Abstract
Fibrillarin, a protein component of C/D box small nucleolar ribonucleoproteins (snoRNPs), directs 2′-O-methylation of rRNA and is also involved in other aspects of rRNA processing. A gene trap screen in embryonic stem (ES) cells resulted in an insertion mutation in the fibrillarin gene. This insertion generated a fusion protein that contained the N-terminal 132 amino acids of fibrillarin fused to a β-galactosidase-neomycin phosphotransferase reporter. As a result, the N-terminal GAR domain was present in the fusion protein but the methyltransferase-like domain was missing. The ES cell line with the targeted fibrillarin allele was transmitted through the mouse germ line, creating heterozygous animals. Western blot analyses showed a reduction in fibrillarin protein levels in the heterozygous knockout animals. Animals homozygous for the mutation were inviable, and massive apoptosis was observed in early Fibrillarin−/− embryos, showing that fibrillarin is essential for development. Fibrillarin+/− live-born mice displayed no obvious growth defect, but heterozygous intercrosses revealed a reduced ratio of +/− to +/+ mice, showing that some of the Fibrillarin heterozygous embryos die in utero. Analyses of tissue samples and cultured embryonic fibroblasts showed no discernible alteration in pre-rRNA processing or the level of the U3 snoRNA. However, the level of the intron-encoded box C/D snoRNA U76 was clearly reduced. This suggests a high requirement for snoRNA synthesis during an early stage in development.
Stable cellular RNAs, such as tRNAs, rRNAs, and small nuclear RNAs (snRNAs), contain a large number of posttranscriptional modifications. The most prevalent modifications in rRNAs and spliceosomal snRNAs are the methylation of the ribose moiety at the 2′-hydroxyl group and the conversion of uridines into pseudouridines, which is directed by guide small nucleolar RNAs (snoRNAs) (reviewed in 6, 15, and 34).
The small nucleolar RNAs (snoRNAs) play a key role in several aspects of pre-rRNA processing, including cleavage and pre-rRNA modification. The snoRNAs are present in the cell as small nucleolar ribonucleoprotein particles (snoRNPs) and can be divided into three groups, based on conserved sequence elements (36). A large family of snoRNAs that share two short sequence elements, called boxes C and D, serve as methylation guides; whereas the H/ACA snoRNAs are the pseudouridylation guide snoRNAs (for a review, see reference 44). The box C/D type snoRNAs have sequences 10 to 22 nucleotides in length of perfect complementarity to sequences within the mature rRNAs; thus, formation of snoRNA-rRNA hybrids positions the conserved box D or D′ element of the snoRNA five base pairs from the nucleotide to be methylated.
Each snoRNP consists of a specific snoRNA and a set of associated proteins common to all box C/D or H/ACA snoRNPs. The box C/D snoRNPs contain four essential proteins, Nop56p, Nop58p, Snu13p, and Nop1p/fibrillarin (17), and it was shown that binding of the 15.5K protein (Snu13p in Saccharomyces cerevisiae) to the box C/D motif is required for the association of the other C/D snoRNP-associated proteins (43). The distribution of these box C/D-associated proteins is asymmetric with the C′ box contacting Nop56 and fibrillarin, the C box interacting with Nop58, and the D and D′ boxes contacting fibrillarin (5).
Human fibrillarin contains an amino-terminal domain that is rich in glycine and arginine residues (termed the GAR domain), a central RNA-binding domain comprising an RNP-2-like consensus sequence, and a C-terminal α-helical domain (3). Crystal structure of a fibrillarin homolog from archaebacteria revealed that the overall fold of its C-terminal domain was similar to the catalytic domain common to many S-adenosylmethionine-dependent methyltransferases (42). The archaebacterial homologs lack the N-terminal GAR domain that is present in eukaryotes (1). Phylogenetic analysis determined a high sequence identity in the C-terminal domain among many fibrillarin homologs, strongly suggesting that they all contain a methyltransferase folding domain (42).
Fibrillarin has been highly conserved throughout evolution; for instance, human fibrillarin is 67% identical to its yeast homolog, termed Nop1p (for nucleolar protein 1) and 81% identical to the Xenopus protein (3, 10). Antifibrillarin autoantibodies are present in patients with the autoimmune disease scleroderma and are able to detect this protein in different organisms (19, 23, 24). A fibrillarin knockout is lethal in S. cerevisiae, and expression of human or Xenopus fibrillarin can functionally replace NOP1 (13). A series of temperature-sensitive lethal point mutations demonstrated that fibrillarin is involved in several functions, namely, pre-rRNA processing and modification and also ribosome assembly (38). Moreover, a point mutation in the putative methyltransferase domain of the yeast fibrillarin, Nop1p, inhibits the overall ribose methylation of rRNAs (38), suggesting that fibrillarin is the methyltransferase in box C/D snoRNPs.
Contrary to snRNAs, which have a cytoplasmic phase of maturation, snoRNAs are restricted to the nucleus (31, 32). Nucleolar localization of box C/D snoRNAs requires the box C/D motif and involves transit through the Cajal bodies (CBs) (18, 25, 41). The protein components of these snoRNPs are essential for this localization, since depletion of the core snoRNP proteins results in failure of box C/D snoRNAs to accumulate in the nucleolus. Metazoan fibrillarin, which localizes to the Cajal bodies and to the nucleoli, has been shown to interact directly with the spinal muscular atrophy disease protein SMN, and this interaction is dependent on transcription by RNA polymerase I (14, 22). Moreover, fibrillarin and SMN exist as a complex in vivo in HeLa cells and colocalize in both nucleoli and Cajal bodies. These results suggested a role for the SMN protein in nucleolar RNA biogenesis (reviewed in reference 33). Inactivation of the SMN gene results in massive cell death in early mouse embryos, demonstrating that the SMN gene product is essential for normal development (26). The recruitment of the SMN protein to Cajal bodies is mediated by dimethyl arginines present in p80 coilin, a protein that is a marker for the Cajal bodies (9). A coilin knockout mouse lacking the C-terminal 487 amino acids of coilin displayed residual Cajal bodies that failed to recruit SMN and spliceosomal Sm snRNP proteins, but did contain fibrillarin (39).
