Abstract
Gene knockout experiments in mice have suggested a hierarchical model of early B cell commitment wherein E2A proteins (E47 and E12) activate early B cell factor (Ebf1), which in turn activates expression of the B cell commitment factor, Pax5. In IL-7 receptor alpha (IL-7Rα) knockout mice, B cell development is blocked before B-lineage commitment at the prepro-B cell stage in adult animals. In IL-7Rα−/− prepro-B cells, E47 is expressed and yet is insufficient to transcriptionally activate the putative downstream target gene, Ebf1. In this study, we show that further increases of E47 expression in IL-7Rα−/− prepro-B cells fails to activate Ebf1, but rather leads to a dramatic induction of the E2A inhibitory factors, Id2 and Id3. In contrast, enforced expression of Ebf1 in IL-7Rα−/− bone marrow potently down-regulates Id2 and Id3 mRNA expression and restores B cell differentiation in vivo. Down-regulation of both Id2 and Id3 during B cell specification is essential in that overexpression of either Id2 or Id3 in wild-type bone marrow blocks B cell specification at the prepro-B cell stage. Collectively, these studies suggest a model where Ebf1 induction specifies the B cell fate by dramatically increasing activity of E47 at the posttranslational level.
Keywords: B cell specification, E2A, Ebf1, Id proteins, IL-7 receptor
The earliest steps in the development of antibody-secreting B lymphocytes from common lymphoid progenitor cells (CLP) in adult mouse bone marrow are orchestrated by the concerted activities of a number of B-lineage transcription factors and signals from growth factor and cytokine receptors. Gene knockout experiments in mice have shown the absolute requirement for the transcription factors PU.1 and Ikaros in the generation of CLP and the requirement for E2A and Ebf1 in the generation of the first specified B cell progenitors referred to as prepro-B cells (1–3). Prepro-B cells can be prospectively identified in murine bone marrow as B220+CD19−IgM−NK1.1− cells that either lack or have just initiated rearrangement of the DH-JH gene segments of the Ig heavy chain (IgH) locus. These cells developmentally progress to B220+CD19+IgM−NK1.1− progenitor B (pro-B/pre-B) cells upon induction of the B cell commitment factor, Pax5, which functions to maintain B cell identity by both positively regulating early B-lineage-associated gene expression and negatively regulating a large number of genes that promote development along alternative hematopoietic cell fates (4–7). B cell development is completely blocked at the pro-B cell stage in the absence of Pax5.
A number of studies have suggested that a complex hierarchical relationship exists between E2A, Ebf1, and Pax5 in early B cell development (8). The E2A gene encodes two alternative mRNA splicing variants, E47 and E12, which can homo- or heterodimerize via their helix–loop–helix domains (9). Both variants are able to rescue B cell development when expressed from a pim-1 promoter/IgH enhancer transgene vector that was crossed onto an E2A knockout background (10). Observations showing that E2A is present (albeit at reduced levels) in Ebf1 knockout animals (3) and that the Ebf1 promoter is bound by E47 in vivo (11, 12) and can be transactivated by overexpression of E47 (13) or E12 (14) in non-B-lineage cell lines, suggest that E2A activity is essential upstream of Ebf1. Similarly, the Pax5 promoter is bound by Ebf1 based on EMSA and Ebf1 can transactivate the Pax5 promoter in transient co-transfection assays (15, 16). Ebf1 is also present in Pax5-deficient pro-B cells derived from Pax5−/− adult mice (17), suggesting that Ebf1 may participate in the activation of Pax5 expression. Complicating the simple hierarchical model where E2A induces expression of Ebf1, which then activates Pax5, are observations showing that up-regulation of E2A protein levels in B cell progenitors are dependent on Ebf1 (18). In addition, expression of Pax5 from the endogenous Ikaros locus resulted in activation of Ebf1 and B cell development in the thymus (19) and in the bone marrow of E2A-deficient mice (20). Pax5 was also shown to directly bind one of two distinct EBF promoters in vivo and could transactivate this promoter in transient transfection assays (12). Collectively, these results demonstrate a complex regulatory circuitry controlling B cell specification and commitment from CLP.
The loss of either the IL-7 receptor alpha (IL-7Rα) chain or the IL-7 ligand also results in a complete block in adult B lymphopoiesis at the prepro-B cell stage, with either modest or no reduction in the numbers of CLP being present in IL-7 knockout animals (21–24). In IL-7Rα− or IL-7-deficient mice, E2A proteins are expressed at wild-type levels and yet Ebf1 and Pax5 are absent, which suggests that other factors participate in Ebf1 induction in addition to E2A, that E2A requires posttranslational modification to activate Ebf1 expression, or that inhibitory proteins suppress the ability of E2A to activate Ebf1 expression in this context (23–25). The activation of Ebf1 seems to be the pivotal event in B cell fate specification in that Ebf1 can surprisingly rescue the B cell developmental program when overexpressed in the context of multiple gene knockout backgrounds that completely lack B-lineage cells including PU.1 (8), E2A (26), IL-7Rα (23), and IL-7 (24).
