Abstract
Wnt signaling stabilizes β-catenin, which in turn influences the transcription of Wnt-responsive genes in conjunction with T-cell factor (TCF) transcription factors. At present, there are two models for the actions of β-catenin. The conventional nuclear model suggests that β-catenin acts in the nucleus to form a heterodimeric transcriptional factor complex with TCF, with TCF providing DNA-specific binding and the C and N termini of β-catenin stimulating transcription. The alternative cytoplasmic model postulates that β-catenin exports TCF from the nucleus to relieve its repressive activity or activates it in the cytoplasm. We have generated modified forms of β-catenin and used RNA interference against endogenous β-catenin to distinguish between these models in cultured mammalian and Drosophila cells. We show that the VP16 transcriptional activation domain can replace the C terminus of β-catenin without loss of function and that the function of β-catenin is compromised by fusion to a transcriptional repressor domain from histone deacetylase, favoring the direct effects of β-catenin in the nucleus. Furthermore, membrane-tethered β-catenin requires interaction with the adenomatous polyposis coli protein but not with TCF for its function, whereas untethered β-catenin requires binding to TCF for its signaling activity. Importantly, by using RNA interference, we show that the signaling activity of membrane-tethered β-catenin, but not free β-catenin, requires the presence of endogenous β-catenin, which is able to accumulate in the nucleus when stabilized by the binding of the β-catenin degradation machinery to the membrane-tethered form. All of these data support a nuclear model for the normal function of β-catenin.
Wnt proteins are closely related secreted glycoproteins that play critical roles in cell proliferation and cell fate determination at many stages of development (37). In addition, deregulation of the Wnt signaling pathway leads to cancer (22). In the absence of a Wnt signal, β-catenin is mostly associated with the plasma membrane, where it associates with E-cadherin and α-catenin and promotes cellular adhesion. Cytosolic β-catenin is normally bound to Axin and the adenomatous polyposis coli (APC) protein, phosphorylated at the N-terminal Ser/Thr residues by casein kinase Iα and glycogen synthase kinase 3 (GSK3), and then degraded by the ubiquitination-proteasome system. In response to a Wnt signal, β-catenin accumulates in the cytoplasm and nucleus and influences the transcription of Wnt-responsive genes in conjunction with the LEF/T-cell factor (TCF) family of transcription factors (2, 19).
Wnt proteins induce the accumulation of nuclear β-catenin, and prominent β-catenin nuclear staining has been found in human cancers with deregulated Wnt signaling. β-Catenin binds to LEF/TCF transcription factors and the C terminus of β-catenin possesses potent transcription activation activity (32). These observations support a model in which β-catenin functions in the nucleus, forming a composite transcription factor, with TCF enabling sequence-specific DNA binding and the C and N termini of β-catenin augmenting the enzymatic activity of RNA polymerase.
However, the nuclear model of β-catenin action has been challenged. It is known that, in the absence of β-catenin, TCF represses Wnt-responsive genes by recruiting Groucho-related transcriptional repressors (5, 23). This has led to the suggestion that the major function of β-catenin might be binding to TCF in the cytoplasm, relieving the repressor activity of TCF (17). This idea has recently been reinforced by a series of experiments performed in Drosophila melanogaster that suggest that membrane-tethered armadillo (Drosophila β-catenin) activates signaling independently of endogenous armadillo and that the C terminus of armadillo provides a nuclear export instead of transcriptional activation function (6). These results support a cytoplasmic model for the function of β-catenin, with β-catenin modulating Wnt signaling through nuclear export or cytoplasmic activation of LEF/TCF transcription factors.
In the present study, we used function-specific mutants and chimeras of β-catenin and small interfering RNA (siRNA) against β-catenin to test whether the function of β-catenin is mediated by nuclear or cytoplasmic β-catenin. Our results suggest that, in our cell culture systems, the function of β-catenin is carried out by nuclear β-catenin and that the signaling activity of membrane-tethered β-catenin requires endogenous β-catenin.
MATERIALS AND METHODS
Expression constructs.
Human β-catenin mutants and LEF1-β-catenin chimeras were tagged with hemagglutinin (HA) epitope fused at the carboxyl termini and cloned into a mammalian expression vector under the control of the cytomegalovirus (CMV) promoter. Site-directed mutagenesis was performed by using PCR, and all constructs were verified by DNA sequencing. A list of the sequences at the joins between different components of various chimeric proteins follows (the joins are indicated by “/”; additional amino acids generated at the joins are indicated in lowercase): VP16-βΔC = EYGG/g/ATQA, HDAC4-β = EPPL/tr/ATQA, CNX-β = TTRA/tr/ATQA, βN-LEFΔN = AGTL/ar/NESE, βC-LEFΔN = DTDL/ar/NESE, and FKBP-LEFΔN = LKLE/tr/NESE. The plasmid encoding CnxPkg has been reported earlier (13). Human β-catenin and plakoglobin mutants, LEF-1, and Renilla luciferase were cloned into the Drosophila expression construct pPac-PL under the control of the Drosophila actin promoter. Human β-catenin and plakoglobin mutants were fused with green fluorescent protein (GFP) at the carboxyl termini. Detailed descriptions of the cloning process and physical maps of the constructs are available upon request.
Mammalian cell culture, transfection, and luciferase assays.
Human kidney epithelial 293 cells and mouse NIH 3T3 cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum or bovine calf serum (HyClone), 1% glutamine, and 1% penicillin-streptomycin (Gibco) at 37°C in 5% CO2. Cells were plated into 12-well plates 24 h before transfections and transfected with Lipofectamine 2000 (Invitrogen) according to the protocol provided by the manufacturer. For 293 cells, each well received 0.01 μg of pCMV-Renilla, 0.2 μg of TOP-FLASH, and 0.5 μg of effector plasmid. For NIH 3T3 cells, each well received 0.02 μg of pCMV-LEF-1, 0.01 μg of pCMV-Renilla, 0.2 μg of LEF-1-luciferase reporter plasmid, and 0.5 μg of effector plasmid. Luciferase assays were performed with the dual luciferase assay kit (Promega) according to the manufacturer's instructions. The luciferase activities were normalized with the Renilla luciferase activities. Each experiment was carried out in triplicate, and error bars represent standard deviations. The activity of the empty vector was set to 1.
RNA interference.
The siRNA targeting human β-catenin was derived from an mRNA sequence (AATGCTTGGTTCACCAGTGGA) of human β-catenin, which contains one underlined mismatch with the corresponding sequence of mouse β-catenin. The siRNA targeting mouse β-catenin was derived from an mRNA sequence (AAACATAATGAGGACCTACAC) of mouse β-catenin, which contains two mismatches (underlined) with the corresponding sequence of human β-catenin mRNA. The siRNA targeting an mRNA sequence (AACAAGATCACCTTCTCCGAG) of human Dvl1 was used as the negative control in siRNA experiments. The siRNA duplexes were prepared by Dharmacon Research (Lafayette, Colo.).
Drosophila S2 cell transfection and dsRNA experiments.
