Abstract
Increasing the immunogenicity to delivered antigens by recombinant attenuated Salmonella vaccines (RASV) has been the subject of intensive study. With this goal in mind, we have designed and constructed a new generation of RASV that exhibit regulated delayed attenuation. These vaccine strains are phenotypically wild type at the time of immunization and become attenuated after colonization of host tissues. The vaccine strains are grown under conditions that allow expression of genes required for optimal invasion and colonization of host tissues. Once established in the host, these virulence genes are turned off, fully attenuating the vaccine strain. In this study, we compared 2 of our newly developed regulated delayed attenuation Salmonella enterica serovar Typhimurium strains χ9088 and χ9558 with the Δcya Δcrp Δasd strain χ8133, for their abilities to express and present a secreted form of the α-helical region of pneumococcal surface protein A (PspA) to the mouse immune system. All 3 strains induced high levels of serum antibodies specific for PspA as well as to Salmonella antigens in orally immunized mice. However, both RASVs expressing delayed attenuation elicited significantly greater anti-PspA immune responses, including serum IgG and T cell secretion of IL-4 and IFN-γ, than other groups. Also, vaccination with delayed attenuation strains resulted in a greater degree of protection against Streptococcus pneumoniae challenge than in mice vaccinated with χ8133 (71–86% vs. 21% survival, P ≤ 0.006). Together, the results demonstrate that the regulated attenuation strategy results in highly immunogenic antigen delivery vectors for oral vaccination.
Keywords: Streptococcus pneumoniae, bacterial vectors
Generating a safe and immunogenic vaccine strain is the biggest challenge in the development of live Salmonella vaccines (1). An ideal Salmonella vaccine strain should exhibit wild-type abilities to withstand stresses (enzymatic, acid, osmotic, ionic, etc.) and host defenses (bile, antibacterial peptides, etc.) encountered after oral or intranasal immunization, and should exhibit wild-type ability to colonize and invade host lymphoid tissues while remaining avirulent. Various attenuated Salmonella strains have been used as live vaccines to induce mucosal and systemic immunity against either the carrier itself or to a vectored antigen (2). Salmonella vaccine strains typically carry defined deletion mutations affecting either metabolic functions or virulence factors (3). Various attenuating mutations have been investigated in the pursuit to develop optimal immune responses (4, 5). Many attenuating mutations were found to either reduce Salmonella survival due to host-induced stresses and/or reduce colonization of lymphoid effector tissues leading to less than ideal immunogenicity (6, 7). To circumvent these problems, we explored ways to achieve regulated delayed attenuation in vivo (8, 9) to create vaccine strains that are phenotypically wild-type at the time of immunization and become attenuated after colonization of host tissues.
One strategy is the deletion of pmi, which encodes 6-phosphomannose isomerase that interconverts fructose-6-P and mannose-6-P (10). Because mannose is required for O-antigen synthesis, Δpmi mutants synthesize complete O-antigen only when grown in the presence of mannose to enable efficient colonization of lymphoid tissues. Synthesis of O-antigen ceases in vivo and O-antigen side chains are lost after ≈7 cell divisions in the absence of mannose. Salmonella enterica serovar Typhimurium pmi mutants are attenuated, even when grown with mannose (11), due to the eventual loss of O-antigen in vivo caused by the lack of nonphosphorylated mannose in host tissues. To ensure that all mannose provided to the vaccine strain during growth is directed toward O-antigen synthesis, the Δ(gmd-fcl)-26 mutation, which deletes 2 structural genes that encode enzymes for the conversion of GDP-mannose to GDP-fucose, was included in our strains. This deletion does not alter attenuation, tissue-colonizing ability, or immunogenicity of a strain with the Δpmi-2426 mutation alone (8).
Another strategy to achieve regulated delayed attenuation relies on the use of the arabinose-regulated araC PBAD activator-promoter (12). Deletion of either fur (13) or crp (14) is attenuating. The promoters, including sequences for activator or repressor protein binding, for the fur and crp genes were deleted and replaced with an araC PBAD cassette to yield Salmonella strains in which the transcription of these 2 genes is regulated by arabinose availability. Growth of such strains in the presence of arabinose leads to transcription of fur and crp, but expression ceases in host tissues, where there is no free arabinose (15). Attenuation develops as Fur and Crp are diluted at each cell division. We have combined 4 of these mutations to construct strain χ9088 [ΔPfur33::TT araC PBADfur Δpmi-2426 Δ(gmd-fcl)-26 ΔasdA33] (8), and a total of 10 mutations to construct strain χ9558 [Δpmi-2426 Δ(gmd-fcl)-26 ΔPfur81::TT araC PBADfur ΔPcrp527::TT araC PBADcrp ΔasdA27::TT araC PBADc2 ΔaraE25 ΔaraBAD23 ΔrelA198::araC PBADlacI TT ΔsopB1925 ΔagfBAC811]. Further details of the rationale for all of the mutations in strain χ9558 [supporting information (SI) Fig. S1 and Table S1] are described elsewhere (16). Here, we focus on the usefulness and immunogenicity of χ9088 and χ9558 as vaccines.
