Abstract
The gene encoding p53 mediates a major tumor suppression pathway that is frequently altered in human cancers. p53 function is kept at a low level during normal cell growth and is activated in response to various cellular stresses. The MDM2 oncoprotein plays a key role in negatively regulating p53 activity by either direct repression of p53 transactivation activity in the nucleus or promotion of p53 degradation in the cytoplasm. DNA damage and oncogenic insults, the two best-characterized p53-dependent checkpoint pathways, both activate p53 through inhibition of MDM2. Here we report that the human homologue of MDM2, HDM2, binds to ribosomal protein L11. L11 binds a central region in HDM2 that is distinct from the ARF binding site. We show that the functional consequence of L11-HDM2 association, like that with ARF, results in the prevention of HDM2-mediated p53 ubiquitination and degradation, subsequently restoring p53-mediated transactivation, accumulating p21 protein levels, and inducing a p53-dependent cell cycle arrest by canceling the inhibitory function of HDM2. Interference with ribosomal biogenesis by a low concentration of actinomycin D is associated with an increased L11-HDM2 interaction and subsequent p53 stabilization. We suggest that L11 functions as a negative regulator of HDM2 and that there might exist in vivo an L11-HDM2-p53 pathway for monitoring ribosomal integrity.
The p53 tumor suppressor gene mediates a major tumor suppression pathway in mammalian cells and is frequently altered in human tumors. The p53 protein induces cell cycle arrest or apoptosis in response to cellular stress by acting as a sequence-specific transcription factor. Various cellular insults, including oncogenic stimulation, DNA damage, nucleotide depletion, and hypoxia, trigger distinct signal transduction cascades, leading to p53 stabilization and activation. Inactivation of this p53-mediated checkpoint function may represent a necessary step for the development of most, if not all, tumors (17, 20, 30).
Accumulating evidence has identified the Mdm2 proto-oncogene as a major regulator of p53 (26, 29). Mdm2 was first cloned as an amplified gene on a murine double-minute chromosome (5) and was subsequently found to be amplified in a portion of human sarcomas (28) and brain tumors (3, 35). Both MDM2 and its human homolog, HDM2, can bind to and inhibit p53 function either by repressing p53's transcriptional activity in the nucleus (26, 29, 45) or by targeting p53 for degradation in the cytoplasm (10, 19; reviewed in reference 47). Deletion of the MDM2 gene in mice results in early embryonic lethality, which can be rescued by the simultaneous deletion of p53, supporting the notion that p53 is the major target of MDM2 during development (16, 22).
A number of cellular factors have been identified that directly bind to or modify MDM2, leading to MDM2 inhibition and p53 activation in response to various cellular stresses (34). The two best-characterized p53-mediated checkpoint pathways are the cellular responses to DNA damage (30) and oncogenic insults (39). Following DNA damage, several kinases, including ATM, Chk1, and Chk2, become activated and phosphorylate p53 and/or MDM2, reducing the binding of MDM2 and p53 (2, 40) and the inhibition of p53 nuclear export (49). Overexpression of various oncogenes, including ras (31), myc (52), E2F1 (1), and E1A (4), activates the transcription of the ARF tumor suppressor, which in turn binds to MDM2 (33, 42, 50), consequently activating p53 by blocking p53 and MDM2 nuclear export and p53 cytoplasmic degradation (43, 48). Oncogenic mutations targeting the components of either the DNA damage-kinase-MDM2-p53 or the oncogenic insult-ARF-MDM2-p53 pathway occur at high frequency in a wide range of human tumors, demonstrating the critical function of these two pathways in preventing tumor development in humans. In addition to ARF and DNA damage-activated kinases, several additional cellular factors, such as Rb tumor suppressor (12, 46), MDMX (14), and p300 histone acetyltransferase (8, 9), have been reported to affect the function of p53 through directly interacting with and regulating the activity of MDM2. More recently, it was shown that mitogen-induced Akt physically associates with and phosphorylates MDM2, leading to an enhanced activity of MDM2 and increased p53 degradation (7, 24, 27, 51). These findings support the notion that MDM2 functions as the primary regulator of p53 and as an integrator for the convergence of different stresses. To further explore the potential of MDM2 in connecting cellular stress to p53, we have undertaken a search for cellular factors that bind to and regulate the function of MDM2.
MATERIALS AND METHODS
Plasmids, cell culture, and actinomycin D treatment.
Mutant HDM2 and L11 constructs were generated by PCR-mediated site-directed mutagenesis and confirmed by direct DNA sequencing. U2OS, Saos-2, SJSA, and WI38 cells were obtained from the American Type Culture Collection, and p53-MDM2 double-deficient mouse embryonic fibroblasts were kindly provided by Steve Jones of the University of Massachusetts (16). All cells were grown in cultures in a 37°C incubator with 5% CO2 in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. Procedures and conditions for cell transfections, immunoprecipitation (IP), and immunoblotting were as previously described (50). For actinomycin D experiments, cells were treated with 5 nM actinomycin D for the indicated time and analyzed for cell cycle progression by flow cytometry or protein distribution by immunofluorescence microscopy.
In vitro transcription and translation and binding assays.
Coupled in vitro transcription and translation reactions were performed using a TNT kit (Promega) and following the manufacturer's instructions. For in vitro binding assays, translated proteins were mixed together and further incubated at 30°C for 30 min in the same reticulocyte lysate. At the end of the incubation, 200 μl of NP-40 lysis buffer was added to each binding reaction followed by IP with appropriate antibodies.
Antibodies.
Affinity-purified rabbit polyclonal antibody to human MDM2 (N-20; Santa Cruz), goat polyclonal antibody to human p53 (FL393; Santa Cruz), mouse monoclonal antibodies to p53 (clone PAb421; Oncogene Science, Uniondale, N.Y.), human MDM2 (clone SMP14; NeoMarkers), tubulin (clone DM1A + DM1B; NeoMarkers), and actin (Sigma) were purchased commercially. Affinity-purified rabbit polyclonal antibody to mouse ARF was raised using a synthetic peptide derived from the C terminus of the mouse ARF protein as an immunogen. Rabbit polyclonal anti-human L11 antibody was produced using a synthetic peptide, CIGAKHRISKEEAMRWFQQK, corresponding to amino acid residues 149 to 168 of human L11.
