Abstract
MicroRNAs play important roles in animal development. Numerous conditional knockout (cKO) studies of Dicer have been performed to interrogate the functions of microRNA during mammalian development. However, because Dicer was recently implicated in the biogenesis of endogenous siRNAs in mammals, it raises the question whether the Dicer cKO defects can be attributable to the loss of microRNAs. Previously, we and others conditionally targeted Dicer and identified its critical roles in embryonic skin morphogenesis. Here, we focus explicitly on microRNAs by taking a parallel strategy with Dgcr8, encoding an essential component of the microprocessor complex that is exclusively required for microRNA biogenesis. With this comparative analysis, we show definitively that the Dicer- and Dgcr8-null skin defects are both striking and indistinguishable. By deep sequencing analysis of microRNA depletion in both Dicer- and Dgcr8-null skin, we demonstrate that most abundantly expressed skin microRNAs are dependent on both Dicer and DGCR8. Our results underscore a specific importance of microRNAs in controlling mammalian skin development.
Keywords: dicer, small RNAs
MicroRNAs are a class of small (19–25nt), noncoding RNAs essential for animal development (1–3). They regulate gene expression post-transcriptionally by guiding the RNA-induced silencing complex (RISC) to their cognate sites at the 3′-untranslated region (UTR) of target mRNAs. Based on bioinformatics predictions, more than one third of mammalian mRNAs are potential targets of microRNAs (4). In the past several years, a two-step model of microRNA biogenesis has been widely validated (5). For most microRNAs, primary microRNA transcripts are first cropped into hairpin intermediates by a nuclear multiprotein Drosha-DGCR8 (Parsha) microprocessor complex (6, 7). Recently, an alternative mirtron pathway was reported where some intronic microRNA precursors can be processed by RNA splicing that bypasses the micropreocessor cleavage (8–10). In either case, the processed microRNA precursors (premiRNA) are transported to the cytoplasm by exportin-5 (11, 12). After the transportation, mature microRNAs are then generated through enzymatic processing by another RNase III enzyme, Dicer (13, 14).
Because of the absolute requirement of Dicer for microRNA biogenesis, numerous conditional knockout (cKO) studies of Dicer have been performed to interrogate microRNA functions in mammalian development (1, 3). At a molecular level, a recent study has shown that in mouse embryonic stem (ES) cells, Dicer ablation results in depletion of all microRNAs (15). However, Dicer has also been reported to function in heterochromatin silencing of these cells (16), and in mouse oocytes, Dicer has been implicated in production of small RNA species other than microRNAs (17, 18).
In contrast to Dicer, DGCR8 function appears to be specific to microRNAs, because it acts in recognizing the premicroRNA hairpin (19, 20). Dgcr8 has also been targeted to mouse ES cells, and although differentiation defects were similar to those of Dicer-null ES cells, Dgcr8-null ES cells appeared to grow faster than Dicer-null cells in the initial stages (16, 21, 22). The underlying basis for the differences in cultured ES cells is still uncertain, and the extent to which Dicer cKO developmental phenotypes in vivo are attributable explicitly to microRNAs has not been explored.
In the present study, we address this possibility directly by conditionally targeting Dgcr8 in skin and comparing these mice to skin-specific Dicer mutant mice that we generated (23) (see also ref. 25). Our findings not only underscore the importance of microRNAs specifically in skin development, but also demonstrate that the striking defects in hair follicle and epidermal differentiation observed by the loss of Dicer function are primarily attributable to Dicer's action on microRNAs.
Results
Dicer and Dgcr8 Skin Conditional Knockout Animals Show Indistinguishable Defects.
