Abstract
DNA palindromes are associated with rearrangement in a variety of organisms. A unique opportunity to examine the impact of a long palindrome in mammals is afforded by the Line 78 strain of mice. Previously it was found that the transgene in Line 78 is likely to be palindromic and that the symmetry of the transgene was responsible for a high level of germ line instability. Here we prove that Line 78 mice harbor a true 15.4-kb palindrome, and through the establishment of cell lines from Line 78 mice we have shown that the palindrome rearranges at the impressive rate of about 0.5% per population doubling. The rearrangements observed to arise from rapid palindrome modification are consistent with a center-break mechanism where double-strand breaks, created through hairpin nicking of an extruded cruciform, are imprecisely rejoined, thus introducing deletions at the palindrome center. Significantly, palindrome rearrangements in somatic tissue culture cells almost completely mirrored the structures generated in vivo in the mouse germ line. The close correspondence between germ line and somatic events indicates the possibility that center-break modification of palindromes is an important mechanism for preventing mutation in both contexts. Permanent cell lines carrying a verified palindrome provide an essential tool for future mechanistic analyses into the consequences of palindromy in the mammalian genome.
In DNA, a “palindrome” exists if a sequence is immediately followed by an inverted copy of itself. True palindromes, with twofold rotational symmetry, are distinctive because they can convert from continuous B-form DNA into a cruciform configuration if subjected to sufficient torsional strain (26). The ability to extrude as a cruciform structure is not a property of random-sequence DNA and is disfavored if even small nonpalindromic spacers intervene between inverted repeats (see, for example, reference 2). The biological consequences of palindromy are varied. On the one hand, key processes can be positively affected by lineform-to-cruciform transitions, as studies of nearly palindromic inverted repeats contained in replication origins or transcriptional control regions have begun to demonstrate (11, 14, 16, 38). On the other hand, palindromes have been implicated in various kinds of illegitimate genome rearrangements in mammalian cells, including gene amplification (12, 32), repeat expansion (39), chromosomal translocations (7, 17), and gross chromosomal deletions (9, 28).
Because long, true palindromes may have genotoxic effects, it is likely that any organism that contains palindromes has also evolved mechanisms to cope with “extracurricular” extrusion events. Few studies have examined this possibility, however. This is because any investigation of long (greater than roughly 200 bp) DNA palindromes quickly runs up against the obstacle presented by the instability and lethality palindromic DNA in Escherichia coli (19, 25). Long, true palindromes are corrupted by cloning-based and PCR-based analyses. Nevertheless, as studies of chromosomal breakpoints in a few human genetic diseases indicate, naturally occurring palindromes may well exist in at least several sites in the human genome. One known palindrome, 160 bp in total length, is located on chromosome 15 downstream of the β-globin gene (9). Other larger palindromes (estimated at somewhere around 230 to 445 bp) are predicted to occur on human chromosomes 11 and 22, and a third nearly palindromic inverted repeat (possessing a 3-bp spacer) is present on chromosome 17 (7, 17, 18, 33). The human studies, though still few in number, all signify an association between palindromy, gross chromosomal aberration, and human genetic disease, and they consistently indicate that the cloning barrier is, above all, responsible for the fact that long true palindromes remain an ill-defined biological phenomenon in higher eukaryotes.
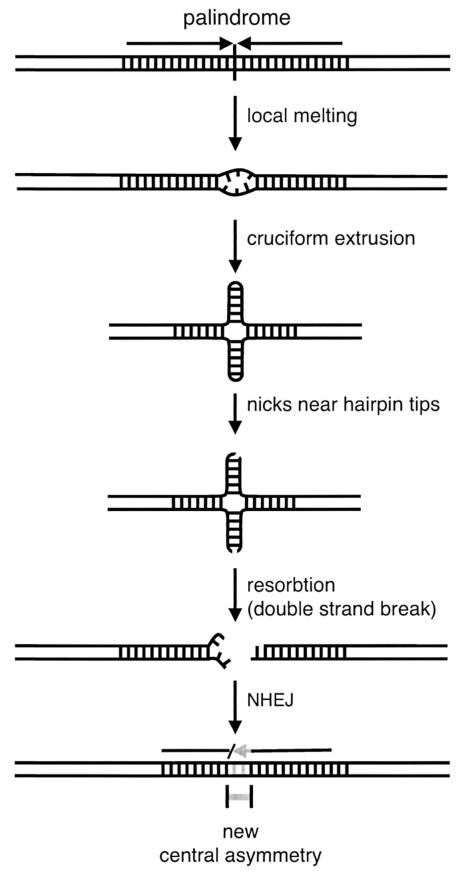
In order to investigate palindrome-mediated rearrangement in mammalian systems, two alternative approaches have been exploited. In one, the germ line instability of transgenes in mice with apparently palindromic integrant structures has been investigated (1, 4, 10). In the second, true palindromic DNA created by in vitro ligation has been transiently transfected into mouse or hamster tissue culture cells (20, 21). In both experimental contexts, introduced palindromic DNA is highly unstable and appears to undergo nonrandom rearrangement (1, 4, 21). A center-break mechanism has been proposed to explain palindrome processing in mammals based upon these observations, as shown in Fig. 1 (1, 21). Palindromic DNA is thought to become a substrate for modification upon cruciform extrusion. The cruciform hairpins are nicked by an endonuclease that cuts one strand near each of the two hairpin tips. The pair of single-strand nicks constitutes a centrally positioned, double-strand break at the center of the palindrome once branch migration has resorbed the extruded structure. The double-strand break is then resealed by nonhomologous end joining (NHEJ). As a result of the center-break-associated modification, an asymmetry is created in the once perfectly palindromic sequence. The new central spacer becomes a barrier to further extrusion events (2, 19).
FIG. 1.
The center-break mechanism of palindrome rearrangement. A cruciform structure is initiated by local underwinding of the DNA. Each extruded hairpin structure is nicked on one strand near the tip. The two nicks comprise a double-strand break at the symmetry center of the palindrome, which becomes reconnected by NHEJ. Overall the process disrupts the perfectly palindromic sequence by introducing central alterations.
The effects of center-break-type alterations have been documented in the germ line of mice bearing chromosomally located transgenes of a probable palindromic structure (1, 4, 22). As reported by several groups, once this type of alteration occurs, very little further rearrangement is observed (1, 4, 10, 22, 39). However, for all of the transgenic studies, it was unknown whether or not the germ line-introduced palindromes were in fact truly palindromic. A possibility that could not be excluded was that the transgenic sequences might initially have been large inverted repeats with small spacers. This uncertainty put limits on any definitive interpretation of the stabilizing effect of central modification and left open a key related question, which is whether long true palindromes confer inviability in mammalian cells just as they do in bacteria (19). The issue of whether or not true palindromes underlie the transgenic observations is central to understanding the impact of palindromic DNA, its structural isomerizations, and the induction of central deletions on genome stability.
