cell polarization (6). Thus, spatial and temporal regulation of microtubule plus-end dynamics is critical for controlling microtubule architecture and function in the cell.
Cellular studies have revealed a specialized class of microtubule-binding proteins, known as plus-end tracking proteins (+TIPs), that accumulate at and track with growing microtubule plus-ends (7). These proteins participate in various processes, including regulation of microtubule dynamics, interactions between microtubules and other cellular structures, and delivery of signaling molecules (8). The cytoplasmic linker protein-170 (CLIP-170) was the first +TIP identified (9, 10). Subsequently, numerous structurally unrelated +TIP proteins have been identified that are evolutionarily conserved in most eukaryotes. Genetic and biochemical studies have revealed that many of these proteins interact with one another and may form higher-order plus-end complexes (11). One member in particular, the end-binding 1 (EB1) protein, interacts directly with most other known +TIPs and, consequently, has been proposed to form the core of the microtubule plus-end complex (8, 12).
Most +TIPs can directly bind to microtubules, but the molecular basis for their dynamic plus-end localization remains unresolved. The copolymerization model for +TIP localization suggests that +TIPs copolymerize with tubulin and thus incorporate preferentially at growing microtubule plus-ends (13–17). Alternatively, the end-recognition model posits that +TIPs have an increased affinity for the microtubule plus-end because of its distinct structural and/or biochemical state (18, 19).
To dissect the mechanisms underlying plus-end tracking, we have developed a cell-free in vitro reconstitution system to observe mammalian EB1 and CLIP-170 proteins and their interactions with microtubules. Using time-lapse total internal reflection fluorescence (TIRF) microscopy, we found that the mammalian EB1 can dynamically track growing microtubule plus-ends. Localization of CLIP-170 specifically to the plus-end requires the presence of mammalian EB1; in the absence of EB1, CLIP-170 moves diffusively along the microtubule lattice. Single-molecule analysis indicates that the CLIP-170–EB1 complex undergoes multiple cycles of binding and dissociation from growing microtubule ends, an observation inconsistent with a copolymerization model. We also found that the plus-end specificity of the CLIP-170-EB1 complex requires GTP hydrolysis. Together, these observations provide new insight into the dynamic and specific localization of these proteins seen in vivo and indicate that EB1 binds preferentially to a distinct conformation at growing microtubule plus-ends.
Results
EB1 Targets Growing Microtubule Plus-Ends Independently of Other Proteins.
To study the +TIP activity of human EB1 protein, we fluorescently tagged recombinant EB1 protein with Alexa 488. In vitro binding experiments demonstrated that the Alexa 488-tagged EB1 protein bound to microtubules to a similar extent as untagged recombinant protein [supporting information (SI) Fig. S1A].
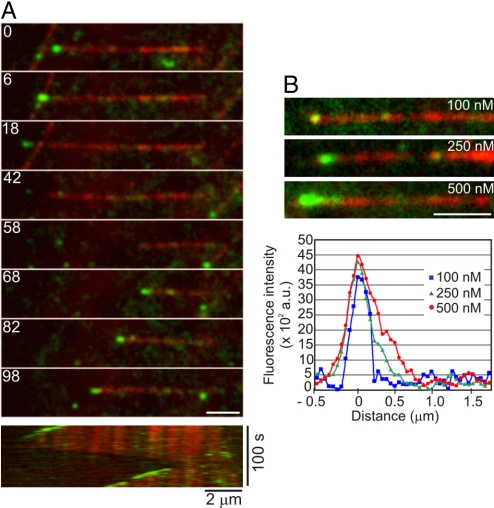
Reconstitution experiments with dynamic microtubules and EB1-Alexa 488 using TIRF microscopy demonstrated that EB1-Alexa 488 alone was capable of +TIP activity. In these experiments, EB1–Alexa 488 labeled all growing microtubule plus-ends; in contrast, we observed no EB1–Alexa 488 labeling at depolymerizing microtubule ends (Fig. 1 and Movie S1). We did not reproducibly detect EB1-Alexa 488 labeling of the more slowly growing microtubule minus-ends in our experiments. Increasing the EB1-Alexa 488 concentration in these experiments resulted in enhanced accumulation of EB1 at growing microtubule plus-ends (Fig. 1B). The robust and specific comet-like labeling of growing microtubule plus-ends indicates that +TIP activity is an intrinsic property of mammalian EB1, similar to previous observations on the Schizosaccharomyces pombe EB1 homolog Mal3 (18).
Fig. 1.
Microtubule plus-end tracking by the human EB1 protein. (A) Montage depicting the specific decoration of growing microtubule plus-ends by 250 nM EB1–Alexa 488. The kymograph at the bottom shows the same microtubule over the duration of the observation period. Microtubule depolymerization is accompanied by loss of EB1–Alexa 488 at the plus-ends, and subsequent rescue of microtubule growth restores plus-end tracking of EB1–Alexa 488. The numbers on each frame represent time in seconds. (B) Images showing the microtubule plus-end decoration pattern at increasing concentrations of EB1-Alexa 488. (Scale bar, 2 μm). The graph shows the corresponding distribution of the fluorescence intensity of EB1–Alexa 488 at the microtubule tip.