In previous work, we demonstrated that oral vaccination of mice with a Δcrp S. Typhimurium vaccine strain expressing a secreted pneumococcal surface protein A (PspA) fusion protein elicited high serum IgG titers against PspA, and those titers were higher than anti-LPS titers against the Salmonella carrier (17). Immunized mice were protected from virulent Streptococcus pneumoniae WU2 challenge (18). In this work, we evaluated the immunogenicity of 2 new S. typhimurium vaccine strains engineered with a regulated delayed attenuation system and synthesizing, as a test antigen, a secreted form of the α-helical region of PspA, similar to the one used previously. Antibody responses, cytokine responses, and protective immunity against S. pneumoniae WU2 challenge were evaluated. The results attained confirm the hypothesis that vaccination with Salmonella vaccine strains with regulated delayed in vivo attenuation elicits strong protective immune responses.
Results
Expression and Stability of rPspA in Salmonella.
The AsdA+ recombinant plasmid pYA3634 (pBR ori) directs the periplasmic secretion of the rPspARx1 (8) (Table S1). Analysis of whole cell lysates from the Δcya Δcrp Δasd strain χ8133, and regulated delayed attenuation strains χ9088 and χ9558 containing pYA3634 all expressed a protein with an approximate molecular mass of 37 kDa, the expected size of the Bla SS-PspA fusion protein that reacted specifically with an anti-PspA polyclonal antibody (Fig. S2). Note that previous observations indicate that ≈95% of the Bla SS-PspA fusion protein is partitioned between the cytoplasm and periplasm (18). Plasmids were maintained and protein expression was shown to be stable when strains were grown under nonselective conditions, in the presence of diaminopimelic acid (DAP), for 50 generations.
Ability to Access the Lymphoid Tissues.
The strategy for regulated delayed attenuation is based on the hypothesis that expression of wild-type characteristics during the initial stage of infection will allow more efficient colonization of host lymphoid tissues, leading to a more robust immune response. To evaluate the effect of regulated delayed attenuation on colonization, we first examined Δpmi strain χ8868 grown with or without mannose added to the growth media. The Δpmi strain colonized Peyer's patches, spleen, and liver significantly better when the cells were grown in the presence of mannose, on days 3 and 7 (Fig. S3A). By day 14, there was no difference in colonization between the 2 groups. We also compared Fur− strain χ9872, which carries the fur-1 mutation (13) with one that carries a regulated fur mutation, strain χ9725. No differences were observed in colonization of Peyer's patches, but there was a 100-fold difference in spleen colonization and a >10-fold difference in liver colonization on day 3 (Fig. S3B). By day 14, there was no difference between groups. Thus, in the case of single mutants, the strains with regulated delayed attenuation have a “jump start” for spleen and liver colonization over the gene knockout mutants. We previously evaluated a fur-1 Δcrp S. Typhimurium mutant in chickens and found it to be nonimmunogenic (Roland and Curtiss, unpublished); therefore, we did not evaluate that combination in mice.
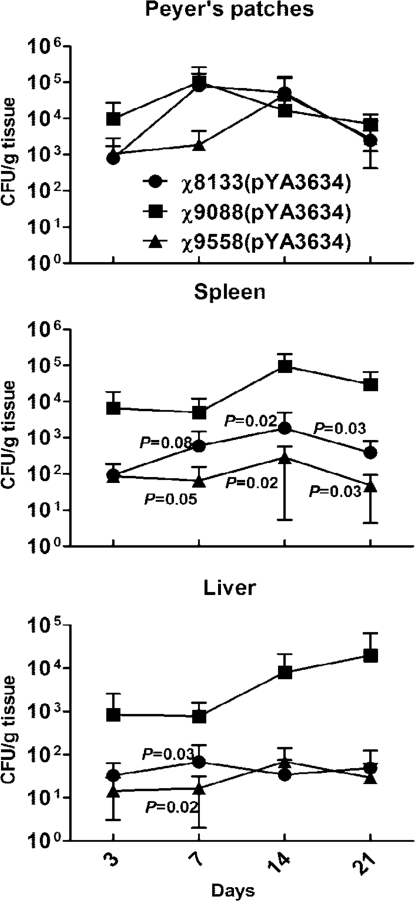
We next evaluated colonization of mouse tissues by Δcya Δcrp strain χ8133(pYA3634), and regulated delayed attenuation strains χ9088(pYA3634) and χ9558(pYA3634). All 3 strains colonized Peyer's patches equally well (Fig. 1). However, strain χ9088(pYA3634) colonized both spleen and liver more efficiently than the other 2 strains (P < 0.05). There was no difference in colonization by χ8133(pYA3634) and χ9558(pYA3634).
Fig. 1.
Comparison of colonization kinetics for the Δcya Δcrp strain χ8133(pYA3634) and regulated attenuation strains χ9088(pYA3634) and χ9558(pYA3634). At each time point, 6 mice per group were euthanized and Peyer's patches, spleen, and liver were collected, homogenized, and plated onto MacConkey plates. Salmonella colonies were counted and expressed as cfu per gram of tissue. P values shown in the representation are compared with χ9088(pYA3634). There were no significant differences between χ8133(pYA3634) and χ9558(pYA3634).
Antibody Responses in Mice After Oral Immunization with Recombinant Salmonella Vaccines.
