Abstract
Despite the attention given to them, the Galápagos have not yet finished offering evolutionary novelties. When Darwin visited the Galápagos, he observed both marine (Amblyrhynchus) and land (Conolophus) iguanas but did not encounter a rare pink black-striped land iguana (herein referred to as “rosada,” meaning “pink” in Spanish), which, surprisingly, remained unseen until 1986. Here, we show that substantial genetic isolation exists between the rosada and syntopic yellow forms and that the rosada is basal to extant taxonomically recognized Galápagos land iguanas. The rosada, whose present distribution is a conundrum, is a relict lineage whose origin dates back to a period when at least some of the present-day islands had not yet formed. So far, this species is the only evidence of ancient diversification along the Galápagos land iguana lineage and documents one of the oldest events of divergence ever recorded in the Galápagos. Conservation efforts are needed to prevent this form, identified by us as a good species, from extinction.
Keywords: genetic isolation, molecular phylogeny, evolution, islands, lizards
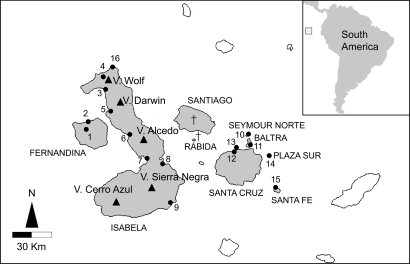
Since Darwin's pioneering work in the archipelago, the Galápagos Islands have been a major scientific resource for evolutionary biologists. This archipelago, currently consisting of about a dozen volcanic islands and more than 100 associated islets, is located on the equator, about 1,000 kilometers west of the South-American coast (Fig. 1). Given their volcanic origin, the Galápagos Islands host unique flora and fauna that have evolved over millions of years in geographic isolation, generating a variety of endemic species with unique and varied ecological, morphological, and behavioral adaptations (1, 2).
Fig. 1.
Galápagos Islands. The islands where land iguanas occur or have occurred in historic times are in gray. The locations of sampling sites are reported in Materials and Methods.
Land iguanas are among the most spectacular representative species of the Galápagos Islands. They once lived in many areas of the Galápagos archipelago (Fig. 1). Currently, many factors contribute to their threatened status (3), one of which may be incomplete taxonomy (4). Two species of Galápagos land iguanas are currently recognized: Conolophus pallidus and Conolophus subcristatus, with the former occurring only on Santa Fe, whereas C. subcristatus occurs on Fernandina, Isabela, Santa Cruz, Plaza Sur, Seymour Norte (a translocated population), and Baltra.
Charles Darwin visited the Galápagos in 1835. During the 5 weeks of his stay in the archipelago, he did not explore Volcan Wolf (the northernmost volcano in Isabela). Thus, although he noticed and commented on both marine and land iguanas (5), he did not encounter a distinct form of land iguana that occurs only on that volcano. Perhaps even more surprising, this form (herein referred to as “rosada,”† meaning “pink” in Spanish) remained unrecorded despite many other scientists having visited Volcan Wolf over the past century. Since it was accidentally seen by some Galápagos National Park rangers in 1986, this form has received no attention. The rosada form is characterized by a distinct phenotype (Fig. 2). It can be clearly distinguished from the syntopic yellow form (C. subcristatus) by evident idiosyncrasies in morphology and color.
Fig. 2.
Adult male iguanas of the yellow (A, Sierra Negra; D, Volcan Wolf) and rosada (B and C, Volcan Wolf) forms (photograph by G.G.)
Earlier genetic studies suggest that the split of the marine and land iguana lineages could have occurred as late as 10.5 million years ago, when the archipelago did not have the current configuration and none of the present islands had yet emerged. Such studies also suggested that the present pattern of diversification of land iguanas originated recently, during the Pleistocene Epoch (6). However, no previous studies included the rosada form. Here, we address its genetic distinctiveness and taxonomic status by means of mtDNA sequencing and microsatellite genotyping.
Results
Phylogenetic Relationships and Genetic Divergence.