In the present report we have proved that the Line 78 strain of mice does indeed contain a long true palindrome. This confirms the conclusions regarding germ line palindrome modification that were earlier based upon studies of the Line 78 transgene (1, 22). A critical next step in understanding the relationship between palindromy and genome stability in mammals is to explore the same Line 78 transgene integrant in a somatic cell context. Permanent cell lines from Line 78 mice were established, making it possible for the first time to determine the rate of palindrome rearrangement in a higher eukaryote. The fine-structure analyses of rearrangements produced in somatic cells, as in the germ line, revealed them to constitute a nonrandom set of outcomes consistent with center-break modification. The rearrangement rate dropped precipitously with the acquisition of central deletions. The concordance between germ line and somatic observations indicates that center-break modification of true palindromes is an important, heretofore disregarded genome maintenance mechanism.
MATERIALS AND METHODS
Mice.
The Line 78 mouse strain has been described previously (1). All cell lines in the present study are heterozygous for the Line 78 transgene and were derived from fetal livers of mixed genetic backgrounds. The 7#7, 7#14, and 18#1 lines (and subclones) were isolated after twice back-crossing a Line 78 heterozygote (1) to BALB/c. Line 630FL7#1 was p53−/− (trp53tm1Ty) (13) on a mixed C57BL/6, CBA, BALB/c, and 129/Sv background.
Establishment of cell lines from Line 78 fetal liver.
Cell suspensions were prepared from day 16 to 19 fetal livers after crossing a Line 78 heterozygous male and a nontransgenic female. Suspensions containing 2 × 106 nucleated cells were infected with Abelson murine leukemia virus (Ab-MLV; stocks were prepared from a p-160 Ab-MLV producer cell line as previously described [3]). Infected liver cells were plated in soft agar, and individual colonies were picked in the case of 7#7, 7#14, and 18#1 (30). Otherwise infected liver cells were maintained in bulk culture; outgrowth of transformants was evident by day 5, and cell lines were isolated by limiting dilution after approximately a month. Lines were routinely carried in RPMI-10% fetal calf serum-50 μM β-mercaptoethanol. Test clones were kept as 10-ml cultures and split 1:50 every 3 days; occasionally (as when the day of passage fell on Thursday) 1:100 split ratios were used.
Sampling of test clones.
Sampling of the test clone culture was performed by limiting dilution. The actual number of wells showing growth was used in the rate calculation (below). Sample subclones were each expanded to about 107 cells, whereupon high-molecular-weight DNA was prepared.
Protocol for determination of transgene structure in sample subclones.
Ten to 12 μg of genomic DNA from each sample subclone was digested, electrophoresed on 0.8% agarose gels, and transferred to membranes in 0.4 M NaOH. Filters were probed with a gel-purified 3.4-kb BamHI fragment from plasmid 1.8 (1). The first-pass analysis was with a BamHI and PstI codigestion. A pair of bands of 3.4 and 2.4 kb (each of an equal, two-copy intensity) was taken as a preliminary indication of an intact palindrome. All other patterns indicated rearrangement as follows. Any banding pattern that included a 2.6- or 3.1-kb signal indicated the possibility of a recombinant lacZ repeat and were confirmed to be homologous recombinants (i) by demonstrating that a 2.6-kb band is also generated with EcoRI digestion or (ii) by showing the absence of the 3.1-kb band when BamHI digestions are performed (one clone from the first time point of 7#7-7-6 was unavailable for analysis). Clones ultimately assigned to the strictly homologous recombinant category (see text) were analyzed by digestion-circularization-PCR (DC-PCR) (see below). Otherwise, a 3.4-kb band appearing with a band of 4.8 kb or smaller indicated a small central deletion spanning the central 6-bp PstI site. A 3.4-kb band with a variable band of up to 5.8 kb indicated a central deletion of larger size. A single band was interpreted as a deletion of even larger extent. (Consistent with this interpretation, the rare lone bands were all smaller than 6.8 kb in size.)
To verify rearranged versus nonrearranged status, all clones that appeared unrearranged according to BamHI and PstI codigestion were tested with EcoRI (DNA was unavailable for day 21 samples). Five clones that revealed a nonintact pattern were reassigned. To uncover all possible examples of mixed cultures, subclones showing more than two bands in the first analysis were digested with ApaI, which is expected to give a single band only if a culture is unmixed. Additionally, EcoRI digests of the intact subclones served to screen this set for mixed cultures. Ten mixed clones in all were identified. The fact that unrearranged cells were present in each of these mixed cultures indicated the possibility that the mixed composition arose as a consequence of rearrangement early after plating. To be conservative, all mixed samples were scored as intact, thus ensuring any error introduced by the 10 ambiguous clones would be in the direction of under- rather than overestimating rearrangement rates.
PCR analysis of Line 78 transgene deletions.
At first the transgene deletions were determined by a trial-and-error approach, testing various pairs of center-directed primers. Seminested PCR (25 cycles each round) was performed with 1.25 U of Platinum Taq DNA polymerase (Gibco BRL) in 25-μl PCRs primed with 200 ng of cellular DNA (initial round) or 1 μl of first-round product (second round). The 174-bp deletion was successfully amplified with Peter1 plus E4 in the first round and Peter2 plus E4 in the second. For the 78-bp deletion, Les1 plus E4 was used in the first round, with E1 plus E4 in the second. The 270-bp deletion was successfully amplified by Les6 plus E4 in the first round and Les5 plus E4 in the second. Later in the study, DC-PCR was used.
DNA sequence analysis was performed after cloning PCR products into the pCR 4-TOPO vector by using a TOPO TA cloning kit (Invitrogen).
DC-PCR analysis of the intact Line 78 transgene.
Genomic DNA was digested with PstI. Repurified samples were self-ligated at a concentration of 7 μg/ml overnight. Fifty nanograms of ligated DNA was used in seminested PCR (25 cycles each round). First-round PCR (2.5 U of Taq polymerase [Invitrogen] in 50-μl PCRs) was with Les1 and ACLF1 for the left side and Les1 and ACRF1 for the right side. One microliter of the PCR was used in the second-round PCR, with E1 in place of Les1.
In the original experiment with transgene-specific primers only, the same ligation protocol was followed, but first-round DC-PCR was performed with Les1 and DC3. Parallel samples were then subjected to second-round PCR with either E1 and DC4 or E1 and DC5.
PCR analysis of the chromosome-transgene junction.
A Line 78 DNA sample was used in nested PCR as follows: left side (first round) was with AC173 plus DC4 and (second round) AC174 plus DC5; right side (first round) was with AC171 plus DC4 and (second round) AC172 plus DC5 (Table 1).
TABLE 1.