Protein microsequencing.
Ten 100-mm-diameter dishes of logarithmically growing U2OS cells were transfected with pCMV-HDM2, and cell lysates were pooled and immunoprecipitated with protein A beads covalently coated with anti-HDM2 antibody (total, 1 mg). After extensive washing, the anti-HDM2 precipitates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gel was silver stained, and the resultant banding patterns were compared with that observed by autoradiography of 35S IPs to identify specific proteins. Proteins of interest were excised and subjected to in-gel protease digestion (lysylendopeptidase; 50 ng/ml). Digested peptide fragments were extracted by acetonitrile and separated by reverse-phase high-pressure liquid chromatography (HPLC) on a Hewlett Packard 1100 HPLC system using a C18 column (Vydac) (1 by 250 mm). Amino acid sequences of individual peptides collected from HPLC were determined on an automated ABI microsequencer.
Luciferase assays.
At 24 h posttransfection, cells were washed in phosphate-buffered saline and harvested by scraping into 250 μl of 1× reporter lysis buffer (Promega). The cells were lysed for 10 min at 4°C with rotation and clarified by centrifugation for 5 min at 4°C in a microcentrifuge at maximum speed. A total of 10 μl of the clarified cell lysate was assayed for luciferase activity as described previously (20). For β-galactosidase assays, 75 μl of the clarified lysate was incubated at 37°C with 500 μl of Z buffer (21 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 40 mM β-mercaptoethanol, pH 7.0) containing chlorophenolred-β-d-galactopyranoside (CPRG; Boehringer Mannheim, Indianapolis, Ind.) at a final concentration of 0.1 mg/ml. The reaction was stopped by adding Na2CO3 to a final concentration of 260 mM, and the optical density at 595 nm was determined on a luminometer (Lumat LB 9501). The luciferase activity for each sample was normalized to β-galactosidase activity to control for transfection efficiency. The normalized luciferase activity of the pGL2-Basic plasmid was set to 1.
Cell transfection and FACS analysis.
Cell transfections were carried out using Lipofectamine reagent (Invitrogen) according to the manufacturer's instructions as described in detail previously (50). All cells were harvested at 24 h posttransfection and analyzed for cell cycle distribution. For fluorescence-activated cell sorter (FACS) analysis, cells were cotransfected with the indicated plasmid, harvested by trypsinization, fixed in 70% ethanol, and stained with propidium iodide (50 mg/ml) containing 50 mg of RNase A/ml. Flow cytometry analysis was conducted using a Becton-Dickinson FACScan. Green fluorescent protein (GFP) was used as a marker for the analysis of transfected cells. DNA content data from at least 10,000 GFP-positive cells are presented in the DNA histograms.
RESULTS
Identification of L11-HDM2 association.
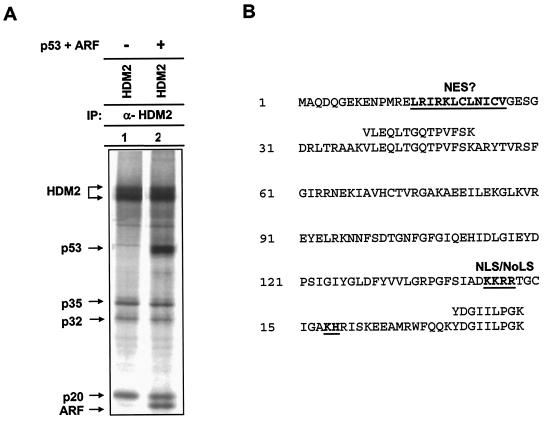
To identify novel HDM2 interacting, and potentially regulatory, molecules, protein complex formation of HDM2 immunocomplexes was determined by coupled metabolic labeling and IP (35S-IP). Analysis of HDM2 complexes revealed three cellular proteins with apparent molecular masses of 35, 32, and 20 kDa that noticeably associated with HDM2 (Fig. 1A). Coexpression of both p53 and ARF did not cause any detectable change of HDM2 association with p35, p32, or p20, suggesting that these proteins do not interact with HDM2 in a competitive manner. To determine the identities of p35, p32, and p20, we purified the proteins in an HDM2 immunocomplex and subjected them to protein microsequencing. Two peptide sequences, VLEQLTGQTPVFSK and YDGIILPGK, matching perfectly with human ribosomal protein L11 (residues 39 to 52 and residues 170 to 178; Fig. 1B), were obtained for p20. Two peptide sequences, GAVDGGLSIPHSTK and RFPGYDSESK, matching perfectly with human ribosomal protein L5 (residues 165 to 178 and residues 179 to 188; accession number U14966), were obtained for p35. Human L11 and L5 proteins contain 178 amino acid residues (20,190 Da) and 297 residues (34,425 Da), respectively, corresponding to the sizes of p20 and p35 detected in the HDM2 immunocomplex. Microsequencing of p32 was not successful. Whether it corresponds to a distinct polypeptide or an alternative translation initiation product of L5 (37) remains to be determined. L5-MDM2 association was previously reported (23), and our finding confirms this binding.
FIG. 1.
Analysis of HDM2 immunocomplex and identification of HDM2-L11 association. (A) HeLa cells were transiently transfected with plasmid DNA expressing HDM2. At 24 h after transfection, cells were metabolically labeled with [35S]methionine and the lysates were immunoprecipitated with antibody to HDM2. Immunoprecipitated proteins were separated by SDS-PAGE and visualized by autoradiography. The molecular identities of L5 and L11 were determined by protein microsequencing following a preparative anti-HDM2 IP. The identity of the 32-kDa protein (p32) has not yet been established. (B) HDM2-associated p20 corresponds to human ribosomal protein L11. L11 protein contains a putative nuclear export signal (NES) and a nuclear and nucleolar localization signal (NLS/NoLS). Two peptide sequences were obtained from protein microsequencing; both matched perfectly with human L11.