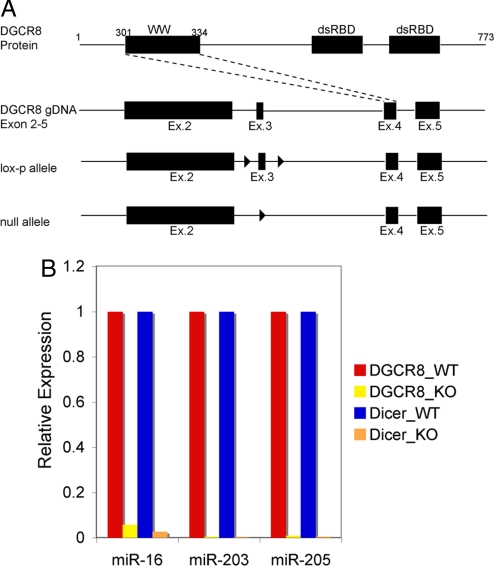
According to microarray gene profiling, Dgcr8, Dicer, and Drosha are expressed throughout all skin lineages, indicating their universal requirement in microRNA biogenesis (data not shown). We therefore generated miceharboring a conditional null Dgcr8 allele as described (22) (Fig. 1A) and crossed them to mice expressing keratin 14-Cre, active in embryonic skin epithelial stem cells (24). To verify that Dgcr8 and microRNA processing were effectively ablated in newborn (P0) skin epidermis, we used real-time quantitative PCR (qPCR) to quantify relative expression levels of three abundant skin microRNAs: miR-16, miR-203 and miR-205 (Fig. 1B). Having confirmed microRNA depletion in Dgcr8 cKO skin, we characterized the mice.
Fig. 1.
Strategy for generating the Dgcr8 floxed allele in mouse genomic DNA (gDNA) and verification that microRNAs were effectively depleted as a result of Dgcr8 skin-specific ablation in mice. (A) Exon 3 within the Dgcr8 coding region was chosen for targeting because its deletion was predicted to generate a frameshift mutation and early truncation of the DGCR8 protein. Arrowheads denote positioning of the Lox sequences, recognized by Cre recombinase. WW, a WW protein–protein interaction module; dsRBD, double-stranded RNA-binding domain. (B) Conditional (K14-Cre) ablation of both Dgcr8 and Dicer in skin resulted in the depletion of microRNAs compared with their WT counterparts. The expression levels of three representative microRNAs highly expressed in skin were chosen for analyses. Values for WT samples were designated as 1. SnoRNA25 served as the internal control.
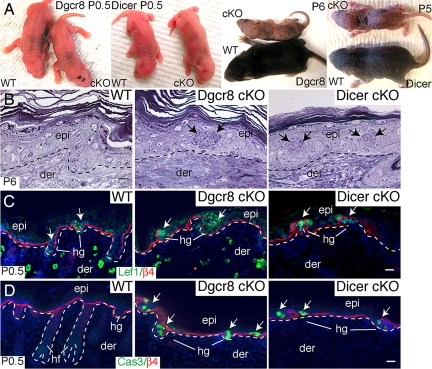
Overall, the phenotype of Dgcr8fl/fl/K14-Cre cKO animals bore a striking resemblance to Dicerfl/fl/K14-Cre cKO mice (23, 25). Although indistinguishable to their WT littermates at birth, both mutants survived up to 5–6 days after birth (P5–P6) with rough skin and failed to gain weight compared with their WT littermates (Fig. 2A). Morphological and histological analyses revealed evaginating hair germs penetrating into and disrupting the epidermis, a hallmark of Dicer cKO skin (23, 25) (Fig. 2B). Immunolocalization confirmed that these structures were positive for Lef1, an essential hair germ marker. The local reduction in β4 integrin expression that classically accompanies hair follicle downgrowth was also seen at these sites (23, 25) (Fig. 2C).
Fig. 2.
Skin phenotypes of Dgcr8 and Dicer cKO mice are strikingly similar. (A) Dgcr8 and Dicer cKO mice can survive up to 5–6 days after birth. Newborn (P0.5) cKO and WT mice are similar in size and appearance. Thereafter, cKO mice fail to gain weight and exhibit taught, flaky skin, a sign of severe dehydration. (B) P6 Dgcr8 and Dicer cKO skins display evaginating hair germs (hg) that appear as balls of undifferentiated cells (arrows) that distort the epidermis (epi). (Scale bar, 10 μm.) (C) Immunofluorescence identifies germs molecularly by transcription factor Lef1 and by hemidesmosomal β4 integrin (nonspecific dermal staining is caused by the secondary antibody). (Scale bar, 20 μm.) (D) Active caspase-3 (Cas3) denotes significant enrichment of apoptotic cells within hair germs in the cKO skin. Dotted lines mark epidermal-dermal boundaries. der, dermis. (Scale bar, 20 μm.)