In this study, we analyzed the effect of depleting fibrillarin in the mouse. A gene trap screen in embryonic stem (ES) cells rendered an insertion mutation in the fibrillarin gene. This line was transmitted through the germ line, and live-born heterozygous animals displayed no obvious phenotype; however, we observed the loss of heterozygous animals. In contrast, homozygous knockout animals were not viable, demonstrating an essential role for fibrillarin in normal development.
MATERIALS AND METHODS
Gene targeting.
The gene trap screen was performed with pGT1, -2, and -3, as previously described (29, 30). The ES cell line E14 (12) was maintained in the presence of leukemia inhibitory factor and electroporated with a mixture of 50 μg each of pGT1, -2, and -3 that had been linearized with HindIII (30). Neomycin-resistant E14 colonies were selected with 200 μg of geneticin/G418 (Gibco BRL) per ml for 10 days, and resistant colonies were differentiated in medium containing 5% fetal calf serum without leukemia inhibitory factor.
Cytological examination of fusion proteins.
Neomycin-resistant colonies were first screened for β-galactosidase/lacZ activity with 5-bromo-4-chloro-3-indoyl-β-d-galactoside (X-Gal). Lines that displayed nuclear X-Gal staining were then analyzed by immunofluorescence with antibodies specific for β-galactosidase (30). Slides were counterstained with 0.5 μg of 4′,6′-diamidino-2-phenylindole (DAPI) and examined on a Zeiss Axioplan II microscope, and images were captured with a Princeton Instruments Pentamax charge-coupled device camera with IPLab software.
Determining the sequence of trapped genes.
Total RNA was prepared with Bio/RNA-X-cell (Bio/Gene Limited), and 5′ rapid amplification of cDNA ends (RACE) products were generated as described previously (29, 30). After dialysis on 0.1 μm nitrocellulose disks (Millipore), 5′-RACE products were sequenced directly with Big Dye terminator cycle sequencing and −40 lacZ primer (USB/Amersham). Databases were searched for sequence matches with the BLAST algorithm (http://www.ncbi.nlm.gov/BLAST/).
Mouse breeding and genotyping.
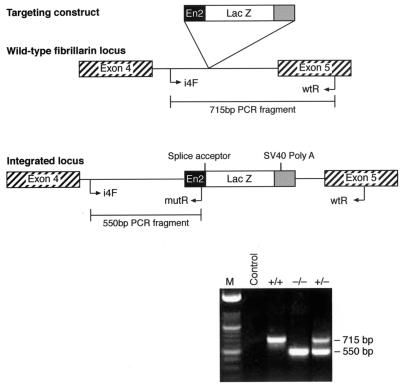
Gene trap integrations were transmitted through the germ line by injection of ES cells into C57BL/6 blastocysts. Chimeric males were then back crossed to MF1 outbred and C57BL/6 inbred females, and offspring were analyzed by PCR. Several backcross generations were performed on the C57BL/6 genetic background prior to setting up heterozygous intercrosses. Mouse genomic tail DNA was prepared according to standard procedures. The wild-type fibrillarin allele was detected with primers corresponding to intron 4 (i4F) and exon 5 (wtR), resulting in the amplification of a 715-bp fragment. The mutant fibrillarin allele was detected with primers corresponding to intron 4 (i4F) and the lacZ cassette (mutR), resulting in the amplification of a 550-bp fragment that diagnoses the targeted allele.
The sequences of the primers were as follows: i4F, GGTTCATATGGTGCCGTGC; wtR CCACTACTCAGTGCAGTGC; and mutR, GGTTCATATGGTGCCGTGC. PCR conditions were 5 min at 94°C, followed by two cycles of 94°C for 45 s,70°C for 45 s, and 72°C for 45 s; then two cycles of 94°C for 45 s, 65°C for 45 s, and 72°C for 45 s; followed by two cycles of 94°C for 45 s, 62°C for 45 s, and 72°C for 45 s; then two cycles of 94°C for 45 s, 58°C for 45 s, and 72°C for 45 s; and finally 25 cycles at 94°C for 45 s, 55°C for 45 s, and 72°C for 45 s and a final 5-min incubation at 72°C.
Analysis of rRNA and snoRNA processing.
Total RNA was extracted from tissue samples and MEF cells with Total RNA Isolation reagent (Advanced Biotechnologies Ltd.) following the manufacturer's instructions. Northern analyses were performed on RNA separated on 1.2% agarose-formaldehyde or 8% polyacrylamide-urea gels as previously described (35).
Hybridization probes were as follows: (064) mITS2-5′ CAA GCG ACG CTC AGA CA; (065) mITS1-5′ TAT TTC GGG TGT GAG CG; (067) m5′ETS/18S AGG TAA GGA AGC GCG AG; (008) 18S CAT GGC TTA ATC TTT GAG AC; (007) 25S CTCCGCTTATTGATATGC; (034) 5.8S GCT AGC TGC GTT CTT CAT C; (200) U3A UUA UGG GAC UUG UU (2′-O-methyl RNA); (283) mU76 CAA GAG TAG CAA ATA TGA TG; and (284) mU83 CAG TCA TGG GTG ATA GAT.