Primer sequences
| Primer | Sequence | 3′ end point |
|---|---|---|
| Transgene specific | ||
| E4 | CTG TAG TTT GCT AAC ACA CC | In, 3 bp to center axis |
| E1 | AGC ATT ATC CTT ATC CAA AAC AGC | In, 99 bp to center axis |
| Les1 | CCT GTG TAG GTT CCA AAA TAT CTA GTG | In, 167 bp to center axis |
| Peter2 | GCA AAA CAG GAG GCA CAT T | In, 201 bp to center axis |
| Peter1 | CCC TGC TCA TCA AGA AGC ACT | In, 242 bp to center axis |
| Les5 | CAG CCG CTA CAG TCC CCA ACA C | In, 262 bp to center axis |
| Les6 | CGA CTT CCA GTT CAA CAT CAG CCG | In, 278 bp to center axis |
| DC3 | CTG ATA GAT AAC CCA AGG CCA GGC | Out, 1,455 bp to edge |
| DC4 | GAA GCT GAA AGG TGG ACA GGA AAC | Out, 1,110 bp to edge |
| DC5 | GTC CAA CAA TCC AGC TTC AGG | Out, 12 bp to edge |
| Chromosome specific | ||
| ACLF1 | CTT TCC AGA TAC GAC GCA GG | Out, 52 bp to left PstI |
| ACRF1 | GGT GAC ATC ACA GTC TAA GGA GG | Out, 53 bp to right PstI |
| AC174 | CCG TGG CAG TAA CCA TTA AGA GC | In, 148 bp to edge |
| AC173 | CTT TCA CAT TCA TGA CGC TGG CC | In, 202 bp to edge |
| AC172 | GGA AAT GTG TCT AGC TCT CAC TGG C | In, 313 bp to edge |
| AC171 | CAT GGT AGG AAG CAT GGC AGC | In, 402 bp to edge |
Rate calculation.
Rearrangement rates (except those in Table 2) were determined by the maximum likelihood estimate (MLE) (see the sample calculation below). The likelihood ratio (LR) test was used to obtain P values for the observed differences. The results of limiting dilutions (number of wells with growth/total at each sampled time point) were incorporated into the calculation. For line 7#7-7-11 on day 21, 29/384 wells showed growth; day 38, 33/768; day 66, 32/768; day 87, 34/768; and day 150, 34/164. For line 7#7-7-6 on day 21, 41/384 wells showed growth; day 39, 47/576; day 64, 51/576; day 80, 54/576; and day 126, 52/576. For 7#14-11, on day 56 we obtained 17/768; day 70, 76/384; and day 92, 57/480.
TABLE 2.
Pilot study
| Cell linea | Day of cultureb | No. of doublingsc | Intact/totald (x/y) | Ratee (p) | 95% CIf |
|---|---|---|---|---|---|
| Intact | |||||
| M630FL7#1 | 43 | 57 | 6/10 | 9.0 | 2.2-23.3 |
| 7#7 | 89 | 119 | 4/12 | 9.0 | 2.7-10.8 |
| 7#7-7 | 68 | 91 | 7/11 | 5.0 | 2.3-8.2 |
| Rearranged | |||||
| 18#1 | 73 | 97 | 13/13 | <0.8 | |
| 18#1-8 | 66 | 88 | 12/12 | <1.0 |
The various lines used in this study are described in Materials and Methods.
Number of days elapsed between single-cell isolation of the test clone and sampling.
Calculated from a population doubling time of 18 h.
Number of subclones exhibiting an intact transgene structure/total number analyzed.
Estimated rearrangement rate calculated by p = 1 − (x/y)1/n. Value listed × 10−3 gives the rate.
Calculated by substituting values for the 95% CI of the ratio x/y into the above equation. Calulation cannot be made for samples giving no rearrangement.
Sample calculation by MLE.
The log likelihood (LL) for a series of estimated rates (R*) was calculated in order to find the MLE. To illustrate, the value for doubling 85 given in Table 3 was calculated as follows (with R* equal to 0.003, n equal to 85, x equal to 24, y equal to 43, the total number of wells with growth being 51, and the total number of wells filled being 576).
TABLE 3.
Rearrangement rate of the Line 78 palindrome in cell line 7#7-7-6
| Day of culture | No. of doublingsa | Intact/totalb (x/y) | Fraction intact | Ratec | 95% CId |
|---|---|---|---|---|---|
| 21 | 28 | 19/25 | 0.76 | 8.6 | 3.6-19.0 |
| 39 | 52 | 31/46 | 0.67 | 7.0 | 4.1-11.8 |
| 64 | 85 | 24/43 | 0.56 | 6.4 | 4.0-10.0 |
| 80 | 106 | 24/45 | 0.53 | 5.8 | 3.5-8.7 |
| 126 | 168 | 22/49 | 0.45 | 5.0 | 3.3-7.0 |
Calculated from a doubling time of 18 h.
Clones with an intact Line 78 transgene/total number of clones analyzed.
Rate of rearrangement per population doubling (value listed × 10−3 gives rate), calculated by the MLE (see Materials and Methods).
The 95% CI (values listed × 10−3 gives interval), calculated by the MLE (see Materials and Methods for details).
(i) The fraction of intact Line 78 transgenes (fraction intact) at a particular doubling (n) is calculated for an estimated rate (R*) according to the following formula: fraction intact = (1 − R*)n, so fraction intact = (1 − 0.003)85, and therefore fraction intact = 0.7746.
(ii) The revised fraction (RF) of intact Line 78 transgenes taking the limiting dilution results into account is calculated by summing the conditional probabilities of a cloning well containing one cell with no mutation, two cells with no mutation, three cells with no mutation, etc. (assuming a Poisson distribution given that there is at least one cell present), according to the following formula: RF = (e−λ)[e(λ)(fraction intact) − 1]/1 − e−λ, so RF = (0.911)[e(0.0932)(0.7746) − 1]/1 − 0.911, and therefore RF = 0.766.
The following formula was used to calculate λ in our example: e−λ = 1 − (total number of wells with growth/total number of wells seeded), so e−λ = 1 − (51/576) = 0.911, therefore λ = 0.0932.
(iii) LL is calculated from the observed number of clones with an intact Line 78 transgene (x) and the total number of clones scored (y): LL = x{ln[RF/(1 − RF)]} + y[ln(1 − RF)], so LL = 24[ln(0.766/0.234)] + 43[ln(0.234)], and therefore LL = −34.0.
LLs calculated for a series of estimated rates (steps i to iii) were plotted against the R* value. In the present example the peak of the curve corresponded to a rate of 6.4 × 10−3. The standard 95% confidence interval (CI) was determined by coming down 2 points from the peak LL.
Data from separate experiments were pooled by adding the LL of each experiment at each estimated rate and graphing as described above. Thus, at an estimated rate of 0.003 the LL for each experiment was determined, added, and then plotted. This was repeated for all R* values.
LR test.