CLIP-170 Requires EB1 for +TIP Activity.
We next examined the interaction of CLIP-170 with microtubules. In CLIP-170, the central coiled-coil domain allows the formation of intramolecular contacts between the N-terminal CAP-Gly microtubule-binding domain and the C-terminal zinc-finger motifs, resulting in a “closed” conformation of CLIP-170 that inhibits microtubule binding (20). Because the amino terminus alone is sufficient for +TIP activity in the cell, to avoid autoinhibition, we used a well-characterized carboxyl-terminal truncation construct of CLIP-170 designated as the H2 fragment (13). The H2 fragment is dimeric, like full-length CLIP-170 (21). We purified recombinant GFP-tagged CLIP-170(H2) and found that CLIP-170(H2)-GFP did not differ significantly from untagged CLIP-170(H2) in microtubule-binding affinity; we found a Kd of 0.9 ± 0.5 μM for unlabeled H2 and 1.5 ± 0.6 μM for H2-GFP under our experimental conditions (Fig. S1A).
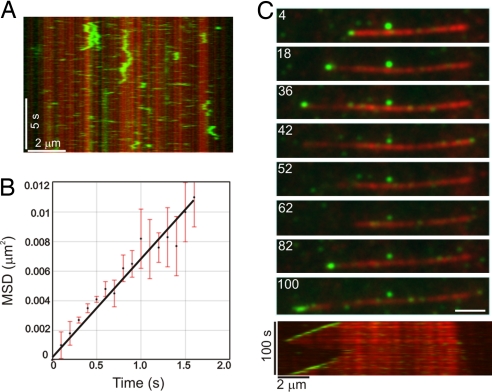
In contrast to EB1, CLIP-170(H2)-GFP alone exhibited no +TIP activity over a broad range of concentrations. Instead, CLIP-170(H2)-GFP bound nonspecifically along the length of the microtubule. At relatively high concentrations (150 nM), CLIP-170(H2) decorated dynamic microtubules uniformly during both the growth and shortening phases (Fig. S1B). Control experiments with GFP alone at similarly high concentrations demonstrated no detectable microtubule binding (Fig. S1C). At lower concentrations (25 nM), CLIP-170(H2) was bound nonspecifically along the microtubule length, and time-lapse (Movie S2) and kymograph analyses (Fig. 2A) indicated that CLIP-170(H2) moved bidirectionally along the microtubule lattice. Mean squared displacement analysis of single CLIP-170(H2)-GFP molecules moving over the microtubule surface indicated that this movement is consistent with a 1-dimensional random walk with an average diffusion coefficient, D, of 3.5 ± 0.1 × 10−11 cm2 s−1 (Fig. 2B). This diffusive motion of CLIP-170 is similar to that of other microtubule end-binding proteins (22–24), but it did not result in plus-end accumulation of CLIP-170(H2)-GFP over time.
Fig. 2.
CLIP-170 requires EB1 for +TIP activity. (A) Kymograph showing the microtubule binding and diffusive movement of CLIP-170(H2)–GFP at 25 nM. Note the transient binding of CLIP-170(H2)–GFP along the length of the microtubule lattice. (B) Plot of the mean squared displacement (MSD) of CLIP-170(H2)–GFP against time. A linear fit to the data yields a diffusion coefficient, D, of 3.5 ± 0.1 × 10−11 cm2 s−1 (MSD = 2Dt). Error bars represent the SEMs of the squared displacement values (n = 38). (C) The montage at the top depicts the specific decoration of growing microtubule plus-ends by 25 nM CLIP-170(H2)–GFP in the presence of 250 nM of unlabeled EB1 protein. The kymograph at the bottom shows the same microtubule over the duration of the observation period. Microtubule depolymerization is accompanied by loss of CLIP-170(H2)2–GFP at the plus-ends and subsequent rescue of microtubule growth restores plus-end tracking of CLIP-170(H2)–GFP. The numbers on each frame represent time (in seconds). (Scale bar, 2 μm.)
Because EB1 and CLIP-170 interact directly with each other (16, 25, 26), we explored whether EB1 could significantly alter the microtubule association of CLIP-170. In the presence of unlabeled EB1, CLIP-170(H2)-GFP dramatically changed localization, from nonspecific lattice labeling to selective targeting of growing microtubule plus-ends (Fig. 2C). Under these conditions, CLIP-170(H2)-GFP preferentially labeled all actively polymerizing microtubule plus-ends in a comet-like pattern, similar to that observed for EB1-Alexa 488 (Movie S3). These results indicate that EB1 is both necessary and sufficient to recruit CLIP-170 to dynamic plus-ends.