Embryonic expression pattern of fibrillarin.
At 9.5 and 12.5 days postcoitus (dpc), embryos dissected from heterozygous intercrosses were fixed for 30 min in 0.5% glutaraldehyde made up in 0.1 M phosphate buffer (pH 7.3)-2 mM MgCl2-5 mM EGTA. Embryos were then washed twice at room temperature in wash buffer (0.1 M phosphate buffer, 2 mM MgCl2, 1% deoxycholate, 2% Nonidet-P40). The embryos were then incubated with an X-Gal staining solution overnight at 37°C. The X-Gal staining solution was prepared by mixing 1 ml of a stock solution of X-Gal (25 mg/ml in dimethyl sulfoxide) with 48 ml of wash buffer containing 5 mM potassium ferrocyanide and 5 mM potassium ferricyanide made up in wash buffer. After staining, the embryos were rinsed with wash buffer and examined on a Leica M2FL III stereo microscope, and images were recorded with a Photometrics CoolSnap color camera.
Histology.
Freshly isolated tissues from 6-month-old heterozygous mutant animals and wild-type littermates were fixed overnight in 10% neutral buffered formalin at 4°C and dehydrated through increasing concentrations of ethanol. Paraffin-embedded sections were stained with hematoxylin and eosin.
Establishment of MEF lines.
At 12.5 dpc, embryos were dissected from heterozygous intercross matings, and spleens were removed and disaggregated with a 20-gauge needle into Dulbecco's modified Eagle's medium containing nonessential amino acids, penicillin, streptomycin, 15% fetal calf serum, 1% β-mercaptoethanol, and 1% l-glutamate. Cultures were left in a 37°C incubator for 2 to 3 days, when fibroblasts could be seen growing out from the tissue mass. Cells were trypsinized at this point and left to settle in a 50-ml Falcon tube for 1 to 2 min. Lumps of tissue sedimented, and the cell suspension was replated and cultured for several passages.
Western blot analysis.
Protein extracts were prepared from wild-type and heterozygous mouse embryonic fibroblast cell lines (MEFs) and mouse NIH 3T3 cells. A 10-cm dish of confluent cells was washed twice with phosphate-buffered saline (PBS), and then cells were resuspended in 200 μl of protein loading dye and left on ice for 5 min. Extracts were boiled at 100°C for 8 min and sonicated before storage at −20°C. Proteins were resolved on sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to a Hybond-P membrane for Western analysis by semidry blotting. Membranes were challenged with 1:1,000 dilutions of either immune serum to fibrillarin (monoclonal antibody D77) (2) or to β-galactosidase (5′-3′ Inc). Bound antibody was visualized with Pierce SuperSignal chemiluminescent substrate by autoradiography. Total proteins were identified by Coomassie blue staining to control for loading differences between samples.
Immunochemical methods and antibodies.
The MEF cell lines were washed with PBS and incubated with 3% paraformaldehyde-0.3% Triton (in PBS) for 5 min at room temperature, followed by incubation with 3% paraformaldehyde for 30 min. The fixed cells were incubated for 1 h at room temperature with 1:1,000 antifibrillarin monoclonal antibody 38F3 (EnCor Biotechnology Inc.), washed three times with PBS, and incubated for 1 h at room temperature with 1:500 fluorescein-conjugated goat anti-mouse IgG (Cappel Laboratories). Samples were observed on a Zeiss Axioplan II microscope and images were acquired with a Princeton Instruments Pentamax charge-coupled device camera with IPLab software.
TUNEL staining.
Embryos derived from Fibrillarin+/− intercrosses were flushed at day 2.5dpc and cultured overnight. At day 3.5dpc, embryos were washed with PBS and fixed in 4% paraformaldehyde-PBS for 5 min at room temperature. They were subsequently washed three times with PBS for 5 min and permeabilized in 0.1% Triton X-100-0.1% sodium citrate for 2 min on ice. Following two rinses with PBS, embryos were incubated with terminal deoxynucleotidyl transferase and fluorescein-labeled dUTP for terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) (In Situ Cell Death Detection kit; Boehringer Mannheim) at 37°C for 1 h. Embryos were subsequently destained three times with PBS for 10 to 15 min before they were analyzed with a Zeiss Axiovert 100 inverted microscope and photographed with a Hamamatsu ORCA-ER charge-coupled device camera with IPLab software. After analysis, the embryos were individually transferred into PCR tubes and processed for genotyping. A multiplex PCR assay was applied with a common forward primer and two reverse primers specific for the wild-type and the mutated fibrillarin gene trap locus, as described for Fig. 2. PCR conditions were identical to those described for Fig. 2.
FIG. 2.
Fibrillarin knockout mice. Genotyping of heterozygous intercross progeny by PCR analysis. The wild-type allele was identified with primers corresponding to intron 4 and exon 5 of fibrillarin, resulting in an amplified product of 715 bp. These two primers are on either side of the gene trap integration site; thus, the mutant allele is interrupted by 3 kb of the lacZ cassette. This results in a lack of amplification and the absence of this PCR product in the targeted allele. The fibrillarin mutant allele was identified with primers corresponding to sequences in intron 4 and a reverse primer in the lacZ construct, giving rise to a 550-bp band, which is not produced in the wild-type allele.
RESULTS AND DISCUSSION
Generation of the mutation.