Rate determinations from different experiments were compared by the LR test. The alternate hypothesis, that all experiments are significantly different, was calculated by adding the LL of the peak rates together (LL1 + LL2). The null hypothesis, that all experiments have the same mutation rate, was determined by calculating LL as though all points were from one experiment (LL1 + LL2). The difference between the LL of the alternate and null hypothesis was then determined with the formula 2[(LL1 + LL2) − (LL1 + LL2)].
The LL difference is multiplied by 2 so that the significance can be tested by using χ2 critical values. The degrees of freedom (df) is the number of experiments minus 1. Where P > 0.05, the null hypothesis (that the two rates are significantly different) is rejected.
RESULTS
The Line 78 transgene is a perfect palindrome.
To investigate effects of true palindromes in mammalian genomes, the central sequence of the Line 78 transgene was determined. Previously, the structure of the suspected palindrome in Line 78 was inferred from Southern blot results, as is the case for all other transgenic palindromes reported to date. That method is not sufficiently sensitive to exclude the possibility that the transgenes in these models were not true palindromes, but instead possessed a very small central asymmetry. Although the transgene restriction maps were consistent with a palindromic structure, and predicted, axis-specific central restriction sites were present (1, 4, 10), we were aware from other studies of Line 78 that it is possible for acquired deletions to spare the nucleotides constituting the PstI site at the exact axis of symmetry (1, 21, 22). Such deletions, which can be as small as 20 bp, do not visibly alter the appearance of the gross restriction pattern on Southern blots and, if present in the Line 78 founder, would not have been detected.
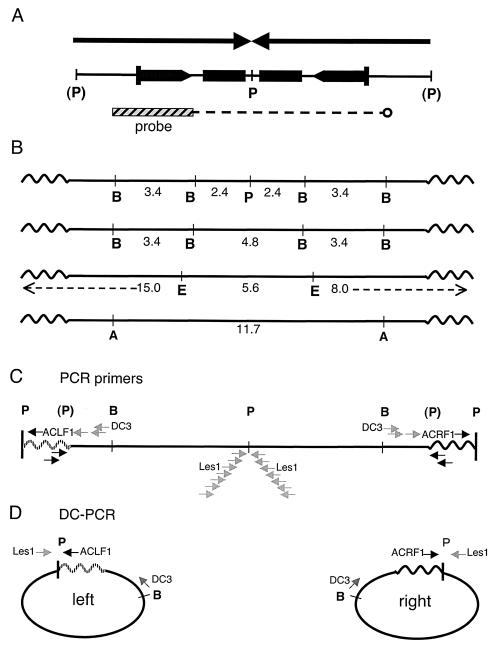
As shown in Fig. 2, an injected PstI-generated DNA fragment integrated as two copies in a tail-to-tail configuration in the Line 78 founder (1). To verify the Line 78 transgene structure at the DNA sequence level, DC-PCR (37), also known as inverse PCR, was performed. DNA prepared from Line 78 cells was digested with PstI, diluted, and self-ligated to form circles. The ligated material was then subjected to PCR. Nested, transgene-specific primers were designed that could amplify across the artificially created junction (in Fig. 2C and D, amplification with primers Les1 and DC3 was followed by a nested set; see Materials and Methods for details). Because earlier studies had suggested that the outer PstI sites were intact, this approach was expected to give either one or two PCR products: a single band of a predicted size would indicate that Line 78 harbors a perfect palindrome. A single band was obtained by DC-PCR, which was consistent with the possibility that the Line 78 sequence is a true palindrome; however, the product was 1 kb longer than expected (data not shown). Two cloned amplicons were subjected to DNA sequence analysis. Each contained, in addition to the transgene sequences, identical, approximately 1-kb nontransgene DNA at the circle joint. A BLAST search of the National Center for Biotechnology Information mouse database located the nontransgene sequence to chromosome 17 (accession no. NT_039658.1). This sequence was verified to be the integration site with new primers designed to span the right and left chromosome-transgene junctions (data not shown) (see Materials and Methods for details).
FIG. 2.
(A) Two copies of the original injected fragment are arranged tail-to-tail in Line 78 (1). (P) indicates that the PstI-generated 5′ overhangs of the fragment had lacked the terminal nucleotide of the PstI recognition site. Rectangles represent lacZ repeats; the two inner copies are truncated. The bar indicates the 3.4-kb BamHI fragment used to probe Southern blots. The dotted line indicates the region of hybridization of this probe within the palindrome. (B) Scale map of the integrated palindrome in Line 78 showing the major diagnostic digests used in the present study. Sizes of probe-positive fragments only are given. Left and right sides are defined according to EcoRI digestion. Wavy lines represent the chromosomal sequences. (C) Location of PCR primers relative to landmark restriction sites (details for each primer are given in Materials and Methods). The chromosomal flanking sequences on the left side are patterned in order to distinguish them from right-side sequences in panels C and D. The primer sequences are given in Materials and Methods; only some of the primers are labeled with their names for orientation. Transgene-specific primers are shown in gray and are mapped as they occur within the two identical palindrome arms. Chromosome-specific primers are in black; left- and right-side primers are not identical. (D) DC-PCR strategies. Only the outside primers used in nested approaches are shown. Paired transgene-specific primers Les1 and DC3 will amplify both sides of the palindrome. The side-specific DC-PCR uses one transgene and one chromosomal primer (Les1 and ACLF1 in the case of the left side and Les1 plus ALRF1 for the right side). A, ApaI; B, BamHI; E, EcoRI; P, PstI; (P), cleaved or former PstI site.
Based upon these results, which provided us with complete information on the integration site, we refined the DC-PCR-based assay through the use of primers specific for the left and right chromosomal flanks (see ACLF1 and ACLR1 in Fig. 2C and D). Both left- and right-side amplification products were separately generated, and the DNA sequences of these were determined. The results revealed that the transgene is symmetric on both sides of the PstI site, establishing that the unrearranged (or intact) Line 78 transgene is, in fact, a true palindrome. According to these analyses, the originally injected PstI fragment had inserted tail to tail between two T's on mouse chromosome 17. Both the incoming fragment and the integration site were completely conserved. Consequently, as diagrammed in Fig. 2, the PstI recognition sites at the outside edges of the palindrome were each destroyed upon integration. The two flanking chromosomal PstI sites happened to occur almost exactly equidistantly, about 1 kb to either side. (This explained why originally only one band was visualized on genomic Southern blots of the Line 78 transgene digested with PstI [1] and why only one band, not two, was generated by DC-PCR amplification with transgene-specific primers [data not shown].) These final DC-PCR results definitively establish palindromy for the Line 78 transgene. Moreover, this analysis formally demonstrates (Fig. 3A) that long true palindromes are not lethal in mice.
