Abstract
The RNA silencing pathway is an intracellular innate response to virus infections and retro-transposons. Many plant viruses counter this host restriction by RNA silencing suppressor (RSS) activity of a double-stranded RNA-binding protein, e.g., tomato bushy stunt virus P19. Here, we demonstrate P19 and HIV-1 Tat function across the plant and animal kingdoms and suppress a common step in RNA silencing that is downstream of small RNA maturation. Our experiments reveal that RNA silencing in HIV-1 infected human cells severely attenuates the translational output of the unspliced HIV-1 gag mRNA, and possibly all HIV-1 transcripts. The attenuation in gag mRNA translation is exacerbated by K51A substitution in the Tat double-stranded RNA-binding domain. Tat, plant virus RSS, or Dicer downregulation rescues robust gag translation and bolsters HIV-1 virion production. The reversal of HIV-1 translation repression by plant RSS supports the recent finding in Arabidopsis that plant miRNAs operate by translational inhibition. Our results identify common features between RNA silencing suppression of plant and animal viruses. We suggest that RNA silencing-mediated translation repression plays a strategic role in determining the viral set-point in a newly HIV-1-infected patient.
Keywords: retrovirus gag RNA, restriction of HIV-1 replication
RNA silencing is a eukaryotic posttranscriptional gene regulation mechanism and innate defense to quell virus infections and genetic damage by retro-transposons. Most plant viruses encode an RNA silencing suppressor (RSS) that counteracts this restriction and drives pathogenesis (reviewed in refs. 1, 2). The importance of RNA silencing suppression in animal retrovirus infection remains controversial (reviewed in refs. 3, 4). Antiviral RNA silencing is initiated when virus-specific double-stranded RNA appears in the cytoplasm and is processed by Dicer endonuclease into 21–25 nucleotide miRNA/miRNA* (guide/passenger) duplexes (5–8). The guide strand and complementary target mRNA is incorporated into RNA-induced silencing complexes (RISC) (9, 10), which coalesce as processing bodies that are sites of target mRNA degradation or translation repression (11, 12).
Physiological expression of HIV-1-encoded miRNA remains controversial (13–16). Evidence that RNA silencing is important for HIV-1 includes the observation that cell-encoded miRNAs dampen virus replication in activated T lymphocytes (17) and contribute to viral latency in resting T lymphocytes (18). HIV-1 restriction of RNA silencing has been attributed to the viral Tat transcriptional transactivator (16). Benasser, et al. (16) determined that Tat RSS activity was genetically separable from Tat transcriptional activity but segregated with the arginine-rich double-stranded RNA-binding domain (16). A similar RNA binding domain is conserved in plant virus RSS and confers interaction with miRNA duplexes in a sequence nonspecific manner that blocks programming of RISC for RNA silencing (19–21). For Tat, tomato bushy stunt virus (TBSV) P19 and other Tombusvirus RSS, mutation of this domain eliminates RNA silencing suppression (16, 22), which posits a common mechanism of activity. The outcome of P19 mutation is reduced TBSV RNA, which culminates in impaired virus propagation and attenuation of disease pathogenesis (19–21). To investigate whether HIV-1 Tat RSS protects against viral RNA degradation, we compared the activity of Tat and P19 in plant protoplasts and animal cells.
Results demonstrate that HIV-1 Tat and TBSV P19 function equivalently in plant protoplasts and animal cells to suppress RNA silencing at a step downstream of the double-stranded RNA (dsRNA) processing, most likely by sequestering mature si/miRNAs. We present evidence that RNA silencing does not affect HIV-1 RNA degradation and instead restricts HIV-1 mRNA translation. HIV-1 Tat and TBSV P19 function equivalently to protect against RNA silencing-mediated suppression of HIV-1 translation.