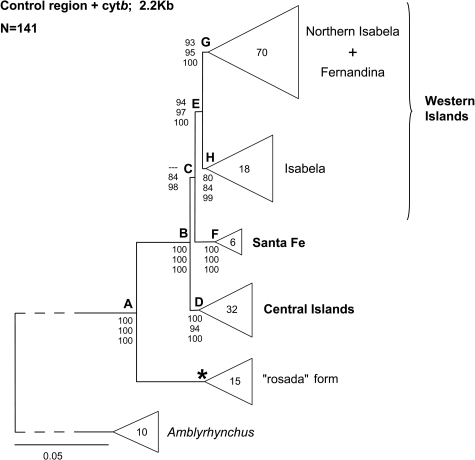
Combined (Pincongruence length difference (ILD) test = 0.17) and separate phylogenetic analyses of the mitochondrial control region (CR) and cytochrome b (cytb) data sets as well as parsimony, maximum likelihood (ML), and Bayesian methods of inference produced very similar results. Eight nodes (Fig. 3 A–H) within the Conolophus clade were strongly supported. Strikingly, the primary split was between the rosada form and all other populations. Not surprisingly, the populations of (i) Santa Fe, (ii) the central islands (Baltra, Santa Cruz, and Plaza Sur), and (iii) the western islands (Fernandina and Isabela) each formed a monophyletic group. All topologies alternative to positioning rosada at the base of the Conolophus tree were rejected, with PShimodaira-Hasegawa (S-H) test ranging from 0.004 to ≪0.001. On the other hand, the S-H test accepted the alternative hypotheses: (i) Santa Fe (central islands, western islands): P = 0.444 and (ii) (Santa Fe, central islands) western islands: P = 0.434.
Fig. 3.
ML phylogenetic tree. The tree is rooted at the midpoint. The branches subtending the node A and the genus Amblyrhynchus have been equally shortened. The asterisk denotes a terminal node. Numbers at the nodes indicate statistical support as follows (from top): MP, ML, Bayesian analysis. For MP and ML, only bootstrap values higher than 70 are shown. For Bayesian analysis numbers indicate posterior probability values. The number of individuals examined is reported inside each terminal triangle.
On average, ML average genetic distances among populations within the central islands group and within the western islands group (excluding the rosada form) were 0.0026 ± 0.0011 (SD) and 0.0044 ± 0.0023, respectively. Higher values were observed between the 2 groups (0.0148 ± 0.0008). The ML average genetic distance between C. pallidus and C. subcristatus was 0.0177 ± 0.0006. The 15 rosada individuals were identical, and their ML average genetic distance from the rest of the land iguanas was 0.0741 ± 0.0018. The ML average genetic distance between Amblyrhynchus and Conolophus was 0.2576 ± 0.0038.
The recombination detection program (RDP3) analysis did not provide any evidence of mtDNA recombination between marine and land iguanas, including the rosada form (all tests were not significant with P set at 0.05 and Bonferroni correction).
Our estimate of the divergence time for the basal split between the rosada and the rest of land iguana lineages (Fig. 3, node A) was ≈5.70 million years (±1.32, SD).
Microsatellite Structure and Variation.
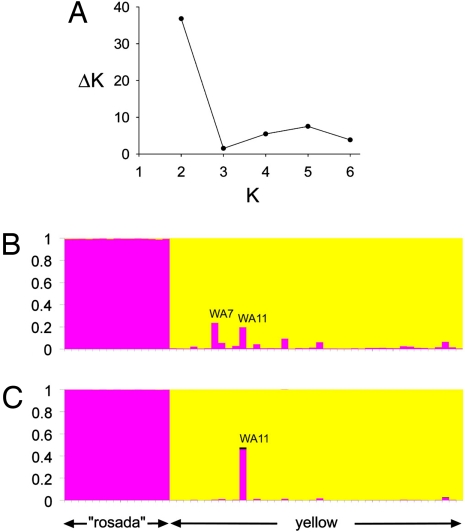
The first STRUCTURE analysis indicated K = 2 as the most appropriate number of clusters (Fig. 4A). The MICRO-CHECKER analysis performed on the 2 clusters did not indicate scoring error attributable to stuttering or allele dropout. Only locus CS8 showed evidence of null alleles in the second cluster (P < 0.001). All rosada individuals were included in the first cluster (rosada group) with q = >0.99. All yellow individuals were assigned to the second cluster (yellow group) with an average q = 0.97 (0.05, SD). Two yellow individuals assigned to the yellow cluster, WA7 and WA11, exhibited q values equal to 0.76 and 0.80, respectively (Fig. 4B).