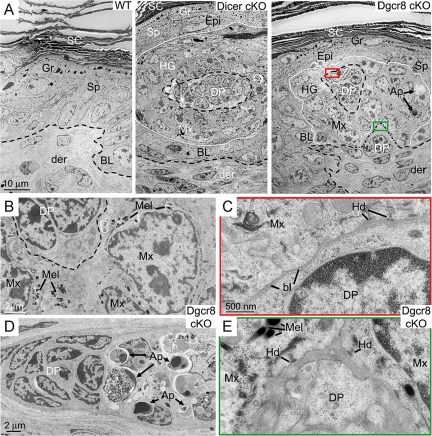
In addition to defects in hair follicle downgrowth, both Dgcr8 and Dicer cKO mutant hair germs and more mature bulbs displayed pervasive signs of apoptosis (23, 25) (Fig. 2D). Morphological features of hair germ evagination, including hemidesmosomes, melanin granules and abnormal apoptosis, were also revealed at the ultrastructural level (Fig. 3). Overall, the skin defects observed in the Dgcr8 cKO mice closely paralleled those of the Dicer cKO mice, suggesting that these defects were caused predominantly by depletion of microRNAs rather than other small RNAs that may depend on Dicer but not DGCR8.
Fig. 3.
Ultrastructural defects of Dicer and Dgcr8 cKO skin. (A) Skins from neonatal WT control, Dicer cKO and Dgcr8 cKO mice were fixed and processed for electron microscopy as described. Shown are regions of the epidermis, where a whorl of evaginated hair germ (hg) cells are readily identified by the presence of dermal papilla (DP) cells at the center of the structure. These aberrations in follicle morphogenesis distorted the surrounding epidermis. (Scale bar, 10 μm.) (B) Cells with hair follicle matrix cell (Mx) morphology were present in the evaginating hair germs of Dgcr8 cKO epidermis. These cells frequently contained melanin granules (Mel). Melanocytes are normally never in mouse skin epidermis, and the appearance of melanin within the whorls was an additional hallmark of the evaginating hair germs. (Scale bar, 2 μm.) (C and E) Closed-up pictures of boxed areas in A. Note the presence of hemidesmosomes (Hd) and a loose basal lamina (bl) at the evaginating hair germ-DP border (C) as well as the epidermal-dermal boundary to what appeared to be an underlying DP (E) in the Dgcr8 cKO skin. (Scale bar, 500 nm.) (D) Enriched apoptotic cells (Ap) in hair germs of Dgcr8 cKO skin. (Scale bar, 2 μm.) Additional abbreviations: SC, stratum corneum; Gr, granular layers; Sp, spinous layers; BL, basal epidermal layer; Der, dermis. Dotted lines mark epidermal-dermal boundaries.
The Most Abundantly Expressed MicroRNAs Are Dependent on both Dicer and DGCR8 in the Skin.
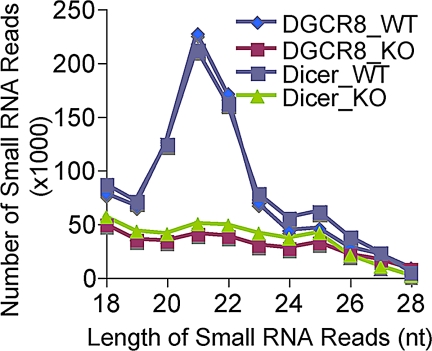
To further investigate the dependence of microRNA biogenesis on DGCR8 and Dicer, we then generated four epidermal small-RNA cDNA libraries from total RNA extracted from skin of P0 Dgcr8 and Dicer cKO mice and their respective WT littermates. Expression of small RNAs in each skin sample was profiled with Solexa deep sequencing as described (26). Overall, the distribution patterns of small RNAs were strikingly different between WT and either Dicer or Dgcr8 cKO samples (Fig. 4). However, the patterns were highly reproducible within either the duplicate WT samples or the Dicer and Dgcr8 cKO samples. Importantly, we observed specific and similar depletion patterns of small RNAs between 19 and 23 nucleotides in both cKO samples (Fig. 4). Because prototypical microRNAs are the only known small RNA species that commonly require both Dicer and DGCR8 processing, this finding suggests that in the skin, Dicer is mainly involved in the production of prototypical microRNAs.
Fig. 4.
Specific and similar depletion of small RNA reads (19–23 nt) in both Dicer and Dgcr8 cKO skin. Small RNA reads from 18 to 28 nucleotides were charted from all four small RNA cDNA libraries. Note the nearly identical distribution patterns of small RNA reads within either two duplicate WT libraries or two cKO libraries.