Results
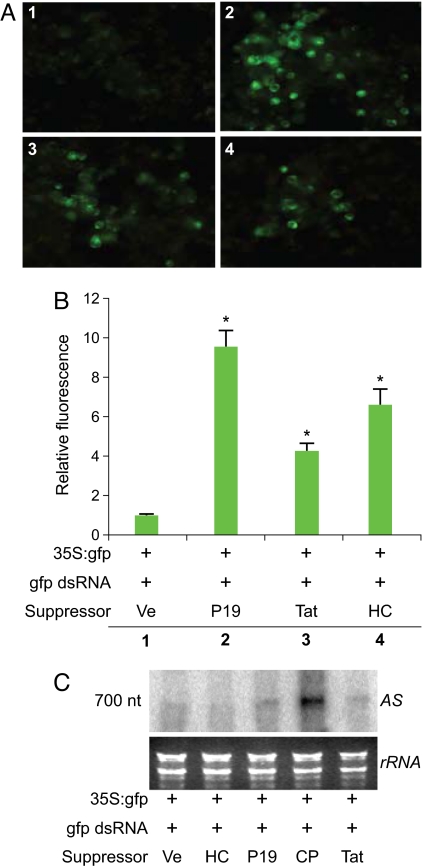
HIV-1 Tat Suppresses RNA Silencing in Plant Cells Downstream of the Maturation Step of dsRNA Duplexes.
We used Nicotiana benthamiana protoplasts to investigate whether or not Tat RSS activity is maintained in the plant kingdom similar to the activity of influenza A virus NS1 RSS (23). Tat was expressed in plant cells downstream of the strong and constitutive 35S promoter derived from cauliflower mosaic virus. The RSS activity of Tat was compared to that of plant viral RSSs by coelectroporation with GFP reporter plasmid and 700 nt GFP-specific dsRNAs that downregulate GFP expression (24). Representative images from 5 independent triplicate transfection assays (Fig. 1A) demonstrated that HIV-1 Tat (Tat), TBSV P19 (P19) and tobacco etch virus helper component-protease (HC-Pro) restored GFP fluorescence compared to the empty vector control. Quantification of GFP fluorescence in the bulk cultures revealed that the RSS activity was statistically significant (p value < 0.0001) (Fig. 1B). Northern blot analysis using a sense strand GFP-specific probe revealed low but detectable levels of the 700 nt GFP effector RNA in the cells electroporated with the empty vector (Ve), HC-Pro, P19, or Tat. By comparison, Turnip crinkle virus coat protein (CP) caused the accumulation of effector dsRNA, consistent with its role in preventing processing of dsRNAs (24, 25). These results confirm the published observation that HC-Pro and P19 do not affect the processing of long dsRNAs into mature siRNAs (19, 21, 22, 26, 27). The results indicated that Tat RSS activity is conserved across kingdoms and functions downstream of dsRNA duplexes.
Fig. 1.
Tat exhibits RSS activity in plant cells that is not attributable to inhibited processing of long dsRNA. (A) N. benthamiana culture cells protoplasts were transfected with GFP reporter plasmid, long GFP-specific dsRNA and the empty vector (Ve) (1) or indicated RSS expression plasmid (2, 3, 4) and monitored for GFP activity 3 days post electroporation. P19, Tat, and HC-Pro restored GFP expression. (B) Quantitative analysis of bulk cultures is summarized from 5 replicate experiments. Expression of indicated RSS increased the GFP fluorescence compared to empty vector control. (C) Northern blot analysis with probe complementary to the antisense strand of gfp effector RNA. HC, P19, and Tat do not induce accumulation of effector RNA. CP induced accumulation of effector RNA. Lower panel is the gel stained with ethidium bromide.
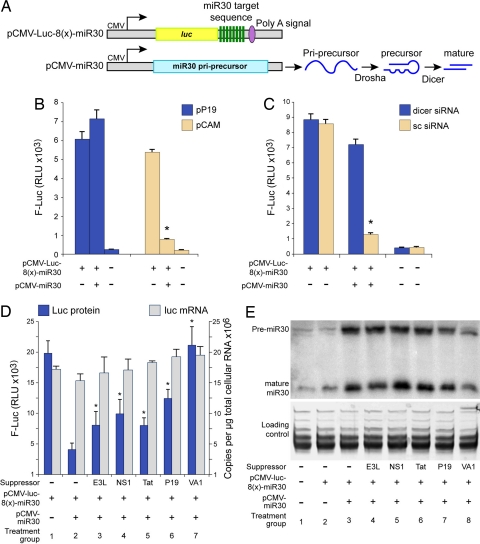
TBSV P19 Suppresses RNA Silencing in Animal Cells Downstream of miRNA Maturation.