EB1 and CLIP-170(H2), alone or together, did not significantly change the microtubule dynamic instability parameters compared with the control values obtained in the absence of any +TIP proteins (Table S1). This indicates that EB1 and CLIP-170(H2) do not significantly affect the dynamics of microtubule assembly and disassembly in this minimal system.
Both EB1 and CLIP-170 Exhibit Rapid Turnover at Microtubule Plus-Ends.
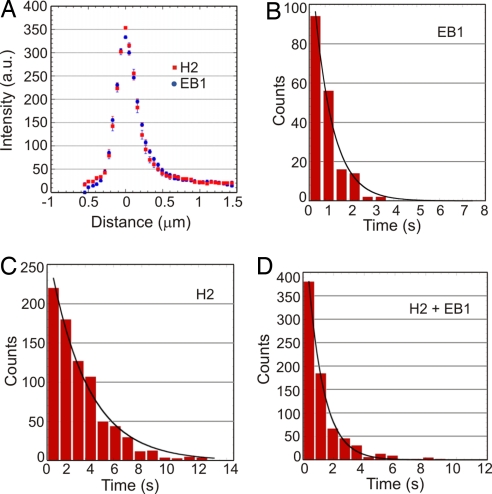
Fluorescence intensity profiles along growing microtubule plus-end structures show that the spatial distribution of labeling are similar for both EB1–Alexa 488 and CLIP-170(H2)–GFP (Fig. 3A). These data are consistent with a model in which EB1 regulates CLIP-170 recruitment and dissociation at microtubule ends (19, 25). Based on the integrated fluorescence intensity in a 1 μm x 1 μm region at growing microtubule plus-ends, we estimate that an average of 10 EB1 molecules and 5 CLIP-170(H2) molecules are present in the comet-like structure at 250 nM of EB1 and 25 nM of CLIP-170(H2). To determine the turnover of individual EB1 and CLIP-170 proteins at microtubule plus-ends, we used single-molecule imaging conditions and greater temporal resolution (Fig. S2). Single EB1–Alexa 488 molecules exchanged rapidly at microtubule plus-ends, with an average dwell time of 0.81 ± 0.06 s (n = 184). In our experiments, the average lifetime of the labeled region (calculated as the ratio of comet tail length to microtubule growth speed) at microtubule plus-ends was 8.3 ± 0.1 s (n = 50); this indicates that single EB1–Alexa 488 molecules go through multiple rounds of binding and unbinding over the lifetime of the microtubule plus-end structure.
Fig. 3.
Rapid turnover of single EB1 and CLIP-170 molecules on growing ends of microtubules. (A) Averaged intensity profiles of EB1–Alexa 488 and CLIP-170(H2)–GFP microtubule plus-end decoration (n = 20 for both). The concentrations of EB1–Alexa 488 and CLIP-170(H2)–GFP were 250 nM and 25 nM, respectively. (B–D) Histograms of dwell times of single binding events of 10 nM EB1–Alexa 488 (B), 5 nM CLIP-170(H2)–GFP in the absence of EB1 (C), or 5 nM CLIP-170(H2)–GFP in the presence of 250 nM unlabeled EB1 (D). Exponential fits to the data yielded mean lifetimes of interactions of 0.81 ± 0.06 s for EB1–Alexa 488 (n = 184), 2.69 ± 0.16 s for CLIP-170(H2)–GFP alone (n = 800), and 1.03 ± 0.03 s for CLIP-170(H2)–GFP in combination with EB1 (n = 753).
In the absence of EB1, single CLIP-170(H2)–GFP molecules had an average dwell time on the microtubule of 2.69 ± 0.16 s (n = 800) as they diffused along its surface. But when these molecules were imaged together with unlabeled EB1, their average dwell time dropped to 1.03 ± 0.03 s (n = 753), similar to that of single EB1–Alexa 488 molecules. The dwell time of the CLIP-170–EB1 complex was not affected by the addition of unlabeled CLIP-170(H2) to a concentration of 25 nM (see Materials and Methods). Thus, EB1 appears to directly influence the turnover of CLIP-170 at microtubule plus-ends. In contrast to the effects of EB1 on CLIP-170 dynamics, the presence of unlabeled CLIP-170(H2) did not significantly affect either the comet tail length or average dwell time of EB1–Alexa 488 at microtubule plus-ends (data not shown). This indicates that the inherent +TIP dynamics of EB1 were not altered by an association with CLIP-170.
Guanosine 5′-[α,β-methylene]triphosphate (GMPCPP) Abolishes +TIP Activity of EB1 and CLIP-170.
During the reconstitution experiments, we observed that the +TIP activity of both EB1–Alexa 488 and CLIP-170(H2)–GFP did not require preincubation with tubulin dimers prior to microtubule polymerization. This observation and the dwell time dynamics discussed above suggest that although EB1 and CLIP-170 can form a complex with free tubulin dimers (16), the formation of such a complex is not required, and such formation does not enhance the targeted concentration of these proteins at plus-ends. Thus, our data support an end-recognition mechanism for +TIP activity, in which these proteins bind with higher affinity to the newly polymerized end of the microtubule because of biochemical and/or structural differences between the new end and the older lattice structure.