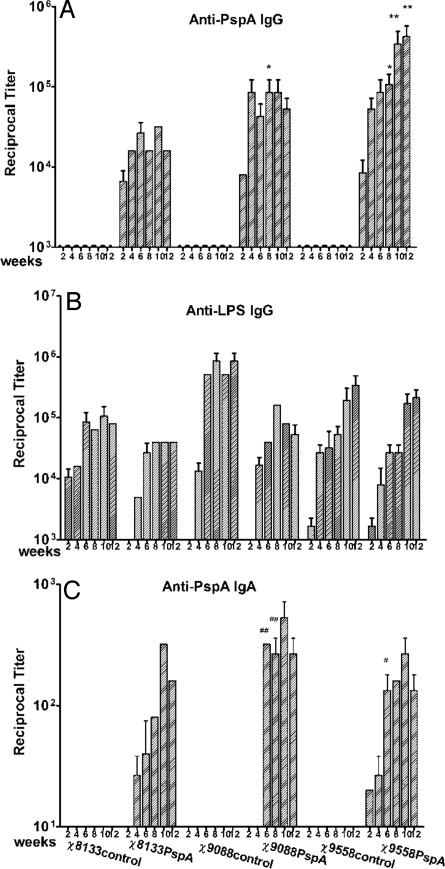
We orally inoculated groups of mice with 1–2 × 109 cfu of either χ8133, χ9088, or χ9558 carrying either PspA expression plasmid pYA3634 or control plasmid pYA3493 (17). Mice were boosted with the same dose of the same strain 8 weeks later. The antibody responses to rPspA and Salmonella LPS in the sera of immunized mice were measured (Fig. 2 A and B). This experiment was performed twice, with each group receiving approximately the same dose of vaccine (+/− 20%). The results from both experiments were similar and have been pooled for analysis. High serum IgG titers against both antigens were observed by 2 weeks after the primary immunization. The maximum preboost anti-rPspA IgG antibody levels were detected by 6 to 8 weeks post immunization. By week 8, the serum anti-rPspA IgG antibody levels of mice immunized with χ9088(pYA3634) or χ9558(pYA3634) was significantly higher than that of the χ8133(pYA3634) immunized mice (P < 0.05), and the anti-rPspA IgG antibody levels in χ9558(pYA3634) immunized mice was significantly higher than mice immunized with either χ9088(pYA3634) or χ8133(pYA3634) (P < 0.05 and P < 0.01, respectively).
Fig. 2.
Serum IgG responses to rPspA (A), to S. Typhimurium LPS (B), and vaginal wash IgA response to rPspA (C) as measured by ELISA. The data represent reciprocal anti-IgG antibody levels in pooled sera from mice orally immunized with attenuated Salmonella carrying either pYA3493 (control) or pYA3634 (rPspA) at the indicated weeks after immunization. Error bars represent variation between triplicate wells. Mice were boosted at week 8. *, P < 0.05 for anti-rPspA serum IgG antibody levels from mice immunized with either of the 2 regulated delayed attenuation strains, χ9558(pYA3634) or χ9088(pYA3634) with that of from mice immunized with the Δcya Δcrp strain χ8133(pYA3634) at week 8. **, P < 0.01 for the anti-rPspA serum IgG antibody levels of χ9558(pYA3634) immunized mice compared with χ8133(pYA3634) immunized mice at weeks 10 and 12. #, P < 0.05 for anti-rPspA vaginal wash IgA antibody levels of χ9558(pYA3634) immunized mice with that of the χ8133(pYA3634) immunized mice at week 6. ##, P < 0.01 for the anti-rPspA vaginal wash IgA antibody levels of χ9088(pYA3634) immunized mice compared with χ8133(pYA3634) immunized mice at week 6 and 8. No immune responses were detected to any antigen tested in mice immunized with buffer only or in preimmune sera from vaccinated mice (reciprocal titer <1:50).
No anti-PspA IgG was detected in mice immunized with strains carrying pYA3493. All strains elicited high antibody titers against LPS; although, by week 12, the titers in mice inoculated with both χ8133 constructs and χ9088(pYA3634) were significantly lower than the other groups (P < 0.05). Similar immune responses were obtained against Salmonella outer membrane proteins (SOMPs) (data not shown).
After the second immunization, significant boosting of serum antibody responses to both PspA and LPS was observed for mice immunized with χ9558(pYA3634). The serum IgG response to PspA was significantly greater than the response in mice immunized with the Δcrp strain χ8133(pYA3634) (P < 0.05).
Detectable mucosal IgA responses were slow to develop, but reached high titers by 6 weeks (Fig. 2C), where the titers from mice immunized with the delayed attenuation strains χ9088(pYA3634) and χ9558(pYA3634), were significantly higher than in mice immunized with the Δcya Δcrp strain χ8133(pYA3634). Maximum mucosal IgA responses for all groups occurred 2 weeks after boosting, at week 10.
IgG Isotype Analyses.
The types of immune responses to rPspA were further examined by measuring the levels of IgG isotype subclasses IgG1 and IgG2a (Fig. S4). Th1-helper cells direct cell-mediated immunity and promote class switching to IgG2a, and Th2 cells provide potent help for B cell antibody production and promote class switching to IgG1 (19, 20). Th1-type dominant immune responses are frequently observed after immunization with attenuated Salmonella (17, 21–23). A Th1- and Th2-type mixed response against PspA was observed for all groups through week 8. After boosting, a strong Th1 response developed for the χ8133(pYA3634) and χ9088(pYA3634) vaccinees, but not for the mice vaccinated with χ9558(pYA3634). This result may be due to the sopB deletion in strain χ9558. Although χ9558 differs from the other strains at several loci, this result is consistent with the known effects of a ΔsopB mutation to dampen the Th1 response against a vectored antigen (24).