To clarify the complex regulatory circuitry between E2A, Ebf1, and Pax5 in the specification and commitment of the B cell lineage, we have performed in vivo genetic complementation assays to address why Ebf1, but not E47, is sufficient to rescue the B cell developmental block in IL-7Rα-knockout animals. Analysis of animals transplanted with IL-7Rα−/− cells expressing either a control retroviral vector or vectors expressing IL-7Rα, E47, Ebf1, or Pax5 showed that B cell development was rescued by Ebf1 in all cases and in 7/11 animals by Pax5. Expression of E47 did not activate Ebf1 mRNA but rather led to increased expression of the E2A-inhibitory proteins Id2 and Id3. Expression of Ebf1 resulted in near complete inactivation of both Id2 and Id3 in prepro-B cells. This suggests that one major function of Ebf1 in B-lineage specification is to down-regulate Id levels to allow for subsequent activation of E2A protein activity. Collectively, these results provide important new insights into the significance of Ebf1 induction as the defining molecular event that determines B-lineage cell fate.
Results
Ebf1 and Pax5 Can Rescue B Cell Development in IL-7Rα-Knockout Mice.
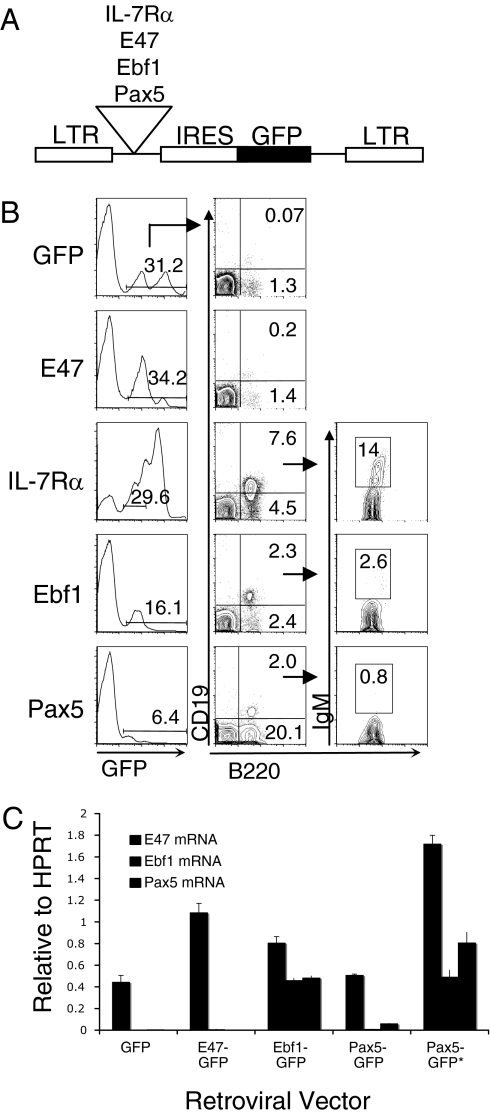
To characterize the mechanism by which Ebf1 can specify the B cell fate, we first transduced IL-7Rα−/− bone marrow cells with retroviruses that co-expressed either E47, Ebf1, Pax5, or the IL-7Rα chain along with a blue-excited green fluorescent protein variant (Bex, hereafter referred to as GFP) (Fig. 1A). Transduced cells were then transplanted into lethally irradiated C57BL/6 recipient mice and analyzed for reconstitution beginning at 3 weeks post-transplantation (PT). As previously noted (23), B cell development in IL-7Rα−/− animals is blocked at the prepro-B cell stage (B220+CD19−NK1.1−IgM−), which was confirmed by analysis of GFP+ control cells in bone marrow of animals reconstituted at 6 weeks PT (n = 15) (Fig. 1B). Expression of E47 failed to promote B cell development to the pro-B/pre-B cell stage (B220+CD19+NK1.1−IgM−) even though E47 mRNA levels were increased in GFP+ prepro-B cells by expression of the retroviral vector (Figs. 1 B and C). Expression of Ebf1 or Pax5 rescued B cell differentiation in IL-7Rα−/− bone marrow cells but failed to restore absolute numbers of pro-B or pre-B lymphocytes due to the absence of IL-7 signaling in rescued cells (Fig. 1B). In contrast to Ebf1, which rescued 100% of reconstituted animals (n = 14), Pax5 only rescued B cell development in 7/11 animals. The ability of Pax5 to rescue B cell development was statistically correlated with the absolute expression levels of Pax5 as determined by the mean fluorescence intensity (MFI) of GFP in prepro-B cells from rescued (MFI = 72.6) versus non-rescued (MFI = 41.7) animals (P = 0.006). We also noted a substantial increase in the frequency of B220+CD19−NK1.1−IgM− cells that expressed Pax5 (Fig. 1B), which we attributed to Pax5 induction of B220 expression on myeloid-lineage cells that expressed Mac-1 and Gr-1 (27) (supporting information (SI) Fig. S1). Because of this, characterization of prepro-B cells in Pax5-rescued animals was always done in B220+CD19−NK1.1−IgM−Mac-1−Gr-1− cells. Further analysis of mRNA expression levels by Real-time PCR using FACS-sorted prepro-B cells from reconstituted mice showed that both Ebf1 and Pax5 up-regulate the B cell-specific genes mb-1, VpreB and λ5 (Fig. S2), further suggesting that rescued B220+CD19+ cells belong to the B lineage. Ebf1 also stimulated a high level of endogenous Pax5 mRNA expression and increased E47 mRNA approximately 1.7-fold compared with E47 levels in GFP control