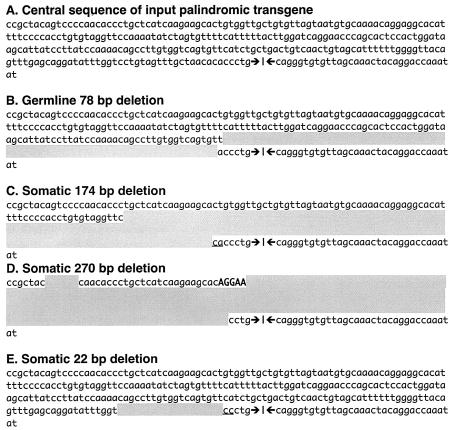
FIG. 3.
Germ line and somatic palindrome modifications are comparable. Sequences were determined by conventional PCR for panels C and D, by DC-PCR for panels A and E, and by both methods for panel B. For comparison, all deletions (shading) are shown on the left side; however, the deletion in panel B is actually on the right side, those of panels C and D are unknown, and that of E is on the left side. Underlining indicates short homologies that that can be assigned to either the left or the right side of a given deletion. Bold typeface indicates junctional insertions of undefined origin (possibly due to terminal deoxynucleotidyl transferase activity in the Ab-MLV cells; however, similar insertions have also been observed in nonlymphoid cells in other studies [21]). (A) Partial sequence of the intact central region of the Line 78 palindrome in test clones 7#7-7-6 and 7#7-7-11. The symmetry axis is indicated by the head-to-head arrows. (B) Inherited deletion arising in our colony (not detected on Southern blots). (C) Deletion in line 18#1. (D) Deletion in test clone 7#14-11. (E) Deletion in the sample subclone 7#7-7-6-22 (day 64). Deletions can also include the central PstI site (see Fig. 6 and examples in reference 22).
Permanent cell lines with the 15.4-kb palindrome.
A determination of palindrome rearrangement rates (rather than frequencies) provides an essential experimental foundation on which to base future mutational studies. In addition, we sought to quantify the stabilizing effect produced by alterations of the palindromic center by center-break-type modification. Permanent cell lines were therefore established by infection of Line 78 fetal livers with Ab-MLV, a procedure that gives rise to pre-B-cell-like transformants (29). Ab-MLV-transformed lymphoid cell lines were chosen specifically because they are stably blocked in differentiation, have a consistent population doubling time, are simple to culture without added growth factors, and typically exhibit stable, diploid chromosome counts (27, 31, 34, 35). A concern with this system is that palindrome modification or hairpin nicking in Ab-MLV transformants might exhibit significant pre-B-cell-specific attributes; however, investigations with transiently transfected palindromes and hairpins previously failed to uncover any major differences between lymphoid and nonlymphoid cells (20, 21). In the early stage of the present study, palindrome rearrangements were evident in spontaneously transformed embryonic fibroblasts arising from a single Line 78 fetus (5). Such cultures were not only heterogeneous but also difficult to subclone and proved quite unsuited to our experimental objectives. In contrast, preliminary experiments with Ab-MLV-transformed cells readily demonstrated ongoing instability through successive rounds of subclonal isolation (Table 2).
Palindrome rearrangement occurs at an extremely high rate in tissue culture cells.
To measure palindrome rearrangement rates, a single cell with an intact Line 78 structure (the “test clone”) is seeded in a cloning well on day 0, expanded, and thereafter carried continuously. At one or more intervals the culture is sampled by a limiting dilution isolation to give single-cell “sample subclones.” The multiple sample subclone cultures, after a brief expansion, are each terminated, and DNA is extracted. Rearrangement in the sample subclones is then evaluated by Southern blot. Rates are estimated according to the relationship x/y = (1 − p)n, where x/y is the fraction of intact Line 78 transgenes in the population, p is the rate of rearrangement of the Line 78 transgene per doubling, and n is the number of population doublings the test clone has undergone (approximated at once per 18 h).
To make a preliminary assessment of the feasibility of the approach outlined above, three pilot studies involving cells established from outcrossed fetal livers were undertaken (Table 2). The rate of palindrome rearrangement was estimated from a single sampling in each case and was calculated according to the above equation at 5 × 10−3 to 9 × 10−3 rearrangements per doubling, a remarkably high figure.
To determine rearrangement rates more precisely, cells descended from one transformant, 7#7, were more intensively analyzed for palindrome instability by a serial sampling protocol. Growth irregularities are associated with an apoptotic crisis that takes place soon after Ab-MLV transformation (35); thus, third-generation subclones of the 7#7 transformant, called 7#7-7-6 and 7#7-7-11, were used. Cultures of each of the intact test clones were initiated from single cells by limiting dilution, expanded, and periodically tested to determine the fraction of unrearranged cells remaining. The palindrome integrity of each of the starting test clones was confirmed by DC-PCR and DNA sequence analysis (data not shown). Up to 49 subclones were isolated and analyzed by Southern blot at each sampled time point.
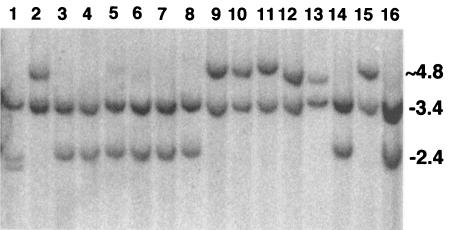
The intact line 7#7-7-6 was tested during a period spanning from 28 to 116 population doublings (days 21, 39, 64, 80, and 126). Figure 4 shows 16 of the 43 subclones analyzed on day 64. It can be seen that after only two months, more than half of the culture was rearranged. Table 3 gives refined rate calculations, based upon MLEs (see Materials and Methods), for the entire experiment. The numbers obtained were fully consistent with the cruder measurements obtained in the pilot studies.
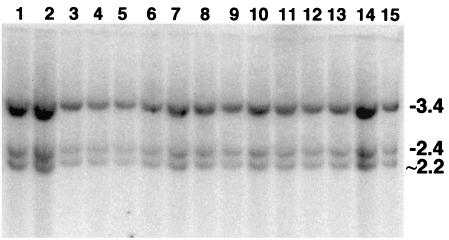
FIG. 4.
Analysis of rearrangements among 7#7-7-6 sample subclones (day 64). Cellular DNA was codigested with BamHI and PstI and was probed as indicated in the legend to Fig. 2. The subclone DNA in lanes 3, 5, 6, 7, 8, and 14 exhibited intact transgenes as indicated by 3.4- and 2.4-kb bands. The remaining subclones were rearranged, as was the subclone in lane 4, as discovered upon a subsequent EcoRI analysis (data not shown) (see Materials and Methods).
A sibling of the cell line 7#7-7-6, clone 7#7-7-11, was also tested, but we used a variant scheme designed to explore the reproducibility of the rate measurements. If reliable, rate measurements should be consistent between parallel cultures and between sibling subclones. At day 21 the 7#7-7-11 culture was sampled and shortly thereafter the test culture was divided in two. 7#7-7-11(A) was sampled on day 87. 7#7-7-11(B) was sampled on days 38 and 66 (Table 4). Three sets of data could thus be compared: (i) 7#7-7-6 as tested on days 21, 39, 64, 80, and 126; (ii) 7#7-7-11(B) tested on days 21, 38, and 66; and (iii) Line 7#7-7-11(A) tested on day 87.
