Abstract
Allelic exclusion of Ig gene expression is necessary to limit the number of functional receptors to one per B cell. The mechanism underlying allelic exclusion is unknown. Because germline transcription of Ig and TCR loci is tightly correlated with rearrangement, we created two novel knock-in mice that report transcriptional activity of the Jκ germline promoters in the Igκ locus. Analysis of these mice revealed that germline transcription is biallelic and occurs in all pre-B cells. Moreover, we found that the two germline promoters in this region are not equivalent but that the distal promoter accounts for the vast majority of observed germline transcript in pre-B cells while the activity of the proximal promoter increases later in development. Allelic exclusion of the Igκ locus thus occurs at the level of rearrangement, but not germline transcription.
Keywords: allelic exclusion, knock-in mouse
The V(D)J recombinase and various DNA repair proteins catalyze the assembly of Ig heavy- and light-chain variable region exons from dispersed gene-segments during B cell development (1). The presence of two alleles for each gene and two light chain loci (Igκ and Igλ), suggests that, in theory, each B cell could produce eight different antibody specificities. However, as postulated by Burnet 50 years ago (2), the vast majority of B cells are monospecific. The underlying basis for the “one cell, one receptor” phenomenon is the allelic and isotypic exclusion of Ig gene expression such that the vast majority of mature B cells possess only one functional heavy and one functional light chain gene rearrangement. Despite intensive effort, the mechanism underlying allelic exclusion is unknown.
Two general models have been proposed to account for allelic exclusion (3). The first is an instructive model in which an event early in development differentially marks the two alleles such that one allele is the preferred substrate for recombinase activity. Once established, this differential mark propagates in a clonal manner. The second model proposes a stochastic mechanism where each allele is equally likely to recombine, but the probability of recombination is low so that the percentage of cells with two functional rearrangements is vanishingly small. In support of the first model, it was observed that the two mouse Igκ alleles replicate asynchronously in S-phase beginning at a very early stage of development (4). However, it has not been proven conclusively that the early replicating allele is the preferential target of recombination, nor what other marks might be clonally propagated to dictate allele-specific recombination. In favor of the second model, we previously reported that a GFP cDNA knocked-in to the mouse Igκ locus is expressed in only 1–5% of pre-B cells (5). As germline transcription is tightly correlated with recombination, we interpreted this to indicate that only a small fraction of unrearranged Igκ alleles are transcriptionally active and therefore suitable targets for the recombinase at any given time. Neither model fully accounts for observed recombination frequencies or patterns, in particular receptor editing of the Igκ locus (6). If the preferred allele is dictated, it is unclear how the second allele becomes a substrate for recombination if the first fails to encode a suitable protein. Similarly, if recombination is rare and stochastic, it is unlikely that a cell would ever recombine both alleles; yet a significant fraction of splenic B cells contain two rearranged Igκ alleles (6).
All rearranging loci undergo sterile or germline transcription. Sterile transcripts originate from promoters upstream of the recombining segments in a developmental and tissue-specific manner that correlates tightly with recombination of the associated loci (7, 8). A recent report demonstrated that germline transcription through a cluster of Jα segments is necessary for their recombination (9). Insertion of a transcription termination sequence into the Jα cluster prevented local Vα-to-Jα rearrangement. These workers concluded that RNA polymerase II transit across the recombining segments recruits chromatin modifications that make the locus accessible to the recombinase. The Jκ cluster of gene segments contains two germline transcript promoters, located approximately 100 bp and 3.5 kb upstream of the Jκ1 gene segment (10–12). Deletion of both promoters abolishes κ locus recombination in AMuLV transformed pro-B cell lines, although the identical experiment has not been done in mice nor has either promoter been deleted individually (13, 14). Although germline transcription appears necessary for recombination, it is unclear if it is sufficient. If germline transcription is sufficient to target the recombinase, monoallelic expression of germline transcripts may underlie antigen receptor allelic exclusion. However, a previous report demonstrated by single cell RT-PCR and RNA FISH that germline transcription of the Jκ cluster is biallelic (15). This is in contrast to our report of variegated, predominantly monoallelic expression of this locus (5). Because these results are at odds with one another, it remains unclear how the Jκ germline transcripts are expressed and how this might regulate locus recombination.
To further probe the mechanism of allelic exclusion and its relation to germline transcription, we created two new knock-in mouse strains that report transcriptional activity of the germline Igκ locus. We found, contrary to expectation, that germline transcription of the Jκ cluster is biallelic and occurs in all pre-B cells. In reconciling this data with previous literature, we discovered that the two Jκ germline promoters in the Igκ locus are not equivalent and that the bulk of germline transcription in this locus derives from the distal promoter in pre-B cells.