Fig. 4.
STRUCTURE analyses. (A) ΔK values are shown for K ranging between 2 and 6. Maximum value is observed when K = 2. (B) Results after the first analysis indicate individuals WA7 and WA11 (yellow morphotypes) as possible hybrids. The second analysis, which was more refined, indicates WA11 as a possible second-generation hybrid (see text).
The second set of STRUCTURE analyses confirmed the assignment of the individual WA7 to the yellow group (q ranging from 0.87 to 0.95, depending on ν values used), whereas the individual WA11 showed mixed ancestry with a substantial proportion of genes derived from a rosada grandparent (q ranging from 0.87 to 0.46, depending on ν values used; Fig. 4C).
Evidence for linkage disequilibrium between loci CS5 and CS9 (P < 0.05, after Bonferroni correction) was found only for the yellow group. This group also showed deviation from Hard-Weinberg Equilibrium (HWE) at locus CS9 attributable to heterozygosity deficiency (P < 0.05). The two clusters showed FST = 0.25 (P ≪ 0.001). The genetic differentiation was confirmed using the stepwise mutation model (RhoST = 0.93, P ≪ 0.001), and a comparison between FST and RhoST showed a significant role for allele size in determining the level of population differentiation (P ≪ 0.001).
The levels of variation differed between the 2 groups, with an observed heterozygosity (HO) value of 0.67 for the yellow group and 0.38 for the rosada group. Similarly, average allelic richness was 6.29 (±2.46) and 3.78 (±2.22) in the groups of yellow and rosada clusters, respectively. Private alleles (i.e., alleles whose occurrence is restricted to only 1 group of individuals) constituted 74% of all alleles. Thirteen and 47 private alleles were observed in the rosada and yellow groups, respectively.
Discussion
The most surprising result was the deep divergence of the rosada lineage at the basis of the Conolophus clade. This species alters the current thinking about the timing of diversification of land iguanas, which was previously supposed to have occurred in the Pleistocene Epoch (6). Although with a large SD, our estimate sets the origin of this relict lineage back to a period when at least some of the present-day islands had not yet formed. In fact, the oldest extant islands in the archipelago, San Cristóbal and Española, are at least 2.35 and 3.3 million years old, respectively, if not older (7). Thus, given its present distribution, the rosada form clearly represents a conundrum because it occurs only on Volcan Wolf, which is considered younger than Volcan Sierra Negra (0.53 million years, the oldest volcano of Isabela) (8) and almost as old as Volcan Cerro Azul (0.35 million years) (9).
The ML average genetic distance between C. subcristatus and C. pallidus is much lower than between the rosada form and each of the 2 named species, supporting the distinctiveness of the taxon. Our preliminary data on the morphology of the rosada and yellow forms also indicate differentiation: in addition to their color pattern and independent of their gender, all rosada individuals investigated are distinguished from the other 2 species by flat dorsal head scales and the prominent adipose nuchal crest with small conic scales. The rosada also shows strong differences in the pattern of the “head-bob” (nodding), a behavior important in territoriality (10) and courtship (11).
The microsatellite data also indicated strong differentiation between the rosada and yellow forms, with mutation and genetic drift (in particular for the rosada form) being important determinants. A similar magnitude of microsatellite differentiation was observed among C. subcristatus populations from other islands (12). Although the 2 forms still share 26% of alleles, none of the rosada individuals investigated incorporated genes from the syntopic yellow iguanas, at least in the past 2 generations, and only 1 yellow individual shows possible mixed ancestry with a rosada grandparent. Thus, introgressive hybridization appears to be rare and not sufficiently strong to have prevented genetic differentiation. In any case, incomplete reproductive isolation between the rosada and syntopic yellow land forms is not surprising, considering that hybridization can still occur between marine and land iguanas (13), 2 genera morphologically, ecologically, behaviorally, and genetically very distant.