cells (Fig. 1C). This suggests that Ebf1 stimulates a feedback loop that reinforces E47 expression. In animals where Pax5 rescued B cell development, endogenous Ebf1 mRNA levels were induced approximately 12-fold in prepro-B cells compared with Ebf1 levels in GFP control prepro-B cells. Ebf1 expression increased an additional10-fold in Pax5-rescued pro-B/pre-B cells to a level that was comparable to Ebf1 expression levels in Ebf1-rescued prepro-B cells (Fig. 1C). This indicates that maximal Ebf1 induction by Pax5 may be occurring with delayed kinetics. The fact that Pax5 can induce Ebf1 expression when expressed at high enough levels suggests that, in addition to the Ebf1-E2A feedback loop, Pax5 may reinforce Ebf1 activation through a positive feedback mechanism by binding the Ebf1 proximal promoter (12, 20). These observations also raise the possibility that Pax5 activation could function as the primary B-lineage specifying event in some developmental contexts through its ability to activate Ebf1.
Fig. 1.
Ebf1 and Pax5 rescue B cell development in IL-7Rα-deficient bone marrow. (A) MSCV retroviral construct coexpressing the cDNA of interest and GFP (IRES = internal ribosome entry site). (B) Flow cytometric analysis of bone marrow from 6-week PT mice (n = 5 each). Plots were gated on GFP+NK1.1− cells. IgM expression was analyzed on B220+CD19+ cells. Numbers indicate percentages of total GFP+ cells from representative animals. (C) Representative Real-time PCR analysis of E47, Ebf1, and Pax5 expression in FACS-sorted prepro-B or pro-B/pre-B (*) cells isolated from recipient mice at 6 weeks PT (n = 3 independent FACS sorts for each retroviral vector). Data show relative expression compared to the internal Hprt control. Error bars indicate the standard deviation for quadruplicate reactions using mRNA isolated from one representative mouse per retroviral construct.
Ebf1 and Pax5 Promote DH-JH Rearrangement in IL-7Rα−/− Bone Marrow Cells.
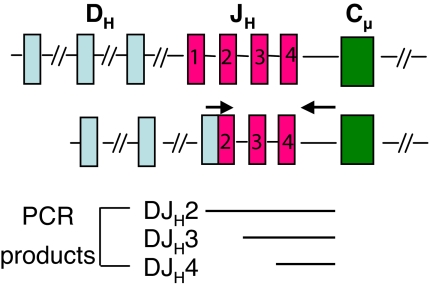
To determine whether Ebf1- or Pax5-rescued cells were undergoing recombination between the Ig heavy chain DH-JH gene segments, we FACS-sorted 500 GFP+ prepro-B or pro-B/pre-B cells from animals reconstituted with retrovirus-expressing cells and used an upstream primer that anneals to the majority of the 10 DH gene segments in C57BL/6 mice and a 3′-primer complementary to a sequence downstream of JH4 (Fig. 2) (28). Using relatively short PCR extension times, this assay detects 3 major PCR products that account for DH rearrangements to either JH2, JH3, or JH4, as determined by direct sequencing of the PCR products (Fig. 2 and data not shown). Results summarized in Table 1 indicate that IL-7Rα-deficient prepro-B cells expressing the GFP control retrovirus show a relatively low incidence of rearrangement (minimally 1 event per 500 cells), with single bands being commonly detected. The low frequency of detection can be explained, in part, by the functional heterogeneity within the prepro-B cell subset, which is likely to be impure for bona fide B-lineage precursors. Similar low frequency rearrangement events were detected in E47- or Pax5-expressing prepro-B cells, which were not statistically different from controls. In contrast, Ebf1 substantially increased the overall frequency of recombination events in prepro-B cells compared with GFP control cells (P < 0.01, Student's t test). Pax5 stimulated recombination in rescued pro-B/pre-B cells (P < 0.05), which reinforces the view that Pax5 rescue of B cell development is occurring with delayed kinetics that correlate with the timing of maximal Ebf1 induction by Pax5 (Table 1 and Fig. 1C).