Results
New Igκ Knock-In Mice Reveal Germline Transcription in All Developing Pre-B Cells.
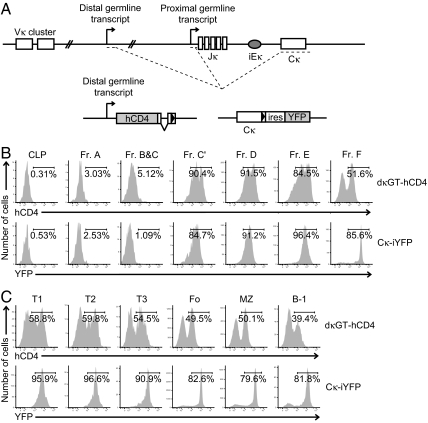
Our previously studied Igκ reporter mouse strain contained a GFP cDNA, heterologous intron and polyadenylation (polyA) sequence inserted into the first Jκ gene segment (referred to as κ0GFP) (5). Although this strain reported proximal promoter activity, the polyA sequence decreased recombination to gene segments 3′ of the knock-in (data not shown) and resulted in skewed allele usage in mature B cells in heterozygous mice. To confirm our previous results in a more rigorous manner, we sought to create additional reporter mice which would not affect recombination of the locus. Because the Jκ cluster contains two germline promoters that were thought to be equivalent, we reasoned that placement of a reporter cassette and polyA sequence downstream of the distal promoter was less likely to affect recombination as the proximal promoter was left intact (the polyA sequence is necessary for proper transcript processing and expression of the reporter cassette). We created a mouse in which a human CD4 (hCD4) cDNA, intron and polyA sequence is inserted downstream of the distal Jκ germline promoter, approximately 3.5 kb upstream of the Jκ1 gene segment (referred to subsequently as dκGT-hCD4, Fig. 1A and supporting information (SI) Fig. S1a). The hCD4 was mutated to prevent intracellular signaling and interaction with class II MHC (16). Because the dκGT-hCD4 insertion could potentially disrupt Igκ control elements, we created a second mouse with an internal ribosome entry site (IRES) and yellow fluorescent protein (YFP) cDNA inserted after the Cκ stop codon and before the endogenous polyA sequence (referred to as Cκ-iYFP, Fig. 1A and Fig. S1b). The Cκ-iYFP insertion has the additional advantage of serving as a reporter for both germline and rearranged transcripts since both include the Cκ exon. Both strains of mice were born in equal proportion to wild-type litter mates and revealed no gross physiological abnormalities (data not shown).
Fig. 1.
Marker protein expression in dκGT-hCD4 and Cκ-iYFP knock-in mice. (A) Schematic of the dκGT-hCD4 and Cκ-iYFP knock-in mutations at the Igκ locus. Both the proximal and distal germline transcript promoters are shown as arrows. Previously defined splicing patterns of both transcripts are depicted as dashed lines. The hCD4 cDNA is followed by an SV40 intron and polyA sequence. Black triangles represent the positions of loxP sites remaining in the locus after Cre recombinase-mediated deletion of a neomycin resistance gene. (B) Flow cytometric analysis of hCD4 or YFP marker expression in developing bone marrow B cells from heterozygous dκGT-hCD4 or Cκ-iYFP knock-in mice. Harvested bone marrow was labeled with antibodies to delineate B cell developmental subsets and mark hCD4-expressing cells (in dκGT-hCD4 mice only). CLP through fraction F are shown. The percentage of marker-positive cells at each developmental stage is shown. Control C57/BL6 mice had no detectable expression of either marker (data not shown). Data are representative of at least five independent experiments analyzing two to four mice per experiment. (C) Flow cytometric analysis of hCD4 or YFP marker expression in splenic B cells from heterozygous dκGT-hCD4 or Cκ-iYFP knock-in mice. Harvested splenocytes were labeled with antibodies to delineate B cell developmental subsets and mark hCD4-expressing cells (in dκGT-hCD4 mice only). Transitional 1 (T1), transitional 2 (T2), transitional 3 (T3), follicular (Fo), marginal zone (MZ), and B-1 B cell subsets are shown along with the percentage of marker positive cells at each developmental stage. Data are representative of at least two independent experiments analyzing two to four mice per experiment.
