Abstract
The eukaryotic genome is packaged into distinct domains of transcriptionally active euchromatin and silent heterochromatin. A hallmark of mammalian heterochromatin is CpG methylation. Lsh, a member of the SNF2 family, is a major regulator of DNA methylation in mice and thus crucial for normal heterochromatin formation. In order to define the molecular function of Lsh, we examined its cellular localization and its association with chromatin. Our studies demonstrate that Lsh is an exclusively nuclear protein, and we define a nuclear localization domain within the N-terminal portion of Lsh. Lsh strongly associates with chromatin and requires the internal and C-terminal regions for this interaction. Lsh accumulates at pericentromeric heterochromatin, suggesting a direct role for Lsh in the methylation of centromeric DNA sequences and the formation of heterochromatin. In search of a signal that is responsible for Lsh recruitment to pericentromeric heterochromatin, we found that histone tail modifications were critical. Prolonged treatment with histone deacetylase inhibitors has been reported to disrupt higher-order heterochromatin organization, and this was accompanied by dissociation of Lsh from pericentromeric heterochromatin. These results are consistent with a model in which Lsh is recruited by intact heterochromatin structure and then assists in maintaining heterochromatin organization by establishing CpG methylation patterns.
The chromatin of eukaryotic cells is organized into two major types: euchromatin and heterochromatin (6, 39). Euchromatin contains the majority of single-copy genes, replicates during early S phase, and contains acetylated histones. Heterochromatin is composed of long stretches of DNA repeats, usually replicates in late S phase, and contains underacetylated histones. A specific methylation mark in heterochromatin in histone 3 at lysine 9 serves as a recognition signal for binding of the heterochromatin protein 1 (HP1) (26). Heterochromatin regulates many diverse nuclear functions, including gene silencing, normal centromere function and nuclear organization.
Another hallmark of mammalian heterochromatin is DNA methylation (5-7, 9, 34, 39). The addition of methyl groups to cytosines within the CpG dinucleotide by DNA methyltransferases is involved in regulating transcription, maintaining genome stability, imprinting, and X chromosome inactivation. In mammalian cells, DNA methylation is catalyzed by two important classes of DNA methyltransferases (5). DNA methyltransferase 1 (Dnmt1) resides at the replication fork and methylates CpG dinucleotides in the newly synthesized daughter strand. The function of Dnmt1 is thought to be essential for maintaining DNA methylation patterns in proliferating cells (30). There are two members of the second class of methyltransferases, Dnmt3a and Dnmt3b, which are required for the initiation of de novo methylation during embryonic development (37).
Although DNA methylation and heterochromatin are closely associated, their precise relationship has not been elucidated. Recently, several members of the SNF2 family of chromatin remodeling enzymes have been found to regulate DNA methylation, suggesting that chromatin structure influences DNA methylation pattern (34).
SNF2 family members are marked by the presence of seven conserved “helicase” motifs involved in ATP binding and hydrolysis (13, 14). Many ATP-dependent chromatin remodeling complexes have been identified, such as SWI/SNF/BRM, ISWI, or Mi-2/NuRD, and each chromatin remodeling complex contains at least one member of the SNF2 superfamily. Chromatin remodeling complexes disrupt histone-DNA interactions and allows for the physical movement, or sliding, of nucleosomes on the DNA utilizing the energy derived from ATP hydrolysis (3). Thus, they alter the accessibility of chromatin to various proteins that regulate DNA-based processes such as DNA replication, transcription, and repair. DDM1, ATRX, and Lsh all belong to the SNF2 superfamily and have been demonstrated to regulate DNA methylation (12, 20, 25). Recombinant DDM1 has been recently demonstrated to induce nucleosome repositioning on a short DNA fragment, thus confirming its membership in the SNF2 family of chromatin remodeling proteins (8). However, neither DDM1, ATR-X, nor Lsh have been reported as yet to interact with large protein complexes, a characteristic of other SNF2 family members.
The DDM1 gene (named for decrease in DNA methylation) deficiency causes a 70% reduction of genomic DNA methylation in the plant Arabidopsis thaliana (27). The loss of DNA methylation is most pronounced at repetitive sequences, followed by hypomethylation at single-copy genes. ATRX deficiency gives rise to changes in the pattern of methylation of several highly repeated sequences, including ribosomal DNA arrays, a Y-specific satellite sequence, and subtelomeric repeats. Mutations in the human ATRX gene result in the inherited disease α-thalassemia (19, 20).
Lymphoid-specific helicase (Lsh) is another member of the SNF2 family of chromatin remodeling proteins (17, 24, 28). Lsh is a nonredundant member of the SNF2 family and is essential for normal murine development and survival. Targeted deletion of Lsh results in perinatal lethality, with abnormal lymphoid development and renal defects (16, 18). DNA derived from Lsh−/− tissue shows a substantial loss of CpG methylaton (12). This suggests that Lsh is an important factor in determining DNA methylation and thus crucial for heterochromatin organization in mammals. However, it is not known whether Lsh controls methylation directly or indirectly, for example, via induction of other enzymes involved in DNA methylation. We have previously reported that Lsh does not influence methyltransferase activity or Dnmt1, Dnmt3a, or Dnmt3b protein levels, suggesting a more direct role of Lsh in establishing or maintaining methylation (12). Here we sought to examine the cellular localization of Lsh and its association with chromatin. We report a close association of Lsh with pericentromeric heterochromatin, suggesting that Lsh via CpG methylation directly participates in the organization of heterochromatin. Disruption of higher-order heterochromatin structure by prolonged exposure to the histone acetylase inhibitor trichostatin A (TSA) is followed by delocalization of Lsh. These results are consistent with a model in which the histone tail modifications that shape higher-order heterochromatin structure provide a signal for Lsh recruitment that in turn is crucial for establishing methylation patterns and the formation of heterochromatin.
MATERIALS AND METHODS
Tissue culture.
For analysis of Lsh expression fetal murine thymus was used (C57BL/6J strain; Jackson Laboratory). PD31, a pre-B-cell line, and EL4, a T-cell line (American Type Culture Collection), were cultured in RPMI medium 1640 (Gibco) supplemented with 2 mM glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (all from Gibco) and 10% fetal calf serum (HyClone). Embryos from crosses between Lsh+/− mice were removed at day 13.5 of gestation and genotyped by PCR as described before (18). Preparation of primary mouse embryonal fibroblast (MEF) cells are described elsewhere (15), and MEFs were grown in Dulbecco modified Eagle medium (Gibco) supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, and antibiotics (Gibco). NIH 3T3 cells (American Type Culture Collection) were maintained in Dulbecco modified Eagle medium with 10% bovine calf serum, and Lsh-expressing GeneSwitch-3T3 cells (Invitrogen) were cultured in the same medium supplemented with 50 μg of hygromycin/ml to select for hygromycin resistance. Cells in exponential growth were treated in the presence of TSA (Sigma) as described previously (31, 42); the drug and medium were renewed regularly. NIH 3T3 cells were incubated at 75 ng of TSA/ml.
Vector construction.