We have used a LEF-luciferase assay in Drosophila S2 cells as described previously (25). S2 cells were grown at room temperature in Schneider's Drosophila medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), 1% glutamine, and 1% penicillin-streptomycin (Gibco). Double-stranded RNA (dsRNA) experiments were performed as described previously (7). PCR was performed with primers containing T7 polymerase binding sites. Complementary single-stranded RNA with a length of ∼700 nucleotides was generated by using a MegaScript T7 transcription kit (Ambion) and annealed to form dsRNA. S2 cells were plated into six-well plates at a density of 106 cells/well. dsRNA was added into S2 cells at 15 μg/well in 1 ml of Drosophila serum-free medium (D-SFM; Gibco). After 1 h of incubation, 1 ml of D-SFM containing 20% fetal calf serum was added. Cells were transfected with the indicated plasmids 24 h after dsRNA treatment. Cells were transfected with CellFECTIN (Invitrogen) according to the protocol provided by the manufacturer. Each well received 0.1 μg of pPac-LEF-1, 0.1 μg of pPac-Renilla, and 0.1 μg of LEF-1-luciferase reporter plasmid and 0.5 μg of effector plasmid. At 36 h after transfection, cells were lysed, and cell lysates were used for luciferase assay. The following genes were targeted with dsRNA: Drosophila armadillo, accession no. X54468; and Drosophila aconitase, accession no. NM_079969. The primers used to generate templates for T7 polymerase were 5′ primer (5′-GGATTAATACGACTCACTATAGGGAGACAGCTAAGCCAGACACGTTC-3′) and 3′ primer (5′-GGATTAATACGACTCACTATAGGGAGAGCTTTCCTGGTTGCCGTAGG-3′) for armadillo and 5′ primer (5′-GGATTAATACGACTCACTATAGGGAGACTCTGTCCAAGTTCGACTCG-3′) and 3′primer (5′-GGATTAATACGACTCACTATAGGGAGAGATAGAGTCAACACCCTGC-3′) for aconitase. Aconitase-dsRNA was used as a negative control in the RNA interference experiments.
Immunoblot analysis.
Cells were lysed with radioimmunoprecipitation assay buffer (10 mM sodium phosphate [pH 7.4], 100 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and protease inhibitors). Equal amounts of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and proteins were transferred onto a nitrocellulose filter. After a blocking step with 5% dry milk in Tris-buffered saline plus 0.1% Tween 20 for 1 h, the membrane was incubated with the primary antibodies overnight at 4°C and with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Signals were detected by an enhanced chemiluminiscence kit (Amersham). Mouse monoclonal anti-β-catenin antibodies were purchased from Transduction Laboratories, and mouse monoclonal anti-α-tubulin antibodies were purchased from Sigma.
Fluoresence microscopy.
COS cells were transfected with HA-tagged β-catenin ΔC. At 36 h after transfection, cells were washed once with phosphate-buffered saline (PBS) and then fixed with 2% paraformaldehyde for 20 min at room temperature. Fixed cells were treated with 0.1% Triton X-100 for 5 min and blocked with 2% fetal bovine serum in PBS for 1 h. Mouse monoclonal anti-β-catenin (Transduction Laboratories) and rabbit anti-HA antibody (HA.11; Covance) were incubated with cells for 1 h at 37°C. Cells were then rinsed and incubated with affinity-purified, fluorescein isothiocyanate (FITC)-conjugated, goat anti-rabbit immunoglobulin (Sigma) and Texas red-conjugated goat anti-mouse immunoglobulin (Jackson Immunoresearch Laboratories) in PBS supplemented with 2% fetal bovine serum for 1 h at 37°C. Cells were then washed with PBS. Nuclei were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) dihydrochloride (Roche) at a final concentration of 1.5 μg/ml in PBS for 30 min in room temperature. Cells were then washed several times with PBS, mounted by using Vetashield mounting medium (Vector), and examined by fluorescence microscopy (Leica).
Analysis of mRNA.
DNA-free RNA was obtained with the RNeasy Minikit (Qiagen) with DNase treatment, and 5 μg of total RNA was reverse transcribed with oligo(dT) and Moloney murine leukemia virus reverse transcriptase. Real-time PCR was done in triplicate with the iCycler iQ thermal cycler and detection system (Bio-Rad) and the PCR core reagents kit (Applied Biosystems) with 500 nM concentrations of primers; the final Mg2+ concentration was adjusted to 4 mM. Fourfold serial dilutions of cDNAs were used to generate curves of log input amount versus threshold cycle, and comparable slopes for a given primer set were obtained for the group of cDNAs being tested (signifying comparable efficiencies of amplification). Induction was normalized for levels of GAPDH (glyceraldehyde-3-phosphate dehydrogenase). When reverse transcriptase was omitted, threshold cycle number increased by at least 10, signifying a lack of genomic DNA contamination or nonspecific amplification. The generation of only correct-size amplification products was confirmed with agarose gel electrophoresis, and the identity of PCR product was confirmed by DNA sequencing. The oligonucleotide primers used were as follows: GAPDH, 5′-GTGAAGGTCGGAGTCAAC-3′ and 5′-TGGAATTTGCCATGGGTG-3′; and DKK1, 5′-AATCTGTCTCGCCTGCAGGAAG-3′ and 5′-ACCCATCCAAGGTGCTATGATC-3′.
RESULTS
Functional substitution of the C terminus of β-catenin by the VP16 transcriptional activation domain.
In the nuclear model, β-catenin is proposed to function as a transcription activator by recruiting nuclear factors to DNA-bound TCF, thereby increasing the enzymatic activity of the RNA polymerase II complex. However, in the cytoplasmic model, β-catenin could function by exporting TCF out of the nucleus, thereby reducing its inhibitory activities, or by activating TCF in the cytoplasm. In order to distinguish between these two models, we first sought to determine whether the function of disabled β-catenin can be compensated for by a fused transcription activation domain and whether a wild-type β-catenin can be compromised by a fused transcription repressor domain.
β-Catenin is composed of an N-terminal region, a central 12- armadillo-repeat region, and a C-terminal region (Fig. 1A). The N-terminal region influences the stability of β-catenin; Ser/Thr residues in the N-terminal region of β-catenin are phosphorylated by GSK3 and targeted by β-TrCP for ubiquitination and degradation; mutations of these phosphorylation sites stabilize the protein. The armadillo repeats contain the binding sites for most β-catenin binding partners, such as E-cadherin, APC, Axin, and TCF. The C-terminal region is required for the normal signaling activity of β-catenin and possesses the capacity to activate transcription when fused to a heterologous DNA-binding domain (32).
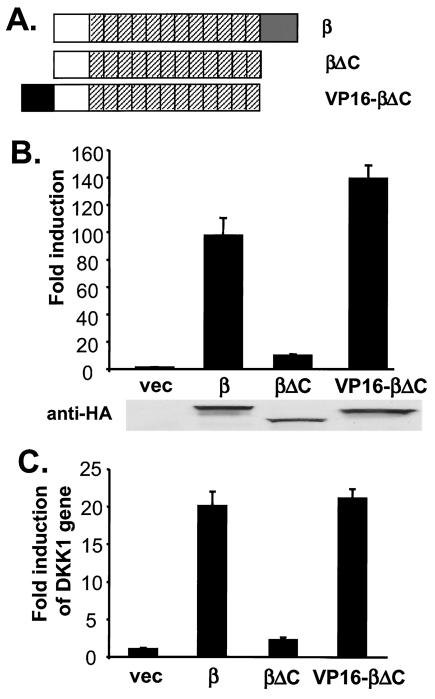
FIG. 1.