We next compared the RSS activity of Tat and plant virus RSS in animal cells. The miR30-based luciferase reporter system (28) contains 8 copies of the mir30 target sequence (pCMV-Luc-8(x)-miR30) (Fig. 2A). Co-transfection of HeLaT4 cells with the primary precursor miR30 (pri-premiR30) expression plasmid pCMV-miR30 and the pCMV-Luc-8(x)-miR30 reporter robustly silenced Luc activity (compare tan bars, reduction by factor of 5, in Fig. 2B). Cotransfection with P19 suppressed miR30 activity by a factor of 7 (compare blue bar in Fig. 2B). By comparison, downregulation of Dicer by Dicer-specific siRNAs likewise eliminated miR30 activity (dicer siRNA, blue bars, Fig. 2C) as compared to treatment with scrambled siRNAs (sc, tan bars, Fig. 2C). Real-time RT PCR to quantify steady state mRNA levels showed that the copy number of luc mRNA did not change in response to P19 expression (data not shown).
Fig. 2.
Tat suppresses miR30 function but does not block miR30 processing in human cells. (A) The luc RNA expressed from reporter plasmid pCMV-Luc-8(x)-miR30 contains 8 miR30 target sites. Co-transfection of pCMV-miR30 is used to over-express miR30 pri-precursor that is processed by Drosha and Dicer to produce precursor miR30 and mature miR30, respectively. (B) Luciferase activity from HelaT4 cells at 48 h posttransfection with indicated plasmids determined that miR30 activity is reduced by P19. (C) Luciferase activity from HelaT4 cells at 48 h posttransfection with indicated siRNA and plasmids determined that miR30 activity is reduced by Dicer downregulation. Firefly Luc (F-Luc) activity from equivalent protein preparations. (D) Luciferase activity and luciferase RNA levels in 293 cells transfected with indicated RSS and miR30 reporter plasmid and miR30 expression plasmid. Equivalent protein preparations were assays for Firefly Luc (F-Luc) activity. (E) Northern blot analysis with antisense miR30-specific RNA probe that detected precursor miR30 (premiR30) and mature miR30. Small RNAs were enriched and separated by 15% urea-PAGE. Lower is gel stained with ethidium bromide.
We used the mirR30 assay to compare suppressor activity of P19 with HIV-1 Tat, vaccinia virus E3L, influenza A virus NS1, and adenovirus VA1. miR30 reduced Luc activity from pCMV-luc8(x)-miR30 by a factor of 5 (blue bars, treatment group 1 and 2, Fig. 2D). Statistically significant partial restoration of Luc activity was observed for E3L, NS1, P19, and Tat and VA1 completely restored Luc activity (blue bars, Fig. 2D, P < 0.05). Real-time RT PCR showed that the steady state level of luc mRNA was similar among all samples (gray bars, Fig. 2D) and indicated that the increased Luc activity was attributable to reversal of translation repression.
Northern blotting with the antisense miR30 RNA probe on size fractionated RNA was used to investigate the amounts of the 71 nt premiR30 and 22 nt mature miR30 (Fig. 2E). Low but detectable endogenous miR30 was detectable (treatment group 1 and 2) and abundant premiR30 and mature miR30 was observed upon transfection of pCMV-miR30 (treatment group 3). Similar levels of premiR30 were detected in E3L, NS1, Tat, and P19 (treatment groups 4–7) and a reduction was observed in response to VA1 (treatment group 8). The ratio of premiR30 to mature miR30 was similar between empty vector control (-suppressor) and E3L (1.2 and 1.4, respectively). The ratio was reduced in response to NS1, Tat, and P19 (range 0.75 to 0.9), indicating no significant block in the processing of premiR30 to mature miR30. VA1 treatment resulted in baseline levels of premiR30 and mature miR30 and the appearance of a smaller species of premiR30 (treatment group 8) and supports the observation that VA1 serves as a decoy substrate for exportin 5, Dicer, and RISC (28, 29). The accumulation of premiR30 and miR30 in the Tat treatment group indicated that, similar to the plant viral RSS P19, Tat does not disrupt the maturation of dsRNA, but reduces the efficiency of a downstream step in the RNA silencing pathway.
RNA Silencing Restricts HIV-1 Gag Protein Synthesis in Human Cells.