Antigen-Specific Stimulation of IL-4 or IFN-γ Production.
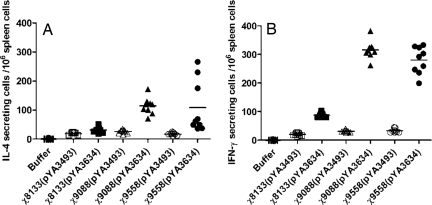
To further evaluate the Th1-Th2 immune response, ELISPOT was used to compare PspA stimulation of IFN-γ (Th1-associated) and IL-4 (Th2-associated) production by spleen cells from immunized and control mice (Fig. 3). Splenic lymphocytes from mice immunized with χ9088(pYA3634) or χ9558(pYA3634) produced similar levels of IFN-γ secreting cells, significantly more than mice immunized with χ8133(pYA3634) (P < 0.001). Similar results were obtained for PspA-specific IL-4 secreting cells, where the numbers were significantly less for χ8133(pYA3634) immunized mice than for the other 2 groups (P < 0.001).
Fig. 3.
Antigen-specific stimulation of IL-4 (A) or IFN-γ (B) Production. Splenectomies were performed on euthanized BALB/c mice 8 weeks after immunization. Buffer controls were also included. Splenocytes were harvested from 3 mice per group, and cells from each spleen were assayed in triplicate. Each symbol represents the results from a single well. ELISPOT analyses were performed as described in Materials and Methods. The results from each well are presented as ELISPOTS per million splenocytes minus any background ELISPOTS from unpulsed mock controls. One-way ANOVA and LSD methods were adopted to compare the secretion levels of IL-4 or IFN-γ between different groups. P < 0.001 for χ9558(pYA3634) and χ9088(pYA3634) versus χ8133(pYA3634) for secretion levels of IL-4 and IFN-γ. P < 0.01 for χ9558(pYA3634) versus χ9088(pYA3634) for secretion levels of IFN-γ.
Status of Systemic Cytokine Environment.
Two weeks after the primary immunization, sera from each group of mice were subjected to Bio-Plex analysis to measure the overall levels of cytokine secretion induced by immunization. This information will help us to further evaluate the effect of vaccination on the immune system. The cytokine secretion profiles were compared (Table S2). The sera from χ8133(pYA3634), χ9088(pYA3634), and χ9558(pYA3634) immunized mice contained higher concentrations of cytokines than the buffer control group (P < 0.01); χ9558(pYA3634) immunized mice had increased levels of both Th1-specific cytokines (IL-2, IL-12, TNF-α) and Th2-specific cytokines (IL-4, IL-5, IL-10), compared with the χ8133(pYA3634) group (P < 0.05). Together, these results indicate that all of the recombinant attenuated Salmonella vaccine (RASV) strains elicited a mixed Th1 and Th2 response and that the regulated delayed attenuation strain χ9558(pYA3634) stimulated stronger cellular immunity and cytokine secretion than χ8133(pYA3634); thus, facilitating antigen presentation and activation of B and T cells.
Evaluation of Protective Immunity.
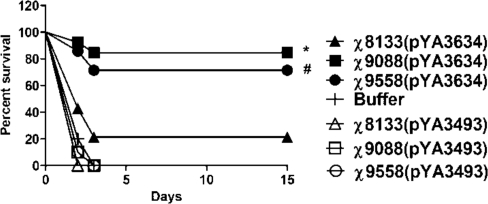
To examine the ability of Salmonella-rPspA vaccines to protect against pneumococcal infection, mice were challenged i.p. with 5 × 104 cfu (250 LD50) of S. pneumoniae WU2 4 weeks after they were boosted (Fig. 4). S. pneumoniae WU2 produces PspA that is cross-reactive with PspARx1 (25). Immunization with any of the pspA-expressing strains provided significant protection against challenge (P < 0.01). However, the protection afforded by χ9088(pYA3634) or χ9558(pYA3634) was significantly greater than χ8133(pYA3634) (P < 0.01). All of the mice that died in these experiments succumbed within 2 or 3 days after challenge.
Fig. 4.
Oral immunization with PspA-expressing Salmonella strains protects BALB/c mice against i.p. challenge with S. pneumoniae WU2; 5 (control) or 7 (vaccine) mice per group were orally immunized twice at 8-weeks intervals with the indicated vaccine strains. Mice were challenged in with ≈5 × 104 cfu of S. pneumoniae WU2 4 weeks after the second oral immunization. The experiment was performed twice. Both experiments gave similar results, and the data have been pooled for presentation and analysis. The average time of death was 2–3 days. *, P = 0.006 vs. survival of mice immunized with Δcya Δcrp strain χ8133(pYA3634); #, P = 0.0063 vs, survival of mice immunized with χ8133(pYA3634).
Isolation of S. pneumoniae from Blood.