Functional substitution of the C terminus of β-catenin by the VP16 transcriptional activation domain. (A) Schematic representation of β-catenin constructs. β-Catenin is composed of the N-terminal domain (□), 12 armadillo repeats (▨), and the C-terminal domain (░⃞). Amino acid residues 675 to 781 were deleted to form β-catenin ΔC, and residues 422 to 490 of VP16 were added to the amino terminus of β-catenin ΔC to form VP16-β-catenin ΔC. All β-catenin derivatives were tagged with HA epitopes at their C termini. (B) Activation of TOP-FLASH by β-catenin (β), β-catenin ΔC (βΔC), and VP16-β-catenin ΔC (VP16-βΔC). Human 293 cells were transfected with indicated plasmids, together with TOP-FLASH and a CMV-Renilla luciferase reporter, and the luciferase activities were determined as described in Materials and Methods. Expression of β-catenin constructs was monitored by immunoblotting with anti-HA antibodies (below the chart). (C) Induction of the DKK1 gene by β-catenin proteins. 293 cells were transfected with indicated plasmids. The abundance of DKK1 mRNA was determined by reverse transcription and quantitative real-time PCR. The results were normalized by the level of GAPDH. vec, Vector.
We first tested whether the C terminus of β-catenin could be replaced by the VP16 transcriptional activation domain. Human β-catenin is used in most of the experiments in the present study, and all β-catenin derivatives that we generated contain the stabilizing S37A mutation and an HA epitope. β-Catenin mutants were ectopically expressed in 293 cells, together with a TOP-FLASH luciferase reporter that contains multiple copies of an optimal TCF-binding site (32). As seen in Fig. 1B, exogenous expression of β-catenin dramatically increased the luciferase activity of TOP-FLASH, and truncating the C-terminal 108 amino acid residues of β-catenin (β-catenin ΔC) decreased the signaling activity of β-catenin by 10-fold.
We reasoned that if the primary function of the C terminus of β-catenin was to activate transcription, it could be functionally substituted with the VP16 transcriptional activation domain. Indeed, fusing the VP16 transcriptional activation domain to the N terminus of β-catenin ΔC fully rescued its signaling activity (Fig. 1B). We initially fused the VP16 activation domain to the N terminus of β-catenin ΔC to avoid potential structural interference with the C-terminal armadillo repeats, but we obtained indistinguishable results when the VP16 transcriptional activation domain was fused to the C terminus of β-catenin ΔC (data not shown). The production of various β-catenin derivatives was similar, as measured by immunoblotting (Fig. 1B), and β-catenin and VP16-β-catenin ΔC had no effect on the FOP-FLASH luciferase reporter, which contains multiple copies of mutant form of TCF binding sites (data not shown).
We confirmed these results with an endogenous gene regulated by β-catenin. Expression profiling has revealed that in 293 cells, DKK1, an inhibitor of the Wnt-1 signaling pathway (16), is upregulated by overexpression of Wnt or β-catenin (M. Chamorro and H. Varmus, unpublished data). Using real-time quantitative PCR, we showed that exogenous expression of β-catenin in 293 cells increased DKK1 expression by 20-fold, whereas expression of β-catenin ΔC had minimal effects (Fig. 1C). Notably, VP16-β-catenin S37A stimulated the expression of DKK1 to a similar extent, as did the full-length β-catenin protein (Fig. 1C).
Together, these results indicate that the C terminus of β-catenin can be functionally replaced by the VP16 transcriptional activation domain, suggesting that the major effects of β-catenin are mediated by transcription activation.
Inhibition of the function of β-catenin by a fused transcriptional repressor domain.
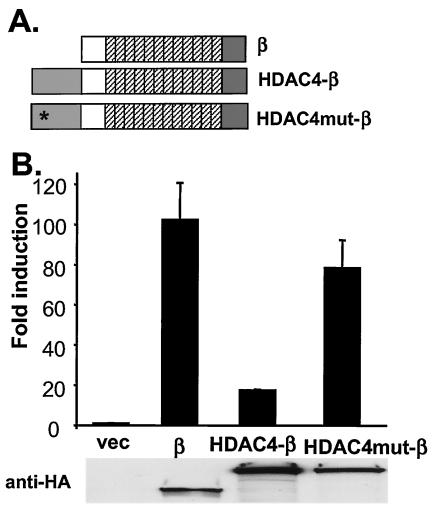
If β-catenin functions as a transcriptional activator in the nucleus, its signaling activity might be compromised by fusion to a transcriptional repressor domain. Histone deacetylases (HDACs) repress transcription by removing acetyl groups from histones and remodeling chromatin into a closed conformation (26). Fusing the catalytic domain of HDAC4 (amino acids 663 to 1083) to the N terminus of β-catenin significantly decreased the activity of β-catenin on the expression of luciferase from the TOP-FLASH plasmid (Fig. 2B). Two residues, His-802 and His-803, in the catalytic domain of HDAC4 are essential for its catalytic activity (35). The HDAC4-β-catenin fusion containing mutated His-802 and His-803 no longer suppressed β-catenin signaling activity (Fig. 2B). Therefore, the effects of the HDAC4 catalytic domain on the signaling activity of β-catenin require the enzymatic activity of HDAC4 and are not due to the steric effects of adding HDAC4 protein to the N terminus of β-catenin. Since HDAC4-β-catenin had no effect on the expression of Renilla luciferase encoded by a cotransfected plasmid (data not shown), it did not have a global inhibitory effect on transcription. Exogenous expression of the HDAC4-β-catenin chimera also had negligible effects on FOP-FLASH (data not shown). These data suggest that the signaling activity of β-catenin is compromised when fused to enzymatically active HDAC4, which can modulate chromatin into a closed conformation, a finding consistent with the view that β-catenin normally functions in the nucleus to promote transcription.
FIG. 2.
Loss of signaling activity by fusing the transcriptional repression domain from an HDAC to β-catenin. (A) Schematic representation of HDAC4-β-catenin chimeras. The catalytic domain (amino acids 663 to 1083) of HDAC4 was fused to the N terminus of β-catenin (HDAC4-β). Two residues (His-802 and His-803) essential for the catalytic activity of HDAC4 were mutated in HDAC4mut-β. All β-catenin derivatives are tagged with HA epitopes at their C termini. (B) Activation of TOP-FLASH by β-catenin and HDAC4-β-catenin chimeras. 293 cells were transfected with TOP-FLASH together with indicated expression constructs. β-Catenin expression was determined by immunoblotting with anti-HA antibodies (below the chart). vec, Vector.
Differential requirements of APC and LEF binding for membrane-tethered and untethered β-catenin.