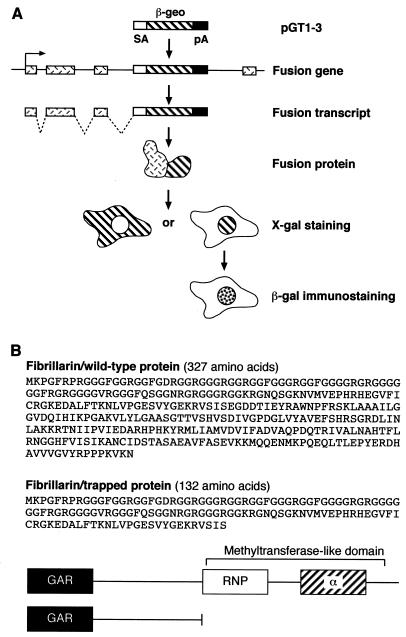
A gene trap screen in mouse embryonic stem cells identified an insertion mutation in the fibrillarin gene. In brief, a β-galactosidase-neomycin phosphotransferase (βgeo) reporter gene, lacking its own promoter and ATG, can be spliced in frame into the gene transcript when integrated into an intron of an expressed gene (Fig. 1A) (30). The sequence of the trapped gene is identified by 5′ RACE from the fusion transcript. This cell line, termed ES 403, was previously reported as part of a gene trap screen (29). The insertion event occurred in intron 4 of the fibrillarin gene and resulted in a fusion protein that contained the N-terminal 132 amino acids of fibrillarin fused to the βgeo reporter. As a result of this, only the N-terminal GAR domain and the spacer 1 region are present in the fusion protein, but the RNP domain and the α-helical domain are missing (Fig. 1B). More importantly, the missing region in the fusion protein comprises the methyltransferase-like domain (Fig. 1B). Whereas endogenous wild-type fibrillarin localized to the nucleoli and to Cajal bodies, the trapped fibrillarin protein in ES cells localized to the nucleoli and also showed a diffuse nuclear localization; however, no localization to the Cajal bodies was observed (data not shown). This is most likely due to the missing sequences in the βgeo-fibrillarin fusion protein. Interestingly, a fibrillarin-green fluorescent protein (GFP) fusion construct comprising the GAR domain and spacer 1 (which resembles exactly the fibrillarin domains present in the fusion protein) also localized to the nucleoplasm and to the nucleoli, but failed to localize to the Cajal bodies (see Fig. 2 in reference 27).
FIG. 1.
Disruption of the murine Fibrillarin gene. (A) Gene trap screening (29, 30). (B) Amino acid sequence of mouse wild-type and trapped fibrillarin proteins (top), and a cartoon depicting the domains present in both proteins (below).
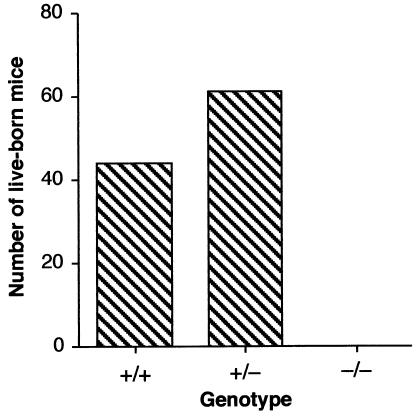
Gene trap events are mutagenic; hence, each ES cell line is heterozygous for what is most likely to be a loss-of-function mutation of the gene in question. The phenotype of heterozygous and homozygous mutations resulting from the fibrillarin gene trap event was investigated in the mouse through the generation of chimeras from a trapped ES cell line (ES 403) and then transmission through the germ line from these animals. Three independent lines of heterozygous mice were generated following blastocyst injection. Genotyping of heterozygous intercross progeny was done by PCR analysis of blastocyst stage embryos and of adult mice. The wild-type allele was amplified with primers corresponding to intron 4 and exon 5 of fibrillarin, resulting in an amplified product of 715 bp, which is not amplified in the targeted allele due to the insertion of approximately 3 kb of the lacZ cassette (Fig. 2). In contrast, a 550-bp band, amplified with the intron 4 forward primer and a reverse primer in the lacZ cassette, is diagnostic of the mutant fibrillarin allele (Fig. 2). Intercrosses of heterozygotes from each line failed to produce homozygous Fibrillarin−/− adult progeny, demonstrating that fibrillarin is an essential protein for development (Fig. 3). Analysis of embryos from heterozygous intercrosses also failed to show any Fibrillarin−/− progeny, even as early as 7.5 dpc (Table 1). The −/− sample shown in Fig. 2 corresponds to an early embryo (2.5 dpc) generated from a heterozygous intercross, since, as described above, no adult −/− progeny was ever detected (Fig. 3 and Table 1, and also see Fig. 7, below).
FIG. 3.
Fibrillarin heterozygous intercross progeny, showing the distribution of +/+, +/−, and −/− mice. Intercrosses of heterozygotes from each line failed to produce homozygous Fibrillarin−/− progeny, and in addition, a significant number of heterozygous mice were lost.
TABLE 1.
PCR analysis of Fibrillarin embryos from heterozygous intercrosses
| dpc | No. of progeny
|
No. of reabsorption sites | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| 7.5 | 4 | 11 | 0 | 0 |
| 8.5 | 6 | 12 | 0 | 0 |
| 9.5 | 6 | 6 | 0 | 13 |
| 10.5 | 5 | 5 | 0 | 15 |
| 11.5 | 4 | 10 | 0 | 5 |
| 12.5 | 5 | 12 | 0 | 16 |
FIG. 7.