L11-HDM2 and L5-HDM2 associations do not require RNA.
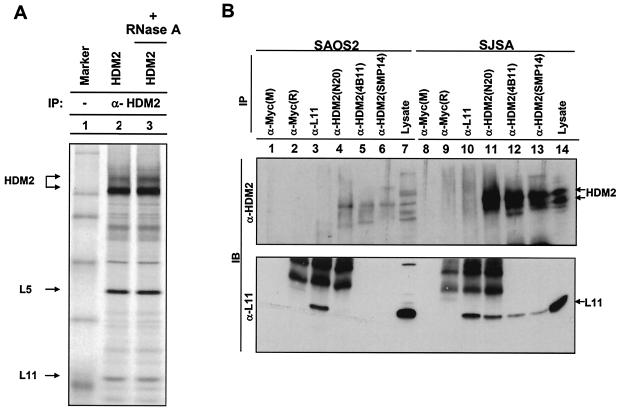
Since L11 and L5 are ribosomal proteins that bind to RNA and 5S and 5.8S RNAs have been reported to associate with HDM2 as well as with the HDM2-p53 complex (23), we determined whether an RNA component is required for the interactions between L11 and L5 with HDM2. Treatment of the cell lysate with RNaseA (10 μg/ml) had no detectable effect on L11-HDM2 and L5-HDM2 associations (Fig. 2A), indicating that RNA, although able to associate HDM2, is not required for HDM2 interaction with either ribosomal protein.
FIG. 2.
HDM2 interacts with ribosomal protein L11. (A) U2OS cells were transiently transfected with plasmid expressing HDM2. Transfected cells were lysed in the presence (+) or absence (−) of RNaseA (10 μg/ml). HDM2-L11/L5 complexes were examined by IP with an α-HDM2 antibody. No obvious effect was observed on L11-Hdm2 association after the RNaseA treatment. (B) L11 associates with HDM2 in vivo. Asynchronously growing human SJSA cells or HDM2-negative Saos-2 cells were lysed and immunoprecipitated with antibodies recognizing L11 and HDM2.
In vivo association of L11 with HDM2.
An in vivo association between L5 and HDM2 was previously demonstrated (23). A rabbit polyclonal antibody specific to L11 was raised and used to examine the in vivo binding of L11 and HDM2. L11 protein was readily detected in HDM2 immunocomplexes precipitated by three different HDM2 antibodies from human SJSA osteosarcoma cells which have HDM2 amplification but not in Saos-2 osteosarcoma cells which do not express detectable levels of HDM2 due to p53 deficiency (Fig. 2B). From these results, we conclude that L11 readily associates with HDM2 in vivo.
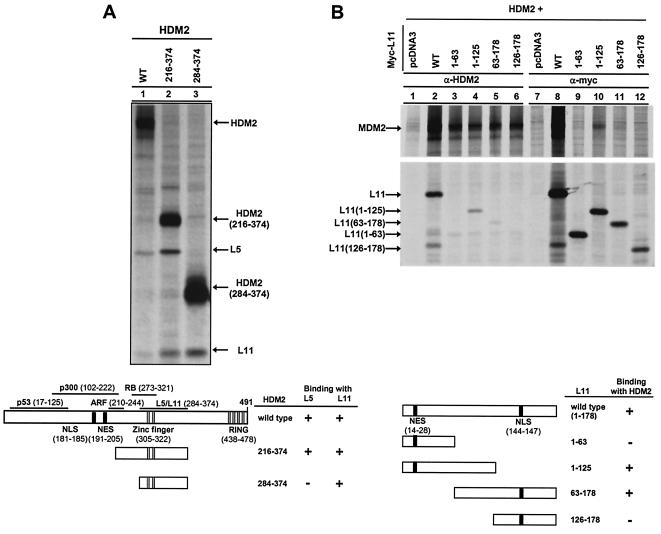
L11 binds to the central domain of HDM2.
A series of HDM2 deletion mutants was generated to map the L5 and L11 binding domains. Deletion of the N-terminal 216 (HDM2216-491) and C-terminal 117 (HDM21-374) residues had no obvious effects on HDM2-L5/L11 association (data not shown), suggesting that a sequence between HDM2 residues 216 and 374 is sufficient for binding L5 and L11. To test this, we generated two smaller HDM2 deletion mutants, HDM2216-374 and HDM2284-374, and assessed their ability to bind with endogenous L5 and L11. HDM2216-374 retained L5 and L11 binding activity (Fig. 3A). A smaller HDM2 fragment, HDM2284-374, lost its ability to bind with L5 but still retained full binding activity with L11. We therefore conclude that L11 can bind with HDM2 independently of L5 and possibly facilitates L5-HDM2 binding.
FIG. 3.
L11 binds to a central domain in HDM2. (A) Mapping of L11 binding domain in HDM2. Wild-type HDM2-expressing plasmids were transiently transfected into U2OS cells. At 24 h after transfection, cells were metabolically labeled with [35S]methionine and the lysates were immunoprecipitated with antibody to HDM2. (B) Mapping of HDM2 binding domain in L11. WT, wild type.
We also generated several L11 deletion mutants to map the HDM2 binding domain in L11 (Fig. 3B). Deletion of either the C-terminal 53 residues (L111-125) or N-terminal 62 residues (L1163-178) reduced, but did not completely disrupt, HDM2 binding, whereas deletion of a central sequence containing residues 63 to 125 completely abolished HDM2 binding. These results indicate that the central region of L11 is required for binding with HDM2.
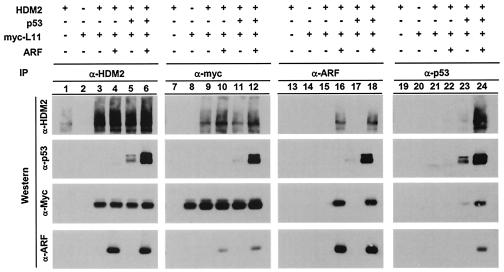
L11 can form a quaternary complex with HDM2, p53 and ARF.