We predicted the pattern of marker protein expression from the new knock-in strains would be similar to the κ0GFP mouse. However, flow cytometric analysis of heterozygous dκGT-hCD4 and Cκ-iYFP mice revealed a very different pattern of marker expression during B cell development (Fig. 1 B and C). Compared to wild-type mice, heterozygous animals from both knock-in strains displayed very little marker expression in the early stages of B cell development (common lymphoid progenitor (CLP) to fraction C). This was expected since the Igκ locus does not begin to rearrange until the pre-B cell stage (fraction D) of development. The small percentage of marker-positive cells in fractions B and C derive mainly from cells transitioning from C to C' (data not shown). At the large cycling and small resting pre-B cell stages (fractions C' and D) almost all B cells became hCD4 or YFP positive. This is in contrast to the κ0GFP mice, in which <1% of small resting pre-B cells from heterozygotes are GFP positive (data not shown). In the dκGT-hCD4 strain, the marker is in the Vκ-Jκ genomic interval and thus is deleted from the locus upon Vκ-to-Jκ rearrangement. As such, we found hCD4 marker expression began to decline in immature B cells (fraction E) and was reduced to approximately 50% in transitional and splenic B cells (Fig. 1C). In contrast, the YFP marker is downstream of Cκ, remains in the locus after rearrangement and continues to be transcribed from the incoming Vκ promoter. Because Vκ promoters are stronger than the germline Jκ promoters, we found the mean fluorescence intensity of YFP to steadily increase as cells progressed from pre-B to splenic B cells. We did detect a fraction of YFP negative cells in IgD+ bone marrow and splenic B cells. These correspond to alleles that have been deleted by recombination to the recombining sequence (RS) element (data not shown).
The κ0GFP knock-in disrupted normal recombination of the mutant allele and led to a 4:1 skew in Igκ usage in favor of the wild-type allele in IgM+ B cells (5). To determine if either dκGT-hCD4 or Cκ-iYFP insertions cause similar recombination defects, we mated both strains to hCκ knock-in mice which carry a human Igκ constant region exon in place of the mouse exon. Analysis of immature B and splenic B cells in hCκ/dκGT-hCD4 heterozygous mice revealed a 2:1 skew in favor of expression of the human Cκ allele (Fig. S2) (17). This agreed with the higher than expected percentage of hCD4 positive splenic B cells (50%) in heterozygous dκGT-hCD4 mice. If the hCD4 knock-in were truly innocuous, approximately 30% of these cells would retain the hCD4 allele in the germline configuration and hence be hCD4 surface positive. In contrast, immature B cells from hCκ/Cκ-iYFP heterozygous mice had a 1:1 ratio of mouse to human Cκ, similar to what is observed in hCκ/C57BL6 heterozygous mice. Homozygous dκGT-hCD4 and Cκ-iYFP mice had normal numbers of B cells at all developmental stages compared to 129/SvJ wild-type littermates (data not shown). Thus, while the hCD4 insertion does cause a slight recombination defect, it does not grossly disrupt B cell development. As predicted, the Cκ-iYFP insertion appeared completely innocuous.
Germline Transcription of the Jκ Cluster Is Bialleic.
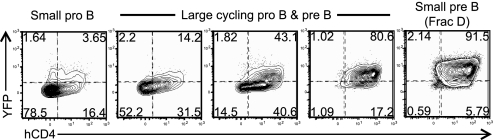
Essentially all small resting pre-B cells appeared positive for either the hCD4 or YFP markers in heterozygotes from the respective knock-in strains. This result suggested that germline transcription may be biallelic. To confirm this interpretation, we mated dκGT-hCD4 and Cκ-iYFP mice and analyzed the resulting compound heterozygotes for expression of both markers during B cell development (Fig. 2). We found that expression of both markers increased as cells progressed from pro-B to the pre-B stage, with almost all small resting pre-B cells being positive for both markers. The hCD4 marker appeared earlier during the pro-to-pre-B cell transition than YFP. This might be due to the fact that the hCD4 marker is a few hundred base pairs from the distal promoter and thus is sensitive to any transcription initiating from this region while the YFP lies at the end of the transcript and requires that RNA polymerase II transcribes the entire 8 kb region; any stalled or aborted transcripts would not give rise to YFP protein. Alternatively, the sensitivity of the assay for hCD4 may exceed that for YFP. Regardless, all pre-B cells were double positive, confirming that on a single cell level, both Igκ alleles are transcriptionally active during recombination of this locus. This is in agreement with a previous report which demonstrated biallelic Jκ transcription by single-cell RT-PCR analysis and FISH (15).
Fig. 2.
Germline transcription at the Jκ cluster is biallelic. Flow cytometric analysis of marker expression in developing B cells from dκGT-hCD4/Cκ-iYFP compound heterozygous mice. Harvested bone marrow was labeled with antibodies to delineate developing B cell subsets and mark hCD4 expression. The progression of cells from the pro-B to the small resting pre-B cell stages is shown. Small pro-B cells were defined as forward scatter low CD43+B220+ while small pre-B cells were forward scatter low CD43−B220+. The intermediate stages represent forward scatter high cells (cycling) with intermediate levels of CD43 progressing from high to low. Data are representative of two independent experiments analyzing three mice each.