To test this mechanism directly, we conducted reconstitution experiments using GMPCPP instead of GTP. GMPCPP is a slowly hydrolyzable analog of GTP that promotes microtubule assembly and results in an overall microtubule lattice structure that mimics the GTP-bound microtubule cap structure at growing plus-ends (27).
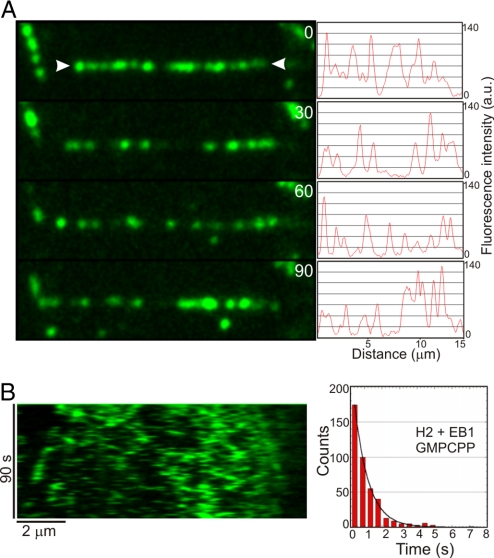
Microtubule polymerization with GMPCPP abolished the +TIP activity of CLIP-170(H2)–GFP in the presence of EB1 and led to nonspecific binding of CLIP-170(H2)–GFP along the entire microtubule lattice (Fig. 4 and Movie S4). Under these conditions, the CLIP-170(H2)–EB1 complex exhibited 1-dimensional diffusion along the microtubule surface with an average dwell time of 0.79 ± 0.02 s (n = 440). Thus, the dwell time measured along the sidewall of the GMPCPP lattice is similar to that observed at microtubule plus-ends formed in the presence of GTP. This similarity indicates that the dynamics of the plus-end complex depend on the nucleotide state of the lattice and do not require copolymerization with tubulin. Thus, we conclude that EB1 and CLIP-170 selectively accumulate at growing microtubule plus-ends because of their ability to recognize a structural or chemical discontinuity along the polymer generated by nucleotide hydrolysis.
Fig. 4.
Effect of GMPCPP on CLIP-170–GFP plus-end tracking activity. (A) Montage showing a dynamic microtubule (arrowheads) polymerized with GMPCPP-tubulin in the presence of 25 nM CLIP-170(H2)–GFP and 250 nM unlabeled EB1. The numbers indicate time (in seconds). Each image is accompanied by an intensity profile of the CLIP-170(H2)-GFP fluorescence along the microtubule length. (B) Kymograph showing the same microtubule over the duration of the observation period. Note that CLIP-170(H2)–GFP does not show plus-end tracking, but rather binds indiscriminately along the entire length of the GMPCPP microtubule. The accompanying histogram shows the distribution of dwell times of single binding events. An exponential fit to the data yields a mean lifetime of interaction of 0.79 ± 0.02 s (n = 440) for the CLIP-170–EB1 complex.
Discussion
In this work, we reconstituted the microtubule plus-end tracking of the +TIPs EB1 and CLIP-170 in vitro in a minimal system and demonstrated an intrinsic ability of mammalian EB1 to bind specifically to the growing plus-end of a microtubule. A similar result was found previously with the S. pombe EB1 homolog Mal3 (18), highlighting the functional conservation of EB1 as a master regulator of +TIP function (7). We also found that an interaction between EB1 and CLIP-170 (16, 25, 26) is sufficient to recruit CLIP-170 to dynamic microtubule plus-ends in vitro, an observation consistent with the findings of RNAi studies in mammalian cells demonstrating the need for EB1 in the localization of CLIP-170 to dynamic plus-ends (25, 28).
In contrast to our observation on these mammalian +TIPs, reconstitution of plus-end dynamics for the yeast homolog of CLIP-170, Tip1, required the presence of a plus-end-directed kinesin, Tea2, in addition to Mal3. These in vitro observations are consistent with genetic data from yeast indicating the need for both Mal3 and Tea2 for the proper cellular localization of Tip1 (29). Thus, whereas CLIP-170 is structurally similar to Tip1, the mammalian and yeast proteins differ significantly in their mechanisms of plus-end localization. CLIP-170 on its own binds directly to the microtubule with an affinity of 0.2–1 μM (15, 16), and our TIRF assays demonstrated diffusion along the microtubule lattice (Fig. 2). In contrast, Tip1 does not bind significantly to microtubules on its own, and Mal3 is not sufficient to recruit Tip1 to plus-ends (18). One potential explanation for this functional difference is the presence of a tandem repeat of the CAP-Gly domain at the N terminus of CLIP-170, as opposed to only a single CAP-Gly domain at the N terminus of Tip1. It has been suggested that at least 2 microtubule-binding domains may be required for +TIP activity (17).