Membrane-tethered β-catenin and plakoglobin (a β-catenin relative) possess signaling activities (13, 17, 38). These observations suggest that β-catenin can function in the cytoplasm. However, since endogenous β-catenin becomes stabilized upon overexpression of membrane-tethered β-catenin, it has been postulated that membrane-tethered β-catenin competes with endogenous β-catenin for its binding partners, such as APC, which promotes the degradation of β-catenin, and that stabilized endogenous β-catenin then mediates the signaling activities of membrane-tethered β-catenin (18).
To investigate these possibilities, we tested the signaling activities of β-catenin mutants with defective binding affinities for LEF/TCF or APC. If membrane-tethered β-catenin functions by titrating components of β-catenin degradation complex and stabilizing endogenous β-catenin, as suggested by the nuclear model, binding to APC, but not to LEF/TCF, might be required for activity. On the other hand, if membrane-tethered β-catenin acts through sequestering repressive LEF/TCF or activating LEF/TCF in the cytoplasm, as suggested by the cytoplasmic model, it should require binding to LEF/TCF, but not to APC, for activity.
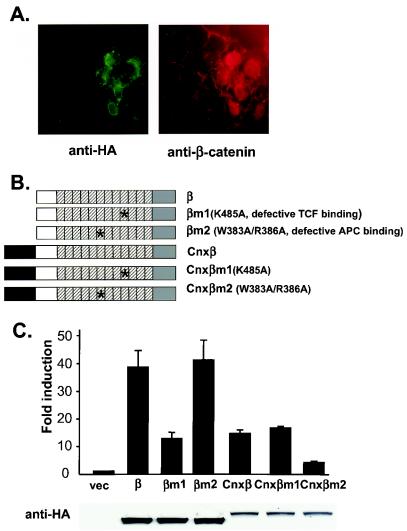
We first examined the effect of membrane-tethered β-catenin on endogenous β-catenin. HA-tagged connexin-β-catenin ΔC (CnxβΔC) was overexpressed in COS cells, and in situ immunofluoresence experiments were performed. Consistent with previous findings, overexpression of CnxβΔC results in stabilization of endogenous β-catenin (Fig. 3A). Similarly, overexpression of connexin-plakoglobin also induces stabilization of endogenous β-catenin (data not shown).
FIG. 3.
Differential requirements of TCF and APC binding for activities of untethered and membrane-tethered β-catenins. (A) Stabilization of endogenous β-catenin by membrane-tethered β-catenin. HA-tagged connexin-β-catenin ΔC (CnxβΔC) was expressed in COS cells. Cells were incubated with rabbit polyclonal anti-HA antibody and mouse monoclonal anti-β-catenin antibody. The epitope recognized by anti-β-catenin antibody was missing in CnxβΔC. Cells were next stained with FITC-conjugated, anti-rabbit immunoglobulin and Texas red-conjugated, anti-mouse immunoglobulin antibodies. Note that β-catenin is localized on the plasma membrane in cells without CnxβΔC (the left side of pictures), whereas β-catenin accumulated in the nucleus in cells with CnxβΔC (the right side of pictures). (B) Schematic representation of β-catenin mutants with defective TCF or APC binding sites. In βm1, Lys 435, crucial for TCF binding, was changed to Ala. In βm2, W383 and R386, crucial for APC binding, were both changed to Ala. Wild-type β-catenin, βm1, and βm2 were also fused to the C terminus of the transmembrane domain of connexin to form Cnxβ, Cnxβm1, and Cnxβm2, respectively. All β-catenin constructs were fused with HA epitopes at their C termini. (C) Differential requirements of TCF and APC binding for untethered and membrane-tethered β-catenin. 293 cells were transfected with TOP-FLASH, and the indicated expression constructs and luciferase activities were assayed. The expression of β-catenin mutants was followed by immunoblotting with anti-HA antibodies (below the chart). vec, Vector.
β-Catenin binds to most of its partners through its armadillo superhelix. β-Catenin mutants that have compromised affinities to various binding partners have been identified by a structure-function analysis of β-catenin mutants (34). Specifically, it has been shown that Lys-435 of β-catenin is essential for binding to LEF/TCF transcription factors, whereas Trp-383 and Arg-386 are important for binding to the 20- and 15-amino-acid repeats of APC. Therefore, mutating Lys-435 would decrease the interaction between β-catenin and LEF/TCF, whereas mutating Trp-383 and Arg-386 would affect the interaction between β-catenin and APC. When β-catenin was fused to the C terminus of the transmembrane domain of connexin and anchored to the cytoplasmic face of intracellular vesicles, it strongly activated TOP-FLASH (Fig. 3C), a finding consistent with previous results with membrane-tethered plakoglobin (13). Next, we generated mutant forms of β-catenin or connexin-β-catenin containing either K435A or W383A/R386A mutations (Fig. 3B). Consistent with the previous study (34), we found by using a coimmunoprecipitation assay that β-catenin K435A bound poorly to LEF-1, whereas β-catenin W383A/R386A bound relatively poorly to APC (data not shown). We then tested these mutants for their signaling activities in 293 cells. Mutating Lys-435, but not Trp-383 and Arg-386, decreased the signaling activities of β-catenin (Fig. 3C). On the other hand, mutating Trp-383 and Arg-386, but not Lys-435, compromised the signaling activities of connexin-β-catenin (Fig. 3C). Therefore, binding to LEF/TCF transcriptional factors is required for the signaling activity of untethered β-catenin but not for that of membrane-tethered β-catenin; conversely, binding to APC is required for the function of membrane-tethered β-catenin but not for that of untethered β-catenin. These results support the hypothesis that membrane-tethered β-catenin activates the signaling pathway by inhibiting the function of the β-catenin destruction complex, possibly by titrating APC, and are inconsistent with the hypothesis that membrane-tethered β-catenin activates the pathway through a direct interaction with LEF/TCF.
Inhibitory RNA confirms that endogenous β-catenin is required for the signaling activity of membrane-tethered β-catenin.
Although membrane-tethered β-catenin induces stabilization of endogenous β-catenin, the question of whether stabilized endogenous β-catenin mediates the signaling activity of membrane-tethered β-catenin remains controversial (6, 8, 31). Because published loss-of-function experiments with armadillo were performed in Drosophila and the membrane function of armadillo is essential for the early development of Drosophila embryos, complete removal of endogenous armadillo precludes a completely satisfying assessment of armadillo signaling. Therefore, only hypomorphic armadillo mutants with various degrees of C-terminal truncations can be used to test the signaling activities of membrane-tethered, full-length armadillo, and interpretations have been complicated by the possibility that the hypomorphic mutants retain signaling activities.
We addressed this question by reducing endogenous β-catenin levels through RNA interference in cultured human, mouse, and Drosophila cells that produce membrane-tethered variants of β-catenin and plakoglobin. The silencing effects of human β-catenin-specific siRNA in 293 cells and mouse β-catenin-specific siRNA in NIH 3T3 cells were documented by immunoblotting (Fig. 4A). The effects of siRNA were highly specific: human β-catenin-specific siRNA, but not mouse β-catenin-specific siRNA, decreased Wnt-1-induced LEF-luciferase in human 293 cells, whereas mouse β-catenin-specific siRNA, but not human β-catenin-specific siRNA, blocked Wnt-1-induced LEF-luciferase in mouse NIH 3T3 cells (data not shown).