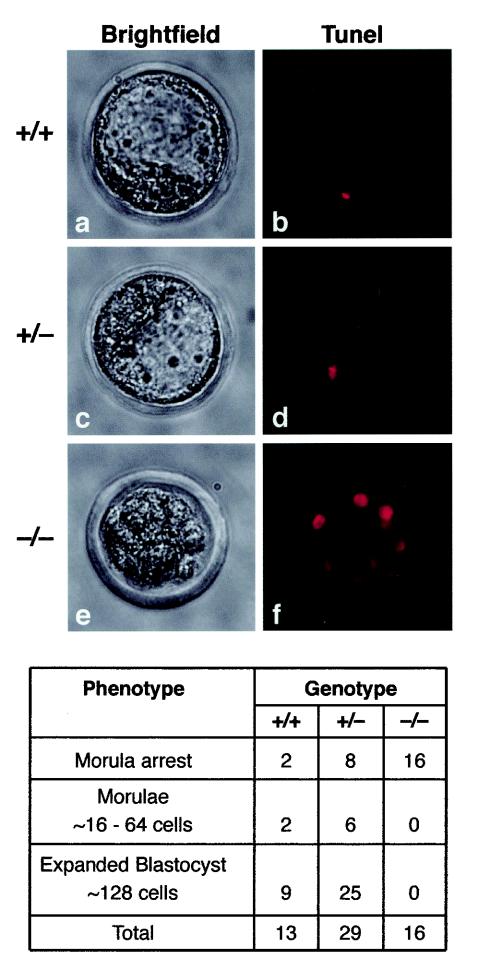
Homozygous embryos die at 2.5 to 3.5 dpc. Morphological alterations and TUNEL staining in control and Fibrillarin mutant embryos. A failure in transition to the blastocyst stage first became apparent at 60 h after mating, with subsequent formation of a disorganized, multicystic structure and finally extensive cellular degeneration at 84 h post coitum. TUNEL staining of Fibrillarin−/− embryos showed strong staining of most cells (f), suggesting apoptotic cell death. In contrast, wild-type and +/− blastocysts (b and d, respectively) showed very few labeled cells.
Fibrillarin heterozygous mutant mice.
We did not observe any discernible phenotype in Fibrillarin+/− live-born mice, despite the fact that a large decrease in the levels of fibrillarin protein was observed in cell lines derived from +/− mice (see below, Fig. 6). This would suggest that an approximate 50% decrease in fibrillarin protein levels is well tolerated.
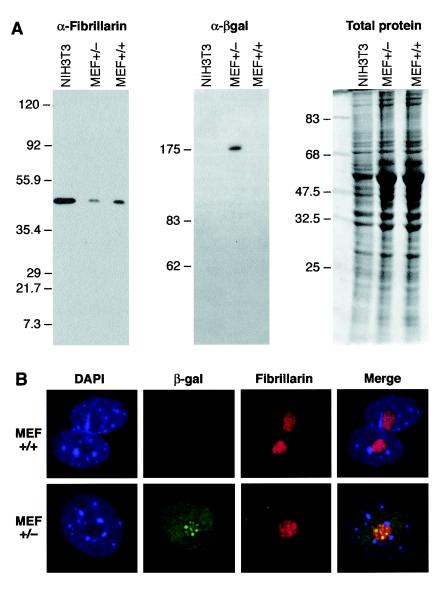
FIG. 6.
(A) Western blots showing reduced fibrillarin protein expression in mouse embryonic fibroblasts derived from Fibrillarin+/− mice. The left panel shows the endogenous fibrillarin protein detected with a specific antibody. The middle panel shows the fibrillarin-βgeo fusion protein detected with an anti-β-galactosidase antibody. The right panel shows total proteins detected by Coomassie blue staining. (B) Localization of fibrillarin-βgeo fusion protein and of endogenous fibrillarin in mouse embryonic fibroblasts derived from +/+ and +/− mice with antibodies specific to β-galactosidase and fibrillarin, respectively.
However, we found that Fibrillarin+/− and Fibrillarin+/+ mice did not segregate in the expected 2:1 ratio; thus, a significant proportion of heterozygous embryos must die during early development (Fig. 3). Thus, it is likely that the level of fibrillarin protein becomes limiting at an early stage of development. Interestingly, there was an indication of increased uterine reasorption starting at 9.5dpc, suggesting that Fibrillarin−/− and some Fibrillarin+/− embryos may implant but do not complete embryogenesis (Table 1).
It is notable that analysis of mice deficient for p80/coilin showed an underrepresentation of −/− mice relative to −/+ and +/+ animals (39), even though the surviving −/− mice showed apparently normal growth. Coilin is a component of the Cajal bodies, in which snoRNP maturation occurs (reviewed in references 7 and 16), and these observations may indicate a particularly sensitive requirement for snoRNP assembly at an early stage in development.
rRNA processing is not affected in Fibrillarin+/− cell lines and/or tissues.
RNA processing was analyzed in brain and liver tissue samples (data not shown), and in mouse embryonic fibroblasts (MEFs) derived from +/+ and +/− mice (Fig. 4A and B). In Saccharomyces cerevisiae, fibrillarin is required for several steps in pre-rRNA processing and subunit assembly (reviewed in reference 40), and the independently transcribed, fibrillarin-associated box C/D snoRNAs U3 (4), U8 (21), and U22 (20) are required for pre-rRNA processing in Xenopus oocytes. However, no clear differences were observed in the levels of any pre-rRNA species, and no alterations were detected in the accumulation of the mature rRNAs in +/− relative to +/+ mice in either the tissue samples or the MEFs. For further description of the pre-rRNA species and processing pathway, see Strezoska et al. (28).