The mtDNA haplotype of the rosada is highly differentiated from those of marine iguanas and the rest of land iguanas. The results of the RDP3 analyses allow us to reject the hypothesis that such differentiation might have occurred by mtDNA recombination after hybridization between land and marine forms. The hypothesis of the origin of the rosada by recent hybridization alone between the 2 forms is not supported either. In fact, a rosada-like haplotype is not found in our sample of yellow iguanas, or in marine iguanas. This is indicated by a phylogenetic analysis that we performed by combining original haplotypes from the present study with those found by Rassmann et al. (6) in their sample of 150 marine iguanas from 21 locations on 14 islands [see supporting information (SI)].
In addition to the taxonomic implications, this form, which we recognize as a good species, is very important because it carries substantial evolutionary legacy, being basal to all other land iguana remnant populations. Thus far, the rosada form is the only evidence of deep diversification along the Galápagos land iguana lineage. No analogous evidence has been found in marine iguanas so far.
These findings call for a conservation program aimed at evaluating the risk of extinction of this newly recognized species, which, based on currently available data, would be assignable to the “critically endangered” category by meeting criteria B and C of the International Union for Conservation of Nature (IUCN) Red List (14).
Materials and Methods
Sampling.
The reader is referred to Fig. 1 for the location of sampling sites. For mtDNA analysis, samples were collected as follows (the number of individuals sampled is in square brackets): C. subcristatus: (1) Roca Limba [5], (2) Cueva Norte [9], (3) Puerto Bravo [27], (4) Piedras Blanca [31], (5) Caleta Tagus [6], (6) Bahia Urbina [10], (7) Bahia Elizabeth [5], (8) Bahia Cartago [8], (9) Villamil [2], (10) Seymour Norte [3], (11) Baltra [8], (12) Cerro Dragón [4], (13) Venecia [7], and (14) Plaza Sur [10]; C. pallidus: (15) Santa Fe [6]; and Amblyrhynchus cristatus: (14) Plaza Sur [5] and (16) P.ta Albemarle [5]. For microsatellite DNA, samples were collected at Volcan Wolf at sites (3) Puerto Bravo [29] and (4) Piedras Blanca [28]. Of these, 15 individuals (11 male and 4 female) were of the rosada form.
Blood Drawing and DNA Extraction.
Blood (≈1 mL) was drawn from the caudal or brachial vein and preserved in 5 mL of lysis buffer (100 mM Tris, 100 mM EDTA, 2% mg/mL SDS). DNA was extracted using the QIAamp DNA Mini Kit (Qiagen).
mtDNA Amplification.
We used the primer pair 12S1984-CB437LD (15) to amplify by PCR a 1,126-bp fragment of the CR positions 1370–2495 in GenBank sequence AY948121. PCR conditions were as described in ref. 14. The primers TGLU14121 (5′-CCGAAAAATCCACCTTGTTATTCAAC-3′) and TTHRREV (5′-GGGGGTGGTTTAATTCCCAGC-3′) were developed and used to amplify by PCR a 1,200-bp fragment that includes the whole cytb gene and small fragments of the tRNAs for glutamic acid and threonine. Sequences were run on an ABI PRISM 3100 (Applied Biosystems) automated sequencer. Conditions for the PCR amplification of the cytb gene are available as supplementary data. Sequences were edited with SEQUENCHER 4.1.2 (Gene Codes). The alignment obtained by using CLUSTAL X (16) was checked by eye. The final data set consists of 1118 bp of CR and 1113 bp of cytb.
Phylogenetic Analyses.
Phylogeny inference was performed using the CR and the cytb data sets both separately and combined. We tested for phylogenetic incongruence of CR and cytb by performing the ILD test (17), after removing the invariant characters (18). Analyses were conducted using maximum parsimony (MP) (19), ML (20), and Bayesian inference (21), as implemented in PAUP* 4.0b10 (22), TREEFINDER 2006 (23), and MRBAYES 3.1.2 (24), respectively. MP heuristic parameters were as follows: starting trees obtained by random-addition (10 replicates) and Tree Bisection and Reconnection (TBR) branch swapping. Gaps in the CR were coded as unordered characters at the end of the data matrix (contiguous gaps were treated as 1 single gap). ML trees were sought via a genetic algorithm, by which method they were less prone to get trapped in local optima (25). Different models were used for the CR and the cytb data. MODELTEST 3.7 (26) was used to select the HKY85+Γ (Ti/Tv rate = 2.464, α = 0.161) for the CR. The GTR+Γ model was instead used for the cytb, with all parameters estimated separately for the first, second, and third positions. The same models were used in the Bayesian analysis. Gaps were recoded as binary data and considered as a separate partition to which a binary (restriction) model of evolution was applied as implemented in MRBAYES. For each partition, parameters' values were estimated during the search. The first, second, and third positions of the cytb were treated as separate partitions. Such partitioned analyses were aimed at modeling the data more accurately to reduce systematic errors that could mislead phylogenetic analyses (27, 28). For MP and ML, nodal support was tested by bootstrapping (29), with 1,000 pseudoreplicates. A. cristatus, the sister taxon of Conolophus (30), was used as an outgroup within the Iguaninae group. Alternative tree topologies were investigated using the S-H test (31). We estimated times of divergence using a nonparametric approach, as implemented in R8S 1.70 (ref. 32; see SI for details).