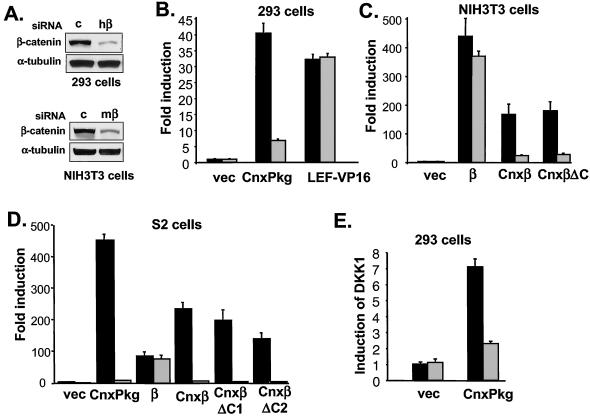
FIG. 4.
Requirement of endogenous β-catenin for the stimulatory effect of membrane-tethered β-catenin/plakoglobin on luciferase reporters. (A) Effect of β-catenin-specific siRNA on the expression of endogenous β-catenin. 293 cells or NIH 3T3 cells were treated with control (c) and human or mouse-specific β-catenin (hβ or mβ) siRNA, and the expression of endogenous β-catenin was determined by immunoblotting with anti-β-catenin antibodies. Equal loading was confirmed by Western blotting with anti-α-tubulin antibodies. (B) Requirement of endogenous β-catenin for the signaling activity of membrane-tethered plakoglobin (CnxPkg) in 293 cells. 293 cells were transfected with control (▪) or human β-catenin-specific (░⃞) siRNA, together with TOP-FLASH and the indicated expression constructs, and the luciferase activities were determined 48 h after transfection. (C) Requirement of endogenous β-catenin for the signaling activity of membrane-tethered β-catenin in NIH 3T3 cells. To achieve high sensitivity in NIH 3T3 cells, β-catenin was cotransfected with a mouse LEF-1 expression plasmid and a LEF-1 luciferase reporter that contains multiple LEF-1 binding sites. NIH 3T3 cells were transfected with control (▪) or mouse β-catenin-specific (░⃞) siRNA, together with the indicated expression constructs, and assayed for luciferase activity. Amino acid residues 675 to 781 of β-catenin were deleted from Cnxβ to form CnxβΔC. (D) Requirement of endogenous armadillo for the signaling activity of membrane-tethered β-catenin in Drosophila S2 cells. S2 cells were grown in six-well plates and treated with control (▪) or armadillo-specific (░⃞) dsRNA at a concentration of 15 μg/well. Cells were then transfected with LEF-luciferase and LEF-1 expression constructs with the indicated plasmids that encode various forms of plakoglobin and β-catenin. Amino acid residues 677 to 781 and 583 to 781 of β-catenin were removed from Cnxβ to form CnxβΔC1 and CnxβΔC2. (E) Requirement of endogenous β-catenin for membrane-tethered plakoglobin-induced DKK1 expression. 293 cells were transfected with control siRNA (▪) or human β-catenin-specific siRNA (░⃞) with the indicated expression plasmids. The expression of DKK1 gene was determined by quantitative real-time PCR after reverse transcription. vec, Vector.
First, we tested the effects of human β-catenin-specific siRNA on the signaling activities of connexin-plakoglobin in human 293 cells by using TOP-FLASH as a readout. Human β-catenin-specific siRNA reduced the signaling activities of connexin-plakoglobin by >80% but had no effect on the signaling activities of LEF-VP16 (Fig. 4B), suggesting that endogenous β-catenin is required for the function of membrane-tethered plakoglobin.
Next, we examined the effects of mouse β-catenin-specific siRNA on LEF-dependent transcription induced by various human β-catenin mutants in mouse NIH 3T3 cells. As shown in Fig. 4C, mouse β-catenin-specific siRNA had only a minimal effect on the induction of luciferase by human β-catenin but significantly decreased the signaling activities of connexin-human β-catenin and connexin-human β-catenin ΔC, presumably by reducing the levels of endogenous murine β-catenin.
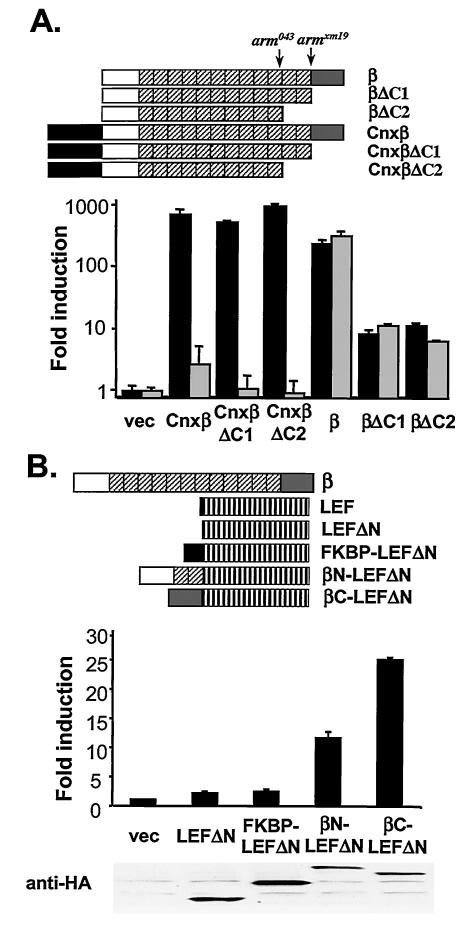
Similar experiments were performed in Drosophila S2 cells. Membrane-tethered connexin-human plakoglobin activated the LEF-luciferase reporter by >400-fold, and connexin-human β-catenin activated the pathway >200-fold (Fig. 4D). Truncation of the C terminus of β-catenin (ΔC1) or removal of the C terminus, together with the last two armadillo repeats (ΔC2) (Fig. 5), did not significantly affect the signaling activities of connexin-β-catenin (Fig. 4D), in contrast to a requirement for the C terminus for the activity of untethered β-catenin in mammalian cells (Fig. 1) or Drosophila cells (Fig. 5). To examine whether these constructs activate the signaling pathway through endogenous armadillo, cells were treated with either control or armadillo-specific inhibitory dsRNA prior to transfection of the indicated expression plasmids. Armadillo-specific dsRNA decreased the signaling activities of all of the membrane-tethered plakoglobin and β-catenin mutants (Fig. 4D), suggesting that membrane-tethered plakoglobin and β-catenin required endogenous armadillo to activate the reporter gene. In contrast, the 100-fold induction of luciferase by untethered β-catenin was not affected by interfering RNA, implying that endogenous armadillo was not required to activate the reporter, presumably because exogenous β-catenin was able to enter the nucleus. Armadillo-specific dsRNA also did not affect LEF-VP16-activated LEF-luciferase (data not shown).
FIG. 5.