The Two Jκ Germline Promoters Are Not Equivalent.
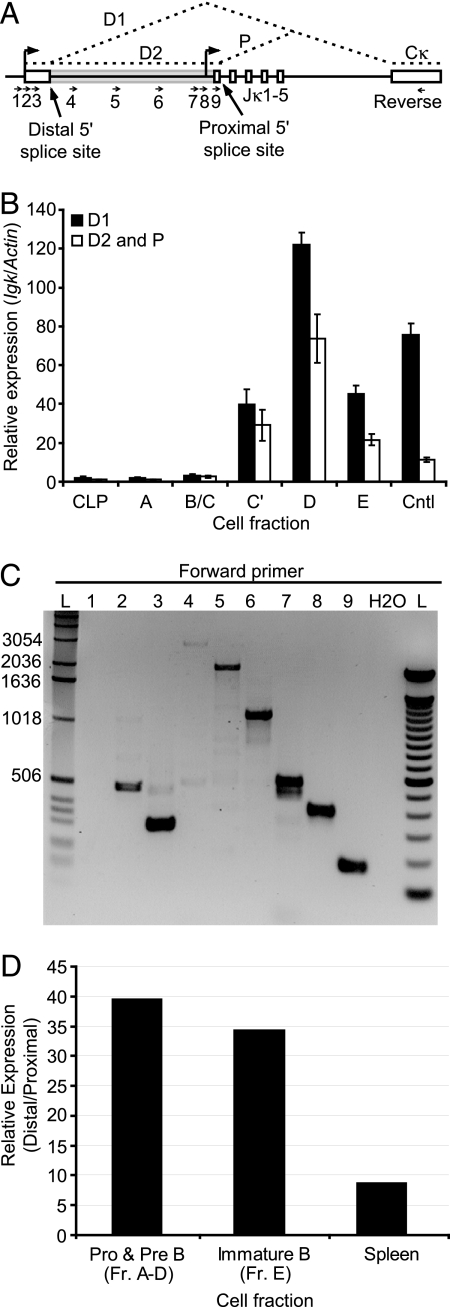
To confirm the validity of the new reporter mice, we fractionated bone marrow B cells from wild-type mice into successive developmental stages by flow cytometry and measured abundance of both the proximal and distal germline transcripts by real-time quantitative RT-PCR (Fig. 3 A and B). In agreement with the reporter mice, the RT-PCR assay demonstrated that Jκ germline transcription begins in fraction C' and continues through the rest of B cell development, reaching a peak in small resting pre-B cells. This is a very different pattern of expression than we observed using our previous κ0GFP knock-in. Although only a small percentage of pre-B cells are GFP positive in κ0GFP mice (5), by conventional RT-PCR the proximal Jκ promoter appeared to be extremely active. It was possible that the κ0GFP mutation is not a faithful reporter of proximal promoter activity, perhaps due to silencing of the targeted allele. However, it is unlikely that a reporter would be systematically silenced early in development only to loose that silencing later (100% of cells that have a germline κ0GFP allele express GFP in peripheral B cells). In other reported cases of transgene silencing, the silencing is stable throughout development (18). Another possibility was that the κ0GFP insertion was accurately reporting proximal promoter activity, but that the RT-PCR analysis used to assess the proximal promoter-derived transcript was flawed. We hypothesized that the transcript originating from the distal Jκ germline promoter may have an alternatively spliced variant that utilizes a splice donor site identical to the proximal transcript splice donor site (Fig. 3A, transcript D2). Such a transcript would wholly encompass the proximal Jκ germline transcript in extent and make it impossible to distinguish the proximal transcript from this distal-promoter-derived transcript by conventional RT-PCR (Fig. 3A). To test if such a transcript exists, we performed RT-PCR on wild-type pre-B cell cDNA using a common anchor primer in the Cκ exon paired with various primers arrayed across the region between the distal germline transcript promoter and the Jκ cluster. As shown in Fig. 3C, we observed bands of the predicted sizes for a transcript that initiates from the distal germline promoter, but that utilizes a 5′ splice site downstream of Jκ1 (lanes 4–6). These were not primary transcripts as the intron between Jκ1 and Cκ was correctly spliced from all observed products. Primers placed upstream of the previously reported distal transcript splice donor site also gave bands of the predicted size for transcript D1 (lanes 2 and 3). A primer placed upstream of the distal germline promoter gave no product, indicating the absence of transcription originating further upstream (lane 1). We confirmed this result by amplifying this novel splice product from a bone marrow pre-B cell cDNA library and subjecting it to DNA sequence analysis (data not shown). In addition, Northern blot analysis revealed transcripts corresponding in size to the unspliced distal promoter transcript and a variant spliced from Jκ1 to Cκ (data not shown). Hence the distal Jκ transcript is alternatively spliced, producing at least two distinct mature transcripts (Fig. 3A and Fig. S3).