Our dwell-time analysis found that neither EB1 nor CLIP-170 remained stably bound to the growing microtubule end. Instead, both on and off rates were relatively high, leading to short dwell times. This in vitro observation closely parallels those reported previously for EB1 binding to microtubules in Xenopus extracts (30) and for the association of CLIP-170 with microtubule ends within mammalian cells (19). Our data demonstrate that EB1 is sufficient to dynamically track microtubule plus-ends, whereas multiple studies have shown that EB1 alone does not bind to tubulin dimers (16, 18). Moreover, current thinking on the GTP cap suggests a depth of ≈40–50 tubulin dimers (31–33), and thus a model in which EB1 copolymerizes specifically with GTP-bound tubulin dimers would not explain the ≈0.5-μm-long EB1 comet tail labeling observed here. Together, these observations argue against a copolymerization model (13, 15, 16) or a polymerization-chaperone model (17).
Our in vitro dynamics instead suggest a model in which EB1 alone, or the CLIP-170–EB1 complex, has a higher binding affinity for the tubulin structure found only at the growing plus-end. Interestingly, Bieling et al. (18) reported a pronounced dependence of Mal3 comet tail length on the rate of microtubule polymerization, consistent with our observations on the comet tail lengths of EB1 and the CLIP-170–EB1 complex. Thus, the structural transition occurring after incorporation of newly polymerized subunits into the microtubule, resulting in significantly decreased affinity of EB1 for the lattice structure, is not tightly linked to subunit incorporation into the polymer, but instead occurs as a subsequent event. In this way, it is similar to the hydrolysis of bound GTP occurring with first-order kinetics on newly incorporated subunits, although whether these events occur at similar rates remains to be determined. The loss of plus-end specificity of binding observed when microtubules were polymerized in the presence of GMPCPP rather than GTP further supports the hypothesis that a conformational change in the microtubule lattice that occurs with, or subsequent to, GTP hydrolysis on incorporated tubulin subunits induces a significant change in the binding affinity of EB1 for the microtubule. These findings indicate that GTP hydrolysis serves as a critical regulatory switch for +TIP tracking.
EB1 enhances microtubule elongation (34), but the mechanisms involved remain unclear. EB1 has been proposed to promote tubulin sheet closure during microtubule polymerization (35), potentially through preferential binding along the seam (36). In vitro, EB1 has been reported to suppress the shortening rate and the frequency of catastrophe (37) or, alternatively, to promote both catastrophes and rescues (35). Much of this experimental variation is likely due to differences in EB1 concentrations used in the assays, because at higher concentrations, EB1 does not localize in a tip-specific manner but instead binds along the microtubule lattice both in vitro (16, 35) and in mammalian cells (34). In vivo, deletion analysis of Mal3 or Tip1 in S. pombe suggests that these proteins act to suppress the frequency of microtubule catastrophes (38–40). In Xenopus extracts, EB1 was shown to decrease the frequency of catastrophes and increase the frequency of rescues (30); in mammalian cells, CLIP-170 functions to promote rescue (41).
In our reconstitution experiments, however, neither EB1 nor CLIP-170, acting alone or in a complex, significantly altered the microtubule dynamic instability parameters that we observed in the presence of tubulin alone. These results suggest that in this minimal system, neither EB1 nor CLIP-170, nor their complex, is sufficient to modulate microtubule dynamics when localized to growing microtubule plus-ends, and that other cellular factors may be needed to drive this activity in the cell.
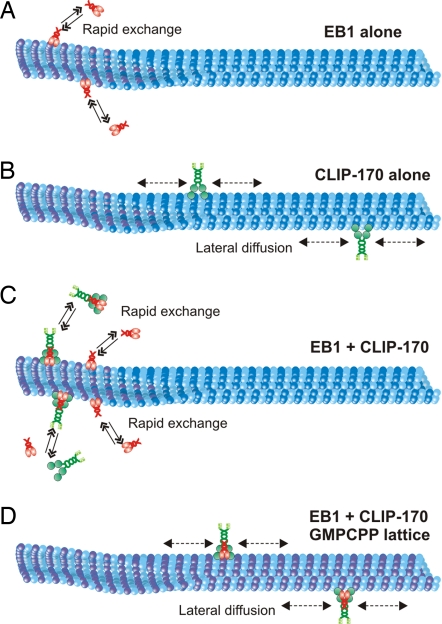
We propose a model in which EB1 dynamically recruits CLIP-170 to growing microtubule plus-ends by recognizing the distinct conformation of the plus-end (Fig. 5). This model is consistent with the increasing perception of EB1 as the master plus-end tracking protein, capable of recruiting multiple distinct +TIPs to the dynamic plus-end (8). The relatively weak binding affinities measured for direct associations among the +TIPs suggest that these proteins function in rapidly changing networks of interactions (7, 8), consistent with observations that +TIPs function in a wide array of cellular processes, including cell polarization, cell migration, and cell division. The enrichment of these +TIPs at the microtubule plus-end likely will facilitate microtubule search and capture, in which dynamic microtubules search the cellular space and become captured by specific targets, such as adherens junctions or other cortical sites of contact (4–6). Future in vitro reconstitution studies modeling the dynamics of these diverse +TIPs are likely to provide further insight into the cellular mechanisms of the plus-end complex.