Based on the mouse Lsh cDNA (U25691) sequence, three primers were designed. The first, a 5′ primer designated LSHN01 (5′-AGAGATGGCCGAACAAACGGAG-3′), encoded the N-terminal of LSH, including the initiation codon. The second, a 3′ primer designated LSHFC1 (5′-CCGGCTAGCTCACTTGTCATCGTCGTCCTTGTAGTCAAATAAACATTCAGCACTGG-3′), encoded the C-terminal sequence of the Lsh peptide, followed by a DNA sequence encoding the eight-amino-acid FLAG tag (DYKDDDDK) and a stop codon. The third was another 3′ primer, LSHFC2 (5′-CCTTGTCATCGTCGTCCTTGAGTCAAATAAACATTCAGCACTGG-3′), that contained a C-terminal sequence for the Lsh protein followed by a DNA sequence encoding the Flag tag without a stop codon. The complete open reading frame of the full-length mouse Lsh cDNA was amplified by reverse transcription-PCR with total RNA purified from PD31 cells as a template and the primer pair LSHN01 and LSHFC1. A 2.5-kb portion of PCR-amplified product was subcloned directly into pCRBLUNT (Invitrogen), forming the vector pCRLSHF1. By using the pCRLSHF1 as a template and two primers, LSHN01 and LSHFC2, the PCR-amplified product was subcloned into pCRBLUNT (Invitrogen), and the new vector was termed pCRLSHF2. The plasmid pCRLSHF1 was cut with EcoRI, and the LSH DNA fragment was subcloned into the EcoRI site of pGene/V5-his (Invitrogen), designating the vector pGeneLSHF. The same EcoRI DNA fragment was subcloned into the EcoRI site of pEGFP-C1 (Clontech), generating the vector pEGFPLSH that encoded the EGFP-tagged Lsh at the C terminus. The pEGFPLSH was partially digested with EcoRI, MunI, and BamHI, respectively. The appropriate DNA fragments were recollected and self-ligated. The serial deletion mutants of Lsh were generated as follows. After EcoRI digestion, the deletion mutant, Δ1-176, did not contain the DNA sequence encoding the 1 to 176 of amino acids of Lsh. The vector generated after MunI digestion had the DNA fragment deleted that encoded amino acids 244 to 469 of Lsh and was thus designated Δ244-469. The vector generated after BamHI digestion did not encode amino acids 475 to 768 of Lsh and was therefore designated Δ475-768. The plasmid pCRLSHF2 was digested with EcoRI and cloned into the EcoRI site of pEGFP-N1 (Clontech), producing the vector pLSHEGFP that encoded the enhanced green fluorescent protein (EGFP)-tagged Lsh at the N terminus. The pLSHEGFP vector was digested with XhoI, and the appropriate DNA fragments were collected and self-ligated. The deletion vector, termed Δ1-31, generated by XhoI digestion did not encode amino acids 1 to 31 of Lsh.
Stable and transient transfections.
The GeneSwitch system (Invitrogen), a mifepristone (Invitrogen)-regulated expression system for mammalian cells, was chosen to produce a cell line with inducible expression of the Lsh gene. The GeneSwitch 3T3 cells express the GeneSwitch regulatory protein from the pSwitch vector. GeneSwitch 3T3 cells were seeded at a concentration of 106 in 10-cm dishes overnight before transfection. Cells were transfected with 2.5 μg of Lsh-inducible vector pGeneLSHF or empty vector pGene/V5-his as a control, by using FuGENE 6 transfection reagent (Roche) according to the manufacturer's recommendations. Stably transfected cells were selected with 500 to 1,000 μg of Zeocin (Invitrogen)/ml. Zeocin-resistant cells were induced with 100 pM mifepristone for induction of the Lsh protein. For EGFP expression NIH 3T3 cells were seeded at a concentration of 1 × 105 to 2 × 105 cells per well in a six-well plate (Costar cluster multiple-well plates) for 15 h before transfection. Cells were transfected with 2 to 4 μg of EGFP-tagged Lsh vector or control vector by using SuperFect transfection reagent (Qiagen) or FuGENE 6 transfection reagent (Roche) according to manufacturers' recommendations. Transiently transfected cells were cultured overnight, and Lsh-GFP expression was observed by fluorescence microcopy or Lsh expression was examined by Western analysis.
Cell cycle analysis.
For cell cycle synchronization, EL4 cells were seeded at ca. 105/ml in a total volume of 10 ml per 25-cm2 flask, with l-mimosine (400 μM; Sigma), aphidicolin (250 ng/ml; Sigma), and nocodazole (400 ng/ml; Sigma). The treated cells were incubated for 24 h in the presence of the blocking reagents, and the drugs were removed for flow cytometric analysis or Western analysis. For cell cycle synchronization, NIH 3T3 cells were incubated at 3 μg of aphidicolin/ml for 18 h and then released into aphidicolin-free medium (10). For cell cycle analysis, cell suspensions were washed twice in phosphate-buffered saline (PBS) and fixed with 70% of ethanol at −20°C for at least 2 h. Cells were washed once with PBS and then incubated in 25 μg of propidium iodide (Sigma)/ml with 100 μg of RNase A/ml in PBS at 37°C for 30 min. Flow cytometric analysis was performed on a Coulter EPICS XL-MCL cytometer. The data were collected and analyzed by using the Coulter System II software provided by the manufacturer.
Indirect immunofluorescence studies.
MEFs were seeded on glass coverslips to 50 to 60% confluency. Cells were fixed in 2% paraformaldehyde solution and permeabilized with 0.5% Triton X-100 (Triton X) solution. Coverslips were blocked with 5% goat serum for 30 min at room temperature and then incubated with primary antibodies in a humidifying chamber at room temperature for 1 h or at 4°C overnight. After the coverslips were washed five times, they were incubated with Cy5 and fluorescein isothiocyanate-conjugated secondary antibodies (Jackson Immunoresearch) in PBS with 1.0 mM MgCl2 at room temperature for 1 h. After an extensive washing, coverslips were mounted onto glass microscope slides by using mounting reagent (Molecular Probes). To immunostain newly synthesized DNA and related proteins, cell staining was performed as previously described (29, 40). The following primary antibodies were used. Dnmt1 was kindly provided by T. Bestor (Department of Genetics and Development, Columbia University, New York, N.Y.) as published before (29), PCNA (PC10; 05-347) was purchased from Upstate Biotechnology, and M2 (anti-Flag) was purchased from Sigma, monoclonal bromodeoxyuridine (BrdU) antibody was from Pharmingen (catalog no. 347580) or Roche (catalog no. 1296736); HP1α (1H5; MAb3584) was purchased from Chemicon, and branched anti-histone 3 lysine 9 methylation antiserum was kindly provided by T. Jenuwein (Research Institute of Molecular Pathology, Vienna, Austria). The cells were observed under a fluorescence microscope (Zeiss) or by three-dimensional confocal microscopy (LSM 510 confocal microscope; Zeiss).
Western analysis and protein fractionation.