For the analyses of small RNAs, we began by specifically focusing on the annotation of microRNAs. In WT samples, we recovered up to 391,102 reads that could be mapped to known small RNA species, including microRNAs, sn/sno-RNAs, tRNAs, and rRNAs as well as ectopically added calibration RNAs (Table 1). Overall, when contrasted against the Dicer and Dgcr8 cKO counterparts, only microRNAs were significantly depleted whereas levels of other noncoding RNAs remained constant (see discussion for sn/snoRNA below) (Table 1). Moreover, the microRNA reads from cKO samples were <5% compared with the WT samples after normalizing to reads from rRNA, tRNA and the spiked-in calibration RNAs that should not be affected by either Dicer or Dgcr8 ablation.
Table 1.
Cloning frequency of small non-coding RNAs from four epidermal libraries revealed the significant depletion only to the microRNA population in both Dicer and Dgcr8 cKO samples
| Sequence type | DGCR8_WT | DGCR8_KO | Dicer_WT | Dicer_5_KO |
|---|---|---|---|---|
| microRNA | 373,467 | 12,071 | 325,707 | 8,070 |
| rRNA | 6,921 | 5,184 | 6,446 | 4,216 |
| tRNA | 9,623 | 8,618 | 10,623 | 6,097 |
| sn/sno-RNA | 990 | 1,012 | 798 | 482 |
| calibration RNA | 101 | 85 | 105 | 86 |
| Total | 391,102 | 26,970 | 343,679 | 18,951 |
The numbers are reads mapped to each small RNA species
Recent reports revealed the existence of endogeneous siRNAs (endo-siRNAs) that are processed from dsRNA by interaction between sense and antisense mRNA transcripts in mouse oocytes (17, 18). To investigate the existence of those endo-siRNAs in the skin, we analyzed small RNA reads in our libraries by examining reads that can be perfectly mapped to mRNAs (Table S1). The number of reads was significantly lower than that of microRNAs. In addition, the biogenesis and/or maintenance of those small RNAs were independent of DGCR8. By contrast, they were depleted by an average of 60% in Dicer cKO samples. Currently, it is unclear whether those small RNAs are functional or simply processed by Dicer as dsRNA. Among them, we noticed the expression of a small RNA derived from a predicted hairpin located in the 5′ region of Dgcr8 mRNA (27) (Table S1, highlighted). However, the low abundance and relatively mild depletion of these types of small RNAs suggests that their role(s) in embryonic skin development may be minor relative to those of microRNAs.
We next analyzed the small RNA reads that were mapped to sn/snoRNA. Although the number of reads was significantly lower than microRNAs, some of the major sn/snoRNA-derived reads showed dramatically differential expression in Dgcr8 and Dicer cKO samples (Table S2). We therefore investigated the top three of the most abundantly expressed small RNAs in this class. All three hosting sn/snoRNAs (SNORD27, SCARNA15, and SNORA58) were predicated to fold into hairpin structures (Fig. S1). Intriguingly, all three cloned small RNA species were derived from one arm of the predicted hairpins, consistent with their dependence on Dicer. However, their independence of DGCR8 suggests that an unknown nuclease activity may be involved in the release of the precursors from their primary transcripts. We also noticed that in SCARNA15, a so-called “Cajal body-specific” RNA, there was evolutionary conservation not only of the small RNA sequence but also of the complementary sequence required for its predicted folding into a hairpin characteristic of microRNA precursors. These findings suggest that there is an evolutionary pressure to maintain the hairpin structure and, potentially, the expression of this small RNA (Fig. S1B).
Currently, we do not know whether this class of small RNAs is functional, because in skin, they appeared to be expressed at very low levels. In addition, we saw no overt phenotypic differences between the skins of our Dgcr8 cKO mice, where this class of small RNAs was still present, and our Dicer cKO mice, where it was largely depleted.
We next examined the Dgcr8- and Dicer-null dependent depletion of specific abundant microRNAs whose cloning frequency in each WT sample was >0.1% of the total number of microRNAs (i.e., ≥373 and 325 reads in the WT libraries, respectively). Although most abundant microRNAs were depleted, miR-21 showed an unusually high residual expression level (≈30%) in both cKO samples. Although this could arise from a very small contamination of nonskin cells robustly expressing miR-21, it is also possible that the mature miR-21 has an unusually long half-life that could result in residual expression at P0 even though K14-Cre is strongly active by E14.5.