Signaling activities of β-catenin C-terminal truncation mutants. (A) Residual signaling activity of β-catenin C-teminal truncation mutants. Human β-catenin C-terminal truncation mutants were generated. The approximate truncation sites in arm043 and armXM19 mutant alleles from Drosophila are labeled (upper panel). In βΔC1, the C terminus of β-catenin (amino acids 677 to 781) was removed. In βΔC2, the C terminus and the last two armdillo repeats (amino acid 583 to 781) were removed. β-Catenin mutants were also fused to the C terminus of the transmembrane domain of connexin. All β-catenin derivatives in Drosophila expression constructs were fused with GFP at their C termini. S2 cells were treated with control (▪) or armadillo-specific (░⃞) dsRNA and transfected with the indicated plasmids as in Fig. 4D. (B) The N terminus of β-catenin contains transcriptional activation activity. β-Catenin-LEF1ΔN chimeric proteins were generated (upper panel). A LEF-1 N-terminal truncation mutant (lacking amino acids 1 to 57) was fused at its N terminus with either the N terminus (amino acids 1 to 218) or C terminus (amino acids 659 to 781) of β-catenin. LEF1-β-catenin chimeras were fused with an HA epitope at the C termini. The effects of LEF1-β-catenin chimera on TOP-FLASH were tested in 293 cells; expression of these chimeras was examined by Western blotting with anti-HA antibodies (below the chart). vec, Vector.
All β-catenin and plakoglobin mutants cloned in a Drosophila expression vector were tagged with GFP at their C termini to show by fluorescence microscopy that they were expressed both in the presence or absence of armadillo dsRNA (data not shown). Further, we have noted that HA-tagged β-catenin has greater signaling activities than GFP-tagged β-catenin protein, presumably due to structural interference by the GFP component (data not shown).
Finally, we tested the contribution of endogenous β-catenin to the signaling activities of membrane-tethered plakoglobin in human 293 cells by using induction of the endogenous DKK1 gene as a readout. Connexin-plakoglobin stimulated the expression of DKK1 by ∼7-fold. Human β-catenin-specific siRNA, but not control siRNA, significantly decreased induction of DKK1 by membrane-tethered plakoglobin (Fig. 4E). Taken together, these results suggest that the signaling function of membrane-tethered β-catenin is mediated by endogenous β-catenin.
Signaling activities of β-catenin C-terminal truncation mutants.
It has been shown that membrane-tethered armadillo has signaling activity in the Drosophila hypomorphic mutants armXM19 (6, 8) and arm043 (6). ArmXM19 lacks the C terminus of armadillo, and Arm043 lacks the C terminus plus the last two armadillo repeats. If membrane-tethered armadillo transduces a signal through endogenous armadillo, both ArmXM19 and Arm043 should retain some signaling activity. To investigate this possibility, we generated human β-catenin C-terminus truncation mutants (ΔC1 and ΔC2), which are truncated at sites homologous to the C terminus of ArmXM19 and Arm043 (Fig. 5A) and tested their signaling activities in S2 cells. S2 cell were pretreated with armadillo-specific dsRNA to reduce or eliminate the contribution of endogenous armadillo. These truncated mutants activated the LEF-luciferase reporter by 10-fold (Fig. 5A). Although their activities are significantly lower than that of full-length β-catenin, they are readily measured, indicating that β-catenin C-terminal truncation mutants retain some signaling activity.
C-terminal truncation mutants of β-catenin contain most of the armadillo repeats and presumably bind to LEF/TCF transcription factors. However, these mutants lack the C-terminal region, a domain responsible for most of the transcriptional activity of β-catenin. The finding that β-catenin C-terminal truncation mutants have residual signaling activities suggests that other regions of β-catenin must also contain transcriptional activation activities. Indeed, it has been suggested that the N terminus of β-catenin contains a transcriptional activation domain (11, 36). To confirm these results, we fused different fragments of β-catenin to the N terminus of LEF-1 ΔN and tested these chimeras for signaling activity. Fusing the C terminus of β-catenin to LEF-1 dramatically increased the stimulatory effect of LEF-1 on TOP-FLASH, a finding consistent with the notion that the C terminus of β-catenin functions as the major transcriptional activation domain (Fig. 5B). Further, fusing the N terminus of β-catenin to LEF-1 also increased the stimulatory effect of LEF-1 on TOP-FLASH, whereas fusing an unrelated protein, FKBP (FK506-binding protein), to LEF-1 had no effect on its signaling activities (Fig. 5B), suggesting that the N terminus of β-catenin contains transcriptional activation activities.
DISCUSSION
β-Catenin accumulates in the nucleus upon activation of the Wnt signaling pathway. It is generally believed that β-catenin functions in the nucleus through formation of a heterodimeric transcription factor with LEF/TCF. However, according to an alternative model, β-catenin exerts its function in the cytoplasm, possibly through nuclear export or cytosolic activation of TCF. There are several key questions in the debate over the nuclear versus the cytoplasmic model. Does membrane-tethered β-catenin exert its effects directly or through stabilized endogenous β-catenin? Does the C terminus of β-catenin serve as a transcriptional activation domain or a nuclear export domain? What is the relationship between the activity of β-catenin and its subcellular locations?
In the present study, we have tried to answer these questions by generating modified forms of β-catenin and by using RNA interference against endogenous β-catenin. Our findings support the model in which β-catenin functions in the nucleus; this was shown most persuasively by our demonstration that, when β-catenin is retained in the cytoplasm by membrane tethering, endogenous β-catenin is required for activity.
One issue in the debate is how membrane-tethered β-catenin or plakoglobin activates the Wnt pathway. Nuclear accumulation of endogenous β-catenin by membrane-tethered β-catenin prompted the hypothesis that membrane-tethered β-catenin signals through endogenous β-catenin (18). Hypomorphic armadillo mutants have been used to test this idea, but the issue has remained controversial, because it is unclear whether the hypomorphic mutants contain any residual activity. Moreover, results have been conflicting: one group found that membrane-tethered armadillo rescues the Wg phenotype in armXM19 and arm043 germ line clone embryos (6), whereas others found that membrane-tethered armadillo exhibits signaling activities in armXM19 but not arm043 embryos (8, 31).
Using RNA interference, we have demonstrated that the signaling activity of membrane-tethered β-catenin relies on endogenous β-catenin in mammalian and Drosophila cell culture systems (Fig. 4). In addition, we have shown that the signaling activities of untethered β-catenin require binding to LEF/TCF, whereas the signaling activities of membrane-tethered β-catenin require binding to APC (Fig. 3). Taken together, these two approaches strongly support the idea that membrane-tethered β-catenin functions by stabilizing endogenous β-catenin, possibly through titration of APC.