Protein extracts were separated on NuPAGE Bis-Tris Pre-Cast gels (for low- to proteins), or NuPAGE Tris-Acetate gels (for high-molecular-weight proteins) by electrophoresis and transferred onto nitrocellulose. Each lane contained 10 μg of protein extracts, with the exception of the cell cycle experiments or fractionation experiments, in which proteins were extracted from equal amounts of cells (i.e., 105). After a blocking step, the membrane was incubated with primary antibodies to distinct proteins, followed by incubation with secondary horseradish peroxidase-coupled antibodies (Amersham Pharmacia). Detection of horseradish peroxidase was performed by using the enhanced chemiluminescence Western blotting detection reagents according to the manufacturer's instructions, followed by visualization on Hyperfilm ECL chemiluminescent film. The following antibodies were used for detection of proteins as the primary antibodies. Lsh antiserum was used as described previously (18). Brg-1 (sc-8749), HDAC1 (sc-7872), MBD1 (sc-10751), CBP (sc-369), and p300 (sc-584) were purchased from Santa Cruz Biotechnology. M2 (anti-Flag; F3165) and Vimentin (LN-6; V2258) were purchased from Sigma. MeCP2 (07-013), PCNA (PC10; 05-347), histone H3 (05-499), acetylated histone 3 (06-599), histone4 (07-108), and acetylated histone 4 (06-598), and dimethyl-histone H3 lysine 9 (MeK9; 07-212) were purchased from Upstate Biotechnology. HP1α (2G9; MAB3446) was purchased from Chemicon, EFGP (JL-8; 8873) was from Clontech, and DNMT3a (64B1446; IMG-268) was from Imgenex. Cell extraction with Triton X, chromatin fractionation, and nuclear matrix isolation were modified as described previously (22, 38). After two washes in PBS, cells were incubated and extracted for 3 min at 4°C in cytoskeleton buffer (CSK; 10 mM PIPES [pH 6.8], 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA) supplemented with leupeptin, aprotinin, and pepstatin (1 μg/ml each), 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and 0.5% (vol/vol) Triton X. The nuclear skeleton that was resistant to Triton X was separated from soluble proteins by centrifugation at 5,000 × g for 3 min. Chromatin proteins were solubilized by DNA digestion with 1 mg of RNase-free DNase I/ml in CSK buffer (with adjusted 50 mM NaCl) in the presence of proteinase inhibitors for 15 min at room temperature, and the supernatant, containing chromatin proteins, was collected by centrifugation. The pellet was washed with 0.25 M ammonium sulfate for 5 min at 4°C and pelleted again. The second pellet was washed with 2 M NaCl in CSK buffer for 5 min at 4°C and then centrifuged. The nuclear matrix was then solubilized in 8 M urea buffer.
RESULTS
Nuclear localization domain of Lsh.
Since Lsh belongs to the SNF2 family of chromatin remodeling proteins and Lsh has been demonstrated to be a major modulator of CpG methylation, Lsh is predicted to have a nuclear localization. By comparing nuclear and cytoplasmic extracts derived from different tissues by Western analysis, we determined that Lsh was only detectable in the nuclear fraction and not in the cytoplasmic preparation (Fig. 1A). Lsh was found to be highly expressed in cells of the lymphoid lineage such as the pre-B-cell line PD31 and embryonic thymocytes but minimally expressed by MEFs (Fig. 1A) and barely detectable in NIH 3T3 fibroblasts (data not shown). As expected, MEFs derived from mice with a targeted deletion of Lsh showed no detectable signal when probed with a specific antiserum raised against the C terminus of Lsh (Fig. 1A). The predominantly nuclear expression of Lsh was further confirmed in 3T3 fibroblasts, when Lsh was tagged with the Flag peptide and the expression was under the control of a mifepristone response element. In this system Lsh was expressed >10-fold above endogenous Lsh levels (Fig. 1B). Furthermore, Lsh localized to the nucleus throughout all stages of the cell cycle (Fig. 1C) even during the S phase, when endogenous Lsh expression was highest. Thus, the nuclear expression profile of Lsh in all cell types examined corresponded well with Lsh's presumed chromatin remodeling activity.
FIG. 1.
Nuclear localization of Lsh. (A) Lsh is localized in nuclear extracts. For analysis of Lsh expression the pre-B-cell line PD31, fetal thymus and embryonal fibroblasts (MEFs) were examined by Western analysis and probed with a specific antibody raised against the C-terminal portion of Lsh. Lanes: C, cytoplasmic extract; N, nuclear extract (equal loading of protein extracts). (B) Lsh is localized in overexpressed nuclear extracts. Lsh was induced in 3T3 cells in the GeneSwitch system and examined by Western analysis. Lanes: C, cytoplasmic extract; N, nuclear extract (equal loading of protein extracts). (C) Lsh is preferentially expressed during S phase. The T-cell line EL4 was synchronized with mimosine (M), aphidicolin (A), or nocodazole (N) or left untreated (Ctrl) and then examined by Western analysis for the presence of Lsh or β-actin. Lanes: C, cytoplasmic extract; N, nuclear extract (extracts were prepared from equal number of cells). The lower panel shows the result of the cell cycle analysis and the distribution of cells at different stages of the cell cycle.
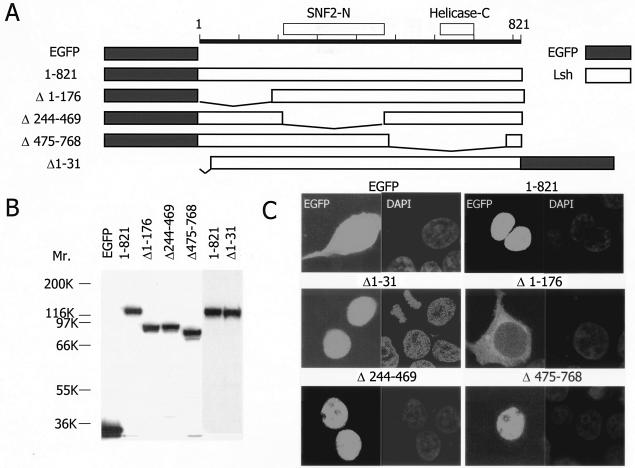
In order to confirm the exclusive nuclear localization and to define the motif within the Lsh protein that is responsible for targeting of Lsh to the nucleus, 3T3 cells were transiently transfected with Lsh expression vectors. A series of LSH deletion mutants were constructed and fused to GFP (Fig. 2A). As shown by Western analysis in Fig. 2B, all constructs resulted in expression of the Lsh protein with the expected molecular size. Expression of wild-type Lsh (amino acids 1 to 821) resulted in an exclusive nuclear expression pattern as shown by fluorescence microscopy, a finding which supported the previous Western analysis of nuclear extracts (Fig. 2C). The position of GFP toward Lsh, either N terminal or C terminal, did not influence the proper nuclear localization (data not shown). In contrast, expression of the GFP protein alone without Lsh revealed a diffuse cytoplasmic distribution (Fig. 2C). In order to determine the nuclear localization domain, mutant Lsh vectors were transiently transfected. Fluorescence analysis revealed that the C-terminal portion of Lsh (amino acids 574 to 768), as well as the internal deletion mutant (amino acids 244 to 469), were dispensable for nuclear localization of Lsh. In contrast the N-terminal deletion (amino acids 1 to 176) completely abolished targeting of Lsh to the nucleus. Since the region from amino acids 1 to 31 was not critical for the nuclear localization of Lsh, these results suggest that the nuclear localization domain of murine Lsh is located between amino acids 31 and 176, a region containing several lysine-rich stretches.
FIG. 2.