Each mouse was marked and bled 24 h after i.p. challenge. S. pneumoniae was recovered from the blood of mice showing significant signs of weakness and listlessness and that ultimately died within 7 days (6833.3 ± 321.5 cfu/mL), but not from mice that appeared to be healthy and that survived past 15 days (P < 0.001).
Passive-Immunization Studies.
A passive-immunization study was conducted to evaluate the roles of antibody and T cell mediated immunity afforded by immunization of mice with the Salmonella vaccines; 100 μL of pooled sera or spleen lymphocytes (1 × 107) taken from immunized mice or controls was administered by tail vein injection into groups of 5 naive mice. This procedure was followed 12 h later by i.p. challenge with 5 × 104 cfu of WU2. Mice receiving sera or cells transferred from χ8133(pYA3493), χ9088(pYA3493), χ9558(pYA3493), or buffer immunized mice died with a mean time of 2 days (Table 1). The sera trans-ferred from χ9088(pYA3634) immunized mice and from χ9558(pYA3634) immunized mice protected all 5 naive mice from challenge. Sera transferred from χ8133(pYA3634) immunized mice protected 4 out of 5 mice from challenge. Passive transfer of spleen lymphocytes, which include B cells and T cells, from χ9088(pYA3634) immunized mice protected all 5 naive mice from challenge; spleen lymphocytes from χ9558(pYA3634) immunized mice protected 3 out of 5 mice from challenge; whereas the spleen cell transfer from χ8133(pYA3634) immunized mice provided no protection.
Table 1.
Passive transfer of pneumococcal immunity by serum or lymphocytes from donors immunized with PspA Salmonella vaccines
| Vaccine strain used to immunize donors | Percentage survival of recipients |
|
|---|---|---|
| Pooled serum | Spleen cells | |
| Buffer | 0 | 0 |
| χ8133(pYA3493) | 0 | 0 |
| χ9088(pYA3493) | 0 | 0 |
| χ9558(pYA3493) | 0 | 0 |
| χ8133(pYA3634) | 80 | 0 |
| χ9088(pYA3634) | 100 | 100* |
| χ9558(pYA3634) | 100 | 60** |
Mice were orally immunized at day 0 and boosted 8 weeks later with the indicated vaccine strains. Serum and cells were collected 4 weeks after boosting and transferred to groups of 5 naive mice. All recipient mice were challenged by i.p with 5 × 104 cfu of WU2 at 12 h after transfer. Survival was calculated 15 days post challenge. Pooled serum, 0.1 mL of serum i.v. Spleen cells, 1 × 107 viable spleen cells i.v.
*, P = 0.002, compared with groups of mice receiving passive transfer from χ8133(pYA3634) immunized or control donors.
**, P = 0.038, compared with groups of mice receiving passive transfer from χ8133(pYA3634) immunized or control donors.
Discussion
Pathogenic bacteria may be attenuated by mutation, so that on infection, host disease symptomology does not occur. However, most means of attenuation make live vaccine strains more susceptible than wild-type strains to environmental stresses encountered after inoculation into the animal or human host. Consequently, fewer bacteria survive to colonize the gut-associated lymphoid tissue (GALT), nasal-ALT, and/or bronchus-ALT with a reduction in the effective immunogenicity of the vaccine. Thus, these attenuation mechanisms hyperattenuate the vaccine, precluding the candidate vaccine from either reaching or persisting in lymphoid tissues to a sufficient extent or duration to permit induction of a protective immune response against the wild-type pathogen of interest. We have addressed this problem by developing methods for regulating the expression of the attenuated phenotype, allowing the live vaccine strain to display abilities similar to a wild-type virulent parental pathogen to successfully colonize effector lymphoid tissues before the display and imposition of the fully attenuated phenotype.
Strains with a single attenuating mutation, Δpmi grown without mannose, or fur-1 were slow to colonize host tissues compared with isogenic strains with regulated delayed attenuation (Fig. S2). By 14 days, a time at which we expect that the regulated strains would be fully attenuated, all strains were being cleared. Similar results were observed for the vaccine strains, except for strain χ9088(pYA3634) in the liver. Although strain χ9558(pYA3634) did not colonize any better than Δcya Δcrp strain χ8133(pYA3634), χ9558 carries a number of additional mutations designed to make it safe in infant mice, making it less able to colonize adult mice than strain χ9088.
The new regulated delayed attenuation strains χ9088(pYA3634) and χ9558(pYA3634) induced stronger immune responses to PspA than Δcya Δcrp strain χ8133(pYA3634), as judged by PspA-specific serum antibody levels, PspA specific lymphocyte cytokine secretion levels, systemic cytokine secretion levels, and protection from virulent S. pneumoniae challenge. These new strains provided a higher degree of protection than we have seen previously. Although we used a 10-fold higher challenge dose of S. pneumoniae WU2 than in a previous study (17), the protection rate was increased >20%.