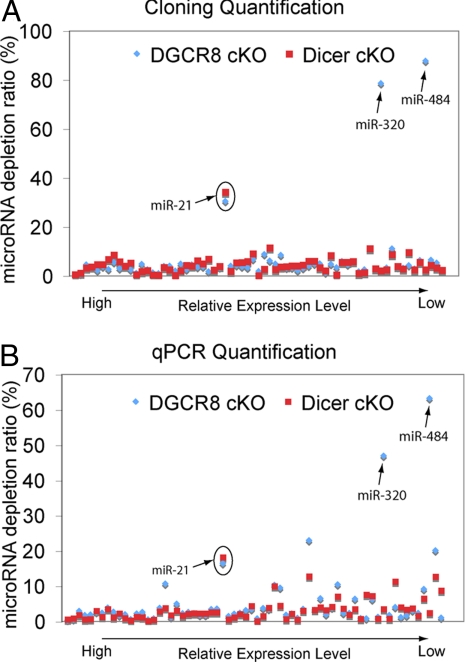
Interestingly, miR-320 and miR-484 were highlyexpressed in the Dgcr8 but not Dicer-deficient samples (Fig. 5A and Table S3). To confirm these distinctions with an alternative approach, we performed real-time qPCR to quantify the relative levels of the 67 most abundantly expressed microRNAs including miR-320 and miR-484 in skin. Consistent with our cloning results, 65 of these 67 microRNAs were depleted similarly in both Dicer and Dgcr8 cKO, whereas miR-320 and miR-484 were depleted only in Dicer but not in the Dgcr8 cKO skin (Fig. 5B). We also noticed that the expression of miR-320 was reported to be independent of DGCR8 in murine ES cells (22).
Fig. 5.
Small RNA cloning and deep sequencing reveals the dependence of microRNA biogenesis on Dicer and DGCR8. (A) Compilation of microRNA sequencing data in Dicer and Dgcr8 cKO samples compared with their WT littermate skins reveal similar dependence of microRNA biogenesis with the exception for miR-320 and miR-484, which require Dicer but not DGCR8. MiR-21 was depleted in both Dicer and Dgcr8 cKO, but only partially (≈70%, circle). MicroRNA depletion ratios were normalized against reads from rRNA, tRNA, and spiked-in calibration RNAs, none of which depends on Dicer or DGCR8. (B) qPCR microRNA expression validation of the cloning/sequencing results. Of 67 skin microRNAs subjected to quantification by qPCR, 65 were dependent similarly on Dicer and DGCR8. MiR-320 and miR-484 displayed dependency on Dicer but not DGCR8.
To probe how the biogenesis of miR-320 and miR-484 does not require DGCR8, we examined the secondary structure of their precursors predicted by miRBase (28). Although premiR-320 appeared to be a bona fide microRNA with the characteristic hairpin structure for the recognition by DGCR8 (19, 20, 29) (Fig. S2A), the annotated flanking sequence of miR-484 did not fold into the typical hairpin structure expected from a prototypical microRNA precursor (Fig. S2B). However, we noticed that the annotated flanking sequences of miR-484 do not completely overlap with the conserved sequences flanking mature miR-484 among vertebrates (Fig. S3A). Interestingly, when we used the conserved sequences flanking mature miR-484 (Fig. S3B) to predict the secondary structure of the miR-484 precursor by mfold (30), the resulted secondary structure showed the characteristic hairpin structure for prototypical microRNAs (Fig. S3C). Based on these data, we surmise that there may be additional features in miR-320 and miR-484 precursors that can allow their independence of DGCR8 recognition whereas the release of mature miR-320 and miR-484 from the hairpin is still dependent on Dicer.
Recently, an alternative mirtron pathway that does not require Drosha/DGCR8 processing was reported (8–10). However, we only recovered miR-877 and miR-1224, two of the most conserved and abundant mirtrons (10) in our libraries. The total reads for these two mirtron microRNAs were only 12 of 719,315 microRNA reads. This observation suggests that the mirtron pathway does not play a major role in microRNA biogenesis in skin.