A series of reciprocal IP-Western blotting assays were carried out to determine the interactions between L11, HDM2, p53, and ARF (Fig. 4). U2OS cells were transiently transfected with various combinations of plasmids expressing these proteins. L11/HDM2 complexes were detected reciprocally in cells expressing both proteins (lanes 3 to 6 and 9 to 12). Coexpression of L11 and HDM2 with ARF, p53, or both did not noticeably affect L11-HDM2 binding (lanes 4 to 6 and 10 to 12) and, importantly, L11 was detected in ARF and p53 immunocomplexes when HDM2 was coexpressed (lanes 13 to 24). Hence, L11, ARF, and p53 do not compete for binding with HDM2 and can simultaneously bind HDM2 to form a multiprotein complex. This conclusion is supported by the deletion analysis results showing that ARF and L11 bind to two separate sequences in HDM2, with amino acids 284 to 374 retaining full binding activity with L11 (Fig. 3A) and ARF binding to the region containing amino acids 210 to 244 of HDM2 (25).
FIG. 4.
L11 can form quaternary complexes with HDM2, p53, and ARF. U2OS cells were transiently transfected with (+) or without (−) plasmid DNA expressing HDM2, p53, and HA-ARF as indicated. At 24 h after transfection, cells were lysed and immunoprecipitated with the indicated antibodies followed by immunoblotting.
L11 reduces HDM2-mediated p53 ubiquitination and stabilizes p53.
HDM2 functions as an E3 ligase to ubiquitinate p53 and promotes p53 degradation (11). To examine whether L11 interferes with HDM2-mediated p53 ubiquitination in vivo, U2OS cells were cotransfected with plasmids expressing hemagglutinin (HA)-epitope-tagged ubiquitin, p53, and HDM2. At 36 h after transfection, proteasome inhibitor MG132 was added to inhibit the degradation of polyubiquitinated proteins. The expression of various proteins was confirmed by direct immunoblotting, and accumulation of ubiquitinated p53 was examined by blotting anti-p53 immunoprecipitates with anti-HA antibody (Fig. 5A). Coexpression of HDM2 with p53 resulted in an accumulation of a high-molecular-weight p53 smear characteristic of polyubiquitinated p53. Ubiquitinated p53 was not detected when either HDM2 or p53 was omitted or when a catalytically inactive RING finger mutant of HDM2 (C464A) was used (data not shown). Coexpression of L11 with HDM2 and p53 (lane 8), like that of ARF (lane 7), almost completely blocked the accumulation of polyubiquitinated p53. Consistent with the decrease of p53 ubiquitination, L11, like ARF, prevented HDM2-mediated p53 degradation, resulting in an increase in the steady-state level of p53 protein in the presence of HDM2 (Fig. 5B). These results demonstrate that L11 protein interferes with HDM2-mediated p53 ubiquitination and subsequent degradation to stabilize p53 protein levels. The steady-state level of HDM2 itself was also increased by the coexpression of either ARF or L11; this is consistent with the idea that L11, like ARF, inhibits both the ubiquitin ligase activity of HDM2 toward its substrate, p53, and HDM2's autoubiquitin ligase activity.
FIG. 5.
L11 inhibits HDM2-mediated p53 ubiquitination and degradation. (A) U2OS cells were transfected with (+) or without (−) the indicated plasmids. At 36 h after transfection, cells were treated with proteasome inhibitor MG132 (50 μM) for 4 h prior to cell lysis. Clarified cell lysate was immunoprecipitated (IP) with anti-p53 antibody, and washed immunoprecipitates were resolved by SDS-PAGE followed by immunoblotting with anti-HA antibody. Expression of individual transfected proteins was determined by direct immunoblotting (bottom panels). WB, Western blotting. (B) Total lysates prepared from cells transfected with (+) or without (−) the indicated plasmids were electrophoretically separated before immunoblotting with the indicated antibodies was performed.
L11 abolishes the inhibition of p53 transactivation by HDM2.
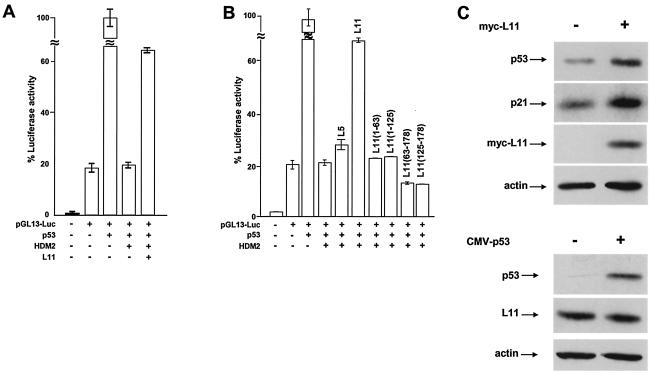
We then determined the effect of the presence of L11 on the HDM2-mediated repression of p53 transactivation activity. Under the conditions in which HDM2 almost completely repressed p53-dependent transactivation from the pGL13-Luc reporter, cotransfection with an L11 expression plasmid restored up to 70% of p53 transactivation activity (Fig. 6A and B).
FIG. 6.
L11 relieves HDM2-mediated repression of p53 transactivation. (A and B) U2OS cells were cotransfected with (+) or without (−) pGL13-Luc reporter plasmids and plasmids expressing indicated proteins. At 24 h after transfection, clarified cell lysates prepared from each transfected cell population were incubated with a luciferase assay buffer and the optical density at 595 nm was determined on a luminometer. The luciferase activity for each sample was normalized to β-galactosidase activity to control for transfection efficiency. The normalized luciferase activity of the pGL2-Basic plasmid was set to 1. (C) Total lysates prepared from cells transfected with the indicated plasmids were electrophoretically separated before immunoblotting with (+) or without (−) the indicated antibodies was performed. CMV, cytomegalovirus.