Fig. 2.
Ebf1 promotes DHJH rearrangement in IL-7Rα-deficient bone marrow. (A) Ig heavy chain locus before and after rearrangement. Arrows indicate locations of the 5′ degenerate primer within the DH gene segments and the 3′ primer downstream of JH4.
Table 1.
DHJH Rearrangement Products
| * Prepro-B | Pro-B/Pre-B | ||||
|---|---|---|---|---|---|
| GFP | E47 | Ebf1‡‡ | Pax5 | Ebf1‡‡ | Pax5‡ |
| 0† | 2 | 3,4 | 3,4 | 2,3,4 | 3,4 |
| 2,3 | 3,4 | 2,4 | 3,4 | 2,3,4 | 3,4 |
| 3,4 | 3 | 2,3,4 | 2 | 2,3,4 | 3,4 |
| 2 | 3 | 2,3,4 | 0 | 2,3,4 | 3,4 |
| 3 | 2,4 | 2,3,4 | 3 | 2,3,4 | 2,3,4 |
*Prepro-B: B220+CD19−NK1.1−IgM−, Pro-B/Pre-B: B220+CD19+NK1.1−IgM−.
† Numbers indicate JH rearrangement product detected. (0 = only germline product detected). Student t test comparing the number of bands detected to the GFP control,
‡P < 0.05,
‡‡ P < 0.01. The number of rearrangement products in prepro-B cells expressing E47 or Pax5 were not statistically significant (P = 0.3 and P = 0.5, respectively)
IgH Transgenes Do Not Rescue B Cell Development in IL-7Rα-Deficient Animals.
Previous studies have shown that Ebf1 functions in coordination with E2A to regulate transcription of the recombinase genes, Rag1 and Rag2, the surrogate light chains λ5 and VpreB, and signaling molecules like Igα/mb-1 in early progenitor B cell subsets (15, 29–32). To test whether the primary function of Ebf1 in B cell specification relates to its role in Ig gene rearrangement, we crossed two different IgH transgene mice (33) to animals deficient in either the IL-7 cytokine or the IL-7Rα chain. Analysis of B cell development in compound transgenic/knockout mice showed that expression of a fully rearranged heavy chain gene could not rescue the B cell developmental block in IL-7- or IL-7Rα-knockout mice (Fig. S3, n = 5 each). Since Ebf1 can rescue B cell development in IL-7- or IL-7Rα-knockout mice, these results indicate that Ebf1 has critical functions in addition to activation of Ig recombination in early B-lineage development.
Ebf1 Potently Downregulates Id2 and Id3 mRNA Levels in Prepro-B Cells.
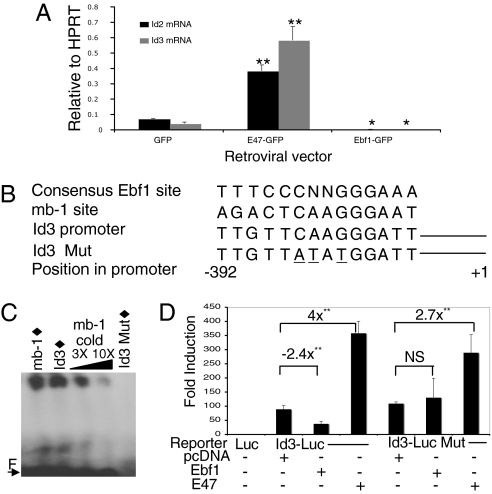
To address why E47 overexpression could not rescue B cell development in IL-7Rα−/− mice (Fig. 1B) or activate Ebf1 transcription (Fig. 1C), we characterized expression of the E2A inhibitory factors, Id2 and Id3, in sorted prepro-B cells that expressed E47 (Fig. 3A). Of the 4 Id proteins expressed in mammalian cells (Id1, Id2, Id3, and Id4), Id2 and Id3 are the only family members expressed in B220+ B-lymphocyte progenitors (34). Real-time PCR analysis confirmed that Id2 and Id3 mRNA are expressed in IL-7Rα-deficient prepro-B cells expressing the GFP control provirus and both factors are significantly induced by E47 (approximately 5- and 7-fold, respectively, compared with GFP control-expressing cells). In contrast to E47, Ebf1 dramatically downregulated both Id2 and Id3 mRNA to essentially undetectable levels in prepro-B cells (P = 0.01 and P = 0.02, respectively). The potent down-regulation of Id2 and Id3 suggests that Ebf1 induction would markedly enhance E2A protein activity, thus providing a molecular basis for co-activation of B cell specification by concerted E2A/Ebf1 activity.
Fig. 3.