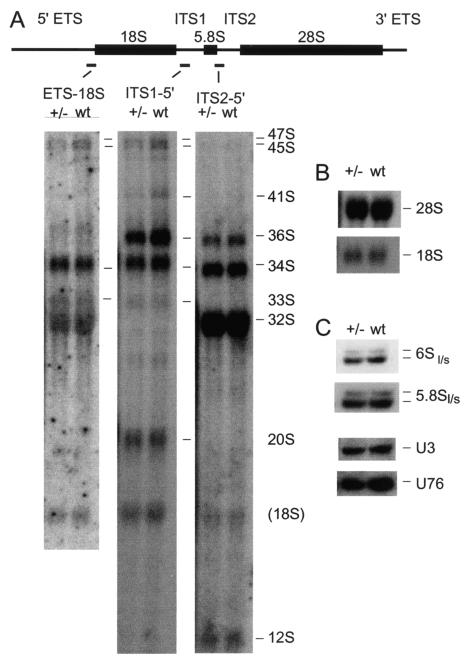
FIG. 4.
Northern analyses of rRNA and snoRNA processing in MEFs derived from Fibrillarin+/− mice. RNA was separated on a 1.2% agarose-/formaldehyde gel to analyze high-molecular-weight pre-rRNAs and rRNAs (panel A) or 8% polyacrylamide-urea gels to analyze low-molecular-weight pre-rRNAs and rRNA (panel B) and snoRNAs (panel C). RNA species identified are indicated on the right of each panel.
An intron-encoded snoRNA is codepleted with fibrillarin.
Yeast Nop1p is dispensable for accumulation of independently transcribed box C/D snoRNAs, including U3, but is required for the maturation and accumulation of intron-encoded box C/D snoRNAs (17, 37). In the case of U18 at least, snoRNA release from the host intron requires the interaction of Nop1p with the Rnt1p endonuclease (8). In mouse cells, fibrillarin binds to intronic pre-snoRNAs prior to processing (11), suggesting a general role for fibrillarin in snoRNA processing (reviewed in reference 36). The independently transcribed U3 and intron-encoded U76 box C/D snoRNAs were analyzed (shown in Fig. 4C; note that all panels in Fig. 4C show hybridization of the same filter, allowing their direct comparison). The level of the U3 snoRNA was not clearly altered in either the tissue sample or MEFs, whereas the level of U76 was markedly reduced. PhosphorImager quantitation standardized to the 5.8S rRNA showed that U76 was reduced 1.9-fold in +/− MEF cells relative to the +/+ control. Together, these results show that a decrease in fibrillarin protein levels results in a corresponding reduction in the level of an intron-encoded snoRNA but has little effect on the independently transcribed U3 snoRNA or rRNA accumulation.
Analysis of fibrillarin expression in mutant animals.
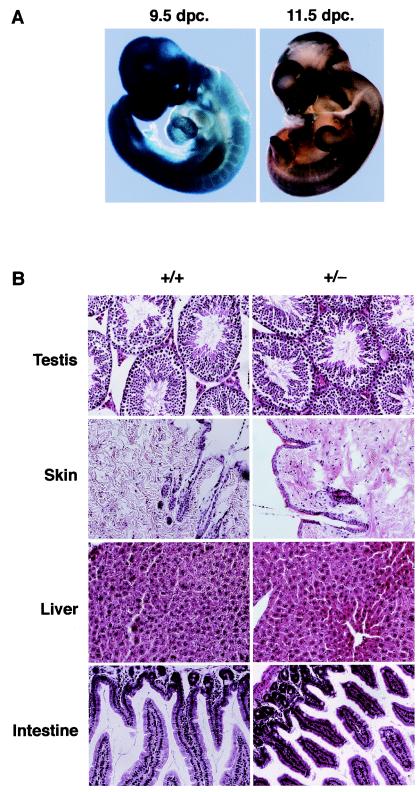
The pattern of fibrillarin gene expression during development was analyzed in whole-mount 9.5- and 11.5-dpc heterozygous embryos. Fibrillarin expression, as measured by β-galactosidase activity, was ubiquitous at 9.5 dpc, but the expression pattern became more restricted to highly proliferating tissues such as neural tube and limb buds at 11.5 dpc (Fig. 5A). The pattern of fibrillarin gene expression was also analyzed in tissue sections of +/− mutant mice. We observed that fibrillarin was highly expressed in the ovary of adult +/− mice (data not shown). This would indicate that the fibrillarin protein is made in oocytes of mothers with an intact gene copy and may be transferred to and used by early embryonic cells during the first rounds of division. The persistence of maternally produced protein in the embryo could explain the normal development of fibrillarin-depleted embryos to the early morula stage despite deletion of the Fibrillarin gene in the embryo (see Fig. 7). Fibrillarin expression was also high in testis and liver, which are both highly proliferating tissues, as expected from a protein involved in ribosome biogenesis (data not shown).
FIG. 5.
(A) Analysis of fibrillarin expression in Fibrillarin+/− embryos. Heterozygous males were mated to heterozygous females to determine fibrillarin gene expression during mouse development. Fibrillarin expression, as measured by β-galactosidase activity, is ubiquitous until 11.5 dpc, when the expression pattern became more restricted to highly proliferating tissues such as neural tube and limb buds. (B) Hematoxylin- and eosin-stained wax sections of testis, skin, liver, and intestine from wild-type (+/+) and heterozygous mutant (+/−) mice. Magnification, ×20.
Histological analysis was performed on tissues isolated from adult wild-type and heterozygous mutant mice, and no discernible differences were observed in highly proliferating tissues such as, testis, skin, liver, and intestine (Fig. 5B).
Fibrillarin expression in wild-type and heterozygote-derived MEFs.