Expression of pspA resulted in lower induction of anti-LPS responses in mice immunized with the Δcya Δcrp strain, χ8133, and the regulated attenuation strain χ9088 (Fig. 2), compared with strains carrying the empty vector, pYA3493, presumably due to antigen burden. In contrast, no antigen burden effect was seen in mice immunized with χ9558 strains, probably because, in addition to regulated delayed attenuation, strain χ9558 also includes a regulated delayed antigen expression system (S.W. and R.C., unpublished results). The basis of this system is similar in principle to regulated delayed attenuation. A chromosomal araC PBAD promoter/activator drives the synthesis of LacI, which represses transcription from the plasmid-borne Ptrc promoter that directs the synthesis of PspA in plasmid pYA3634. Thus, in vitro, in the presence of arabinose, PspA is not synthesized. Once the vaccine reaches the arabinose-free environment in host tissues, PspA synthesis is initiated. A detailed explanation of this feature will be described elsewhere.
Mixed Th1- and Th2-type immune responses were observed for rPspA until after the boost, when a strong Th1 response developed. Strain χ9558(pYA3634) stimulated a much more balanced Th1 and Th2 response, likely due to the inclusion of the sopB mutation (24). Passive transfer of serum from mice immunized with any of the 3 vaccine strains expressing PspA were protected from challenge, but transfer of spleen cells containing both B and T cells provided protective immunity only when derived from mice immunized with the new generation vaccines. Together, the results of this study demonstrated that the Salmonella vaccine strains χ9088(pYA3634) and χ9558(pYA3634) featuring a novel regulated delayed in vivo attenuation system are superior to χ8133(pYA3634) not only in inducing PspA specific antibody responses, but also in eliciting specific cytokine secretion, resulting in significant protection of mice against pneumococcal challenge.
Materials and Methods
Bacterial Strains, Plasmids, Media, and Growth Conditions.
The bacterial strains and plasmids used in this study are listed in Table S1. S. typhimurium cultures were grown at 37 °C in LB broth or on LB agar (26). For animal experiments, plasmid-containing χ9088 and χ9558 cultures were supplemented with 0.2% mannose or 0.2% mannose, and 0.05% arabinose, respectively. No additions were made to the media for growing plasmid-containing χ8133 cultures. DAP was added (50 μg/mL) for the growth of Asd− strains (27). S. pneumoniae WU2 was cultured on brain heart infusion (BHI) agar containing 5% sheep blood or in Todd–Hewitt broth plus 0.5% yeast extract (25). Plasmid pYA3493 is an expression vector for constructing antigen fusions to the first 23 aa of β-lactamase (17). Plasmid pYA3634 carries amino acids 3–257 of codon-optimized pspA RX-1 fused to the first 35 aa of β-lactamase (8).
Strain Construction and Characterization.
The mutations carried by strains χ9088 and χ9558 are shown in Fig. S1. Details of the construction of strain χ9558 will be described elsewhere. MacConkey agar supplemented with 1% maltose was used to confirm the phenotype of crp mutants (17). Chrome Azurol S (CAS) plates were used to confirm the constitutive synthesis of siderophores characteristic of fur mutants (28). The presence of the ΔasdA33 and ΔasdA16 mutations in Salmonella was confirmed by inability of the strain to grow on media without DAP (27). LPS profiles of Salmonella strains were examined as described (29). Plasmid stability was determined as previously described (30).
SDS/PAGE and Immunoblot Analyses.
For details, see SI Materials and Methods.
Immunization of Mice.
Female BALB/c mice, 6–7 weeks old, were obtained from Charles River Laboratories. All animal procedures were approved by the Arizona State University Animal Care and Use Committee. Mice were acclimated for 7 days before starting the experiments. The animals were housed directly on bedding for all experiments. RASV strains were grown statically overnight in LB broth containing the appropriate supplements at 37 °C. The day after, an overnight culture of 1 mL was inoculated into 100 mL of LB broth containing the appropriate supplements and grown with aeration at 37 °C to an OD600 of 0.8 to 0.9. Cells were pelleted by centrifugation at room temperature (6,000 × g for 15 min), and the pellet resuspended in 1 mL of buffered saline with gelatin (BSG). Dilutions of the vaccine strains were plated onto LB agar to determine bacterial titers. Groups of 5 to 7 mice were orally inoculated with 20 μL of BSG alone or BSG containing 1 × 109 cfu of Salmonella on day 0 and again 8 weeks later. At days 3, 7, 14, and 21 after the primary dose, spleen, Peyer's patches, and liver samples were collected and homogenized to a total volume of 1 mL in BSG, and dilutions of 10−1 to 10−6 (depending on the tissue) were plated onto MacConkey agar and LB agar to determine the numbers of viable bacteria. Samples that were positive by enrichment in selenite cysteine broth were recorded as >2 cfu, and negative samples as were recorded as <2 cfu. Blood was obtained by mandibular vein puncture at biweekly intervals. After centrifugation, the serum was removed from the whole blood and stored at −20 °C.
Antigen Preparation.
We purified rPspA and S. typhimurium outer membrane proteins (SOMPs) as described in ref. 16. S. typhimurium LPS was obtained from Sigma. The rPspA clone was kind gift from Susan Hollingshead at the University of Alabama at Birmingham (31).
ELISA.
Sera from all mice in a group were pooled for analysis. ELISA was used to assay antibodies in serum to S. typhimurium LPS, SOMPs, and to rPspA, and in vaginal wash to rPspA as previously described (17). A brief summary can be found in SI Materials and Methods.
Passive Transfer of Cells and Sera.