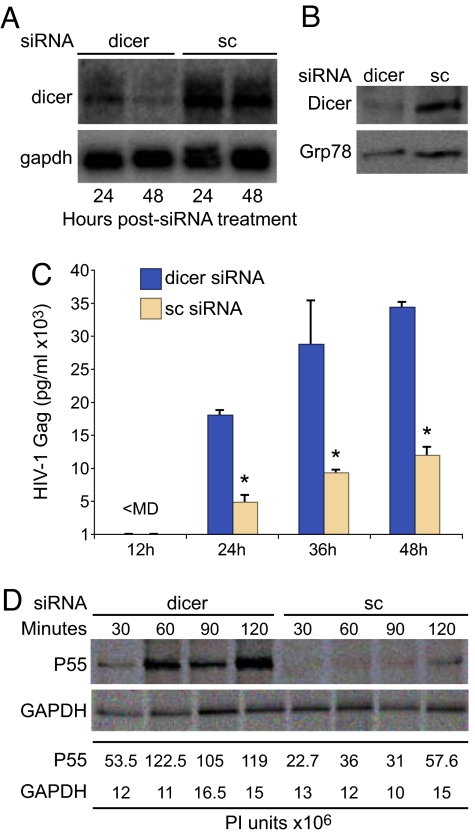
To determine whether the RNA silencing pathway inhibits gag mRNA translation, Dicer was downregulated in HeLa T4 cells by transfection with Dicer-specific siRNAs (30). Northern and Western blotting detected significant Dicer mRNA downregulation by 24 h and 48 h posttransfection (Fig. 3A), and Dicer protein was downregulated in comparison to treatment with non-silencing scrambled control (sc) (Fig. 3B, 48 h sample). These cells were transfected with HIV-1NL4–3 and virion production was measured at 24 h, 36 h, and 48 h post transfection. Dicer downregulation increased virion production 3- to 4-fold (Fig. 3C). Pulse labeling and Gag IP showed that the rate of Gag protein synthesis was increased significantly by Dicer downregulation (Fig. 3D, P < 0.05). By comparison, gapdh translation was not affected, as determined by GAPDH immunoprecipitation (IP). Analysis of CEMx174 lymphocytes infected with HIV-1NL4–3 yielded a similar increase in Gag protein synthesis upon Dicer knockdown (data not shown). Meanwhile, TCA-precipitable counts measuring [35S]cysteine/methionine incorporation were similar between the samples treated with the Dicer-specific siRNAs (3.3 × 106 ± 8 × 105 cpm) and the sc siRNAs (3 × 106 ± 6 × 105 cpm). The results demonstrate that HIV-1 replication in human cells is attenuated by RNA silencing and that downregulation of the RNA silencing pathway significantly increases de novo Gag protein synthesis independently of general effects on cellular protein synthesis.
Fig. 3.
Down-regulation of Dicer enhances the production of the HIV-1 structural protein in human cells. (A) Northern blot of total cellular RNA from HelaT4 cells transfected with dicer siRNA (dicer) or scrambled siRNA (sc) determined downregulation of dicer mRNA at both 24 and 48 h posttreatment. (B) Immunoblot with Dicer and Grp78 antiserum determined downregulation of Dicer protein at 48 h. (C) Down-regulation of Dicer enhanced Gag production in HeLaT4 cells transfected with HIV-1NL4–3. Gag ELISA was performed on cell-free medium from 3 independent transfections. Gag levels were normalized to cotransfected Luciferase. (D) IP assay determined that Dicer downregulation increases rate of synthesis of Gag P55 but not GAPDH.
RNA Silencing-Mediated Restriction of the HIV-1 Gag Protein Synthesis Is Suppressed Equivalently by Tat and P19.