TABLE 4.
Rearrangement rate of the Line 78 palindrome for cell line 7#7-7-11
| Day of culture | No. of doublingsa | Intact/totalb (x/y) | Fraction intact | Ratec | 95% CId |
|---|---|---|---|---|---|
| 21 | 28 | 19/19 | 1.00 | 0.0 | 0-3.6 |
| 38 (Ae) | 50 | 23/31 | 0.74 | 6.0 | 2.7-10.8 |
| 66 (A) | 88 | 20/30 | 0.67 | 4.7 | 2.3-8.2 |
| 87 (B) | 116 | 21/32 | 0.66 | 3.9 | 1.9-6.2 |
| Pooledf | 5.5 | 4.5-6.7 |
See footnote a of Table 3.
See footnote b of Table 3.
See footnote c of Table 3.
See footnote d of Table 3.
A and B identify two subcultures established after day 21.
Rate of palindrome rearrangement given by pooling all 7#7-7, -6, and -11 data except one time point (see the text) by the MLE (see Materials and Methods).
All of the data for the 7#7-7-6 and 7#7-7-11 test clones are given in Tables 3 and 4. By the LR test (see Materials and Methods), individual rate determinations for the test clones were not significantly different from one another (P > 0.1). Therefore, data were pooled from both test clones (7#7-7-6 and 7#7-7-11), giving an overall rearrangement rate of 5.5 × 10−3 per doubling (95% CI, by the MLE, of 4.5 × 10−3 to 6.7 × 10−3). This more extensive data set is again consistent with results of the smaller pilot study (Table 2) and show that (i) the rate of palindrome rearrangement can be measured in a mammalian cell culture system, and (ii) rates are reproducibly very high in wild-type cells.
Limitation of the rate measurement.
It was anticipated that over the long term, the serial sampling method might be compromised due to vagaries of clonal outgrowth. A single very late sample of 7#7-7-11 was taken on day 150 (corresponding to about 200 population doublings) specifically to determine whether this was a real concern. We found that at day 150, only 1 of 30 sample subclones was still unrearranged (intact). This gave a calculated rate of 15 × 10−3, which, compared to rates obtained for both 7#7-7-6 and 7#7-7-11 by the LR test (see Materials and Methods), was highly significantly different (P < 0.0005). Independent confirmation of a confounding factor associated with this one late sample was independently obtained by the observation that all 29 rearranged sample subclones exhibited one of only two different structures. The pauciclonal composition had to have arisen by subclonal outgrowth in the culture, because it was not present in earlier samples. We conclude that subclonal outgrowth can develop, and if so, it will bias the ratios of rearranged to unrearranged cells in cultures that have been carried over prolonged periods. The 150-day sample was excluded from the rate calculation. These results indicate that the possibility of subclonal outgrowth in a given culture must be accommodated in the experimental design, and monitored by specific tests, as just described.
Central deletion stabilizes a palindrome more than 25-fold.
The line 18#1 had been included in our pilot study to investigate whether or not a transgene with a small deletion of the palindrome symmetry center would be stable in somatic cells (Fig. 3C). When examined by subcloning, the deleted transgene in 18#1 did in fact appear to have been stabilized (Table 2) as previously described for germ line deletions (1, 4). To extend the preliminary analysis, the rate of transgene rearrangement in the test clone 7#14-11 was measured. The 7#14 transformant originated from the same fetal liver as transformant 7#7, thus its central deletion of several hundred base pairs (Fig. 3E) was somatically acquired. The deletion, located just adjacent to the central PstI site, had been accompanied by a small insertion so that 7#14 contained a large perfect inverted repeat separated by a 300-bp unique spacer.
The test clone 7#14-11 was sampled on days 56, 70, and 92. As shown for a number of day 92 subclones (Fig. 5), all of the sample subclones collected throughout the course of the experiment had the same structure as the original 7#14 transformant. Pooled, the 7#14-11 data give a rearrangement rate of 0 with an upper 95% confidence limit of 1.8 × 10−4 rearrangements per doubling (Table 5). This amounts to a greater than 25-fold reduction (P value by the LR test was <0.0005) below the lower bound of the 95% CI for the perfect palindrome (Table 4). Because we never observed rearrangement, the magnitude of the stabilizing effect may well be even greater. We conclude that the new central asymmetry, here generated by a somatic center-break event, stabilizes the transgene.
FIG. 5.
Lack of further rearrangement in 7#14-11. Some day 92 subclones from a culture of 7#14-11 are shown. No further rearrangements in any of the 105 subclones sampled from 7#14-11 were seen.
TABLE 5.
Rearrangement rate of the Line 78 palindrome for cell line 7#14-11
The structures of spontaneously generated variants indicate a center-break mechanism.
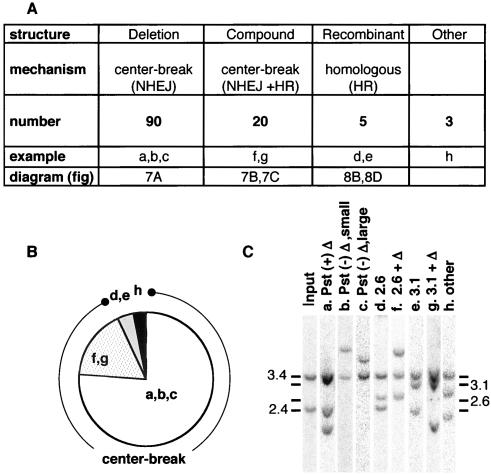
One-hundred eighteen rearrangements in total were recovered in the serial subcloning experiments. Each was characterized in order to elucidate the underlying mechanism and in order to compare somatic versus germ line events (1, 22, 39). Results are summarized in Fig. 6. Rearrangements fell into three categories according to their structural properties: those that had acquired non-homology-based deletions within the palindrome, those indicating homologous recombination between lacZ repeats, and composite structures that contained both a nonhomologous deletion as well as a recombinant lacZ repeat.
FIG. 6.
Rearrangement outcomes. (A) Three classes of outcome are defined according to structure: deletion subclones have restriction maps indicative of a central deletion, compound subclones have restriction maps indicative of a central deletion and also contain a homologously recombined lacZ copy (one subclone listed in this category had a central deletion undetected on Southern blots but revealed by DC-PCR), and recombinant subclones have restriction maps predicted for a straightforward homologous recombination and were confirmed in three of the five examples to lack central deletions by DC-PCR. The deduced mechanism and the number of subclones in each category as well as a reference to the appropriate diagrams in Fig. 7 and 8 are also given (202 other subclones were intact). Deletion subclones were subgrouped (a, b, c) according to the size of the central deletion as well as whether the central PstI site was or was not preserved. The compound and recombinant categories are each subgrouped according to whether the lacZ recombinant is 5′-inner-to-3′-outer (2.6-kb band, d and f) or 5′-outer-to-3′-inner (3.1-kb band, e and g.) Three subclones (other) had banding patterns that did not fit the criteria for any category (see the text for details). (B) Pie chart showing the proportions of each type of structure among the 118 rearranged subclones. (C) Examples of different types of patterns obtained with BamHI and PstI codigestion. Lanes are from four different gels. Adjustments were made in the composite so that reference intact lanes from each image (cropped out) would overlie.