Nuclear localization domain of Lsh. (A) Construction map of Lsh deletional mutants. (B) Expression of deletion mutants. Nuclear extracts were examined 24 h after transfection by Western analysis with specific antiserum raised against the GFP peptide (equal loading of protein extracts). (C) The nuclear localization domain is N terminal. At 24 h after transfection with indicated plasmids, 3T3 cells were examined under a fluorescence microscope.
Association of Lsh with Triton X-resistant fractions.
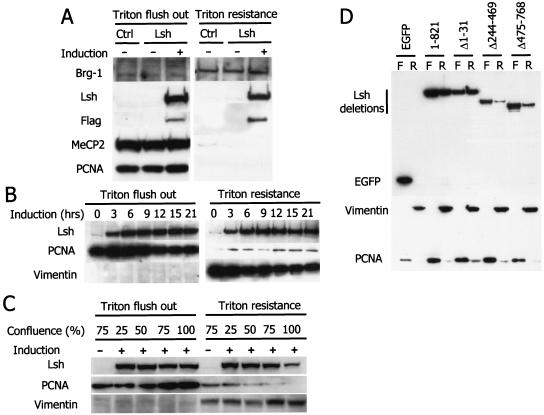
In order to determine the nuclear function of Lsh, we further investigated the nuclear compartment of Lsh localization. We first examined whether Lsh was present in a soluble and/or extractable form (as many DNA-binding factors are) or whether it was closely bound to nuclear components, as has been reported for other SNF2 family members (4, 36, 38, 46). Lsh was induced as a Flag-tagged fusion protein in 3T3 cells (Fig. 1B), and the nuclei were then washed with high concentrations of Triton X detergent (Fig. 3A). Western analysis of flushed material and the Triton X-resistant fraction demonstrated that as much as about half of the induced Lsh was found to be Triton resistant, indicating a close association of Lsh with chromatin or the nuclear matrix. By comparison, most transcription factors or nuclear proteins, such as MECP2 and PCNA, can be efficiently extracted by Triton X washes, whereas chromatin-associated proteins such as BRG1 and hBRM, are fairly Triton X resistant (38, 46) (Fig. 3A). In order to determine whether overexpression of Lsh “saturates” potential chromatin-binding sites and thus is responsible for the extractable form of Lsh, we performed a time kinetic experiment to induce low amounts of Lsh. However, even at 0 and 3 h of induction, when low levels of Lsh were detectable, the relationship of free and bound Lsh was unaltered, and the ratio of bound to free Lsh remained at ca. 50:50 (Fig. 3B). This suggested that both forms may play a physiological role and are not simply a by-product of overexpression. Since Lsh expression peaked during S phase (Fig. 1C), we tested whether the resistance of Lsh to Triton X washes is correlated with the replication phase. At confluency, when 3T3 cells are arrested at G1, Lsh was found to be predominantly Triton X extractable, whereas during the growth phase the ratio of free to bound Lsh shifted back to 50:50 (Fig. 3C). As a control, Vimentin, a nuclear scaffolding protein, was examined that was contained within the Triton X-resistant fraction throughout the cell cycle. In contrast, PCNA (proliferation cellular nuclear antigen) shifted slightly within the Triton X-resistant fraction from chromatin association at 25% confluency to dissociation at 100% confluency. The shift in Lsh's Triton X resistance during cell cycle progression suggests that the ability of Lsh to bind to chromatin is best in proliferating cells.
FIG. 3.
Lsh is associated with chromatin. (A) Lsh is present in the Triton X-resistant fraction. After Lsh induction in 3T3 cells that were transfected with the Lsh expression vector (Lsh) or transfected with the empty control vector (Ctrl) cells were extracted with Triton X-100, and the resistant and soluble fractions were examined by Western analysis for the presence of Lsh with antibodies against the C-terminal portion of Lsh (Lsh), the Flag epitope (Flag), Brg-1, MeCP2, and PCNA (extracts were prepared from equal number of cells). (B) Kinetics of Lsh expression. At different time points after Lsh induction 3T3 cells were extracted with Triton X, and the different fractions were examined by Western analysis for the presence of Lsh, PCNA, or Vimentin (extracts were prepared from equal number of cells). (C) Association with Triton X-resistant fraction is dependent on cell cycle. 3T3 cells at different stages of confluency were extracted with Triton X, and the different fractions were examined by Western analysis for the presence of Lsh or PCNA and Vimentin serving as controls (extracts were prepared from equal number of cells).(D) The C-terminal and internal domain are required for Triton X resistance. After transfection with the indicated deletion mutants, Triton X-resistant (lanes R) and soluble (Triton flush out [lanes F]) fractions were examined by Western analysis for the presence of GFP, Vimentin, and PCNA (extracts were prepared from equal number of cells).
In order to define which domains of the Lsh protein are responsible for Triton X resistance, we transfected GFP-tagged Lsh mutants into 3T3 cells and extracted them with Triton X. A deletion of 31 amino acids at the N-terminal portion of Lsh did not affect the ratio of free to bound Lsh and was similar to that of wild-type Lsh (Fig. 3D). However, deletion of the internal region comprising amino acids 244 to 469, as well as deletion of the C-terminal located portion (amino acids 574 to 768), shifted the distribution of Lsh toward the free form. This suggests that the entire Lsh protein downstream of amino acid 244 is critical for efficient chromatin binding. In addition, these domains may contain protein interaction or protein folding functions.
Lsh localizes to pericentromeric heterochromatin.
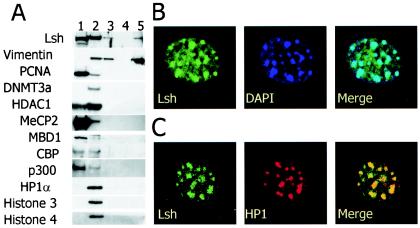
In order to examine to which nuclear structure Lsh is recruited, the Triton X-resistant fractions were further eluted by digestion with DNase I and high-salt washes. Under these conditions chromatin-bound fractions are separated from the nuclear matrix (22). Vimentin, for example, a component of the nuclear matrix scaffold, resists elution of high-salt concentrations (Fig. 4A). DNA-binding proteins (MecP2 and MBD1) or transcriptional regulators (p300) or the replication factor PCNA are only loosely associated with the nuclear components and are readily eluted by detergent. In contrast, about half of Lsh resists Triton X washes, and most of this fraction is detectable in the “chromatin” fraction which also contains histones, HP1α and Dnmt3a (Fig. 4A) (10). This suggests that the bound Lsh protein primarily associates with chromatin, as opposed to binding to the nuclear scaffold (for comparison, see lanes 2 and 5 in Fig. 4A). However, a small portion of induced Lsh appears in the scaffolding fraction and, at present, it cannot be determined whether this is of physiologic relevance or an artifact due to overexpression of Lsh.
FIG. 4.
Lsh localizes to pericentromeric heterochromatin. (A) Lsh predominantly associates with chromatin. 3T3 cells were extracted with Triton X, and different fractions examined by Western analysis with the indicated antibodies: lane 1, flushed fraction; lane 2, Triton X-resistant fraction after DNase digestion; lane 3, ammonium sulfate wash; lane 4, 2 M NaCl wash; lane 5, solubilized pellet. (B) Lsh colocalizes with DAPI. At 24 h after transfection, GFP-tagged Lsh was examined by fluorescence microscopy. (C) Lsh colocalizes with HP1α. At 24 h after transfection, GFP-tagged Lsh was immunostained for detection of HP1α.