Fig. 5.
Model for plus-end tracking activity of mammalian EB1 and CLIP-170. (A) EB1 (red) is an intrinsic +TIP protein that specifically decorates growing microtubule plus-ends by targeting the plus-end structure (purple). (B) In the absence of EB1, CLIP-170 (green) binds nonspecifically along the length of the microtubule and exhibits 1-dimensional diffusion on the microtubule surface. (C) EB1 is required for +TIP activity of CLIP-170, either by binding to the microtubule plus-end as a preformed complex with CLIP-170 or by recruiting CLIP-170 to the microtubule plus-end. +TIP activity of both proteins is characterized by rapid exchange of these proteins at the growing microtubule tips. (D) GMPCPP (purple microtubule lattice) eliminates plus-end tracking of the CLIP-170–EB1 complex and results in its binding along the length of the microtubule, as well as 1-dimensional diffusion along the lattice.
Materials and Methods
Protein Purification and Labeling.
His-tagged recombinant GFP-labeled CLIP-170(H2) was affinity-purified using a nickel column and then desalted using a PD-10 column (Amersham Biosciences) and exchanged into BRB80 buffer (80 mM piperazine-1,4-bis(2-ethanesulfonic acid), 1 mM MgCl2, and 1 mM EGTA [pH 6.8]) supplemented with 50 mM NaCl. Recombinant GST-tagged full-length EB1 protein was purified using the PreScission expression system (GE Healthcare). In brief, the fusion protein was purified using glutathione Sepharose beads, and the GST tag subsequently cleaved through site-specific cleavage with the PreScission protease. The released EB1 protein was further purified using a NAP-10 gel filtration column and exchanged into BRB80 buffer supplemented with 50 mM NaCl. Purified proteins were flash frozen and stored at −80 °C until use. Purified EB1 protein was fluorescently labeled at cysteine residues using Alexa–Fluor 488 maleimide (Invitrogen) in BRB80 buffer. The degree of labeling was estimated as ≈70% by spectroscopic analysis.
Microtubule-Binding Assays.
Microtubule-binding assays were conducted by coincubating increasing concentrations of rhodamine-labeled microtubules with purified recombinant CLIP-170(H2), CLIP-170(H2)–GFP, EB1, or EB1–Alexa 488 proteins for 30 min at 37 °C. The samples were then centrifuged at 35,000 × g for 20 min, after which the resultant supernatant and pellet fractions were analyzed by SDS/PAGE and densitometry. Under our experimental conditions, both tagged and untagged EB1 were bound relatively weakly to microtubules and did not reach saturation.
Microtubule Seeds.
Rhodamine-labeled and biotinylated GMPCPP microtubule seeds were prepared by polymerizing 50 μM tubulin (at a bovine tubulin: rhodamine-tubulin: biotin-tubulin ratio of 22.5: 1.5: 1) in the presence of 1 mM GMPCPP (Jena Bioscience) at 37 °C for 30 min. The assembled microtubules were harvested by centrifuging at 35,000 × g for 20 min and then resuspended in BRB80 buffer supplemented with 1 mM GMPCPP. These microtubules were sheared with a 100-μL Hamilton syringe to generate short seeds before use.
Microtubule Plus-End Tracking Assays.
The in vitro reconstitution experiments were conducted in flow cells (15 μL volume) constructed using slides and silanized coverslips (Repel silane, Amersham Biosciences) attached with double-sided adhesive tape. The flow cell was coated with 20% monoclonal antibiotin antibody (clone BN-34, Sigma) and then blocked with 5% Pluronic F-127 (Sigma). Then 125 nM sheared GMPCPP microtubule seeds were introduced into the flow cell. Tubulin polymerization was initiated by introducing 22 μM 1:30 rhodamine-labeled bovine tubulin in BRB80 buffer supplemented with 0.1% methyl cellulose (4000 cP, Sigma), an oxygen-scavenging system containing glucose oxidase, catalase, glucose, 100 mM DTT, 1 mM Mg-GTP, and +TIP proteins. Unless stated otherwise, 250 nM EB1–Alexa 488 and 25 nM CLIP-170(H2)–GFP were used. Microtubule dynamics were visualized at 22 °C using a TIRF microscope fitted on an inverted microscope (Nikon TE-2000U). TIRF excitation was achieved using a 488-nm optically pumped semiconductor laser (Sapphire, Coherent) to visualize Alexa 488 and a 532-nm diode-pumped solid-state laser (CrystaLaser) to visualize rhodamine. The laser light intensities used for imaging were kept low, so that the photobleaching rates were slow compared with the dwell times. Images were captured with a back-illuminated electron-multiplying CCD camera (Cascade-512B, Photometrics) using either time-lapse capture at 2-sec intervals for 2-color microscopy or burst mode at 10 frames per second for the analysis of +TIP dynamics at the single-molecule resolution. Control experiments showed that the dwell times of single binding events of the CLIP-170–EB1 complex at 5 nM CLIP-170(H2)–GFP in the presence of 250 nM unlabeled EB1 or at 5 nM CLIP-170(H2)–GFP in the presence of both 25 nM unlabeled CLIP-170(H2) and 250 nM unlabeled EB1 were statistically indistinguishable.