Cotransfection of a plasmid expressing L5 had a negligible effect on the HDM2-mediated repression of p53 transactivation (Fig. 6B), indicating that even when overexpressed, L5 alone does not significantly affect the ability of HDM2 to repress p53. This result is consistent with our observations that L11 can bind with HDM2 independently of L5, supporting the notion that L5 is not required for the function of L11 in HDM2 regulation. Deletion of either N-terminal (L1163-178 and L11126-178) or C-terminal (L111-63 and L111-125) sequences eliminated the ability of L11 to repress HDM2 inhibition of p53 (Fig. 6B). Both L111-125 and L1163-178 mutants retained a partial HDM2 binding activity (Fig. 3B). These results suggest that additional sequences in L11 other than those simply required for HDM2 binding are necessary to block HDM2 function.
Ectopic expression of L11 resulted in a dose-dependent increase in the steady-state level of endogenous p53 protein in U2OS cells, as previously observed, while overexpression of p53 did not appear to significantly affect L11 protein level (Fig. 6C). Notably, this was coupled with a parallel increase in p21 levels, suggesting that expression of L11 can stabilize endogenous p53 to activate p21.
Ectopic expression of L11 blocks S-phase entry dependent on p53.
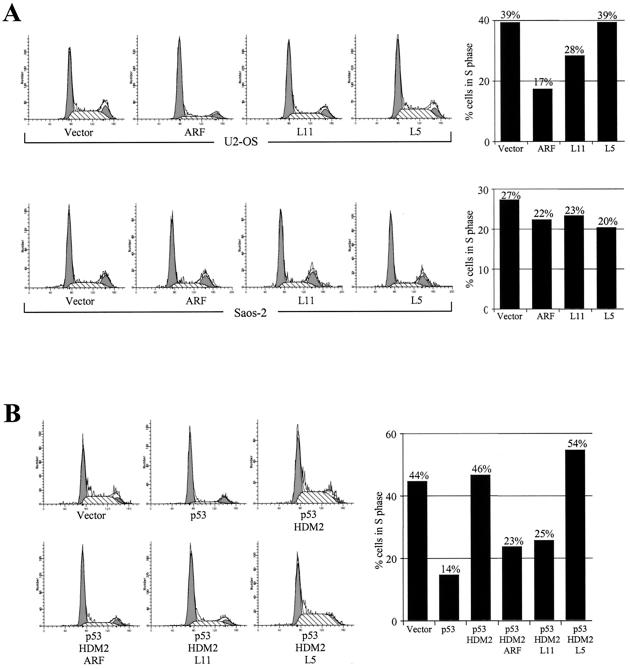
To determine whether overexpression of L11 is able to induce a p53-dependent cell cycle arrest, L11 was transiently transfected into U2OS (which contains functional p53) and Saos-2 (p53-defective) cells and the cell cycle distribution was determined by flow cytometry analysis (FACS). Cells were simultaneously transfected with a plasmid expressing GFP for gating the positively transfected cells. As shown in Fig. 7A, overexpression of L11 blocked S-phase entry in U2OS but not in Saos-2 cells, indicating that L11 induces a cell cycle arrest dependent on the presence of p53. Previous studies have shown that ARF can reverse the MDM2-inhibited p53 function to induce cell cycle arrest. To address whether L11 has a similar function, we carried out FACS analysis with U2OS cells transiently transfected with L11, HDM2, and p53 (Fig. 7B). As shown previously, expression of p53 blocked cell cycle progression into the S phase and this was abolished by coexpression of HDM2. In cells transfected with p53 and MDM2, cotransfection with L11, but not with another HDM2-binding ribosomal protein, L5 (23), restored S-phase entry to a level comparable to that seen in cells cotransfected with ARF, indicating a specific function of L11 in reversing HDM2 inhibition of p53 activity.
FIG. 7.
L11 induces a p53-dependent cell cycle arrest. (A) U2OS or Saos-2 cells were transfected with or without (Vector) a plasmid expressing a GFP marker and the indicated proteins. (B) U2OS cells were cotransfected with or without (Vector) a plasmid expressing a GFP marker and the indicated proteins. Transfected cells were sorted, and their cell cycle distribution characteristics were determined by flow cytometry at 24 h after transfection. The proportions of cells in S phase in each transfected cell population were compared using bar graphs. A minimum of 10,000 GFP-positive cells were analyzed for each transfection.
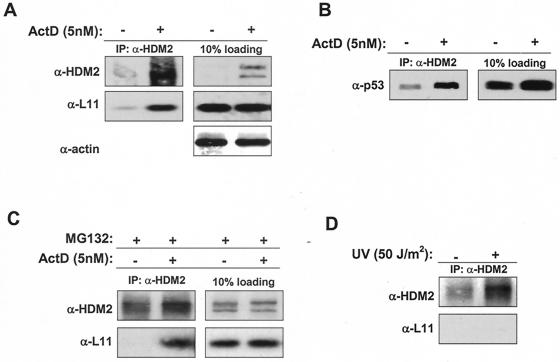
Ribosomal stress induced by a low concentration of actinomycin D increases L11-MDM2 interaction and stabilizes p53.
In search of biological functions of the L11-HDM2-p53 pathway, we hypothesized that since it is a ribosomal protein, L11 functions in monitoring ribosomal biogenesis and cell growth. To test this idea, we first determined the nature of HDM2-L11 interactions in mediating ribosomal biogenesis by treating cells with low concentrations of actinomycin D. At high concentrations (e.g., >30 nM) actinomycin D causes DNA damage and inhibits transcription from all three classes of RNA polymerase promoters, whereas at low concentrations (e.g., <10 nM) actinomycin D selectively inhibits RNA pol I-dependent transcription and thus ribosomal biogenesis (13, 32). The binding of L11 with HDM2 was barely detectable under normal conditions, presumably because the majority of L11 was bound to 60S ribosomal subunits either in the nucleolus or in the cytoplasm (Fig. 8A). Treating WI38 cells with 5 nM of actinomycin D apparently did not affect the level of endogenous L11 but clearly increased the levels of endogenous HDM2 and the L11-HDM2 complex (Fig. 8A). Consequently, upon actinomycin D treatment the levels of p53 and the HDM2-bound p53 were also increased (Fig. 8B). To rule out the possibility that the increase in the HDM2-L11 interaction was solely due to an increase of HDM2, cells were treated with the proteasome inhibitor MG132, alone or in combination with actinomycin D treatment, to stabilize and accumulate HDM2 protein before lysis. Although both cell populations accumulated HDM2 protein to similar levels, L11-HDM2 complex was readily detected in the cells treated with combination of MG132 and actinomycin D but was almost undetectable in cells treated with MG132 alone (Fig. 8C). Hence, perturbation of ribosomal assembly by actinomycin D enhances L11-HDM2 association.