The first category, simple nonhomologous deletion, was the largest. Seventy-six percent of the rearranged subclones had sustained deletions that extended from the center outward to one or both sides of the symmetry axis. Depending upon its extent, a central deletion will give the following band patterns with BamHI and PstI codigestion: (i) three bands, one 3.4 kb, one 2.4 kb, and a third variably sized band of less than 2.4 kb; (ii) two bands, one of 3.4 kb plus a variably sized band of less than 5.8 kb; or (iii) one band of less than 6.8 kb. Any deviation from the upper limit for the variable bands in each case would have constituted evidence of a deletion that extended beyond the palindrome proper, but no such examples came to light. All structures in this category fit the above criteria and are simply explained by the center-break scheme (1, 21) (Fig. 1 and 7A).
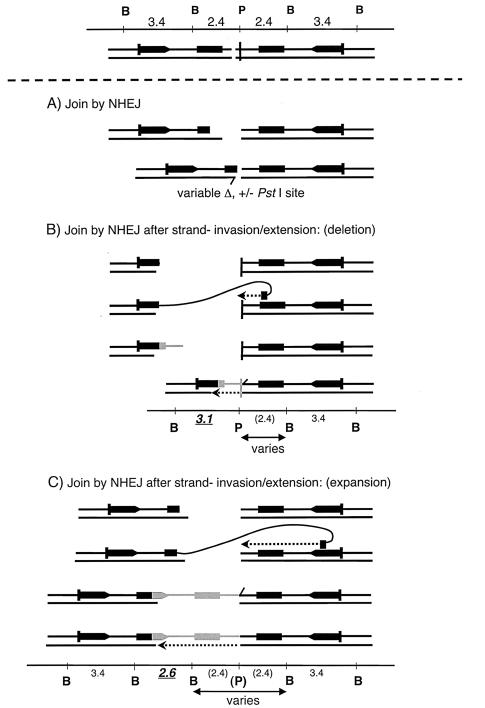
FIG. 7.
Center-break activity can generate recombinant lacZ structures as well as simple deletions. Top, a center-break is created at the palindrome symmetry center. (A) The break is enlarged and then reconnected by NHEJ. Alignment of broken ends can sometimes be mediated by microhomologies at the joined termini followed by limited gap filling (indicated by the short displaced top strand and dashed arrow); however, events that occur completely independently of any microhomology are also seen, as in Fig. 3. (B) NHEJ with strand invasion (expansion). The terminus to one side of the center-break is degraded to within the inner lacZ repeat. The 3′ end can then invade an intact outer lacZ repeat on the other side of the break. Following extension, the newly elongated end peels off and is joined to its partner by NHEJ (with gap filling). (C) NHEJ with strand invasion (deletion). The terminus to one side of the center-break is degraded to within the outer lacZ repeat; thereafter, strand invasion, extension, and joining take place as described for panel B. The predicted restriction map is given for each outcome.
Rearranged subclones with a recombinant lacZ repeat comprised about 20% of the total and were distinguished according to the presence of a 2.6- or a 3.1-kb band on Southern blots. Such clones were subjected to additional digests as described in Materials and Methods. According to other bands occurring together with the 2.6- or 3.1-kb bands, clones with recombinant lacZ repeats belonged to two distinct structural categories (Fig. 7B and C). In one category, only the additional bands predicted for straightforward homologous recombination were seen (Fig. 8); however, in the other, a banding pattern indicated compound events where homologous recombination had been accompanied by NHEJ (Fig. 7B and C). Subclones were categorized as follows.
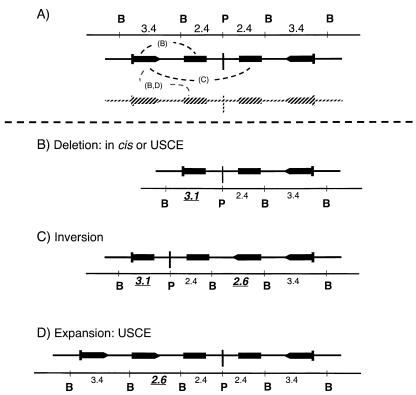
FIG. 8.
Structures generated by homologous exchange. (A) Three recombinant products from crossing over between inner and outer lacZ repeats (dashed lines) are theoretically detectable. (B) Recombination between lacZ repeats located on the same arm of the palindrome is deletional and is indicated by the appearance of a 3.1-kb band in addition to the 2.4- and 3.4-kb bands. (C) Recombination between inner and outer lacZ repeats on opposite arms of the palindrome results in inversion and yields a four-band pattern: 3.1- and 2.6-kb bands (recombinant) plus 2.4- and 3.4-kb bands (nonrecombinant). (D) Reciprocal exchange between sister chromatids (as diagrammed by the gray copy in panel A) would give one expanded chromosome with an increased number of lacZ repeats (and a 3.4-, 3.1-, 2.6-, 2.4-kb banding pattern) and one deleted product (with a 3.4-, 3.1-, 2.4-kb banding pattern) as diagrammed in panel B.
A small number of subclones exhibited one of three strictly homologous patterns and for simplicity will be described first. In principle, strictly homologous recombination of the Line 78 transgene, as illustrated in Fig. 8, might take place either in cis or between sister chromatids and might be deletional or inversional. The three detectable outcomes are shown in Fig. 8B to D. Following BamHI and PstI codigestion, straightforward inversion (involving one of the inner copies and one of the outer copies of the lacZ repeat) is predicted to generate two new diagnostic bands of 3.1 and 2.6 kb while retaining the 3.4- and 2.4-kb bands associated with the original intact structure. As diagrammed in Fig. 8C, the 3.1-kb band indicates a recombinant possessing the 5′ end of the outer lacZ repeat and the 3′ end of the inner, truncated lacZ copy. The 2.6-kb band indicates the reciprocal: the 3′ end of the outer lacZ repeat combined with the 5′ end of the inner copy. Homologous deletion, depicted in Fig. 8B, gives the diagnostic 3.1-kb band, together with the 3.4- and 2.4-kb intact bands. Homologous expansion, occurring through sister chromatid exchange, would give the diagnostic 2.6-kb band, and as for the other outcomes, it would also show the 3.4- and 2.6-kb bands (Fig. 8D). Altogether, we found no inversion clones, four deletion clones, and three expansion clones. Examples of Southern blots showing the deletion- and expansion-type homologous recombination patterns are given in Fig. 6C, lanes d and f.