Western blot analysis of embyronic protein with an antifibrillarin antibody confirmed that there was a large decrease in fibrillarin protein levels in mouse embryonic fibroblasts derived from 12.5-dpc +/− embryos compared to MEFs derived from wild-type embryos (Fig. 6A, left panel). As expected, the β-galactosidase antibody revealed the presence of a fusion protein only in MEFs derived from heterozygous embryos (Fig. 6A, middle panel).
We analyzed the subcellular distribution of endogenous fibrillarin and of the fibrillarin-βgeo fusion protein in mouse embryonic fibroblasts derived from fibrillarin +/+ and +/− mice with antibodies specific for fibrillarin and β-galactosidase, respectively. As observed in ES cells (data not shown), endogenous wild-type fibrillarin localizes primarily to nucleoli in MEF cell lines, whereas the trapped fibrillarin fusion protein localized to the nucleoli but also showed a diffuse nuclear localization (Fig. 6B). We also found that the subcellular localization of other nuclear proteins that localize to the Cajal bodies or to the nucleolus, such as SMN, UBF, nucleolin, and the spliceosomal Sm proteins, was not altered in heterozygous MEFs (data not shown).
Massive cell death in early Fibrillarin−/− embryos.
Progeny from heterozygous intercrosses were isolated as uncompacted five to seven-cell morulae 56 h post coitum. The embryos were maintained overnight in culture and subsequently genotyped by PCR, as described for Fig. 2. During this time period, normal morulae first compact (8- to 16-cell stage), then differentiate to cavitating blastocysts (64 cells), and finally expand to form an inner cell mass and the outer trophoblast cell layer (128 cells). Homozygous mutant embryos initially were indistinguishable from wild-type and heterozygous embryos. However, after overnight culture these mutant embryos showed signs of aberrant development; for instance, morulae failed to compact or form expanded blastocysts in all mutant embryos (morula arrest) (Fig. 7). At later time points the −/− embryos became progressively more disorganized and fragmented with extensive cellular degeneration.
TUNEL staining of such embryos revealed a high number of intensely labeled cells, which is diagnostic of apoptotic cell death (Fig. 7, upper panel [f]). In contrast, heterozygous and wild-type blastocysts revealed very few apoptotic cells (Fig. 7, upper panel [b and d]). Abnormal cleavage stage embryos with degenerating cells are commonly found in laboratory mice, particularly following superovulation. However, a significant difference was found in the number of abnormal cleavage stage embryos between homozygous mutant and heterozygous/wild-type embryos [X2 = 18.92; P < 0.00001]. This indicates that embryonic lethality at premorula stages is specifically associated with the loss of the fibrillarin gene. The association of the morula arrest phenotype with a homozygous mutant genotype was highly significant (P < 0.01; +/+: 1; +/−: 1; −/−: 0).
These experiments confirmed that homozygous disruption of fibrillarin in the mouse leads to developmental arrest and death prior to implantation. Thus, the death of mutant embryos may coincide with depletion of maternal fibrillarin in these embryos. In summary, we have shown that homozygous depletion of mouse fibrillarin leads to embryonic lethality, which occurs at an early stage of development, and that a significant proportion of +/− embryos die during early development. These results demonstrate that fibrillarin is essential for development.
Acknowledgments
We thank Michael Terns (University of Georgia, Athens) and Wendy Bickmore (MRC HGU) for comments and critical reading of the manuscript. We are grateful to Edouard Bertrand (Montpellier) for communicating unpublished results. We thank Ian Jackson (MRC HGU) for helpful discussions and David Brownstein (University of Edinburgh) for mouse pathology.
We acknowledge support from the Medical Research Council (K.N. and J.F.C.) and from the Wellcome Trust (E.P. and D.T.).
REFERENCES
- 1.Amiri, K. A. 1994. Fibrillarin-like proteins occur in the domain Archaea. J. Bacteriol. 176:2124-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aris, J. P., and G. Blobel. 1988. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 107:17-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aris, J. P., and G. Blobel. 1991. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc. Natl. Acad. Sci. 88:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borovjagin, A. V., and S. A. Gerbi. 1999. U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J. Mol. Biol. 286:1347-1363. [DOI] [PubMed] [Google Scholar]
- 5.Cahill, N. M., K. Friend, W. Speckmann, Z. H. Li, R. M. Terns, M. P. Terns, and J. A. Steitz. 2002. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 21:3816-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decatur, W. A., and M. J. Fournier. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278:695-698. [DOI] [PubMed] [Google Scholar]
- 7.Filipowicz, W., and V. Pogacic. 2002. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14:319-327. [DOI] [PubMed] [Google Scholar]
- 8.Giorgi, C., A. Fatica, R. Nagel, and I. Bozzoni. 2001. Release of U18 snoRNA from its host intron requires interaction of Nop1p with the Rnt1p endonuclease. EMBO J. 20:6856-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hebert, M. D., K. B. Shpargel, J. K. Ospina, K. E. Tucker, and A. G. Matera. 2002. Coilin methylation regulates nuclear body formation. Dev. Cell. 3:329-337. [DOI] [PubMed] [Google Scholar]
- 10.Henriquez, R., G. Blobel, and J. P. Aris. 1990. Isolation and sequencing of NOP1. A yeast gene encoding a nucleolar protein homologous to a human autoimmune antigen. J. Biol. Chem. 265:2209-2215. [PubMed] [Google Scholar]
- 11.Hirose, T., M. D. Shu, and J. A. Steitz. 2003. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol. Cell. 12:113-123. [DOI] [PubMed] [Google Scholar]