To determine whether Lsh binds to euchromatin or heterochromatin, we transiently expressed Lsh as a GFP-tagged fusion protein. As revealed by confocal immunofluorescence analysis, Lsh colocalized with DAPI (4′,6′-diamidino-2-phenylindole) spots that are indicative for pericentromeric heterochromatin in murine cells (Fig. 4B). Pericentromeric heterochromatin DNA consists primarily of minor and major satellite sequences and large blocks of repetitive sequences and presents the bulk of heterochromatin visualized by immunefluorescence analysis. Pericentromeric heterochromatin is thought to contribute to normal centromere functions during mitosis such as spindle formation and chromosome segregation. The colocalization of Lsh with DAPI was confirmed in 3T3 cells that overexpressed Flag-tagged Lsh (data not shown). Furthermore, Lsh colocalized with HP1α, a major component of pericentromeric heterochromatin (Fig. 4C). In addition, we have demonstrated that chromatin immunoprecipitated with anti-Lsh/Flag antibodies in 3T3 cells showed a close association of Lsh with minor and major satellite sequences that are the main components of pericentromeric DNA (Q. Yan and K. Muegge, unpublished data). Thus, our data show that Lsh is localized at pericentromeric heterochromatin, and previous work indicated that Lsh is crucial for CpG methylation at pericentromeric DNA (12). Taken together, these results support the idea that Lsh plays a direct role in the formation of pericentromeric heterochromatin.
Lsh is not required for targeting of Dnmt1 to replication foci.
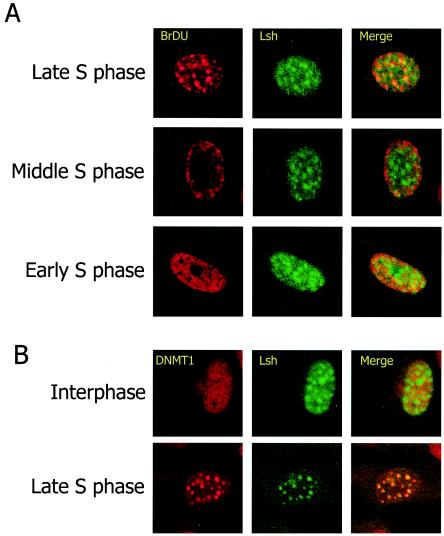
Lsh-deficient cells show a global methylation defect that is similar to the one reported in Dnmt1−/− embryos. Thus, we hypothysed that Lsh and its presumed chromatin remodeling activity maybe important for targeting Dnmt1 to sites of methylation. Dnmt1 has a significant preference for hemimethylated DNA relative to unmethylated DNA in vitro. Dnmt1 associates with sites of DNA synthesis, the replication foci, throughout the S phase but shows a diffuse nucleoplasmic distribution pattern during other phases of the cell cycle. The direct interaction with PCNA allows Dnmt1 to track along the newly synthesized strands and to ensure proper copying of the methylation “code.” Thus, Dnmt1 is thought to play a role in the maintenance of CpG methylation (5). Based on Lsh's nuclear expression pattern, we tested whether Lsh could directly participate in the maintenance of genomic methylation. Therefore, we first determined whether Lsh could associate with replication foci, the site of newly synthesized DNA. Lsh-induced 3T3 fibroblasts were synchronized with a high concentration of the cell cycle inhibitor aphidicolin. Cells were then released into normal cell cycle and at different time points pulsed with BrdU in order to detect newly synthesized DNA. Costaining for detection of incorporated BrdU, as well as Lsh, allowed the identification of different stages of the cell cycle. As shown in Fig. 5A, Lsh associated with replication foci only during late S phase, when few, large foci are visible by BrdU incorporation. Lsh did not colocalize with replication foci at early S phase, characterized by a fine punctate staining pattern, or mid-phase with a staining pattern of BrdU at the periphery of the nucleus and perinucleolar sites (29). Lsh also colocalized with Dnmt1 in late S phase (Fig. 5B). Thus, Lsh potentially may facilitate maintenance of methylation during the late phase of replication.
FIG. 5.
Lsh colocalizes with late replication foci. (A) Lsh colocalizes with late replication foci. Lsh was induced in 3T3 cells and immunostained with anti-Flag tag antibody and with anti-BrdU antibody for detection of incorporated BrdU. (B) Lsh colocalizes with Dnmt1. Lsh-induced 3T3 cells were immunostained with anti-Flag tag antibody and immunostained for detection of Dnmt1.
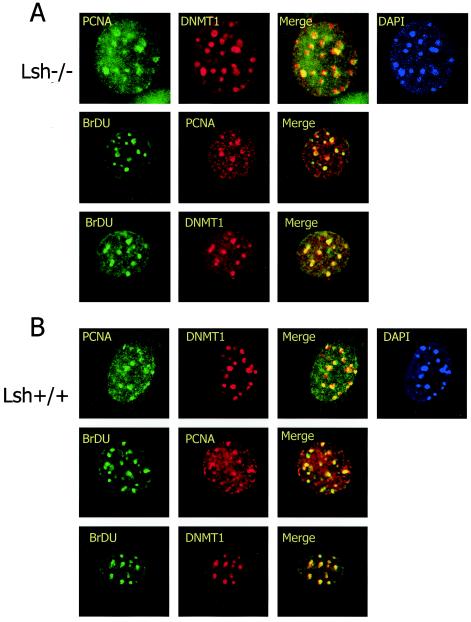
In order to test the possibility that Lsh is crucial for recruitment of Dnmt1 to late replication foci, MEFs derived from Lsh-deficient mice or wild-type controls were stained for BrdU incorporation and probed for PCNA or Dnmt1 to detect replication foci. As shown in Fig. 6A and B, PCNA, Dnmt1, and BrdU colocalized with each other in wild-type cultures, indicating the formation of normal replication foci. The same colocalization was detectable in Lsh−/− MEFs, suggesting that Lsh is dispensable for proper localization of Dnmt1 during late S phase. Thus, Lsh, although present at replication foci, does not alter targeting of Dnmt1 to replication foci.
FIG. 6.
Lsh is not required for targeting of Dnmt1 to replication foci. Lsh−/− (A) or Lsh+/+ (B) MEFs were immunostained for detection of PCNA, Dnmt1, and incorporated BrdU.
Delocalization of Lsh follows disruption of higher-order heterochromatin organization.
Next, we sought to determine the signal that is required to target Lsh to pericentromeric heterochromatin. Targeted deletion of Lsh in MEFs leads to hypomethylation at pericentromeric minor and major satellite sequences (12). Since Lsh is crucial for methylation at centromeric DNA, it may also require CpG methylation as a signal for targeting, and thus Lsh could participate in a self-reinforcing loop of DNA methylation. To test this idea, we sought to compare the ability of Lsh to associate with heterochromatin in Lsh wild-type and Lsh-deficient MEFs. When GFP-tagged Lsh was transiently expressed in Lsh−/− hypomethylated MEFs, Lsh still retained the ability to colocalize with DAPI (Fig. 7A). This suggests that Lsh does not require DNA methylation for heterochromatic localization.
FIG. 7.