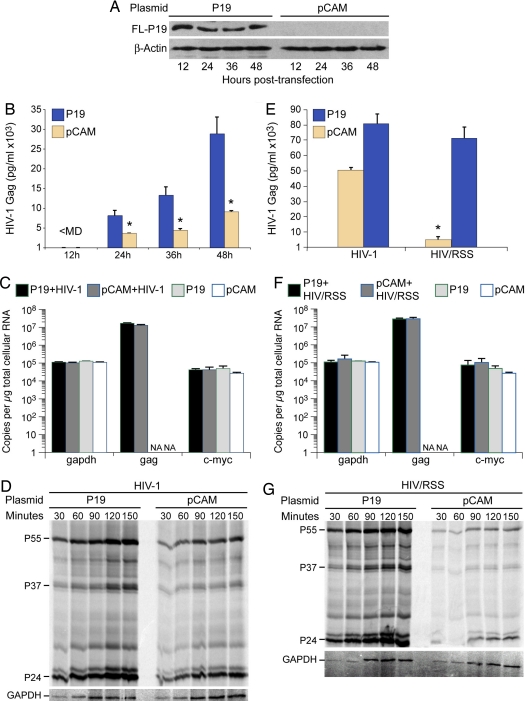
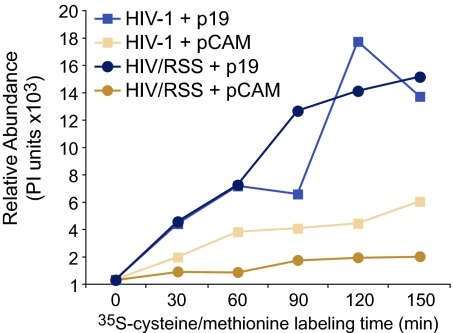
We next evaluated whether Tat and P19 RSS rescue gag mRNA translation. Flag-tagged P19 was expressed in HelaT4 cells and P19 was consecutively verified by Western blotting at 12, 24, 36, and 48 h (Fig. 4A). At the 24 h time point, the cells were transfected with HIV-1NL4–3and virion levels were measured by Gag p24 ELISA on supernatant medium. P19 produced a significant increase in virion level at each time point (Fig. 4B) but did not alter the copy number of gag mRNA (Fig. 4C). Real-time RT PCR results revealed no change in gapdh or c-myc mRNA in response to P19 or HIV-1 expression (Fig. 4C). At the 48 h time point, the rate of Gag protein synthesis was assessed by pulse labeling and Gag IP experiments. The increase in virion level by P19 expression was attributable to increased Gag protein synthesis (Fig. 4D). We next compared virion production between HIV-1NL4–3 and the derivative provirus HIV-1/RSS that contains the K51A mutation in the tat gene, which eliminates RSS activity but retains transcriptional transactivation activity (16). Gag ELISA results showed a significant reduction in virion production by a factor of ten in response to K51A (tan bars, Fig. 4E). Expression of P19 was sufficient to complement the defect in virion production from HIV-1/RSS and bolstered virion production from HIV-1 (blue bars, Fig. 4E). Real-time RT PCR data showed no difference in the copy number of HIV-1/RSS gag mRNA nor gapdh and c-myc RNA loading controls in response to P19 (Fig. 4F). These levels were also were unchanged in relation to HIV-1 (Fig. 4C). Pulse labeling and Gag IP demonstrated that HIV-1/RSS leads to a significant reduction in the rate of Gag protein synthesis (Fig. 4G Right) and that P19 expression significantly increased the rate of Gag protein synthesis (Fig. 4G Left). As summarized in Fig. 5, these IP data indicated that the K51A mutation reduced Gag protein synthesis by a factor of 3 (tan squares, HIV-1 + pCAM; tan circles, HIV-1/RSS + pCAM). Similar to the Gag ELISA results, P19 expression bolstered Gag protein synthesis to above the level of HIV-1 + pCAM (compare to blue squares and circles). Measurement of TCA-precipitable counts revealed similar levels of [35S]cysteine/methionine incorporation in the P19 treated cells (4.2 × 106 ± 1 × 106) and the pCAM control (3.2 × 106 ± 3.8 × 105 cpm). Statistical analysis by Dunnett's method determined no significant difference in de novo cellular protein production (P = 0.2273). Likewise, metabolic labeling with 3H–uridine demonstrated no significant difference in total cellular RNA synthesis during the 1 h labeling period (P19 treatment: 5.2 × 105 ± 1.8 × 104 and the pCAM control: 6 × 105 ± 1.3 × 104 cpm). The results demonstrated that Tat and P19 increase the translatability of HIV-1 gag mRNA.
Fig. 4.