At week 12, sera and spleen cells were harvested from 5 mice per group. The sera were pooled and Spleen cells (1 × 107) were suspended in PBS and injected into the lateral tail veins of naive, syngeneic BALB/c mice. Naive syngeneic BALB/c mice received 100 μL of serum from different groups of mice through tail vein. All groups were challenged i.p. after 12h with 5 × 104 cfu of S. pneumoniae in 200 μL of BSG.
IL-4 and IFN-γ ELISPOTs.
At week 8, spleen cells were harvested from 3 mice per group. Cells from each spleen were assayed by ELISPOT in triplicate wells as previously described (31). A brief summary of this procedure can be found in SI Materials and Methods.
Measurement of Cytokine Concentrations.
Cytokine concentrations were determined by using the Bio-Plex Protein Array System (Bio-Rad), as previously described (32). A brief summary of this procedure can be found in SI Materials and Methods.
Pneumococcal Challenge.
At week 12, the ability of the Salmonella-PspA vaccine to protect immunized mice against S. pneumoniae was assessed by i.p. challenge with 5 × 104 cfu of S. pneumoniae WU2 in 200 μL of BSG (34). The LD50 (LD50) of S. pneumoniae WU2 in BALB/c mice was 2 × 102 cfu by i.p. administration (data not shown). Mice were marked 24 h after i.p. challenge, and blood samples were taken. Dilutions were made into saline and plated on BHI agar containing 5% sheep blood. Bacterial colonies were enumerated after overnight incubation at 37 °C.
Statistical Analysis.
Numerical data were expressed as means ± SE. An ANOVA (SPSS Software) analysis, followed by least significant difference (LSD) method, were used to evaluate differences in Ab titer and cytokine-secreting cells response discerned to 95% confidence intervals. The Kaplan–Meier method (SPSS Software) was applied to obtain the survival fractions after i.p. challenge of orally immunized mice. By using the log-rank test, the P value for statistical differences between surviving pneumococcal challenges and Salmonella-vaccinated groups or BSG was discerned at the 95% confidence interval. The Pearson χ2 test was used to analysis the data of passive transfer. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments.
We thank Susan Hollingshead (University of Alabama, Birmingham, AL) for providing WU2 strain, PspA polyclonal antibody, and purified rPspA; Rebin Kader (Arizona State University) for expert technical assistance; and Erika Arch (Arizona State University) for her assistance with the manuscript. This work was supported by National Institutes of Health Grants R01 AI24533, R01 AI057885, and R01 AI056289, and the Bill and Melinda Gates Foundation Grant 37863.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811697106/DCSupplemental.
References
- 1.Kwon YM, Cox MM, Calhoun LN. Salmonella-based vaccines for infectious diseases. Expert Rev Vaccines. 2007;6:147–152. doi: 10.1586/14760584.6.2.147. [DOI] [PubMed] [Google Scholar]
- 2.Medina E, Guzman CA. Use of live bacterial vaccine vectors for antigen delivery: Potential and limitations. Vaccine. 2001;19:1573–1580. doi: 10.1016/s0264-410x(00)00354-6. [DOI] [PubMed] [Google Scholar]
- 3.Raupach B, Kaufmann SHE. Bacterial virulence, proinflammatory cytokines and host immunity: How to choose the appropriate Salmonella vaccine strain? Microbes Infect. 2001;3:1261–1269. doi: 10.1016/s1286-4579(01)01486-1. [DOI] [PubMed] [Google Scholar]
- 4.Dunstan SJ, Simmons CP, Strugnell RA. Comparison of the abilities of different attenuated Salmonella typhimurium strains to elicit humoral immune responses against a heterologous antigen. Infect Immun. 1998;66:732–740. doi: 10.1128/iai.66.2.732-740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garmory HS, et al. The use of live attenuated bacteria as a delivery system for heterologous antigens. J Drug Target. 2003;11:471–479. doi: 10.1080/10611860410001670008. [DOI] [PubMed] [Google Scholar]
- 6.Hohmann EL, Oletta CA, Miller SI. Evaluation of a phoP/phoQ-deleted, aroA-deleted live oral Salmonella typhi vaccine strain in human volunteers Vaccine. 1996;14:19–24. doi: 10.1016/0264-410x(95)00173-x. [DOI] [PubMed] [Google Scholar]
- 7.Tacket CO, et al. Safety and immunogenicity in humans of an attenuated Salmonella typhi vaccine vector strain expressing plasmid-encoded hepatitis B antigens stabilized by the asd-balanced lethal vector system. Infect Immun. 1997;65:3381–3385. doi: 10.1128/iai.65.8.3381-3385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtiss R, III, et al. In: Virulence Mechanisms of Bacterial Pathogens. Brogden K, et al., editors. Washington, DC: American Society for Microbiology Press; 2007. pp. 297–313. [Google Scholar]
- 9.Curtiss R, III, Wanda SY, Zhang X, Gunn B. Salmonella vaccine vectors displaying regulated delayed in vivo attenuation to enhance immunogenicity. 107th General Meeting American Society for Microbiology; Toronto, Canada. 2007. p. 278. abstr E-061. [Google Scholar]
- 10.Stocker BA, Wilkinson RG, Mäkelä PH. Genetic aspects of biosynthesis and structure of Salmonella somatic polysaccharide. Ann N Y Acad Sci. 1966;133:334–348. doi: 10.1111/j.1749-6632.1966.tb52375.x. [DOI] [PubMed] [Google Scholar]