Lsh dissociates from chromatin after TSA treatment. (A) Lsh associates with DAPI in hypomethylated MEFs. Lsh wild-type and Lsh−/− MEFs were transfected with a GFP-tagged Lsh expression vector and examined after 24 h. (B) TSA induces hyperacteylation. 3T3 cells were treated in the presence of 75 ng of TSA/ml, and extracts were examined by Western analysis with antibodies raised against histone 3 (H3), acetylated H3 (AcH3), or acetylated H4 (AcH4; equal loading of protein extracts). (C) Lsh dissociates from chromatin after TSA treatment and reassociates after recovery. After TSA treatment or after an additional 24 h of recovery, 3T3 cells were extracted with Triton X, and the different fractions were examined by Western analysis for the presence of induced Lsh; Vimentin served as a control. Triton X-resistant (lanes R) and soluble (Triton flush out [lanes F]) (extracts were prepared from equal number of cells) fractions are shown.
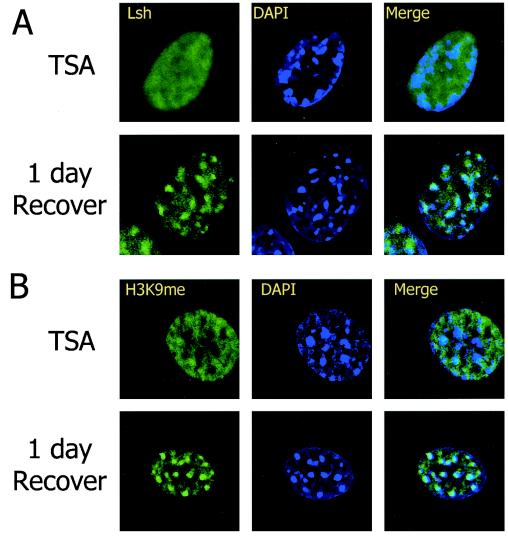
Many SNF2 family members have been reported to participate in large protein complexes, a hallmark of SNF/SWI chromatin remodeling complexes but, to date, no direct interactions of Lsh with other proteins have been found. During chromatin immunoprecipitation, Lsh can precipitate HP1α after cross-linking (Yan and Muegge, unpublished), suggesting a close association of Lsh with HP1 on heterochromatic nucleosomes. However, Lsh is not directly interacting with HP1α, since Flag-tagged Lsh cannot directly precipitate HP1α without cross-linking. In order to understand better the signal that recruits Lsh to pericentromeric heterochromatin, we examined whether Lsh requires higher-order heterochromatin organization for its localization. The pericentromeric heterochromatin structure depends on the presence of underacetylated histones. Prolonged treatment with the histone deacetylase inhibitor TSA results in disruption of pericentromric heterochromatin structure and centromere function (42). HP1α, which is recognized by methylated lysine 9 residue of histone 3, is no longer retained at centromeric heterochromatin. Also, the binding of the H3-K9me branched peptide antibody, recognizing a close configuration of methylated lysine residues at histone 3 tails, is disturbed (31, 42). To test whether Lsh's pericentric localization was dependent on higher-order heterochromatin organization, we treated 3T3 cells with TSA to increase global levels for histone 3 and 4 acetylation (Fig. 7B). After induction of hyperacetylation, 3T3 cells displayed an altered DAPI staining pattern with relocation of centromeric regions toward the nuclear periphery (Fig. 8), as has been previously reported (42). Similarly, staining with the branched anti-H3-K9 methylation antibody was disturbed, indicating a successful disruption of the heterochromatin structure (31) (Fig. 8B). Using anti-Flag antibody as a probe, indirect immunofluorescence staining revealed a diffuse staining pattern for Lsh and a failure of Lsh to associate with DAPI. This indicated that Lsh localization requires a higher-order heterochromatin structure (Fig. 8A). In order to confirm that Lsh binding to pericentromeric heterochromatin depends on a higher-order structure, we examined the Triton X resistance of the Lsh protein. As shown in Fig. 7C, almost all Lsh was extractable with Triton X, indicating a loss of the ability to bind to chromatin. Under the same conditions HP1α was flushed by Triton X and no longer retained by heterochromatin. To demonstrate that cells did not simply succumb to histone hyperacteylation after TSA treatment, cells were washed with PBS and cultured for 24 to 48 h to allow for recovery. The association of either Lsh (Fig. 7C) or HP1 (data not shown) with heterochromatin was reversible and returned to levels of untreated cells within a day, as judged by Western analysis. The colocalization of Lsh with DAPI could also be restored within a day of recovery (Fig. 8A). Similarly, the higher-order structure as indicated by staining with the branched H3-K9me antibody reversed after 1 day of recovery (Fig. 8B). Therefore, the signal that is required for recruitment of Lsh was not erased even after TSA treatment, and removal of the drug resulted in the relocalization of Lsh. These results suggest that binding of Lsh to pericentromeric heterochromatin is dependent on histone modifications and closely associated with normal higher-order structure of heterochromatin.
FIG. 8.
Lsh requires higher-order heterochromatin structure for localization. (A) Lsh dissociates from DAPI after TSA treatment and reassociates after recovery. After TSA treatment of 3T3 cells, Lsh was induced and cells were immunostained with anti-Flag antibody. (B) Dissociation of branched anti-H3-K9me from DAPI after TSA treatment and reassociation after recovery. The cells were also immunostained with the antibody raised against the branched H3-K9 methylated peptide.
In order to define whether Lsh plays a role in maintenance versus initiation of pericentromeric heterochromatin, we examined the time course of HP1 association in the absence of Lsh (Table 1). We have recently found that HP1α association with pericentromeric heterochromatin is not altered in the absence of Lsh, despite changes in histone methylation levels (45). In order to determine whether HP1 reassembly is delayed in the absence of Lsh, we disrupted HP1α association with pericentromeric chromatin by TSA treatment in wild-type and Lsh-deficient cells. Table 1 demonstrates that the dissociation of HP1 with DAPI occurs in ca. 77 to 85% of cells in both cultures. The recovery rate of HP1 localization in Lsh-deficient cells after 3, 6, or 12 h resembles that of wild-type cells, further supporting the idea that Lsh plays a role in maintaining rather than initiating heterochromatin formation, as further discussed below.
TABLE 1.
HP1 recovery after TSA treatmenta
| Time (h) | % HP1 recovery
|
|
|---|---|---|
| LSH+/+ cells | LSH−/− cells | |
| 0 | 25.2 | 33.0 |
| 3 | 58.5 | 62.5 |
| 6 | 71.9 | 74.5 |
| 12 | 78.9 | 86.9 |
MEFs derived from Lsh+/+ or Lsh−/− embryos were treated for 5 days with TSA (75 ng/ml), washed, and cultured for the indicated time periods. Cells were then stained with HP1α antibody and DAPI in order to monitor the recovery of higher-order chromatin structure in the absence of Lsh. The percentage indicates the percentage of cells in which HP1 colocalized with DAPI.