Expression of P19 enhanced production from HIV-1 and HIV/RSS. (A) Immunoblot of HelaT4 cells transfected with indicated plasmids with Flag or beta-actin antiserum determined expression of P19-Flag fusion protein at indicated times posttransfection. (B) Gag production from HeLaT4 cells transfected with HIV-1NL4–3 was reduced by P19 RSS. Gag ELISA was performed on cell-free medium from 3 independent cotransfections of HIV-1NL4–3 and pCMV-p19FL (P19) or empty vector (pCAM). Gag levels were normalized to cotransfected Luciferase. (C) P19 expression did not change steady state levels of HIV-1 gag, c-myc, or gapdh transcripts. Evaluation of total cellular RNA preparations from 3 replicate transfections by reverse transcription and real-time PCR with HIV-1 gag, c-myc, and gapdh specific primers. (D) IP assay demonstrated that P19 expression increases rate of synthesis of HIV-1 Gag P55 but not GAPDH. (E) Gag production from HeLaT4 cells transfected with HIV-1NL4–3 or HIV/RSS was increased by coexpression of P19 RSS. Gag ELISA was performed on cell-free medium from 3 independent cotransfections of indicated provirus and pCMV-p19FL (P19) or empty vector (pCAM). Gag levels were normalized to cotransfected Luciferase. (F) P19 expression did not change steady state levels of HIV/RSS gag, c-myc, or gapdh transcripts. Evaluation of total cellular RNA preparations from 3 replicate transfections by reverse transcription and real-time PCR with HIV-1 gag, c-myc, and gapdh specific primers. (G) IP assay determined that P19 expression increases rate of synthesis of HIV/RSS Gag P55 but not GAPDH.
Fig. 5.
Tat is the viral RSS and the plant viral RSS P19 can replace Tat RSS activity. Quantification of Gag IP assay results of Fig. 4 C and F. Introduction of K51A mutation in HIV/RSS reduces Gag production and P19 expression increases rate of HIV-1 and HIV/RSS Gag protein synthesis to similar levels.
Conclusion
Our data indicate that the production of HIV-1 Gag protein, and thereby production of virus particles, is restricted by RNA silencing, which confirms the results of Triboulet, et al. (17) and de Vries, et al.(31). Viral strategies to counter RNA silencing include RNA protection, silencing suppression, evasion, modulation, and adaptation (4). Our results indicate that HIV-1 Tat confers RNA silencing suppression, which counters host-mediated inhibition of HIV-1 translation that is attributable to cell-encoded miRNAs (17, 18). The observation that virion production from an HIV-1 strain lacking RSS activity is severely attenuated indicates that Tat RSS promotes acute HIV-1 infection. Heterologous plant virus P19 RSS is sufficient to overcome translational RNA silencing and provides a mechanistic explanation for the observation that Ebola V35 can replace Tat RSS activity (32). HIV-1 Tat and TBSV P19 also cosegregate in their loss of activity by point mutation of the dsRNA-binding domain.
Our results demonstrate that the RSS activity of Tat and P19 is modest in comparison to adenovirus VA1 RNA. While modest in our system, P19 RSS activity is paramount to prevent host attenuation of TBSV infection and pathogenesis (reviewed in refs. 1, 2). Given our observation that P19 expression does not reduce global cellular translation and that Tat RSS activity was not recapitulated in a shRNA reporter assay (13), viral RSS activity is not a global phenomenon and is targeted to select small RNAs.
The evidence demonstrating reversal of HIV-1 translation repression by a plant RSS support the recent finding in Arabidopsis that plant miRNAs operate by translational inhibition (33). Our IP results, quantitative RNA analyses in human cells, and functional analysis in N. benthamiana cells indicate that the underlying mechanism of RNA silencing suppression by human virus Tat and plant virus P19 are related. The demonstration that Tat and P19 do not reduce maturation of dsRNA duplexes indicates a common mechanism to prevent guide strand programming of RISC, and agrees with research by Lin and Cullen (13) that Tat does not block premiRNA processing by Dicer. The results indicate that influenza A virus NS1 RSS also relies on si/miRNA-binding, whereas vaccinia virus E3L functions upstream and relies on binding to long dsRNAs, thereby preventing their processing into mature si/miRNAs.
Materials and Methods
Plasmids.