Fig. 3.
The distal germline Jκ promoter is far more active than the proximal germline Jκ promoter in developing pre-B cells. (A) Schematic of the Jκ cluster. The transcription start sites for both the distal and proximal promoters are shown as arrows above the diagram along with the deduced splicing pattern of the germline transcripts in dashed lines. The unfilled box at the left represents the first exon of one of the distal-promoter transcripts (D1), and the gray box the first exon of the other (D2). The two different splice donor sites are indicated. Approximate primer locations for both real-time RT-PCR (B) and gel-based RT-PCR (C) of Jκ transcripts are shown as small arrows. (B) Quantitative real-time PCR analysis of spliced Jκ germline transcripts in sorted B cell subsets from wild-type mice. Distal promoter transcripts were quantified using primer #3 (transcript D1) or primer #9 (transcript D2) paired with the reverse primer in Cκ. Proximal promoter derived transcripts (transcript P) were quantified using the same primer pair as for the alternatively spliced distal transcript D2. Harvested bone marrow from C57/BL6 mice was labeled as in (Fig. 1B) and sorted by flow cytometry into the indicated fractions before harvesting cells for RNA isolation. Cntl represents a positive control RNA sample from AMuLV transformed pro-B cells. Values were normalized to Actin transcript abundance (± SD). Data are representative of two independent experiments. (C) RT-PCR analysis of Jκ germline transcripts in primary pre-B cells. Pre-B cells from wild-type mice were sorted and processed for RNA isolation and subsequent RT-PCR. Various forward primers (numbered arrows) were paired with a common reverse primer in the Cκ exon. An ethidium bromide-stained agarose gel analysis of the resultant products is shown. Lane marked L is a standard DNA size ladder. Numbers above the lanes indicate the forward primer used according to the diagram in (A). Data are representative of two independent experiments. (D) 5′RLM-RACE analysis of germline Igκ transcripts in developing primary B cells. Primary cells were harvested and sorted from wild-type C57/BL6 mice and processed for RNA isolation and subsequent 5′RLM-RACE (strategy shown in Fig. S3). The graph represents the ratio of transcripts initiating from the distal and the proximal Igκ germline transcript promoters. No significant quantities of either transcript were detected in thymocytes or water controls. Data are representative of two independent experiments, with each PCR done in triplicate.
Because of the presence of this alternatively spliced distal transcript, previous analyses of distal and proximal germline transcript abundance and promoter activity may be incorrect. To better analyze the relative amounts of transcription initiation from the distal and proximal promoters, we designed assays based on the 5′ RNA ligase mediated rapid amplification of cDNA ends (5′ RLM-RACE) protocol (Fig. S3) (19). RNA ligase is used to attach an adapter oligonucleotide to the decapped 5′ end of mRNA which is then analyzed using a Taqman-based RT-PCR assay using start-site specific and adapter oligonucleotides, allowing the discrimination of transcripts based on their start site. As shown in Fig. 3D, the vast majority of transcription at each developmental stage is triggered by the distal promoter. Comparing this data with data obtained by conventional RT-PCR (Fig. 3B) allows us to conclude that most of the transcripts generally ascribed to the proximal promoter are actually due to an alternatively spliced form of the distal Jκ germline transcript. Northern blot analysis confirmed that bone marrow pre-B cells express relatively little proximal promoter driven germline transcript (data not shown).
The Proximal Jκ Promoter Is Only Weakly Active in Small Pre-B Cells.
Although the distal germline Jκ promoter is active in all pre-B cells, the proximal promoter is active in only a small fraction. Given the low abundance of proximal germline transcript detected by the RLM-RACE assay in pre-B cells, we re-evaluated the expression of GFP in heterozygous κ0GFP knock-in mice. Using a stricter gating scheme we found, in contradiction to our previous report, fewer than 1% GFP positive cells amongst small pre-B cells (data not shown). This low percentage is in line with the RLM-RACE assay. Expression of GFP in all other B cell developmental subsets was similar to what we found previously (5).