E47 and Ebf1 differentially regulate Id2/Id3 expression during early B cell development. (A) Real-time PCR analysis of Id2 and Id3 expression levels in FACS-sorted GFP+ prepro-B cells from recipient mice at 3–6 weeks PT. Data shown are representative of one reconstituted mouse per retroviral construct out of three that were analyzed in quadruplicate. (B) Putative Ebf1 binding site at position −392 in the Id3 promoter. Point mutations introduced into the site are underlined. (C) EMSA using 10 ng of recombinant Ebf1 protein and radiolabeled sequences (filled diamonds) shown in (B). Increasing amounts of unlabeled mb-1 cold probe were used as competitor. “F” indicates free probe. (D) Luciferase assay using extracts from K562 cells co-transfected with either the wild-type or mutant Id3 promoter-reporter and Ebf1, E47, or control pcDNA expression plasmids. Luciferase activity was normalized to a co-transfected Renilla expression plasmid. Results are shown as fold-induction over the promoterless luciferase vector in all experiments. Error bars indicate the SEM from four different experiments performed in triplicate. (*, P < 0.05; **, P < 0.01) NS = not significant.
Ebf1 Directly Binds and Negatively Regulates Id3 Promoter Activity.
Inspection of the Id3 promoter sequence revealed a consensus binding site for Ebf1 positioned about 390 bp upstream of the transcription start site (Fig. 3B). EMSA analysis using recombinant Ebf1 protein (35) revealed that Ebf1 binds this sequence in vitro (Fig. 3C, lane 2). The band co-migrates with a shifted complex containing recombinant Ebf1 and the known Ebf1 binding site within the mb-1 promoter and is competed away by a cold mb-1 probe. To determine whether Ebf1 could regulate expression of an Id3-driven reporter gene through this sequence, we then co-transfected an Ebf1 expression plasmid and an Id3 promoter-driven luciferase reporter construct into K562 cells. After normalization of luciferase activity to Renilla luciferase, the results showed that Ebf1 could significantly repress luciferase expression in a binding-site-dependent manner (Fig. 3D, P < 0.001). Co-transfection of two additional cell lines, BOSC23 and NIH 3T3, with the same constructs resulted in similar reductions in luciferase activity (3.2- and 3.6-fold, respectively, data not shown). Each of these lines express the endogenous Id3 gene as demonstrated by Western blot analysis using an Id3-specific monoclonal antibody (clone 6–1, Cal Bioreagents) that recognizes both murine and human Id3 protein (Fig. S4). Conversely, E47 expression activated the Id3 promoter-reporter 4-fold (Fig. 3D, P < 0.001), which was likely dependent on the presence of 6 consensus E-box sequences located within the upstream Id3 promoter region. E47 also activated the mutant Id3 promoter construct, which indicates that E47 activation of Id3 is not dependent on Ebf1. A recent study showed that Ebf1 could directly bind the Id2 promoter by ChIP analysis, which supports the hypothesis that Ebf1 may negatively regulate Id2 gene expression (36).
Id2 and Id3 Down-Regulation Are Essential for B Cell Specification.
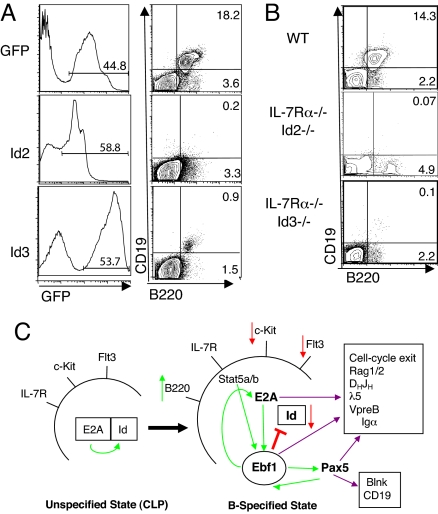
To address whether Id2 and/or Id3 down-regulation is necessary for B cell specification, we overexpressed Id2 or Id3 in wild-type C57BL/6 bone marrow cells using a retroviral vector linked with GFP. Transduced cells were transplanted into lethally irradiated syngeneic recipient animals and B cell development within total bone marrow GFP+ cells was then assessed at 6 weeks PT (Fig. 4A). Analysis of Id2- or Id3-expressing cells revealed that either factor blocked B cell development at the B220+CD19−NK-1.1−IgM− prepro-B cell stage (n ≥ 4 each). These results indicate that down-regulation of both Id2 and Id3 is an essential event in B-lineage specification.
Fig. 4.