HIV/RSS was constructed by PCR-based site-directed mutagenesis of pNL4–3 to introduce K51A (16). P19 eukaryotic expression plasmid pCMVp19FL 9 was constructed by PCR of the P19 ORF from template pRTL2p19 (34) with primers containing terminal ClaI and BamHI restriction sites [supporting information (SI) Table S1]. Plant expression plasmid pRTL2Tat was constructed by SacI and BamHI restriction of pRTL2 (35) and pCMV-Tat-1 (36) (a gift of Andrew Rice) isolation from agarose and ligation. All constructions were verified by sequencing. Previously described plasmids are pRTL2:smGFP, HC-Pro, and CP (24), VA1 (28); NS1 and E3L (23, 32). The pCMV-Luc-8(x)-miR30(p) and pCMV-miR30 was a gift of Bryan Cullen (28).
Cells Culture, Transfection and Infection.
Monolayer human HEK293 embryonic kidney cells and HeLaT4 (CD4+ HeLa) cells were cultured in DMEM with 10% FBS at 37 °C and 5% CO2. Human CEMx174 lymphocytes were cultured in RPMI medium 1640/10% FBS. Transient transfections of 1 × 105 HelaT4 cells in triplicate in 6-well plates featured 1 μg of p19FL or empty pCAM vector and 0.2 μg of pGL3 firefly luciferase transfection control in Fugene6. After 2 days, transfectants were subcultured and transfected with 1 μg of HIV-1NL4–3 or HIV/RSS proviral plasmid. Culture media were harvested for HIV-1 Gag P24 ELISA (Zeptrometrix). Cells lysates were harvested in parallel in 100 μl Nonidet P-40 lysis buffer (20 mM Tris·HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, and 1% Nonidet P-40). Ten μl of lysate was assayed in Luciferase reagent (Promega) and relative light units were used to standardize minor differences in transfection efficiency.
Virus stocks for infections were generated by transfection of 1 × 106 HEK293 cells with HIV-1NL4–3 or HIV-1/RSS and infection of stock culture of CEMx174 T cells. After 48 h, cells were harvested on Ficoll Hypaque and cultured with naïve CEMx174 cells at a 1:10 ratio. Fluorescence-activated cell sorting (FACS) analysis of intra-cellular Gag expression was performed with anti-p24 KC57-FITC antibody (Beckman-Coulter) and Fix and Perm (CALTAG).
Protein Analysis.
Cells were lysed in RIPA buffer (50 mM Tris pH 8.0, 0.1% SDS, 1% Triton-X, 150 mM NaCl, 1% deoxycholic acid, 2 mM PMSF) and 50 μg protein was subjected to SDS/PAGE and transferred to nitrocellulose membrane. Immunoblotting antibodies detected Flag and β-actin (Abcam). Visualization was performed with Luminol reagent (Santa Cruz Biotechnology). The IP and TCA precipitation protocols are described previously (37).
Statistical Analysis.
One-way analysis of variance model was applied to log base 2-transformed data. Dunnett's method analyzed the mean difference among multiple groups.
N. benthamiana Protoplast Assays.
Isolation of N. benthamiana cultured cell protoplasts and electroporation are described in detail by Qi, et al. (24). One-million protoplasts were electroporated with 5 μg of pRTL2:smGFP, in the presence or absence of 5 μg of double stranded GFP effector RNA by electroporation. The assays were performed in triplicate wells of 6-well plates with 5 μg of suppressor plasmid. At 3 days postelectroporation, GFP fluorescence intensity was measured by CytofluorTM 2350 Fluorescence Measurement System with the plate reader software (Millipore).
Northern Blot Analyses.
For detection of plant GFP effector RNA, 5 μg of total RNA was separated on 5% PAGE with 8M urea and 0.5X TBE. For detection of miR30 precursor miRNA and mature miRNA, 10 μg of enriched small RNA were separated by 15% PAGE/8M urea/0.5X TBE. The small RNA preparations were isolated from total cellular RNA with differential ethanol precipitation using miRvana protocol (Ambion). The RNAs were transferred to Hybond-XL nylon membrane (Amersham Biosciences) and subjected to UV crosslinking. The membranes were hybridized in ULTRAhyb Ultrasensitive Hybridization Buffer (Ambion) overnight, washed twice in 2× SSC/0.1% SDS for 15 min and twice in 0.2× SSC/0.1% SDS for 15 min. Hybridization and washing were performed at 65 °C and at 37 °C for detection of large RNA species and small RNA species, respectively, and visualized by PhosphorImaging. To generate 32P-UTP-labeled antisense miR30 probe, 1 μg of PCR product containing T7 promoter was in vitro transcribed by T7 RNA polymerase.