Tip-to-tip distances along the ML tree were averaged to calculate ML average genetic distances between populations within and between each group resulting from the phylogenetic analysis.
Recombination Analysis.
We tested for possible mtDNA recombination between Amblyrhynchus and Conolophus by applying the following methods: (i) RDP (33), (ii) GENECONV (34), and (iii) CHIMAERA (35), as implemented in RDP3 (ref. 33; see SI for details).
Microsatellite Characterization.
The extracted DNA was genotyped at 9 microsatellite loci as in Tzika et al. (12).
Microsatellite Structure and Variation.
MICRO-CHECKER 2.2.3 (36) was used to check for possible typing errors, null alleles, large allele dropout, and errors attributable to stutter peaks. Population structure and individual assignment were performed using a Bayesian method implemented in STRUCTURE 2.2 (37). We first performed a STRUCTURE analysis to assess the number of groups (K). We used a model that assumed admixture and uncorrelated allele frequencies and used no prior population information. The K value that maximized the statistic ΔK (38) was chosen as the optimal K value. Based on the assignment obtained, once the K value was assessed, we reran STRUCTURE using the number of distinct clusters and the phenotype of individuals (yellow vs. rosada) as prior population information. This was aimed to infer the ancestry of all individuals that, from the previous run, could potentially have mixed ancestry. In this run, we used a model that accounted for the occurrence of null alleles (39), as the MICRO-CHECKER analysis suggested. Aware of the resolution limit of the number of loci used (40, 41), we set the parameter GENSBACK = 2 to investigate the hypothesis that each individual belongs to the alternative phenotype or has 1 parent or grandparent with the alternative phenotype. To test for sensitivity of the data to ν (in our case, the probability that an individual may be misclassified or has mixed ancestry within the past 2 generations), we conducted 3 runs by setting the parameter MIGRPRIOR = 0.1, 0.05, and 0.01, as suggested by Pritchard et al. (37). For all STRUCTURE analyses, membership coefficients (q) were calculated using 1.0 × 106 repetitions of a Markov chain Monte Carlo simulation, after an initial “burnin” of 5.0 × 105 repetitions. Tests for significant deviations from HWE and genotyping disequilibrium were performed as implemented in GENEPOP 3.3 (42). The level of polymorphism was measured as the mean number of alleles per locus and both HO and expected heterozygosity, respectively, using the program GENETIX 4.05 (43). Because the observed number of alleles in a sample is dependent on sample size, allelic richness was also estimated with the program FSTAT 2.9.3 (44). Population differentiation was investigated taking into consideration both the infinite and stepwise mutation models. Assuming an infinite mutation model, the Wright's fixation index FST was assessed by the estimator θ with the program FSTAT 2.9.3. The estimator RhoST of the RST statistic, which is based on a stepwise mutation model, was calculated using the program RSTCALC (45). To determine whether stepwise-like mutations contributed to genetic differentiation, we performed a statistical test based on randomization of allele size (46). The test, implemented in the program SPAGeDi (47), can be interpreted as testing whether FST = RhoST. In case of tests with multiple comparisons, the sequential method of Holm (48), also known as “sequential Bonferroni,” was applied as implemented in the MULTIPLICITY PROGRAM 2 (49).
Supplementary Material
Acknowledgments.