DISCUSSION
CpG methylation is a major hallmark in mammalian heterochromatin (5-7, 9, 34, 39). It plays an important role in gene suppression and is thought to control chromatin structure, for example, by modulating histone tail modifications. We have previously reported that Lsh is a crucial regulator of CpG methylation in mice (12). Furthermore, we have recently observed abnormal H3-K4 methylation patterns at heterochromatin in the absence of Lsh and reactivation of retroviral gene expression (45). Thus, Lsh modifies some (although not all, see below) characteristics of heterochromatin in mice. In the present study, we investigated the possibility that Lsh participates directly in the configuration of heterochromatin as opposed to indirectly inducing other protein factors. We demonstrated here a nuclear localization of Lsh, a strong association with chromatin, and a specific accumulation of Lsh at pericentromeric heterochromatin. These results provide an important link with previous results that demonstrated an effect of Lsh on heterochromatin structure and support a model in which Lsh is a direct participant with pericentromeric heterochromatin and plays an important role in normal heterochromatin formation.
Lsh deletion strongly affects DNA methylation at many other repetitive sequences that are not pericentromeric. The effect of DNA hypomethylation on single-copy genes is less pronounced (only five of nine genes show hypomethylation by Southern analysis [see also reference 12]), a phenomen also reported in DDM1 mutants. These pronounced Lsh effects on repetitive sequences are consistent with a model in which Lsh primarily guards heterochromatin at repetitive sites, and alterations of chromatin structure after Lsh deletion may then spread into single-copy genes.
Since Lsh-deficient cells have residual CpG methylation, functional homologues for Lsh may exist that can also localize to heterochromatin. ATRX, another member of the SNF2 chromatin remodeling family, has been reported to localize to pericentromeric heterochromatin (4, 33), and ATRX mutations cause DNA methylation defects in patients; however, these defects do not occur at pericentromeric DNA sequences but at other repetitive elements (20). Possibly, ATRX and Lsh may have redundant roles with respect to DNA methylation at heterochromatic sites such that Lsh substitutes for ATRX function. Another SNF2 family member, SNF2H, is associated with heterochromatin during late S phase of the cell cycle (11). SNF2H is thought to play a role at heterochromatin by enabling DNA replication through highly condensed regions during S phase. An effect on CpG methylation has not yet been demonstrated. DNA methyltransferases also localize to heterochromatic regions (1, 10, 32). Dnmt1, thought to be the major “maintenance” methyltransferase, colocalizes to heterochromatin during the late S phase of replication (29, 40). Since the global DNA methylation deficiency in Lsh−/− mutants is similar to the one reported for Dnmt1−/− mutants (12, 30), we hypothesized cooperative effects of Lsh with Dnmt1. Thus, we tested for the role of Lsh in targeting of Dnmt1 to replication foci. Although we did not see any difference in the composition of replication foci with or without Lsh, this does not exclude a possible role for Lsh in promoting the efficiency of Dnmt1 methylation. However, these questions have to be resolved in an in vitro system utilizing recombinant Lsh.
Heterochromatin has various properties, and it is a current challenge to understand the relationship and sequential requirements of distinct chromatin modifications and chromatin components (39). A major concern has been the interplay of histone modifications and CpG methylation. Several lines of evidence suggest that histone modifications provide a key signal for DNA methylation in lower organisms. For example, mutants of H3-K9 methyltransferase (such as dim-5 or KYP) lower cytosine methylation in Neurospora crassa or Arabidopsis thaliana (23, 43). Hyperacetylation after treatment with the histone deacetylase inhibitor TSA affects CpG methylation in Neurospora, a phenomenon not reported for mammalian species (42). On the other hand, there have been reports supporting the reverse relationship in which DNA methylation affects histone modifications. For example, mutants of met1 (a Dnmt1-like DNA methyltransferase in Arabidopsis) can reduce H3-K9 methylation (41). Although treatment with the demethylation agent 5-azacytidine does not alter HP1 binding or staining with the branched H3-K9me antibody (42, 45), it does alter H3-K4 methylation levels at heterochromatin (45). In human cancer cells, 5-azacytidine treatment or deletion of DNMT1 and DNMT3b can alter the H3-K9 methylation status at the specific promoter region of tumor suppressor genes, thus linking CpG methylation to chromatin modifications in mammals (2, 35). In the present study we observed that the reverse relationship also exists: disruption of higher-order heterochromatin structure by histone hyperacteylation correlated with dissociation of Lsh from pericentromeric heterochromatin. This suggests a model in which heterochromatin organization constitutes the signal for Lsh localization (and CpG methylation), and Lsh (and CpG methylation) plays a role in perpetuating heterochromatin formation rather than initiating it (7).
Pericentromeric heterochromatin plays an important role in spindle fiber attachment and sister chromatid cohesion during mitosis. The association of Lsh at pericentric regions and the effect on the DNA methylation of pericentric satellite sequences suggest a role in mitosis. We have recently found a severe growth defect in MEF derived from Lsh−/− embryos with signs of abnormal mitosis, including centrosome hyperamplification, abnormal spindle formation, and micronuclei formation (15). That treatment with the DNA-demethylating agent azacytidine mimicked the absence of Lsh (15) suggests that Lsh localization to pericentromeric heterochromatin is crucial to ensuring normal DNA methylation patterns that are required for normal mitosis.
At present, we do not know the precise heterochromatic signal that is required for Lsh localization. A three-dimensional configuration may be recognized by Lsh either directly or indirectly through other heterochromatic components. H3-K9 methylation or HP1 may contribute to Lsh recruitment, a hypothesis awaiting further testing. Recently, short RNA molecules have been proposed to be involved in the formation of heterochromatin structure, since deletion of RNA interference machinery inhibits heterochromatin structure in fission yeast (44). RNA interference leads to short double-stranded RNA that inhibits the accumulation of homologous transcripts. The process appears to be involved in the regulation of H3-K9 methylation and heterochromatic gene silencing (44). DNA repeats that allow bidirectional transcription may lead to the formation of double-stranded RNA and have been suggested as a nucleation center for heterochromatin (21). The pairing of RNA molecules with corresponding sequences may serve as target for Suv39h histone methyltransferases (26). After successful methylation of histone 3 at lysine 9, HP1 is recruited and contributes to establishment of a higher-order heterochromatin structure. This in turn may signal Lsh recruitment, which then facilitates CpG methylation. DNA methylation, occurring at repetitive sequences and at centromeric sites may then fortify heterochromatin structure, thus granting its stable propagation through multiple rounds of cell division. Thus, our results are consistent with a model in which Lsh, although important for heterochromatin formation, serves as a link in maintaining rather than initiating heterochromatin structure.
Acknowledgments
We are grateful to Joost Oppenheim, Zack Howard, and Doug Ferris for critical review of the manuscript. We thank Timothy Bestor and Thomas Jenuwein for the generous gifts of the Dnmt1 and the branched H3-K9me antiserum.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
This project was funded in whole or in part with federal funds from the National Cancer Institute and the Alternative Medicine Program, National Institutes of Health, under contract no. NO1-CO-12400.