- 11.Collins LV, Attridge S, Hackett J. Mutations at rfc or pmi attenuate Salmonella typhimurium virulence for mice. Infect Immun. 1991;59:1079–1085. doi: 10.1128/iai.59.3.1079-1085.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-del Portillo F, Foster JW, Finlay BB. Role of acid tolerance response genes in Salmonella typhimurium virulence. Infect Immun. 1993;61:4489–4492. doi: 10.1128/iai.61.10.4489-4492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curtiss R, III, Kelly SM. Salmonella typhimurium deletion mutants lacking adenylate cyclase and cyclic AMP receptor protein are avirulent and immunogenic. Infect Immun. 1987;55:3035–3043. doi: 10.1128/iai.55.12.3035-3043.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katzman RL, Lisowska E, Jeanloz RW. Invertebrate connective tissue. Isolation of D-arabinose from sponge acidic polysaccharide. Biochem J. 1970;119:17–19. doi: 10.1042/bj1190017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curtiss R, III, et al. Salmonella vaccine strains with regulated delayed attenuation in vivo. Infect Immun. 2009 doi: 10.1128/IAI.00693-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang HY, Srinivasan J, Curtiss R., III Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar Typhimurium vaccine. Infect Immun. 2002;70:1739–1749. doi: 10.1128/IAI.70.4.1739-1749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xin W, et al. Analysis of type II secretion of recombinant pneumococcal PspA and PspC in a Salmonella enterica serovar Typhimurium vaccine with regulated delayed antigen synthesis. Infect Immun. 2008;76:3241–3254. doi: 10.1128/IAI.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeKruyff RH, Rizzo LV, Umetsu DT. Induction of immunoglobulin synthesis by CD4+ T cell clones. Semin Immunol. 1993;5:421–430. doi: 10.1006/smim.1993.1048. [DOI] [PubMed] [Google Scholar]
- 20.Gor DO, Rose NR, Greenspan NS. Th1-Th2: A procrustean paradigm. Nat Immunol. 2003;4:503–505. doi: 10.1038/ni0603-503. [DOI] [PubMed] [Google Scholar]
- 21.Pashine A, et al. Th1 dominance in the immune response to live Salmonella typhimurium requires bacterial invasiveness but not persistence. Int Immunol. 1999;11:481–489. doi: 10.1093/intimm/11.4.481. [DOI] [PubMed] [Google Scholar]
- 22.Pascual DW, et al. Expression of recombinant enterotoxigenic Escherichia coli colonization factor antigen I by Salmonella typhimurium elicits a biphasic T helper cell response. Infect Immun. 1999;67:6249–6256. doi: 10.1128/iai.67.12.6249-6256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramarathinam L, Niesel DW, Klimpel GR. Salmonella typhimurium induces IFN-γ production in murine splenocytes. Role of natural killer cells and macrophages. J Immunol. 1993;150:3973–3981. [PubMed] [Google Scholar]
- 24.Link C, et al. An SopB-mediated immune escape mechanism of Salmonella enterica can be subverted to optimize the performance of live attenuated vaccine carrier strains. Microbes Infect. 2006;8:2262–2269. doi: 10.1016/j.micinf.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Briles DE, et al. PspA, a protection-eliciting pneumococcal protein: Immunogenicity of isolated native PspA in mice. Vaccine. 1996;14:858–867. doi: 10.1016/0264-410x(96)82948-3. [DOI] [PubMed] [Google Scholar]
- 26.Bertani G. Studies on lysogenesis I: The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakayama K, Kelly SM, Curtiss R., III Construction of an asd+ expression-cloning vector: Stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Nat Biotechnol. 1988;6:693–697. [Google Scholar]
- 28.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 29.Hitchcock PJ, Brown TM. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konjufca V, Wanda S-Y, Jenkins MC, Curtiss R., III A recombinant attenuated Salmonella enterica serovar Typhimurium vaccine encoding Eimeria acervulina antigen offers protection against E. acervulina challenge. Infect Immun. 2006;74:6785–6796. doi: 10.1128/IAI.00851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nabors GS, et al. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies to heterologous PspA molecules. Vaccine. 2000;18:1743–1754. doi: 10.1016/s0264-410x(99)00530-7. [DOI] [PubMed] [Google Scholar]
- 32.Sedgwick JD, Holt PG. A solid-phase immunoenzymatic technique for the enumeration of specific antibody-secreting cells. J Immunol Methods. 1983;57:301–309. doi: 10.1016/0022-1759(83)90091-1. [DOI] [PubMed] [Google Scholar]
- 33.Jonathan RK, et al. Circulating cytokines and chemokines in acute symptomatic parvovirus B19 infection: Negative association between levels of pro-inflammatory cytokines and development of B19-associated arthritis. J Med Virol. 2004;74:147–155. doi: 10.1002/jmv.20158. [DOI] [PubMed] [Google Scholar]
- 34.Nayak AR, et al. A live recombinant avirulent oral Salmonella vaccine expressing pneumococcal surface protein A induces protective responses against Streptococcus pneumoniae. Infect Immun. 1998;66:3744–3751. doi: 10.1128/iai.66.8.3744-3751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.