A second issue in the debate is the function of the C terminus of β-catenin. The conventional nuclear model of β-catenin proposes that the C terminus of β-catenin serves as a transcriptional activation domain (32). However, the cytoplasmic model proposes that it functions as a nuclear export domain (6). This idea is based, in part, on the observation that a motif at the N-terminal portion of the armadillo C terminus may be related to the corresponding region of pendulin, the Drosophila α-importin homologue, which supposedly binds to karyopherin CAS (named for cellular apoptosis susceptibility) to mediate its nuclear export. Two glutamic acid residues conserved between armadillo and pendulin appear to be important for the signaling activity of armadillo in Drosophila, and the C terminus of armadillo can be functionally substituted with the C terminus of pendulin but not with a mutant version of the C terminus with these two Glu residues mutated (6). However, we have not been able to observe a critical role of these Glu residues for the localization and signaling activities of armadillo; mutating the two Glu residues to either Asp or Ala did not alter the subcellular localization of the mutant proteins and had no effect on the stimulatory activities of armadillo on luciferase reporters in 293 or S2 cells (data not shown). Therefore, our results support a transcriptional activational function instead of a nuclear export function for the C terminus of β-catenin.
Third, what is the relationship between the signaling activity of β-catenin and its subcellular localization? Chan and Struhl have observed that adding a nuclear localization signal (NLS) to the C terminus of armadillo decreased its signaling activity in Drosophila, a finding that argues against a nuclear function for β-catenin (6). Consistent with this observation, we found that adding the NLS of simian virus 40 large T antigen to the C terminus of β-catenin caused a significant redistribution of β-catenin toward the nucleus in COS cells and reduced the signaling activities of β-catenin by threefold in 293 cells (data not shown). However, introducing positive charges into the C terminus of β-catenin could have affected the transcriptional activity of the C-terminal domain independent of its effect on location. We have also found that adding the NLS to the N terminus of β-catenin did not alter the subcellular localization and signaling activities of β-catenin (data not shown).
If endogenous ArmXM19 can mediate the signaling activity of membrane-tethered armadillo, why does ectopically expressed ArmXM19 stabilized by an N-terminal truncation only have low or no ectopic activity (6)? It is conceivable that the signaling activity of ArmXM19 stabilized by either membrane-tethered armadillo or N-terminal truncation can be blocked by endogenous E-cadherin or by other β-catenin inhibitors, such as ICAT (27) or Chibby (29). However, membrane-tethered armadillo in sufficient amounts might titrate such inhibitors of armadillo, thus promoting the signaling activity of endogenous ArmXM19. It is also possible that membrane-tethered armadillo titrates inhibitory TCF and provides a sensitized environment for stabilized endogenous ArmXM19. Further experiments are needed to address this issue.
Our experiments with β-catenin chimeras strongly suggest that β-catenin primarily functions as a transcriptional activator. We have shown that fusing the VP16 transcriptional activation domain to a β-catenin C-terminal truncation mutant completely restored its signaling activity, whereas fusion of the catalytic domain of HDAC4 to β-catenin suppressed its signaling activity. In agreement with our observations, a chimeric protein in which the C-terminal domain of β-catenin is replaced with the transcriptional repression domain of Drosophila engrailed shows a strong dominant-negative effect in Xenopus embryos (20). However, in Drosophila, the C terminus of armadillo cannot be replaced by the VP16 transcriptional activation domain, and fusion of the engrailed transcriptional repressor to armadillo does not interfere with the signaling activity of armadillo (6). The causes of these discrepancies are not yet known but could reflect different experimental systems with different concentrations of components.
The discrepancies between our results and those of Chan and Struhl might also be partially explained by the possibility that TCF has a repressor function in Drosophila embryos (5) but primarily functions as an activator in the cell culture systems that we have used. Somatic mutation of TCF in Drosophila wing imaginal disk does not show any repressor function for TCF (24). It is likely that TCF transcription factors function predominantly as transcriptional activators in most Wnt signaling processes. This would explain why losses of TCF in Drosophila or mammals produce phenotypes resembling the loss Wnt signaling (4, 9, 14, 33).
The conclusions we have drawn from our experiments are strongly reinforced by other evidence that β-catenin acts as a transcriptional activator. β-Catenin binds to transcriptional coactivators such as CBP/p300 (10, 28) and Brahma/Brg-1 (1). Significantly, Pygopus and Legless, two potential chromatin-remodeling factors dedicated to the activation of Wnt-responsive genes, are nuclear proteins and form a trimeric complex with β-catenin (3, 15, 21, 30). By using chromatin immunoprecipitation methods, we have demonstrated that β-catenin binds to DKK1 promoter in vivo (Chamorro and Varmus, unpublished). Similarly, chromatin immunoprecipitation has been used to show that β-catenin binds to the pitx2 promoter (12). All of these data support a model in which the primary role of β-catenin in the Wnt signaling pathway is to enhance transcription by direct actions in the nucleus, most likely recruiting different transcriptional coactivators to the TCF binding sites of Wnt-responsive genes.
Acknowledgments
We thank Dianqing Wu, Rudolf Grosschedl, Peter Vogt, and Po Chen for providing reagents and William Pao for critically reading the manuscript.
F.C. was supported by a Susan G. Komen Breast Cancer Foundation postdoctoral fellowship.
REFERENCES
- 1.Barker, N., A. Hurlstone, H. Musisi, A. Miles, M. Bienz, and H. Clevers. 2001. The chromatin remodeling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 20:4935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behrens, J., J. P. von Kries, M. Kuhl, L. Bruhn, D. Wedlich, R. Grosschedl, and W. Birchmeier. 1996. Functional interaction of β-catenin with the transcription factor LEF-1. Nature 382:638-642. [DOI] [PubMed] [Google Scholar]
- 3.Belenkaya, T. Y., C. Han, H. J. Standley, X. Lin, D. W. Houston, and J. Heasman. 2002. pygopus Encodes a nuclear protein essential for wingless/Wnt signaling. Development 129:4089-4101. [DOI] [PubMed] [Google Scholar]
- 4.Brunner, E., O. Peter, L. Schweizer, and K. Basler. 1997. pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature 385:829-833. [DOI] [PubMed] [Google Scholar]
- 5.Cavallo, R. A., R. T. Cox, M. M. Moline, J. Roose, G. A. Polevoy, H. Clevers, M. Peifer, and A. Bejsovec. 1998. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395:604-608. [DOI] [PubMed] [Google Scholar]