REFERENCES
- 1.Bachman, K. E., M. R. Rountree, and S. B. Baylin. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276:32282-32287. [DOI] [PubMed] [Google Scholar]
- 2.Bachman, K. E., B. H. Park, I. Rhee, H. Rajagopalan, J. G. Herman, S. B. Baylin, K. W. Kinzler, and B. Vogelstein. 2003. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell 3:89-95. [DOI] [PubMed] [Google Scholar]
- 3.Becker, P. B., and W. Hörz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 4.Bérubé, N. G., C. A. Smeenk, and D. J. Picketts. 2000. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum. Mol. Genet. 9:539-547. [DOI] [PubMed] [Google Scholar]
- 5.Bestor, T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9:2395-2402. [DOI] [PubMed] [Google Scholar]
- 6.Bird, A. P., and A. P. Wolffe. 1999. Methylation-induced repression: belts, braces, and chromatin. Cell 99:451-454. [DOI] [PubMed] [Google Scholar]
- 7.Bird, A. P. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 8.Brzeski, J., and A. Jerzmanowski. 2003. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J. Biol. Chem. 278:823-828. [DOI] [PubMed] [Google Scholar]
- 9.Burgers, W. A., F. Fuks, and T. Kouzarides. 2002. DNA methyltransferases get connected to chromatin. Trends Genet. 18:275-277. [DOI] [PubMed] [Google Scholar]
- 10.Chen, T., Y. Ueda, S. Xie, and E. Li. 2002. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 277:38746-38754. [DOI] [PubMed] [Google Scholar]
- 11.Collins, N., R. A. Poot, I. Kukimoto, C. Garcia-Jimenez, G. Dellaire, and P. D. Varga-Weisz. 2002. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat. Genet. 32:627-632. [DOI] [PubMed] [Google Scholar]
- 12.Dennis, K., T. Fan, T. M. Geiman, Q. Yan, and K. Muegge. 2001. Lsh, a member of the SNF2 family, is required for genome wide methylation. Genes Dev. 15:2940-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisen, J. 1999. Proteins in the SNF2 family of proteins. [Online.] http://www.tigr.org/∼jeisen/SNF2/snf2.html.
- 14.Eisen, J. A., K. S. Sweder, and P. C. Hanawalt. 1995. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 14:2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan, T., Q. Yan, J. Huang, S. Austin, E. Cho, D. Ferris, and K. Muegge. 2002. Lsh-deficient murine embryonal fibroblasts show reduced proliferation with signs of abnormal mitosis. Cancer Res. 63:4677-4683. [PubMed]
- 16.Geiman, T. M., and K. Muegge. 2000. Lsh, an SNF2/helicase family member, is required for proliferation of mature T lymphocytes. Proc. Natl. Acad. Sci. USA 97:4772-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geiman, T. M., S. K. Durum, and K. Muegge. 1998. Characterisation of gene expression, genomic structure and chromosomal localisation of Hells (Lsh). Genomics 54:477-483. [DOI] [PubMed] [Google Scholar]
- 18.Geiman, T. M., L. Tessarollo, M. R. Anver, J. B. Kopp, J. M. Ward, and K. Muegge. 2001. Lsh, an SNF2 family member, is required for normal murine development. Biochim. Biophys. Acta 1526:211-220. [DOI] [PubMed] [Google Scholar]
- 19.Gibbons, R. J., D. J. Picketts, L. Villard, and D. R. Higgs. 1995. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome). Cell 80:837-845. [DOI] [PubMed] [Google Scholar]
- 20.Gibbons, R. J., T. L. McDowell, S. Raman, D. M. O'Rourke, D. Garrick, H. Ayyub, and D. R. Higgs. 2000. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat. Genet. 24:368-371. [DOI] [PubMed] [Google Scholar]
- 21.Hall, I. M., G. D. Shankaranarayana, K. Noma, N. Ayoub, A. Cohen, and S. I. Grewal. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232-2237. [DOI] [PubMed] [Google Scholar]
- 22.He, D., A. N. Nickerson, and S. Penman. 1990. Core filaments of the nuclear matrix. J. Cell Biol. 110:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson, J. P., A. M. Lindroth, X. Cao, and S. E. Jacobsen. 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416:556-560. [DOI] [PubMed] [Google Scholar]
- 24.Jarvis, C. D., T. Geiman, M. P. Vial-Storm, O. Osipovich, U. Akella, S. Candeias, I. Nathan, S. K. Durum, and K. Muegge. 1996. A novel putative helicase produced in early murine lymphocytes. Gene 169:203-207. [DOI] [PubMed] [Google Scholar]
- 25.Jeddeloh, J. A., T. L. Stokes, and E. J. Richards. 1999. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22:94-97. [DOI] [PubMed] [Google Scholar]
- 26.Jenuwein, T. 2002. An RNA-guided pathway for the epigenome. Science 297:2215-2218. [DOI] [PubMed] [Google Scholar]
- 27.Kakutani, T., J. A. Jeddeloh, and E. J. Richards. 1995. Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res. 23:130-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, D. W., K. Zhang, Z. Q. Ning, E. H. Raabe, S. Tintner, R. Wieland, B. J. Wilkins, J. M. Kim, R. I. Blough, and R. J. Arceci. 2000. Proliferation-associated SNF2-like gene (PASG): a SNF2 family member altered in leukemia. Cancer Res. 60:3612-3622. [PubMed] [Google Scholar]
- 29.Leonhardt, H., A. W. Page, H. U. Weier, and T. H. Bestor. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865-873. [DOI] [PubMed] [Google Scholar]
- 30.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 31.Maison, C., D. Bailly, A. H. Peters, J. P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329-334. [DOI] [PubMed] [Google Scholar]
- 32.Margot, J. B., M. C. Cardoso, and H. Leonhardt. 2001. Mammalian DNA methyltransferases show different subnuclear distributions. J. Cell Biochem. 83:373-379. [DOI] [PubMed] [Google Scholar]
- 33.McDowell, T. L., R. J. Gibbons, H. Sutherland, D. M. O'Rourke, W. A. Bickmore, A. Pombo, H. Turley, K. Gatter, D. J. Picketts, V. J. Buckle, L. Chapman, D. Rhodes, and D. R. Higgs. 1999. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc. Natl. Acad. Sci. USA 96:13983-13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meehan, R. R., S. Pennings, and I. Stancheva. 2001. Lashings of DNA methylation, forkfuls of chromatin remodeling. Genes Dev. 15:3231-3236. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen, C. T., D. J. Weisenberger, M. Velicescu, F. A. Gonzales, J. C. Lin, G. Liang, and P. A. Jones. 2002. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 62:6456-6461. [PubMed] [Google Scholar]
- 36.Nielsen, A. L., C. Sanchez, H. Ichinose, M. Cervino, T. Lerouge, P. Chambon, and R. Losson. 2002. Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1α. EMBO J. 21:5797-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 38.Reyes, J. C., C. Muchardt, and M. Yaniv. 1997. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol. 137:263-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richards, E. J., and S. C. Elgin. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108:489-500. [DOI] [PubMed] [Google Scholar]
- 40.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 41.Soppe, W. J., Z. Jasencakova, A. Houben, T. Kakutani, A. Meister, M. S. Huang, S. E. Jacobsen, I. Schubert, and P. F. Fransz. 2002. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21:6549-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taddei, A., C. Maison, D. Roche, and G. Almouzni. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3:114-120. [DOI] [PubMed] [Google Scholar]
- 43.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277-283. [DOI] [PubMed] [Google Scholar]
- 44.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 45.Yan, Q., J. Huang, T. Fan, H. Zhu, and K. Muegge. 2003. Lsh, a modulator of CpG methylation, is crucial for normal histone methylation. EMBO J. 22:5154-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao, K., W. Wang, O. J. Rando, Y. Xue, K. Swiderek, A. Kuo, and G. R. Crabtree. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95:625-636. [DOI] [PubMed] [Google Scholar]