FIG. 8.
Perturbation of ribosomal biogenesis by actinomycin D (ActD) increases L11-HDM2-p53 complex levels. (A and B) WI38 cells were treated with 5 nM actinomycin D for 24 h (+) or left untreated (−) before being lysed for analysis for the expression of various proteins and protein complex formation. Approximately 500 ug of total protein was utilized for each IP. (C) WI38 cells were treated with the proteasome inhibitor MG132 for 6 h before lysis to stabilize HDM2 in the presence (+) or absence (−) of 5 nM actinomycin D. A total of 200 ug of protein was used in each IP. (D) WI38 cells were treated with UV (50 J/m2) (+) at 24 h before harvesting or left untreated (−), and IP was performed as described above.
To further substantiate the notion that actinomycin D-induced ribosomal stress, but not other types of stress, specifically increased L11-HDM2 binding, we have examined L11-HDM2 interaction in cells that sustained several additional stresses. Exposure of WI38 cells to UV irradiation accumulated HDM2 but did not increase L11-HDM2 association (Fig. 8D). Likewise, we found that overexpression of E2F1, which, like overexpression of several other growth-promoting oncogenes, stimulated the transcription of ARF and accumulated HDM2 and p53, did not appreciably affect the level of either L11 protein or L11-HDM2 association (data not shown). These results suggest that L11 does not play a major role in cellular response to either DNA damage or hyperproliferation induced by oncogene overexpression and instead specifically regulates a HDM2-p53 response to ribosomal perturbation.
DISCUSSION
In this study, we provide evidence for a physical interaction and functional regulation between ribosomal protein L11, a component of cell growth machinery, and HDM2, a key regulator of p53-mediated checkpoint pathways. First, L11 directly binds with HDM2 via its central domain and can form a ternary complex with p53. Second, L11 inhibits HDM2-promoted p53 ubiquitination to subsequently stabilize p53 protein levels. Third, L11 expression relieves HDM2-mediated repression of p53 transactivation and can activate endogenous p53.
The 491 residues of HDM2 can be divided into three functional domains: an N-terminal region which binds p53, a C-terminal RING finger domain that promotes p53 ubiquitination, and a central region of approximately 300 residues that contains multiple sites for binding and regulation by different proteins, including RB (12, 46), ARF (33, 42, 50), p300 histone acetyltransferase (9) Akt (in phosphorylation) (7, 24, 27, 51), and now L11 (see Fig. 3A for a schematic summary). These findings attest to the notion that the central 300-amino-acid domain of MDM2/HDM2 contains multiple sites for the binding of various cellular factors to channel multiple stress pathways into p53 control.
The activity residing in this region has long been elusive. We have found that L11 ribosomal protein binds HDM2 via this central domain (Fig. 3), providing the first functional assignment to this region. It is intriguing that L11 regulates HDM2 and p53 in a manner similar to the regulation of HDM2 and p53 by ARF. Both L11 and ARF normally localize to the nucleolus and relocalize to interact with HDM2 in the nucleoplasm (21, 48), both can bind directly with HDM2 and form ternary complexes with p53 and HDM2, and both inhibit HDM2-promoted p53 ubiquitination and restore p53's transactivating activity in the presence of HDM2. There is, however, one notable distinction between L11- and ARF-mediated HDM2 regulation mechanisms. ARF inhibits HDM2's nuclear export, whereas L11 does not (reference 48 and our unpublished results). We suspect that binding of ARF and L11 to two different, nonoverlapping sequences in HDM2 can contribute to this mechanistic difference. ARF has been suggested to link oncogene-induced hyperproliferative signals to p53-dependent cell cycle arrest (39). By transfection and co-IP assays, we observed the formation of a p53-HDM2-ARF-L11 quaternary complex (Fig. 4). Assembly of such a quaternary complex raises the intriguing possibility that in cells experiencing two insults at the same time, one causing a ribosomal perturbation or growth inhibition and one stimulating cells to hyperproliferate, L11 and ARF can in theory simultaneously bind to HDM2 to additively inhibit HDM2 and induce a more rapid and effective cell cycle arrest.
How L11 inhibits HDM2 ubiquitin ligase activity, as in the case of ARF-mediated inhibition of MDM2 ubiquitin ligase activity, is not clear. In vitro, the RING finger domain of HDM2 alone is sufficient to catalyze the synthesis of polyubiquitin chain (autoubiquitination) and association of L11 with HDM2 does not inhibit HDM2-mediated p53 ubiquitination (M. Furukawa, J. McCarville, and Y. Xiong, unpublished data); this suggests that an additional factor(s) is involved in L11 inhibition of HDM2-mediated p53 ubiquitination in vivo. Further investigation of the mechanism of L11 inhibition will provide insights into inhibition of HDM2 activity and ubiquitin ligase activity in general.