Discussion
Our previous analysis of germline Jκ transcription, based on the κ0GFP knock-in mouse, concluded that transcription was rare and stochastic in the pre-B cell population and that those rare transcriptionally active alleles were preferentially rearranged (5). However, the GFP insertion disrupted recombination of the locus and expression of the marker was not consistent with another report of biallelic germline transcription (15). We created the dκGT-hCD4 and Cκ-iYFP germline reporter mice in an attempt to reconcile these observations. Unexpectedly, the new mice revealed that germline Jκ transcription is biallelic in all pre-B cells. In reconciling the new results with our previous report, we discovered that the two Jκ germline promoters are not equivalent and that the vast majority of germline transcription in this locus derives from the distal germline promoter, 3.5 kb upstream of Jκ1. Analysis of the reporter mice in conjunction with 5′ RLM-RACE revealed that, whereas the distal germline promoter becomes highly active in pre-B cells (fractions C' and D), the proximal germline promoter is only weakly active in pre-B cells and becomes somewhat more active as cells progress in development. The difference in activity between the two promoters has not been appreciated until now because the distal promoter generates two alternatively spliced transcripts, one of which completely overlaps with the proximal germline transcript (Fig. 3A). Hence transcripts from the distal and proximal promoters cannot be distinguished from one another by conventional RT-PCR.
The relative activity of the distal and proximal Jκ germline transcript promoters (as determined by the 5′ RLM-RACE assay and RT-PCR) matched well with the observed pattern of marker expression in both the dκGT-hCD4 and κ0GFP mice, and the overall amount of both germline transcripts is reflected in the Cκ-iYFP strain. Although the alternatively spliced distal transcript encompasses the GFP cDNA in κ0GFP mice, we believe that the unusually long 5′ untranslated region (UTR) in the resulting mRNA prevents efficient translation of GFP, and, as a result, GFP is only translated from the proximal promoter-derived transcript. These three Igκ knock-in mice are thus faithful reporters of the abundance of germline transcript originating from either promoter or from the locus as a whole.
Successful heavy chain locus rearrangement results in pre-B cell receptor (pre-BCR) signaling that induces several rounds of mitotic division and retargeting of recombinase activity to the light chain loci (20). Our new mice reveal that pre-BCR signaling results in the rapid onset of germline transcription of both Igκ alleles in all pre-B cells. It is unclear how allelic exclusion is maintained under such circumstances. It is possible that germline transcription is a requisite for recombination, but not sufficient, such that other modifications in addition to transcription are necessary for efficient rearrangement. Previous studies have demonstrated that locus accessibility increases in conjunction with increases in histone acetylation and methylation of histone H3K4 as cells transition from the pro-B to pre-B stage (20–23). However, it is not clear if any of these locus modifications occurs in an allele-specific manner to dictate preferential recombination. Similarly, although the Igκ locus undergoes monoallelic DNA demethylation during B-cell development, this process occurs as a late step in the progressive modification of chromatin structure of the locus and it is unclear if the demethylation occurs before the onset of recombination (both Igκ alleles are largely methylated in pre-B cells) (22–24). The two Igκ alleles do re-localize in pre-B cells such that one allele tends to associate with heterochromatin (23, 25). However, given that germline transcription is biallelic and ubiquitous among pre-B cells, association with heterochromatin appears to have little effect on transcriptional competence. Although heterochromatin association is thought to be repressive, instances of transcriptional activity within this compartment have been noted previously (26, 27). Rather than serving to repress transcription, such localization may lead to preferential allele recombination by sequestering one allele away from the recombinase, chromatin modifying enzymes, or other nuclear compartments. Maintenance of transcriptional competence of the heterochromatin-associated allele may play a role in allowing this allele to recombine if the first fails to encode a suitable light chain. Another possibility is that germline transcription and accessibility of the Vκ gene segments is regulated on an allelic basis, perhaps by heterochromatin association. If the Jκ cluster is in an “always on” state in pre-B cells, the regulated portion of Igκ recombination may be accessibility of the Vκ segments. Whether Vκ germline transcription is allelically biased has not been explored.
Our results demonstrate that the distal Jκ promoter gives rise to the majority of germline transcript in this region. This promoter is approximately 3.5 kb upstream of the Jκ cluster. This arrangement is very different from the germline promoters associated with other recombining loci that are within several hundred base pairs of the relevant gene segment (7). Recent experiments using the TCRα locus as a model found that blocking transcription from a germline promoter prevented rearrangement to downstream Jα segments (9). However, the effect was local and only involved segments in close proximity to the transcriptional blockade; recombination to segments further downstream was unaffected. The distance from the distal germline Igκ promoter to the Jκ cluster suggests that it might only weakly promote recombination at the Jκ cluster. We propose that biallelic transcription from the distal promoter results in a modest increase in accessibility at the Jκ cluster, resulting in infrequent, stochastic recombination. Once recombination occurs on either allele, the incoming Vκ promoter greatly increases accessibility to remaining Jκ segments downstream, promoting further replacement rearrangements on the already rearranged allele until the recombinase is inactivated. In favor of this model, we found that the dκGT-hCD4 knock-in resulted in a slight recombination defect. We suggest that this defect is a result of the polyA transcription termination signal positioned between the distal promoter and the Jκ gene segments (9). The polyA sequence prevents polymerase transit across the locus and further reduces the modest level of accessibility at the Jκ cluster. However, because we cannot remove the polyA sequence from our targeted allele, we cannot test this formally. After a certain time period in the absence of a productive rearrangement (perhaps as levels of the pre-BCR decay or certain transcription factors increase), the proximal promoter might become increasingly active, enhancing local accessibility and rearrangement. Alternatively, proximal promoter activity, and thus Jκ accessibility, may increase in the setting of a receptor editing signal, a possibility we are currently exploring.