Discussion
Together, our phenotypic and small RNA profiling studies confirmed critical roles of microRNAs during mammalian skin development. Our deep sequencing analysis suggests that Dicer but not DGCR8 (the microprocessor complex) may be involved in the biogenesis of small RNAs, for example, endo-siRNA and sn/snoRNA-derived small RNAs in the skin. However, the indistinguishable phenotypic consequences of targeting Dgcr8 or Dicer during embryonic skin development support the view that the primary function of both proteins is in processing of microRNAs. It is difficult to distinguish whether the skin differs in this respect from ES cells. It is formally possible that the reported growth-related differences between Dicer and Dgcr8 null ES cells may be attributable not to differences in Dicer and DGCR8 function, but rather to variations in cell culture conditions and/or variations in procedures used by different groups to derive these mES lines (16, 21, 22).
Taken together our findings provide valuable insights into the relative importance of microRNAs and other small RNAs in a mammalian tissue in vivo. When contrasted against studies in mouse oocytes and ES cells, the lack of marked skin differences between Dicer and Dgcr8 mutant mice suggest that different cell types and/or tissues may differ in this regard. Future comparative studies of Dicer and Dgcr8 cKO in other developmental systems, for example, hematopoiesis, myogenesis, and neurogenesis, should help in ascertaining the extent to which microRNAs dominate among other classes of small RNAs in regulating mammalian development.
Materials and Methods
Small RNA Cloning, Sequencing, and Annotation.
Total RNAs were isolated from newborn skin as described (23). Fifteen micrograms of total RNAs were used for the preparation of each library for Solexa sequencing as described (26). For each library, 0.01 fmol of each of the internal control oligonucleotides were added into the total RNA. Small RNA reads were matched against the known transcripts of known noncoding RNAs as described (31, 32). For the mapping of small RNA reads to mRNAs, small RNA reads that were perfectly matched to mouse mRNA (75,876 entries retrieved from GenBank) were scored. Data were shown when their total hits are >10 times in four libraries.
Immunofluorescence and Antibodies.
OCT sections were fixed for 10 min in 4% PFA in PBS and washed three times for 5 min in PBS at RT. Immunofluorescence was performed as described (23). The primary antibodies used as the indicated concentrations were: Lef1 (1:100, Fuchs Lab), β4 Integrin (β4, 1:100, BD Biosciences), Active Caspase-3 (1:1000, R&D Systems).
Real-Time PCR Gene Expression Analysis.
MicroRNA real-time PCR quantification were performed by using the miScript system (Qiagen) according to the manufacture's instruction. SnoRNA25 RNA were served as the internal control. The LightCycler 480 system was used for real-time PCR. Differences between samples and controls were calculated based on the 2−ΔΔCP method.
Electron Microscopy.
Tissues were fixed for greater or equal than 1 h in 2% glutaraldehyde, 4% formaldehyde, and 2 mM CaCl2 in 0.05 M sodium cacodylate buffer, and then processed for Epon embedding. Samples were visualized with a Tecnai G2 transmission electron microscope.
Supplementary Material
Acknowledgments.
We thank E. Lai for comments and suggestions, N. Stokes, L. Polak, and D. Oristian for assistance in the mouse facility, S. Dewell for assistance with Solexa sequencing and data analysis, and A. Tarakhovsky (The Rockefeller University, New York) for Dicerfl/fl mice. This work was supported by grants from the National Institutes of Health (NIH) and the Howard Hughes Medical Institute (E.F., T.T., and M.S.) and NIH Pathway to Independence Award K99AR054704 (to R.Y.). E.F. and T.T. are investigators of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810766105/DCSupplemental.
References
- 1.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y, et al. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denli AM, et al. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 7.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 8.Okamura K, et al. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berezikov E, et al. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lund E, et al. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 14.Hutvagner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 15.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 19.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 20.Sohn SY, et al. Crystal structure of human DGCR8 core. Nat Struct Mol Biol. 2007;14:847–853. doi: 10.1038/nsmb1294. [DOI] [PubMed] [Google Scholar]
- 21.Murchison EP, et al. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, et al. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi R, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 24.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andl T, et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner M, et al. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedersen JS, et al. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berninger P, Gaidatzis D, van Nimwegen E, Zavolan M. Computational analysis of small RNA cloning data. Methods. 2008;44:13–21. doi: 10.1016/j.ymeth.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.