Image Analysis and Statistics.
Plus-end tracking of EB1–Alexa 488 and CLIP-170(H2)–GFP was analyzed using kymographs generated with the “Multiple Kymograph” plug-in for ImageJ submitted by J. Rietdorf and A. Seitz (European Molecular Biology Laboratory, Heidelberg, Germany). Microtubule growth rates and +TIP dwell times were extracted from these kymographs. The lengths of the +TIP comet tails, defined as the distances from the peak fluorescence intensity at the microtubule tips to the baseline lattice intensity, were measured from line profiles of fluorescence intensities of EB1–Alexa 488 and CLIP-170(H2)–GFP. To analyze the diffusional movement of CLIP-170(H2)–GFP, manual tracking was used to trace the displacement of single molecules on the microtubule lattice. Dwell time and mean squared displacement data were fitted using KaleidaGraph (Synergy Software), and statistical significance was determined using the Student t test.
Supplementary Material
Acknowledgments.
We thank Yujie Sun for assistance with the TIRF microscopy and Lee Ligon and Jennifer Ross for helpful discussions. This work was supported by National Institutes of Health Grants R01 GM068591 (to E.H.) and P01 AR051174 (to Y.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807614106/DCSupplemental.
References
- 1.Dammermann A, Desai A, Oegema K. The minus end in sight. Curr Biol. 2003;13:R614–R624. doi: 10.1016/s0960-9822(03)00530-x. [DOI] [PubMed] [Google Scholar]
- 2.Howard J, Hyman AA. Dynamics and mechanics of the microtubule plus-end. Nature. 2003;422:753–758. doi: 10.1038/nature01600. [DOI] [PubMed] [Google Scholar]
- 3.Kirschner M, Mitchison T. Beyond self-assembly: From microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- 4.Siegrist SE, Doe CQ. Microtubule-induced cortical cell polarity. Genes Dev. 2007;21:483–496. doi: 10.1101/gad.1511207. [DOI] [PubMed] [Google Scholar]
- 5.Jaworski J, Hoogenraad CC, Akhmanova A. Microtubule plus-end tracking proteins in differentiated mammalian cells. Int J Biochem Cell Biol. 2008;40:619–637. doi: 10.1016/j.biocel.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 6.Levy JR, Holzbaur EL. Special delivery: Dynamic targeting via cortical capture of microtubules. Dev Cell. 2007;12:320–322. doi: 10.1016/j.devcel.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Akhmanova A, Steinmetz MO. Tracking the ends: A dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 8.Lansbergen G, Akhmanova A. Microtubule plus-end: A hub of cellular activities. Traffic. 2006;7:499–507. doi: 10.1111/j.1600-0854.2006.00400.x. [DOI] [PubMed] [Google Scholar]
- 9.Pierre P, Scheel J, Rickard JE, Kreis TE. CLIP-170 links endocytic vesicles to microtubules. Cell. 1992;70:887–900. doi: 10.1016/0092-8674(92)90240-d. [DOI] [PubMed] [Google Scholar]
- 10.Perez F, Diamantopoulos GS, Stalder R, Kreis TE. CLIP-170 highlights growing microtubule ends in vivo. Cell. 1999;96:517–527. doi: 10.1016/s0092-8674(00)80656-x. [DOI] [PubMed] [Google Scholar]
- 11.Akhmanova A, Hoogenraad CC. Microtubule plus-end-tracking proteins: Mechanisms and functions. Curr Opin Cell Biol. 2005;17:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Vaughan KT. TIP maker and TIP marker: EB1 as a master controller of microtubule plus-ends. J Cell Biol. 2005;171:197–200. doi: 10.1083/jcb.200509150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diamantopoulos GS, et al. Dynamic localization of CLIP-170 to microtubule plus-ends is coupled to microtubule assembly. J Cell Biol. 1999;144:99–112. doi: 10.1083/jcb.144.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnal I, Heichette C, Diamantopoulos GS, Chretien D. CLIP-170/tubulin–curved oligomers coassemble at microtubule ends and promote rescues. Curr Biol. 2004;14:2086–2095. doi: 10.1016/j.cub.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 15.Folker ES, Baker BM, Goodson HV. Interactions between CLIP-170, tubulin, and microtubules: Implications for the mechanism of Clip-170 plus-end tracking behavior. Mol Biol Cell. 2005;16:5373–5384. doi: 10.1091/mbc.E04-12-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ligon LA, Shelly SS, Tokito MK, Holzbaur EL. Microtubule binding proteins CLIP-170, EB1, and p150Glued form distinct plus-end complexes. FEBS Lett. 2006;580:1327–1332. doi: 10.1016/j.febslet.2006.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slep KC, Vale RD. Structural basis of microtubule plus-end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bieling P, et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 19.Dragestein KA, et al. Dynamic behavior of GFP-CLIP–170 reveals fast protein turnover on microtubule plus-ends. J Cell Biol. 2008;180:729–737. doi: 10.1083/jcb.200707203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lansbergen G, et al. Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization. J Cell Biol. 2004;166:1003–1014. doi: 10.1083/jcb.200402082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheel J, et al. Purification and analysis of authentic CLIP-170 and recombinant fragments. J Biol Chem. 1999;274:25883–25891. doi: 10.1074/jbc.274.36.25883. [DOI] [PubMed] [Google Scholar]