Methods
Generation of Knock-In Mice.
Jκ-hCD4: A 400 bp fragment surrounding the reported distal Jκ transcription start site and upstream of the 5′ distal transcript splice site, was found to have greater than 60% homology between human, mouse, and dog and had robust promoter activity in luciferase assays. A QuickSite Mutagenesis kit (Stratagene) was used to introduce an AvrII restriction site 250 bp downstream of the transcription start site and mutate all ATG codons to alternate codons between the start site and AvrII site. A human CD4 cDNA lacking the 33 C-terminal amino acids (intracellular domain) and possessing a Y43I mutation (thought to abolish interaction with MHC class II) along with a SV40 intron/poly A and a floxed neomycin resistance gene were cloned into the AvrII site. This region was extended by 2 kb and 5 kb 5′ and 3′ homology arms and a diphtheria toxin subunit A (DTA) cassette to generate the final targeting construct (Fig. S1).
Cκ-iYFP.
A floxed neomycin resistance gene followed by an IRES-YFP cassette (gift of Richard Locksley, University of California, San Francisco, CA) was cloned into the BsmBI site located 9 bp downstream of the stop codon of Cκ. The fragment containing the Cκ exon was extended by 2 kb and 5.5 kb 5′ and 3′ homology arms and a DTA cassette to generate the final targeting construct.
Targeting constructs were linearized and electroporated into Prm1-Cre ES cells followed by two weeks of neomycin selection. Neomycin resistant ES cell colonies were picked and initially screened for either knock-in by long distance PCR. PCR positive clones were further verified by Southern blot using probes outside of the homology arms. Two successfully targeted ES clones per construct were used for subsequent blastocyst injection. Both clones for each construct successfully gave rise to germline transmitting chimeric males. Males were subsequently mated to 129/SvImJ females and the pups assayed for deletion of the neomycin resistance gene by PCR. Further breeding was used to remove the Prm1-Cre transgene from the background.
Flow Cytometry and Cell Sorting.
Flow cytometric analysis of bone marrow and splenic B cell development in either strain was done as previously described (28, 29). For hCD4 knock-in mice, cells were additionally stained with human CD4 antibody (clone RPA-T4, eBiosciences). Cell sorting was done by flow cytometry or magnetic beads (Miltenyi). The identity of all antibodies is available upon request.
Transcript Analysis.
Reverse transcription was done using SuperScript II or MoMLV-RT (Invitrogen) according to manufacturers instructions. Subsequent real time or conventional PCR was done using JumpStart Taq polymerase (Sigma) and Taqman labeled probes (for real time only). PCR cycling conditions were 95 °C for 4 min followed by 25 to 45 cycles of 95 °C for 30 sec, 60 °C for 30 sec, and 72 °C for1 min. Primer sequences are provided in Table S1. RLM-RACE was performed using a kit from Ambion. Linker-ligated RNA was converted to first-strand cDNA using reverse transcriptase. This cDNA was analyzed using a nested PCR strategy that involved 14 cycles of amplification with linker and promoter-specific primers followed by a second round of amplification on an aliquot of the primary product using internal primers and a taqman probe (Fig. S3C). RLM-RACE products were cloned and subjected to sequence analysis to verify their identities. Individual cloned products served as absolute standards for quantification of distal and proximal promoter initiated transcript abundance by real-time taqman PCR.
Supplementary Material
Acknowledgments.
We acknowledge Dr. Richard Locksley (UCSF) for the gift of a cloned neoR-IRES-YFP cassette. We thank Dr. Christian Vettermann and Hong-Erh Liang for suggestions and insightful comments and Benjamin Taylor and Matthias Merkenschlager (Imperial College, London, United Kingdom) for discussions regarding unpublished data. This work was supported by a grant from the National Institutes of Health to M.S.S. (R01 HL48702).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808895106/DCSupplemental.