We thank the Parque Nacional Galápagos and the Charles Darwin Research Station, G. dell'Omo, S. Rosa, R. Palozzi, M. Cruz, and all the people who supported this study in the collection of samples. We acknowledge D. Tautz, K. Rassmann, and A. Caccone for making their samples available for our study. We thank to A. Caccone and M. Milinkovitch for their criticism on an early version of this paper. Samples were collected under CITES permits IT/IM/2003/MCE/02328, IT/IM/2004/MCE/00041, and IT/IM/2005/MCE/01819 granted to the first author. This work was supported by the Italian Ministry of Education, University, and Research (MIUR), with a grant to G.G.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. FJ536155–FJ536164 (CR) and FJ536145–FJ536154 (cytb) for Amblyrhynchus and FJ536004–FJ536144 (CR) and FJ535863–FJ536003 (cytb) for Conolophus].
This paper and the name “rosada” used herein are disclaimed for nomenclatural purpose [Articles 8.2, 8.3 in ICZN International Code of Zoological Nomenclature. Fourth Edition (ITZN, Padova, 1999)]. We postpone a formal description.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806339106/DCSupplemental.
References
- 1.Darwin C. The Origin of Species by Means of Natural Selection. London: John Murray; 1859. [Google Scholar]
- 2.Bowman RI. In: Patterns of Evolution in Galápagos Organisms. Bowman RI, Berson M, Leviton AE, editors. Pacific Division, San Francisco: American Association for the Advancement of Science; 1983. pp. 237–537. [Google Scholar]
- 3.Snell HL, Snell MH, Tracy CR. Variation among populations of Galápagos land iguanas (Conolophus)—Contrasts of phylogeny and ecology. Biol J Linn Soc. 1984;21:185–207. [Google Scholar]
- 4.Daugherty C, Cree A, Hay JM, Thompson MB. Neglected taxonomy and continued extinctions in Tuatara. Nature. 1990;347:177–178. [Google Scholar]
- 5.Darwin C. The Voyage of the Beagle. New York: Dutton; 1950. [Google Scholar]
- 6.Rassmann K, Markmann M, Trillmich F, Tautz D. In: Iguanas, Biology and Conservation. Alberts AC, Carter RL, Hayes WK, Martins EP, editors. Berkeley: Univ California Press; 2004. pp. 71–83. [Google Scholar]
- 7.White WM, McBirney AR, Duncan RA. Petrology and geochemistry of the Galápagos Islands: Portrait of a pathological mantle plume. J Geophys Res-Solid Earth. 1993;98:533–563. [Google Scholar]
- 8.Nordlie BE. Morphology and structure of the western Galápagos volcanoes and a model for their origin. Geol Soc Am Bull. 1973;84:2931–2956. [Google Scholar]
- 9.Naumann T, Geist D. Physical volcanology and structural development of Cerro Azul Volcano, Isabela Island, Galápagos: Implications for the development of Galápagos-type shield volcanoes. Bull Volcanol. 2000;61:497–514. [Google Scholar]
- 10.Carpenter CC. Behavioral and ecological notes on the Galápagos land iguanas. Herpetologica. 1969;25:155–164. [Google Scholar]
- 11.Christian KA, Tracy CR. In: Iguanas of the World. Burghardt GM, Rand AS, editors. Park Ridge, NJ: Noyes Publications; 1982. pp. 366–379. [Google Scholar]
- 12.Tzika AC, et al. Population genetics of Galápagos land iguana (genus Conolophus) remnant populations. Mol Ecol. 2008;17:4943–4952. doi: 10.1111/j.1365-294X.2008.03967.x. [DOI] [PubMed] [Google Scholar]
- 13.Rassmann K, Trillmich F, Tautz D. Hybridization between the Galapagos land and marine iguana (Conolophus subcristatus and Amblyrhynchus cristatus) on Plaza Sur. J Zool Lond. 1997;242:729–739. [Google Scholar]
- 14.International Union for Conservation of Nature. IUCN Red List Categories and Criteria. Gland, Switzerland: IUCN; 2001. [Accessed June 2008]. Version 3.1 (IUCN Species Survival Commission) http://www.iucnredlist.org/info/categories_criteria2001. [Google Scholar]
- 15.Hanley TC, Caccone A. Development of primers for the characterization of the mitochondrial control region of Galápagos land and marine iguanas (Conolophus and Amblyrhynchus) Mol Ecol Notes. 2005;5:599–601. [Google Scholar]
- 16.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farris JS, Kallersjo M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1994;10:315–319. [Google Scholar]
- 18.Cunningham CW. Can three incongruence tests predict when data should be combined? Mol Biol Evol. 1997;14:733–740. doi: 10.1093/oxfordjournals.molbev.a025813. [DOI] [PubMed] [Google Scholar]