Real-Time RT PCR.
The RT-PCR protocol (37) used random hexamers and Sensiscript reverse transcriptase (Qiagen) and 100 ng RNA. Ten percent of the cDNA was used for real-time PCR with primers complementary to luc mRNA or actin (Table S1) in Lightcycler (Roche).
Acknowledgments.
We thank members of the K.B.-L. laboratory for critical comments on the manuscript and Tim Vojt for assistance with illustrations. This work was supported by National Institutes of Health National Cancer Institute R01CA108882; P01CA16058; P30CA100730.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806822106/DCSupplemental.
References
- 1.Li WX, Ding SW. Viral suppressors of RNA silencing. Curr Opin Biotechnol. 2001;12:150–154. doi: 10.1016/s0958-1669(00)00190-7. [DOI] [PubMed] [Google Scholar]
- 2.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cullen BR. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat Immunol. 2006;7:563–567. doi: 10.1038/ni1352. [DOI] [PubMed] [Google Scholar]
- 4.Yeung ML, Benkirane M, Jeang KT. Small non-coding RNAs, mammalian cells, and viruses: Regulatory interactions? Retrovirology. 2007;4:74. doi: 10.1186/1742-4690-4-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullivan CS, Ganem D. MicroRNAs and viral infection. Mol Cell. 2005;20:3–7. doi: 10.1016/j.molcel.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Voinnet O. Induction and suppression of RNA silencing: Insights from viral infections. Nat Rev Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- 7.Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38(Suppl):S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 8.Yeung ML, Bennasser Y, Jeang KT. miRNAs in the biology of cancers and viral infections. Curr Med Chem. 2007;14:191–197. doi: 10.2174/092986707779313417. [DOI] [PubMed] [Google Scholar]
- 9.Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 12.Jagannath A, Wood MJ. Localization of Double-stranded siRNA to Cytoplasmic P-Bodies Is Ago2-dependent and Results in Upregulation of GW182 and Ago2. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol. 2007;81:12218–12226. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 15.Omoto S, et al. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Triboulet R, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4(+) T lymphocytes. Nat Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 19.Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 20.Omarov RT, Ciomperlik JJ, Scholthof HB. RNAi-associated ssRNA-specific ribonucleases in Tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex. Proc Natl Acad Sci USA. 2007;104:1714–1719. doi: 10.1073/pnas.0608117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silhavy D, et al. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bucher E, Hemmes H, de Haan P, Goldbach R, Prins M. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol. 2004;85:983–991. doi: 10.1099/vir.0.19734-0. [DOI] [PubMed] [Google Scholar]
- 24.Qi Y, Zhong X, Itaya A, Ding B. Dissecting RNA silencing in protoplasts uncovers novel effects of viral suppressors on the silencing pathway at the cellular level. Nucleic Acids Res. 2004;32:e179. doi: 10.1093/nar/gnh180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu F, Ren T, Morris TJ. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J Virol. 2003;77:511–522. doi: 10.1128/JVI.77.1.511-522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasschau KD, Carrington JC. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 27.Llave C, Kasschau KD, Carrington JC. Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc Natl Acad Sci USA. 2000;97:13401–13406. doi: 10.1073/pnas.230334397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersson MG, et al. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutvagner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 31.de Vries W, Berkhout B. RNAi suppressors encoded by pathogenic human viruses. Int J Biochem Cell Biol. 2008 doi: 10.1016/j.biocel.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Haasnoot J, et al. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brodersen P, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 34.Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Restrepo MA, Freed DD, Carrington JC. Nuclear transport of plant potyviral proteins. Plant Cell. 1990;2:987–998. doi: 10.1105/tpc.2.10.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice AP, Carlotti F. Mutational analysis of the conserved cysteine-rich region of the human immunodeficiency virus type 1 Tat protein. J Virol. 1990;64:1864–1868. doi: 10.1128/jvi.64.4.1864-1868.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartman TR, et al. RNA helicase A is necessary for translation of selected messenger RNAs Nat Struct Mol. Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]