References
- 1.Schlissel MS. Regulating antigen-receptor gene assembly. Nat Rev Immunol. 2003;3:890–899. doi: 10.1038/nri1225. [DOI] [PubMed] [Google Scholar]
- 2.Burnet FM, Fenner F. The Production of Antibodies. 2nd Ed. Melbourne: Macmillan; 1949. p. viii. [Google Scholar]
- 3.Corcoran AE. Immunoglobulin locus silencing and allelic exclusion. Semin Immunol. 2005;17:141–154. doi: 10.1016/j.smim.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Mostoslavsky R, et al. Asynchronous replication and allelic exclusion in the immune system. Nature. 2001;414:221–225. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- 5.Liang HE, Hsu LY, Cado D, Schlissel MS. Variegated transcriptional activation of the immunoglobulin kappa locus in pre-b cells contributes to the allelic exclusion of light-chain expression. Cell. 2004;118:19–29. doi: 10.1016/j.cell.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 6.Mostoslavsky R, Alt FW, Rajewsky K. The lingering enigma of the allelic exclusion mechanism. Cell. 2004;118:539–544. doi: 10.1016/j.cell.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 7.Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- 8.Schlissel MS, Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 1989;58:1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- 9.Abarrategui I, Krangel MS. Regulation of T cell receptor-alpha gene recombination by transcription. Nat Immunol. 2006;7:1109–1115. doi: 10.1038/ni1379. [DOI] [PubMed] [Google Scholar]
- 10.Leclercq L, Butkeraitis P, Reth M. A novel germ-line JK transcript starting immediately upstream of JK1. Nucleic Acids Res. 1989;17:6809–6819. doi: 10.1093/nar/17.17.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin DJ, van Ness BG. Initiation and processing of two kappa immunoglobulin germ line transcripts in mouse B cells. Mol Cell Biol. 1990;10:1950–1958. doi: 10.1128/mcb.10.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Ness BG, et al. Transcription of the unrearranged mouse C kappa locus: Sequence of the initiation region and comparison of activity with a rearranged V kappa-C kappa gene. Cell. 1981;27:593–602. doi: 10.1016/0092-8674(81)90401-3. [DOI] [PubMed] [Google Scholar]
- 13.Cocea L, et al. A targeted deletion of a region upstream from the Jkappa cluster impairs kappa chain rearrangement in cis in mice and in the 103/bcl2 cell line. J Exp Med. 1999;189:1443–1450. doi: 10.1084/jem.189.9.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferradini L, et al. Rearrangement-enhancing element upstream of the mouse immunoglobulin kappa chain J cluster. Science. 1996;271:1416–1420. doi: 10.1126/science.271.5254.1416. [DOI] [PubMed] [Google Scholar]
- 15.Singh N, Bergman Y, Cedar H, Chess A. Biallelic germline transcription at the kappa immunoglobulin locus. J Exp Med. 2003;197:743–750. doi: 10.1084/jem.20021392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moebius U, Pallai P, Harrison SC, Reinherz EL. Delineation of an extended surface contact area on human CD4 involved in class II major histocompatibility complex binding. Proc Natl Acad Sci USA. 1993;90:8259–8263. doi: 10.1073/pnas.90.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casellas R, et al. Contribution of receptor editing to the antibody repertoire. Science. 2001;291:1541–1544. doi: 10.1126/science.1056600. [DOI] [PubMed] [Google Scholar]
- 18.Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer BC. Revolutions in rapid amplification of cDNA ends: New strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal Biochem. 1995;227:255–273. doi: 10.1006/abio.1995.1279. [DOI] [PubMed] [Google Scholar]
- 20.Schlissel MS. Regulation of activation and recombination of the murine Igkappa locus. Immunol Rev. 2004;200:215–223. doi: 10.1111/j.0105-2896.2004.00157.x. [DOI] [PubMed] [Google Scholar]
- 21.Stanhope-Baker P, Hudson KM, Shaffer AL, Constantinescu A, Schlissel MS. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85:887–897. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- 22.Goldmit M, Schlissel M, Cedar H, Bergman Y. Differential accessibility at the kappa chain locus plays a role in allelic exclusion. EMBO J. 2002;21:5255–5261. doi: 10.1093/emboj/cdf518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldmit M, et al. Epigenetic ontogeny of the Igk locus during B cell development. Nat Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- 24.Mostoslavsky R, et al. Kappa chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 1998;12:1801–1811. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosak ST, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert N, et al. Chromatin architecture of the human genome: Gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 27.Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- 28.Rumfelt LL, Zhou Y, Roweley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19- early B cell precursors. J Exp Med. 2006;203:675–687. doi: 10.1084/jem.20052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardy RR, Shinton SA. Characterization of B lymphopoiesis in mouse bone marrow and spleen. Methods Mol Biol. 2004;271:1–24. doi: 10.1385/1-59259-796-3:001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.