- 19.Farris JS. Methods for computing Wagner trees. Syst Zool. 1970;18:374–385. [Google Scholar]
- 20.Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 21.Bayes T. Studies in the history of probability and statistics: IX. Thomas Bayes's essay towards solving a problem in the doctrine of chances. Biometrika. 1763/1958;45:296–315. [Google Scholar]
- 22.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods) Sunderland, MA: Sinauer Associates; 2002. Version 4. [Google Scholar]
- 23.Jobb G. TREEFINDER. Germany: Munich; 2006. [Accessed May 2006]. Version of May 2006. Distributed by the author at www.treefinder.de. [Google Scholar]
- 24.Ronquist F, Huelsenbeck JP. In: Statistical Methods in Molecular Evolution. Nielsen R, editor. New York: Springer; 2005. pp. 183–233. [Google Scholar]
- 25.Jobb G, von Haeseler A, Strimmer K. TREEFINDER: A powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471–2148-4–18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 27.Reeder TW. A phylogeny of the Australian Sphenomorphus group (Scincidae: Squamata) and the phylogenetic placement of the crocodile skinks (Tribolonotus): Bayesian approaches to assessing congruence and obtaining confidence in maximum likelihood inferred relationships. Mol Phylogenet Evol. 2003;27:384–397. doi: 10.1016/s1055-7903(02)00448-7. [DOI] [PubMed] [Google Scholar]
- 28.Brandley MC, Schmitz A, Reeder TW. Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst Biol. 2005;54:373–390. doi: 10.1080/10635150590946808. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 30.Wiens JJ, Hollingsworth BD. War of the iguanas: Conflicting molecular and morphological phylogenies and long-branch attraction in iguanid lizards. Syst Biol. 2000;49:143–159. doi: 10.1080/10635150050207447. [DOI] [PubMed] [Google Scholar]
- 31.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 32.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 33.Martin DP, Williamson C, Posada D. RPD2: Recombination detection and analysis from sequence alignments. Bioinformatics. 2005;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
- 34.Padidam M, Sawyer S, Fauquet CM. Possible emergence of new geminiviruses by frequent recombination. Virology. 1999;265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- 35.Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: Computer simulation. Proc Natl Acad Sci USA. 2001;98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Oosterhout C, Hutchinson WF, Derek PM, Wills DPM, Shipley P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- 37.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;55:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 39.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boecklen WJ, Howard DJ. Genetic analysis of hybrid zones: Numbers of markers and power of resolution. Ecology. 1997;78:2611–2616. [Google Scholar]
- 41.Vähä JP, Primmer CR. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol Ecol. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- 42.Raymond M, Rousset F. An exact test for population differentiations. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- 43.Belkhir K, Borsa P, Chikhi L, Raufaste N, Catch F. Laboratoire Génome, Populations, Interactions CNRS UMR 5000. Montpellier, France: Univ de Montpellier II; 2004. [Google Scholar]
- 44.Goudet J. Fstat version 1.2: A computer program to calculate F-statistics. J Hered. 1995;86:485–486. [Google Scholar]
- 45.Goodman SJ. RSTCalc: A collection of computer programs for calculating estimates of genetic differentiations from microsatellite data and determining their significance. Mol Ecol. 1997;6:881–885. [Google Scholar]
- 46.Hardy OJ, Charbonnel N, Fréville H, Heuertz M. Microsatellite allele sizes: A simple test to assess their significance on genetic differentiation. Genetics. 2003;163:1467–1482. doi: 10.1093/genetics/163.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hardy OJ, Vekemans X. SPAGeDi: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2002;2:618–620. [Google Scholar]
- 48.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 49.Brown BW, Russell K. Multiplicity Program. Houston: Univ of Texas; 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.