Enforced expression of Id2 or Id3 inhibits B-lineage specification. (A) Flow cytometric analysis of bone marrow from mice reconstituted with GFP- (n = 5), Id2- (n = 4), or Id3-expressing (n = 5) cells at 3–6 weeks PT. Plots are gated on GFP+ cells. (B) Loss of Id3 (n = 3) or Id2 (n = 1) does not rescue B cell development in IL-7Rα-knockout mice. (C) Model for Ebf1-induced activation of B cell specification from unspecified CLP. Red arrows indicate inhibition or down-regulation, while green arrows indicate activating events or up-regulation.
Loss of Id2 or Id3 Fails to Rescue B Cell Development in IL-7Rα−/− Mice.
In IL-7Rα-deficient prepro-B cells, Id2, Id3, and E2A are all expressed at high levels while Ebf1 expression is absent. To test whether loss of Id2 or Id3 might allow E2A to stimulate endogenous Ebf1 transcription and rescue B cell development in IL-7Rα-knockout mice in the absence of retroviral Ebf1 expression, we generated IL-7Rα−/−Id2−/− and IL-7Rα−/−Id3−/− double-deficient mice. Analysis of IL-7Rα−/−Id2−/− or IL-7Rα−/−Id3−/− mice showed that loss of individual Id proteins was not sufficient to promote B cell development in the context of IL-7Rα−/− bone marrow cells (Fig. 4B). We were unable to generate IL-7Rα−/−Id2−/−Id3−/− mice since the loss of both Id2 and Id3 results in an embryonic lethal phenotype (R. Benezra, personal communication). These results indicate that inactivation of both Id proteins by Ebf1 expression is necessary for B cell fate specification to occur in IL-7Rα-deficient animals.
Discussion
The results described above demonstrate that Id2 and Id3 function as repressors of B-lineage fate specification and that Ebf1 overcomes this inhibition through direct or indirect down-regulation of Id2 and Id3 gene transcription. These data suggest a model (Fig. 4C) wherein activation of Ebf1 expression promotes a substantial increase in E2A activity at both the transcriptional level, through up-regulation of E47 mRNA (Fig. 1C), and at the posttranslational level, by Ebf1 down-regulation of E2A inhibitory protein activity (Fig. 3A). High levels of E2A in cooperation with Ebf1 would then stimulate cell-cycle exit and activation of multiple E2A/Ebf1 target genes involved in sterile IgH transcription and recombination at DH-JH gene segments, as well as activation of Pax5. In support of this model, high levels of E2A protein activity are known to repress lymphocyte proliferation (37) and recombinase activity is negatively regulated in cycling cells (38). In addition, E2A-GFP knock-in mice crossed with Ebf1-deficient animals showed that up-regulation of E2A was dependent on Ebf1 at the onset of B cell specification (18). This scenario suggests a complex interplay between key regulatory factors that function to both reinforce early B cell developmental decisions through positive feedback loops that further up-regulate expression of differentiation-promoting factors and inactivation of inhibitory pathways that dampen activity of select factors like E2A in a temporally-specific manner. The primary effector molecule driving B-lineage specification and development in this process seems to be Ebf1 through its ability to potently activate E2A, which would explain how Ebf1 can rescue B cell development in multiple gene knockout contexts that lack the earliest B-lineage cells.
One lingering question related to B cell specification is how Ebf1 becomes activated in the first place if increased levels of E47 only lead to increased expression of the E2A inhibitors, Id2 and Id3. The observation that Id2/Id3 mRNA levels increase in E47-expressing prepro-B cells suggest that some level of E47 must be transcriptionally competent and free from Id protein inhibition to transcriptionally activate these genes, although this level of “free” E2A is not sufficient to activate Ebf1 transcription. One possibility is that E47 may be posttranslationally modified by phosphorylation to stimulate homodimerization in a manner that is not inhibitable by Id proteins (25). Alternatively, E2A activation alone may not be sufficient to induce Ebf1 without cooperation from other regulatory factors stimulated by IL-7 receptor signaling. Another attractive possibility is that Ebf1 is induced by removal of repressive signals like Flt3 (39) or TGFβ signaling (34) that may restrain CLP from differentiation along the B cell pathway. Removal of repressive signals via daughter cell migration away from the CLP niche in the bone marrow may be sufficient to allow IL-7 receptor signaling to maximally activate Ebf1 expression. Further work will be necessary to clarify these issues.
Methods
Mice.
IL-7Rα- and IL-7-deficient mice were all on a C57BL/6 background. Transgenic lines Vh81x and m167 were obtained from Dr. J. Kearney (University of Alabama-Birmingham, Birmingham, AL). All mice were bred and maintained at University of Alabama-Birmingham under a protocol approved by the Institutional Animal Care and Use Committee in compliance with all state and federal regulations governing the use of experimental animals.
Retroviral Transductions.