- 12.Hooper, M., K. Hardy, A. Handyside, S. Hunter, and M. Monk. 1987. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature 326:292-295. [DOI] [PubMed] [Google Scholar]
- 13.Jansen, R. P., E. C. Hurt, H. Kern, H. Lehtonen, M. Carmo-Fonseca, B. Lapeyre, and D. Tollervey. 1991. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J. Cell Biol. 113:715-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, K. W., K. Gorzynski, C. M. Hales, U. Fischer, F. Badbanchi, R. M. Terns, and M. P. Terns. 2001. Direct interaction of the spinal muscular atrophy disease protein smn with the small nucleolar rna-associated protein fibrillarin. J. Biol. Chem. 276:38645-38651. [DOI] [PubMed] [Google Scholar]
- 15.Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20:3617-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 109:145-148. [DOI] [PubMed] [Google Scholar]
- 17.Lafontaine, D. L., and D. Tollervey. 2000. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol. 20:2650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narayanan, A., W. Speckmann, R. Terns, and M. P. Terns. 1999. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell. 10:2131-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochs, R. L., M. A. Lischwe, W. H. Spohn, and H. Busch. 1985. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol. Cell. 54:123-133. [DOI] [PubMed] [Google Scholar]
- 20.Peculis, B. A. 2001. snoRNA nuclear import and potential for cotranscriptional function in pre-rRNA processing. RNA. 7:207-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peculis, B. A., and J. A. Steitz. 1993. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell 73:1233-1245. [DOI] [PubMed] [Google Scholar]
- 22.Pellizzoni, L., J. Baccon, B. Charroux, and G. Dreyfuss. 2001. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 11:1079-1088. [DOI] [PubMed] [Google Scholar]
- 23.Pollard, K. M., G. Reimer, and E. M. Tan. 1989. Autoantibodies in scleroderma. Clin. Exp. Rheumatol. 7(Suppl. 3):S57-S62. [PubMed] [Google Scholar]
- 24.Reimer, G., V. D. Steen, C. A. Penning, T. A. Medsger, Jr., and E. M. Tan. 1988. Correlates between autoantibodies to nucleolar antigens and clinical features in patients with systemic sclerosis (scleroderma). Arthritis Rheum. 31:525-532. [DOI] [PubMed] [Google Scholar]
- 25.Samarsky, D. A., M. J. Fournier, R. H. Singer, and E. Bertrand. 1998. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 17:3747-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schrank, B., R. Gotz, J. M. Gunnersen, J. M. Ure, K. V. Toyka, A. G. Smith, and M. Sendtner. 1997. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. 94:9920-9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snaar, S., K. Wiesmeijer, A. G. Jochemsen, H. J. Tanke, and R. W. Dirks. 2000. Mutational analysis of fibrillarin and its mobility in living human cells. J. Cell Biol. 151:653-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strezoska, Z., D. G. Pestov, and L. F. Lau. 2000. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S RRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 20:5516-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutherland, H. G., G. K. Mumford, K. Newton, L. V. Ford, R. Farrall, G. Dellaire, J. F. Caceres, and W. A. Bickmore. 2001. Large-scale identification of mammalian proteins localized to nuclear sub-compartments. Hum. Mol. Genet. 10:1995-2011. [DOI] [PubMed] [Google Scholar]
- 30.Tate, P., M. Lee, S. Tweedie, W. C. Skarnes, and W. A. Bickmore. 1998. Capturing novel mouse genes encoding chromosomal and other nuclear proteins. J. Cell Sci. 111:2575-2585. [DOI] [PubMed] [Google Scholar]
- 31.Terns, M. P., and J. E. Dahlberg. 1994. Retention and 5′ cap trimethylation of U3 snRNA in the nucleus. Science 264:959-961. [DOI] [PubMed] [Google Scholar]
- 32.Terns, M. P., C. Grimm, E. Lund, and J. E. Dahlberg. 1995. A common maturation pathway for small nucleolar RNAs. EMBO J. 14:4860-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terns, M. P., and R. M. Terns. 2001. Macromolecular complexes: SMN-the master assembler. Curr. Biol. 11:R862-R864. [DOI] [PubMed] [Google Scholar]
- 34.Terns, M. P., and R. M. Terns. 2002. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 10:17-39. [PMC free article] [PubMed] [Google Scholar]
- 35.Tollervey, D. 1987. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 6:4169-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tollervey, D., and T. Kiss. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9:337-342. [DOI] [PubMed] [Google Scholar]
- 37.Tollervey, D., H. Lehtonen, M. Carmo-Fonseca, and E. C. Hurt. 1991. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre- rRNA processing in yeast. EMBO J. 10:573-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tollervey, D., H. Lehtonen, R. Jansen, H. Kern, and E. C. Hurt. 1993. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72:443-457. [DOI] [PubMed] [Google Scholar]
- 39.Tucker, K. E., M. T. Berciano, E. Y. Jacobs, D. F. LePage, K. B. Shpargel, J. J. Rossire, E. K. Chan, M. Lafarga, R. A. Conlon, and A. G. Matera. 2001. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol. 154:293-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 41.Verheggen, C., J. Mouaikel, M. Thiry, J. M. Blanchard, D. Tollervey, R. Bordonne, D. L. Lafontaine, and E. Bertrand. 2001. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 20:5480-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, H., D. Boisvert, K. K. Kim, R. Kim, and S. H. Kim. 2000. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution. EMBO J. 19:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watkins, N. J., A. Dickmanns, and R. Luhrmann. 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 22:8342-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinstein, L. B., and J. A. Steitz. 1999. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 11:378-384. [DOI] [PubMed] [Google Scholar]