- 6.Chan, S. K., and G. Struhl. 2002. Evidence that Armadillo transduces wingless by mediating nuclear export or cytosolic activation of Pangolin. Cell 111:265-280. [DOI] [PubMed] [Google Scholar]
- 7.Clemens, J. C., C. A. Worby, N. Simonson-Leff, M. Muda, T. Maehama, B. A. Hemmings, and J. E. Dixon. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 97:6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox, R. T., L. M. Pai, J. R. Miller, S. Orsulic, J. Stein, C. A. McCormick, Y. Audeh, W. Wang, R. T. Moon, and M. Peifer. 1999. Membrane-tethered Drosophila Armadillo cannot transduce Wingless signal on its own. Development 126:1327-1335. [DOI] [PubMed] [Google Scholar]
- 9.Galceran, J., I. Farinas, M. J. Depew, H. Clevers, and R. Grosschedl. 1999. Wnt3a−/−-like phenotype and limb deficiency in Lef1−/− Tcf1−/− mice. Genes Dev. 13:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of β-catenin in vertebrates. EMBO J. 19:1839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu, S. C., J. Galceran, and R. Grosschedl. 1998. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with β-catenin. Mol. Cell. Biol. 18:4807-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kioussi, C., P. Briata, S. H. Baek, D. W. Rose, N. S. Hamblet, T. Herman, K. A. Ohgi, C. Lin, A. Gleiberman, J. Wang, V. Brault, P. Ruiz-Lozano, H. D. Nguyen, R. Kemler, C. K. Glass, A. Wynshaw-Boris, and M. G. Rosenfeld. 2002. Identification of a Wnt/Dvl/β-catenin → Pitx2 pathway mediating cell-type-specific proliferation during development. Cell 111:673-685. [DOI] [PubMed] [Google Scholar]
- 13.Klymkowsky, M. W., B. O. Williams, G. D. Barish, H. E. Varmus, and Y. E. Vourgourakis. 1999. Membrane-anchored plakoglobins have multiple mechanisms of action in Wnt signaling. Mol. Biol. Cell 10:3151-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korinek, V., N. Barker, K. Willert, M. Molenaar, J. Roose, G. Wagenaar, M. Markman, W. Lamers, O. Destree, and H. Clevers. 1998. Two members of the Tcf family implicated in Wnt/β-catenin signaling during embryogenesis in the mouse. Mol. Cell. Biol. 18:1248-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kramps, T., O. Peter, E. Brunner, D. Nellen, B. Froesch, S. Chatterjee, M. Murone, S. Zullig, and K. Basler. 2002. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear β-catenin-TCF complex. Cell 109:47-60. [DOI] [PubMed] [Google Scholar]
- 16.Mao, B., W. Wu, Y. Li, D. Hoppe, P. Stannek, A. Glinka, and C. Niehrs. 2001. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature 411:321-325. [DOI] [PubMed] [Google Scholar]
- 17.Merriam, J. M., A. B. Rubenstein, and M. W. Klymkowsky. 1997. Cytoplasmically anchored plakoglobin induces a WNT-like phenotype in Xenopus. Dev. Biol. 185:67-81. [DOI] [PubMed] [Google Scholar]
- 18.Miller, J. R., and R. T. Moon. 1997. Analysis of the signaling activities of localization mutants of β-catenin during axis specification in Xenopus. J. Cell Biol. 139:229-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molenaar, M., M. van de Wetering, M. Oosterwegel, J. Peterson-Maduro, S. Godsave, V. Korinek, J. Roose, O. Destree, and H. Clevers. 1996. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell 86:391-399. [DOI] [PubMed] [Google Scholar]
- 20.Montross, W. T., H. Ji, and P. D. McCrea. 2000. A β-catenin/engrailed chimera selectively suppresses Wnt signaling. J. Cell Sci. 113(Pt. 10):1759-1770. [DOI] [PubMed] [Google Scholar]
- 21.Parker, D. S., J. Jemison, and K. M. Cadigan. 2002. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development 129:2565-2576. [DOI] [PubMed] [Google Scholar]
- 22.Polakis, P. 2000. Wnt signaling and cancer. Genes Dev. 14:1837-1851. [PubMed] [Google Scholar]
- 23.Roose, J., M. Molenaar, J. Peterson, J. Hurenkamp, H. Brantjes, P. Moerer, M. van de Wetering, O. Destree, and H. Clevers. 1998. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395:608-612. [DOI] [PubMed] [Google Scholar]
- 24.Schweizer, L., D. Nellen, and K. Basler. 2003. Requirement for Pangolin/dTCF in Drosophila Wingless signaling. Proc. Natl. Acad. Sci. USA 100:5846-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schweizer, L., and H. Varmus. 2003. Wnt/Wingless signaling through beta-catenin requires the function of both LRP/Arrow and frizzled classes of receptors. BMC Cell Biol. 4:4. [DOI] [PMC free article] [PubMed]
- 26.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 27.Tago, K., T. Nakamura, M. Nishita, J. Hyodo, S. Nagai, Y. Murata, S. Adachi, S. Ohwada, Y. Morishita, H. Shibuya, and T. Akiyama. 2000. Inhibition of Wnt signaling by ICAT, a novel β-catenin-interacting protein. Genes Dev. 14:1741-1749. [PMC free article] [PubMed] [Google Scholar]
- 28.Takemaru, K. I., and R. T. Moon. 2000. The transcriptional coactivator CBP interacts with β-catenin to activate gene expression. J. Cell Biol. 149:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takemaru, K. I., S. Yamaguchi, Y. S. Lee, Y. Zhang, R. W. Carthew, and R. T. Moon. 2003. Chibby, a nuclear β-catenin-associated antagonist of the Wnt/Wingless pathway. Nature 422:905-909. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, B., F. Townsley, R. Rosin-Arbesfeld, H. Musisi, and M. Bienz. 2002. A new nuclear component of the Wnt signalling pathway. Nat. Cell Biol. 4:367-373. [DOI] [PubMed] [Google Scholar]
- 31.Tolwinski, N. S., and E. Wieschaus. 2001. Armadillo nuclear import is regulated by cytoplasmic anchor Axin and nuclear anchor dTCF/Pan. Development 128:2107-2117. [DOI] [PubMed] [Google Scholar]
- 32.van de Wetering, M., R. Cavallo, D. Dooijes, M. van Beest, J. van Es, J. Loureiro, A. Ypma, D. Hursh, T. Jones, A. Bejsovec, M. Peifer, M. Mortin, and H. Clevers. 1997. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88:789-799. [DOI] [PubMed] [Google Scholar]
- 33.van de Wetering, M., E. Sancho, C. Verweij, W. de Lau, I. Oving, A. Hurlstone, K. van der Horn, E. Batlle, D. Coudreuse, A. P. Haramis, M. Tjon-Pon-Fong, P. Moerer, M. van den Born, G. Soete, S. Pals, M. Eilers, R. Medema, and H. Clevers. 2002. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111:241-250. [DOI] [PubMed] [Google Scholar]
- 34.von Kries, J. P., G. Winbeck, C. Asbrand, T. Schwarz-Romond, N. Sochnikova, A. Dell'Oro, J. Behrens, and W. Birchmeier. 2000. Hot spots in beta-catenin for interactions with LEF-1, conductin, and APC. Nat. Struct. Biol. 7:800-807. [DOI] [PubMed] [Google Scholar]
- 35.Wang, A. H., N. R. Bertos, M. Vezmar, N. Pelletier, M. Crosato, H. H. Heng, J. Th'ng, J. Han, and X. J. Yang. 1999. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol. 19:7816-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams, B. O., G. D. Barish, M. W. Klymkowsky, and H. E. Varmus. 2000. A comparative evaluation of β-catenin and plakoglobin signaling activity. Oncogene 19:5720-5728. [DOI] [PubMed] [Google Scholar]
- 37.Wodarz, A., and R. Nusse. 1998. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 14:59-88. [DOI] [PubMed] [Google Scholar]
- 38.Zecca, M., K. Basler, and G. Struhl. 1996. Direct and long-range action of a wingless morphogen gradient. Cell 87:833-844. [DOI] [PubMed] [Google Scholar]