All cDNA sequences were cloned into the EcoRI site of the murine stem cell virus (MSCV) retroviral vector upstream of an IRES element and GFP. cDNA constructs were kind gifts from L. Park (IL-7Rα chain, Amgen), C. Murre (E47, University of California San Diego, La Jolla, CA), R. Grosschedl (Ebf1, Max Planck Institute, Leipzig, Germany), M. Busslinger (Pax5, Institute of Molecular Pathology, Vienna, Austria) and X.H. Sun (Id2 and Id3, Oklahoma Medical Research Foundation, Oklahoma City, OK). Retrovirus preparation and transplantation was performed as described previously (40).
Flow Cytometry.
Bone marrow extracted from the femurs and tibiae of mice 3–6 weeks PT were analyzed using labeled antibodies against B220(RA3–6B2)Pacific Blue, CD19 (ID3)Texas Red, NK1.1 (PK136)PE, IgMAPC, Mac-1(M1/70)PE−Cy7, and Gr-1(RB6–8C5)PE−Cy7. Prepro-B (B220+CD19−NK1.1−IgM−) and pro-B/pre-B cell (B220+CD19+NK1.1−IgM−) populations were analyzed and sorted using LSRII (BD Biosciences) or a triple-laser Mo-Flo cell sorter (Cytomation).
Real-time PCR.
GFP+ prepro-B and pro-B/pre-B cells from transplanted mice were sorted and resuspended in RNA STAT-60 (Tel-Test B,). RNA was purified using the MicroRNAeasy kit (Qiagen). cDNA was made using the High Capacity cDNA Archive Kit (Applied Biosystems). Real-time PCR was performed with SYBR Green PCR Master Mix using the ABI Prism 7900HT sequence detection system (Applied Biosystems). Reactions containing 2 μl cDNA were performed in a 20 μl total volume. HPRT was used as an internal control. All primer sequences are available upon request.
DH-JH Rearrangement.
Five hundred transduced prepro-B (B220+CD19−NK1.1−IgM−) or pro-B/pre-B (B220+CD19+NK1.1−IgM−) cells were sorted into complete DMEM media with 10% FBS. All FACS-sorted populations from mice reconstituted with Pax5-expressing cells were also negative for Mac-1 and Gr-1. Cells were spun at 2000 rpm for 5 min and then resuspended in 10 μl 1X PCR buffer (Roche) with 0.1% Triton-X. Nested PCR to detect DH-JH rearrangement products were performed using primers as previously described (28).
EMSA.
The Id3 probe was a 20 bp fragment containing the putative Ebf1 binding site in the Id3 promoter (GTTGTTCAAGGGATTTATGA). The Id3 mutant probe has three point mutations in the core of the Ebf1 site (GTTGTTATATGGATTTATGA). The Ebf1 binding site within the mb-1 promoter was used as a positive control. Probes were labeled with [32P]-dCTP using Klenow. 10 ng of recombinant Ebf1 protein (provided by J. Hagman, National Jewish Medical Research Center, Denver, CO) was incubated with the labeled probe in the presence of 20 mM Hepes pH 7.9, 70 mM KCl, 1 mM DTT, 2.4 mM MgCl2, 2% glycerol, 0.1% Nonidet P-40, and 0.1 mg/ml BSA for 30 min. Samples were separated on a 1X TGE/6% acrylamide gel at 4 °C.
Luciferase Reporter Assay.
The Id3 promoter region from −1319 to +14 was cloned into the pGL2-Basic vector (Promega). The Dual-Luciferase Reporter Assay System (Promega) was used to detect both firefly and Renilla luciferase activity in Lipofectamine 2000 (Invitrogen)-transfected K562, BOSC23, or NIH 3T3 cells. Ebf1 and E47 cDNAs were cloned into the EcoRI site of pcDNA3.1 (Invitrogen).
Supplementary Material
Acknowledgments.
We thank Dr. Robert Stephan for Real-time PCR primers, Dr. John Kearney for transgenic mice and helpful discussions, and Drs. C. Scott Swindle and Rose Ko for assistance with transplants. This work was supported by R01AI055667 (C.A.K), R01AI14782–30 (T.L.C.), R01 AI054661 (H.G. and J.H.) and T32 AI007051 (M.A.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802550106/DCSupplemental.