- 22.Culver-Hanlon TL, Lex SA, Stephens AD, Quintyne NJ, King SJ. A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat Cell Biol. 2006;8:264–270. doi: 10.1038/ncb1370. [DOI] [PubMed] [Google Scholar]
- 23.Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 24.Brouhard GJ, et al. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komarova Y, et al. EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol Biol Cell. 2005;16:5334–5345. doi: 10.1091/mbc.E05-07-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishima M, et al. Structural basis for tubulin recognition by cytoplasmic linker protein 170 and its autoinhibition. Proc Natl Acad Sci U S A. 2007;104:10346–10351. doi: 10.1073/pnas.0703876104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caplow M, Ruhlen RL, Shanks J. The free energy for hydrolysis of a microtubule-bound nucleotide triphosphate is near zero: All of the free energy for hydrolysis is stored in the microtubule lattice. J Cell Biol. 1994;127:779–788. doi: 10.1083/jcb.127.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watson P, Stephens DJ. Microtubule plus-end loading of p150(Glued) is mediated by EB1 and CLIP-170 but is not required for intracellular membrane traffic in mammalian cells. J Cell Sci. 2006;119:2758–2767. doi: 10.1242/jcs.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browning H, Hackney DD, Nurse P. Targeted movement of cell end factors in fission yeast. Nat Cell Biol. 2003;5:812–818. doi: 10.1038/ncb1034. [DOI] [PubMed] [Google Scholar]
- 30.Tirnauer JS, Grego S, Salmon ED, Mitchison TJ. EB1–microtubule interactions in Xenopus egg extracts: Role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol Biol Cell. 2002;13:3614–3626. doi: 10.1091/mbc.E02-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voter WA, O'Brien ET, Erickson HP. Dilution-induced disassembly of microtubules: relation to dynamic instability and the GTP cap. Cell Motil Cytoskeleton. 1991;18:55–62. doi: 10.1002/cm.970180106. [DOI] [PubMed] [Google Scholar]
- 32.VanBuren V, Cassimeris L, Odde DJ. Mechanochemical model of microtubule structure and self-assembly kinetics. Biophys J. 2005;89:2911–2926. doi: 10.1529/biophysj.105.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schek HT, III, Gardner MK, Cheng J, Odde DJ, Hunt AJ. Microtubule assembly dynamics at the nanoscale. Curr Biol. 2007;17:1445–1455. doi: 10.1016/j.cub.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ligon LA, Shelly SS, Tokito M, Holzbaur EL. The microtubule plus-end proteins EB1 and dynactin have differential effects on microtubule polymerization. Mol Biol Cell. 2003;14:1405–1417. doi: 10.1091/mbc.E02-03-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitre B, et al. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat Cell Biol. 2008;10:415–421. doi: 10.1038/ncb1703. [DOI] [PubMed] [Google Scholar]
- 36.Sandblad L, et al. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 2006;127:1415–1424. doi: 10.1016/j.cell.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 37.Manna T, Honnappa S, Steinmetz MO, Wilson L. Suppression of microtubule dynamic instability by the +TIP protein EB1 and its modulation by the CAP-Gly domain of p150glued. Biochemistry. 2008;47:779–786. doi: 10.1021/bi701912g. [DOI] [PubMed] [Google Scholar]
- 38.Browning H, et al. Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J Cell Biol. 2000;151:15–28. doi: 10.1083/jcb.151.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brunner D, Nurse P. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell. 2000;102:695–704. doi: 10.1016/s0092-8674(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 40.Busch KE, Brunner D. The microtubule plus-end tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr Biol. 2004;14:548–559. doi: 10.1016/j.cub.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 41.Komarova YA, Akhmanova AS, Kojima S, Galjart N, Borisy GG. Cytoplasmic linker proteins promote microtubule rescue in vivo. J Cell Biol. 2002;159:589–599. doi: 10.1083/jcb.200208058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.