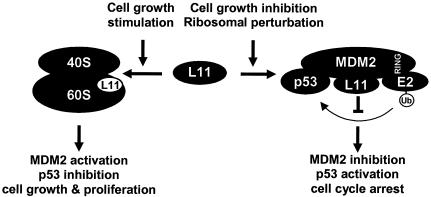
In yeast species such as Saccharomyces cerevisiae, cell cycle arrest results in response to increased cellular size while nutrient deprivation coordinately blocks both cell growth and cell cycle, indicating that sufficient cell growth is required for and must regulate the progression of the cell cycle (15). Similar observations have also recently been reported regarding mammalian cells (6). Ribosomal biogenesis includes the expression of approximately 150 rRNA genes and the synthesis of nearly 80 ribosomal proteins and a large number of assembly factors and consumes up to 80% of the energy of a proliferating cell (reviewed in references 18 and 44). Ribosomal assembly must therefore be tightly regulated for the economy of the cell and rapidly responsive to various environmental and intracellular growth conditions or insults. As the major component of cell growth, ribosome biogenesis could conceivably be the target of a checkpoint pathway for monitoring cell growth and coupling a growth condition change or insult to the cell cycle. A mammalian ribosome contains 77 ribosomal proteins, and the function of most ribosomal protein subunits has not been investigated individually. We suggest that the L11-HDM2-p53 pathway functions in monitoring ribosome biogenesis and couples cell growth to the cell cycle (Fig. 9). We postulate that L11 is constantly swinging between binding with HDM2 in the nucleoplasm and being assembled into ribosomes, with the latter activity exhibiting higher affinity and being more prevalent than L11's association with HDM2. A signal stimulating cell growth would promote L11 assembly into functional ribosomes, allowing HDM2 to repress p53 and thereby linking increased cell growth (protein synthesis) with a reduced threshold (e.g., p21 level) for entering the S phase. An inhibition of cell growth that decreases ribosome biogenesis or a direct perturbation of ribosomal biogenesis would release L11 to bind with HDM2, leading to increased p53 activity and lowering the threshold of entry into a proliferative cycle.
FIG. 9.
A schematic model for the function of the L11-MDM2-p53 pathway. (See text for discussion.)
The findings reported here raise a number of other questions. Given that L11 is highly conserved during evolution, the p53 gene is conserved in Drosophila, and Mdm2 is present in all known vertebrates (e.g., frogs, zebra fish, and mammals), could L11's regulatory effect toward p53 (and thus, coupling of cellular growth to the cell cycle) have evolved as early as or prior to the divergence of vertebrates? In mammals, MDM2 has a closely related homologue, MDMX, that also binds to and negatively regulates p53 (41). MDMX shares with MDM2 similar overall structural organizations including the conserved central zinc finger domain. Does L11 similarly regulate MDMX? MDMX functions in part by interacting with MDM2 (38). Could L11 influence the MDM2-MDMX interaction or MDMX affect L11-mediated regulation on MDM2? Lastly, both Rb family proteins and p53 have been reported to negatively regulate Pol I-dependent rRNA transcription (recently reviewed by Ruggero and Pandolfi) (36). Could the L11-Mdm2-p53 pathway and the Rb/p53-Pol I pathway be intertwined on a feedback loop to positively regulate each other? Studies of these questions could help lead us to a better understanding of how cell growth control is coordinated with cell cycle regulation and are experimentally possible.
Acknowledgments
We thank John Cogswell and Cathy Finlay (GlaxoSmithKline) for providing the Hdm2 and p53 adenoviruses, Bert Vogelstein for providing pGL13-Luc plasmid, Karen Vousden for communicating their unpublished results, and Y. Joe He for helping figure preparation.
Y.Z. is a recipient of a Career Award in Biomedical Science from the Burroughs Wellcome Fund and a Howard Temin Award from National Cancer Institute. Y.X. is a recipient of a Career Development Award from the United States Department of Army Breast Cancer Research Program. This study was supported by the M. D. Anderson Research Trust (Y.Z.) and NIH grant CA65572 (Y.X.).
REFERENCES
- 1.Bates, S., A. C. Phillips, P. Clark, F. Stott, G. Peters, R. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumor suppressors RB and p53. Nature 395:124-125. [DOI] [PubMed] [Google Scholar]
- 2.Chehab, N. H., A. Malikzay, M. Appel, and T. D. Halazonetis. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev. 14:278-288. [PMC free article] [PubMed] [Google Scholar]
- 3.Corvi, R., L. Savelyeva, S. Breit, A. Wenzel, R. Handgretinger, J. Barak, M. Oren, L. Amler, and M. Schwab. 1995. Non-syntenic amplification of MDM2 and MYCN in human neuroblastoma. Oncogene 10:1081-1086. [PubMed] [Google Scholar]
- 4.de Stanchina, E., M. E. McCurrach, F. Zindy, S.-Y. Shieh, G. Ferbeyre, A. V. Samuelson, C. Prives, M. F. Roussel, C. J. Sherr, and S. W. Lowe. 1998. E1A signaling to p53 involves the p19ARF tumor suppressor. Genes Dev. 12:2434-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fakharzadeh, S. S., S. P. Trusko, and D. L. George. 1991. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 10:1565-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fingar, D. C., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16:1472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottlieb, T. M., J. F. Leal, R. Seger, Y. Taya, and M. Oren. 2002. Cross-talk between Akt, p53 and Mdm2: possible implications for the regulation of apoptosis. Oncogene 21:1299-1303. [DOI] [PubMed] [Google Scholar]
- 8.Grossman, S. R., M. E. Deato, C. Brignone, H. M. Chan, A. L. Kung, H. Tagami, Y. Nakatani, and D. M. Livingston. 2003. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300:342-344. [DOI] [PubMed] [Google Scholar]
- 9.Grossman, S. R., M. Perez, A. L. Kung, M. Joseph, C. Mansur, Z. X. Xiao, S. Kumar, P. M. Howley, and D. M. Livingston. 1998. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol. Cell 2:405-415. [DOI] [PubMed] [Google Scholar]
- 10.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 11.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh, J.-K., F. S. G. Chan, D. J. O'Connor, S. Mittnacht, S. Zhong, and X. Lu. 1999. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol. Cell 3:181-193. [DOI] [PubMed] [Google Scholar]