References
- 1.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 2.Bain G, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 3.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 4.Delogu A, et al. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–281. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: The guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 6.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 8.Medina KL, et al. Assembling a gene regulatory network for specification of the B cell fate. Dev Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Kee BL, Quong MW, Murre C. E2A proteins: essential regulators at multiple stages of B-cell development. Immunol Rev. 2000;175:138–149. [PubMed] [Google Scholar]
- 10.Bain G, et al. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- 11.Greenbaum S, Zhuang Y. Identification of E2A target genes in B lymphocyte development by using a gene tagging-based chromatin immunoprecipitation system. Proc Natl Acad Sci USA. 2002;99:15030–15035. doi: 10.1073/pnas.232299999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roessler S, et al. Distinct promoters mediate the regulation of ebf1 gene expression by interleukin-7 and pax5. Mol Cell Biol. 2007;27:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith EM, Gisler R, Sigvardsson M. Cloning and characterization of a promoter flanking the early B cell factor (EBF) gene indicates roles for E-proteins and autoregulation in the control of EBF expression. J Immunol. 2002;169:261–270. doi: 10.4049/jimmunol.169.1.261. [DOI] [PubMed] [Google Scholar]
- 14.Kee BL, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 16.Hirokawa S, Sato H, Kato I, Kudon A. EBF-regulating Pax5 transcription is enhanced by STAT5 in the early stage of B cells. Eur J Immunol. 2003;33:1824–1829. doi: 10.1002/eji.200323974. [DOI] [PubMed] [Google Scholar]
- 17.Nutt SL, Urbanek P, Rolink A, Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell development: Difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 1997;11:476–491. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- 18.Zhuang Y, Jackson A, Pan L, Shen K, Dai M. Regulation of E2A gene expression in B-lymphocyte development. Mol Immunol. 2004;40:1165–1177. doi: 10.1016/j.molimm.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 19.Fuxa M, et al. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon K, et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 21.Peschon JJ, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller JP, et al. The earliest step in B lineage differentiation from common lymphoid progenitors is critically dependent upon interleukin 7. J Exp Med. 2002;196:705–711. doi: 10.1084/jem.20020784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikuchi K, Lai AY, Hsu CL, Kondo M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J Exp Med. 2005;201:1197–1203. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dias S, Silva H, Jr., Cumano A, Vieira P. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J Exp Med. 2005;201:971–979. doi: 10.1084/jem.20042393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sloan SR, Shen CP, McCarrick-Walmsley R, Kadesch T. Phosphorylation of E47 as a potential determinant of B-cell-specific activity. Mol Cell Biol. 1996;16:6900–6908. doi: 10.1128/mcb.16.12.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seet CS, Brumbaugh RL, Kee BL. Early B Cell Factor Promotes B Lymphopoiesis with Reduced Interleukin 7 Responsiveness in the Absence of E2A. J Exp Med. 2004;199:1689–1700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson K, et al. Ectopic expression of PAX5 promotes maintenance of biphenotypic myeloid progenitors coexpressing myeloid and B-cell lineage-associated genes. Blood. 2007;109:3697–3705. doi: 10.1182/blood-2006-05-026021. [DOI] [PubMed] [Google Scholar]
- 28.Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19-early B cell precursors. J Exp Med. 2006;203:675–687. doi: 10.1084/jem.20052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sigvardsson M, O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 30.Sigvardsson M, et al. Early B-cell factor, E2A, and Pax-5 cooperate to activate the early B cell-specific mb-1 promoter. Mol Cell Biol. 2002;22:8539–8551. doi: 10.1128/MCB.22.24.8539-8551.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martensson A, Martensson IL. Early B cell factor binds to a site critical for lambda5 core enhancer activity. Eur J Immunol. 1997;27:315–320. doi: 10.1002/eji.1830270145. [DOI] [PubMed] [Google Scholar]
- 32.Gisler R, Sigvardsson M. The human V-preB promoter is a target for coordinated activation by early B cell factor and E47. J Immunol. 2002;168:5130–5138. doi: 10.4049/jimmunol.168.10.5130. [DOI] [PubMed] [Google Scholar]
- 33.Chen X, Martin F, Forbush KA, Perlmutter RM, Kearney JF. Evidence for selection of a population of multi-reactive B cells into the splenic marginal zone. Int Immunol. 1997;9:27–41. doi: 10.1093/intimm/9.1.27. [DOI] [PubMed] [Google Scholar]
- 34.Kee BL, Rivera RR, Murre C. Id3 inhibits B lymphocyte progenitor growth and survival in response to TGF-beta. Nat Immunol. 2001;2:242–247. doi: 10.1038/85303. [DOI] [PubMed] [Google Scholar]
- 35.Fields S, et al. The “zinc knuckle” motif of Early B cell Factor is required for transcriptional activation of B cell-specific genes. Mol Immunol. 2008;45:3786–3796. doi: 10.1016/j.molimm.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pongubala JM, et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 37.Engel I, Murre C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J. 2004;23:202–211. doi: 10.1038/sj.emboj.7600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang H, et al. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol Cell. 2005;18:699–709. doi: 10.1016/j.molcel.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 39.Holmes ML, Carotta S, Corcoran LM, Nutt SL. Repression of Flt3 by Pax5 is crucial for B-cell lineage commitment. Genes Dev. 2006;20:933–938. doi: 10.1101/gad.1396206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cotta CV, Zhang Z, Kim HG, Klug CA. Pax5 determines B- versus T-cell fate and does not block early myeloid-lineage development. Blood. 2003;101:4342–4346. doi: 10.1182/blood-2002-10-3139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.