- 13.Iapalucci-Espinoza, S., and M. T. Franze-Fernandez. 1979. Effect of protein synthesis inhibitors and low concentrations of actinomycin D on ribosomal RNA synthesis. FEBS Lett. 107:281-284. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, M. W., and S. J. Berberich. 2000. MdmX protects p53 from Mdm2-mediated degradation. Mol. Cell. Biol. 20:1001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston, G. C., J. R. Pringle, and L. H. Hartwell. 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105:79-98. [DOI] [PubMed] [Google Scholar]
- 16.Jones, S. N., A. E. Roe, L. A. Donehower, and A. Bradley. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378:206-208. [DOI] [PubMed] [Google Scholar]
- 17.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 10:1054-1072. [DOI] [PubMed] [Google Scholar]
- 18.Kressler, D., P. Linder, and J. de La Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubbutat, M. H. G., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 20.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 21.Llanos, S., P. A. Clark, J. Rowe, and G. Peters. 2001. Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat. Cell Biol. 3:445-452. [DOI] [PubMed] [Google Scholar]
- 22.Luna, R. M., D. S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203-206. [DOI] [PubMed] [Google Scholar]
- 23.Marechal, V., B. Elenbaas, J. Piette, J.-C. Nicolas, and A. J. Levine. 1994. The ribosomal protein L5 is associated with mdm-2 and mdm-2-p53 complexes. Mol. Cell. Biol. 14:7414-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayo, L. D., and D. B. Donner. 2001. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl. Acad. Sci. USA 98:11598-11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Midgley, C. A., J. M. Desterro, M. K. Saville, S. Howard, A. Sparks, R. T. Hay, and D. P. Lane. 2000. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 19:1312-2323. [DOI] [PubMed] [Google Scholar]
- 26.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237-1245. [DOI] [PubMed] [Google Scholar]
- 27.Ogawara, Y., S. Kishishita, T. Obata, Y. Isazawa, T. Suzuki, K. Tanaka, N. Masuyama, and Y. Gotoh. 2002. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J. Biol. Chem. 277:21843-21850. [DOI] [PubMed] [Google Scholar]
- 28.Oliner, J. D., K. W. Kinzler, P. S. Meltzer, D. L. George, and B. Vogelstein. 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358:80-83. [DOI] [PubMed] [Google Scholar]
- 29.Oliner, J. D., J. A. Pietenpol, S. Thiagalingam, J. Gyuris, K. W. Kinzler, and B. Vogelstein. 1993. Oncoprotein MDM2 conceals the activation domain of tumor suppressor p53. Nature 362:857-860. [DOI] [PubMed] [Google Scholar]
- 30.Oren, M. 2000. Regulation of the p53 tumor suppressor protein. J. Biol. Chem. 274:36031-36034. [DOI] [PubMed] [Google Scholar]
- 31.Palmero, I., C. Pantoja, and M. Serrano. 1998. p19ARF links the tumor suppressor p53 and Ras. Nature 395:125-126. [DOI] [PubMed] [Google Scholar]
- 32.Perry, R. P., and D. E. Kelley. 1970. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J. Cell. Physiol. 76:127-139. [DOI] [PubMed] [Google Scholar]
- 33.Pomerantz, J., N. Schreiber-Agus, N. J. Liegeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, and R. A. DePinho. 1998. The INK4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92:713-723. [DOI] [PubMed] [Google Scholar]
- 34.Prives, C. 1998. Signaling to p53: breaking the MDM2-p53 circuit. Cell 95:5-8. [DOI] [PubMed] [Google Scholar]
- 35.Reifenberger, G., L. Liu, K. Ichimura, E. E. Schmidt, and V. P. Collins. 1993. Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res. 53:2736-2739. [PubMed] [Google Scholar]
- 36.Ruggero, D., and P. P. Pandolfi. 2003. Does the ribosome translate cancer? Nat. Rev. Cancer 3:179-192. [DOI] [PubMed] [Google Scholar]
- 37.Schatz, O., M. Oft, C. Dascher, M. Schebesta, O. Rosorius, H. Jaksche, M. Dobrovnik, D. Bevec, and J. Hauber. 1998. Interaction of the HIV-1 rev cofactor eukaryotic initiation factor 5A with ribosomal protein L5. Proc. Natl. Acad. Sci. USA 95:1607-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharp, D. A., S. A. Kratowicz, M. J. Sank, and D. L. George. 1999. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J. Biol. Chem. 274:38189-38196. [DOI] [PubMed] [Google Scholar]
- 39.Sherr, C. J. 1998. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 12:2984-2991. [DOI] [PubMed] [Google Scholar]
- 40.Shieh, S.-Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 41.Shvarts, A., W. T. Steegenga, N. Riteco, T. van Laar, P. Dekker, M. Bazuine, R. C. van Ham, W. van der Houven van Oordt, G. Hateboer, A. J. van der Eb, and A. G. Jochemsen. 1996. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 15:5349-5357. [PMC free article] [PubMed] [Google Scholar]
- 42.Stott, F. J., S. Bates, M. C. James, B. B. McConnell, M. Starborg, S. Brookes, I. Palmero, K. Ryan, E. Hara, K. H. Vousden, and G. Peters. 1998. The alternative product from the human CDK2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17:5001-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao, W., and A. J. Levine. 1999. p19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. USA 96:6937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas, G. 2000. An encore for ribosome biogenesis in the control of cell proliferation. Nat. Cell Biol. 2:E71-E2. [DOI] [PubMed] [Google Scholar]
- 45.Thut, C. J., J. A. Goodrich, and R. Tjian. 1997. Repression of p53-mediated transcription by MDM2, a dual mechanism. Genes Dev. 11:1974-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao, Z., J. Chen, A. J. Levine, N. Modjtahedi, J. Xing, W. R. Sellers, and D. M. Livingston. 1995. Interaction between the retinoblastima protein and the oncoprotein MDM2. Nature 375:694-698. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Y., and Y. Xiong. 2001. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 12:175-186. [PubMed] [Google Scholar]
- 48.Zhang, Y., and Y. Xiong. 1999. Mutation in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol. Cell 3:579-591. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, Y., and Y. Xiong. 2001. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 292:1910-1915. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Y., Y. Xiong, and W. G. Yarbrough. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725-734. [DOI] [PubMed] [Google Scholar]
- 51.Zhou, B. P., Y. Liao, W. Xia, Y. Zou, B. Spohn, and M. C. Hung. 2001. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat. Cell Biol. 3:973-982. [DOI] [PubMed] [Google Scholar]
- 52.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]