Although, according to Southern blot results, the BamHI- and PstI-generated banding patterns of the seven subclones just described indicated be uncomplicated homologous recombinants, it was of interest to determine whether some of these might nonetheless have sustained a small, central alteration. We performed DC-PCR on five extant DNA samples, and in fact two of these were clearly not the result of homologous recombination only: in one case no PCR product was generated with the right-side primer (the exceptional nature of this subclone was confirmed by Southern blot analysis with other restriction enzymes), and in the second case a shortened DC-PCR product was generated with the left-side primer. The DNA sequence of this subclone revealed a 20-bp deletion (Fig. 3E). The tiny deletion underscores the necessity of establishing the central integrity of the original transgenic palindrome in these and similar studies. DNA from the remaining two recombinants was unavailable. To sum up, at least 3 (confirmed), and no more than 5 total, of the 118 rearranged outcomes were the products of an uncomplicated homologous exchange.
The 22 remaining subclones with recombinant lacZ repeats were all found to belong to the compound category, where both homology-dependent and homology-independent processes clearly played a role. Their banding patterns were inconsistent with homologous exchange but could be explained on the basis of the scheme diagrammed in Fig. 7B and C. A scheme essentially like that in Fig. 7C was earlier proposed to explain germ line Line 78 transgene expansions (39). In brief, following the generation of a central double-strand break the ends are not rejoined directly by NHEJ. Instead, resection of one of the ends to within lacZ homology occurs, generating a 3′ end that can invade an intact lacZ copy. The broken end is extended by polymerization, expelled from the temporary template, and only then rejoined via NHEJ to the other terminus created by the initial double-strand break (39). There can be an overall decrease (Fig. 7B) or an increase (Fig. 7C) in the number of lacZ repeats depending upon which lacZ repeat (an inner or an outer copy) is gene converted by invasion-extension and upon the extent to which ends are shortened by degradation prior to NHEJ. For simplicity, Fig. 7 illustrates compound events arising from strand invasions-extensions in cis. The same types of products can be generated by strand invasion of an unbroken sister chromatid (15, 39) (for some recombinants, trans events provide an economical model with fewer strand degradation steps). Clones with patterns consistent with the schemes in Fig. 7B and C were taken to be compound events. Specifically, a 2.6-kb band had to appear in conjunction with a 3.4-kb band (always) and with or without a third variably sized band of ≤5.8 kb (Fig. 7C). A 3.1-kb band had to appear in conjunction with a 2.4-kb band or a 3.4-kb band, and in the latter case it could also have a third band less than 2.4 kb in size (Fig. 7B). Southern blot results for representative 2.6- and 3.1-kb compound events are illustrated in Fig. 6C, lanes f and g.
Three subclones could not be assigned to any of the above structural groups; that is, they were not consistent with simple deletion, homologous recombination, or the compound events depicted in Fig. 7. These were classified as “other” in Fig. 6. Despite their exceptional structure, none of the three clones was grossly aberrant. One of these, as already mentioned, was initially taken as a 2.6-kb homologous recombinant, but on further analysis it gave an inconsistent DC-PCR result along with an anomalous pattern with BamHI only (5.7, 3.4, and 2.4 kb). A second (with 3.4-, 2.9-, and 2.2-kb BamHI- and PstI-generated bands) is shown in Fig. 6C, lane h, and a third had a 4.7-, 3.4-, and 1.9-kb pattern (data not shown). On paper, these band patterns could result from two separate rearrangement events or perhaps from multiple episodes of strand invasion-extension (involving one or both ends generated by a central break). Patchy, multiorigin insertions are observed for NHEJ products in plant cells and have also been seen in extrachromosomal palindrome rearrangement products as well as in some transgene rearrangements (4, 8, 21). No definitive experiment can elucidate which of these possibilities is likely to pertain, and the three exceptional recombinants were not analyzed further. Regardless, apart from the sizes of the bands exhibited, the rearrangements classed as “other” had no obviously unusual feature. That is, there were no more than three bands total, no single band lay outside the size ranges of other variants, no band was unusually intense, and all three clones had apparently acquired nonhomologous central deletions.
In short, almost all (93%) of the outcomes produced by rapid palindrome rearrangement in tissue culture cells belong to one of three delimited categories, consistent with a center-break mechanism.
DISCUSSION
Consistent, rapid palindrome modification in germ line and somatic cell contexts.
DNA sequence level analysis of the transgene in Line 78 has proved it to be a perfect 15.4-kb palindrome, providing conclusive evidence that giant, true palindromes are not lethal in mice. The Line 78 model is the only one available to date for any organism for which the symmetry axis of a palindrome is demonstrated where the palindrome is large enough to lie outside the range of what can be molecularly cloned. This circumstance provides a unique opportunity to investigate the effects of a true palindrome in somatic cells and, in particular, to focus on the properties of the mammalian machinery that modifies palindromes through central break and join events (1, 4, 21, 22, 39).
The basic features of the center-break mechanism observed in germ line rearrangement events are apparently recapitulated in tissue culture, with the advantage that it is now possible to measure rates and to survey large numbers of rearrangement outcomes. In tissue culture cells, the Line 78 palindrome rearranged at the extremely high rate of about 0.5% per population doubling. Where central asymmetry had been introduced into the Line 78 transgene through center-break modification, the rate of rearrangement dropped at least 25-fold. The possibility that the measured differences in stability could have been influenced by background gene effects is excluded by the fact that the intact or centrally deleted Line 78 cells are isogenic, having been isolated from the same fetal liver. Other confounding factors were minimized by a redundant sampling protocol, by monitoring the consistency of the results obtained thereby, and by using an approach based upon screening rather than selecting for variants. Although palindrome instability was most precisely measured in a single (mixed) genotypic background, pilot experiments conducted on cell lines derived from other crosses confirm the generality and reproducibility of observations made with Ab-MLV-transformed cells.
Nearly 120 transgene rearrangements were examined. Virtually all (93%) palindrome modifications were simple deletion or deletion with gene conversion, and along with a minor component of pure homologous recombinants (no more than 4%), the outcomes were consistent with those seen in the mouse germ line (1, 39). Thus, the outcome of palindrome rearrangement is remarkably similar between germ line and somatic cells. Moreover, the almost universal observation of central deletion implicates the center-break mechanism in both contexts (1, 21, 22).
Notably, in these studies there was no evidence for any type of palindrome-provoked genetic disaster. For example, although homologous recombination between lacZ copies in the tissue culture system gave rise to an expansion in the size of the transgene, as with the germ line rearrangements (39), such expansions were incremental rather than rampant amplifications. Overall, there was no indication (according to hybridization intensity or the appearance of supernumerary bands) of increased copies of the transgene sequences in any subclone. Deletions occurred in a delimited fashion; complete loss of the transgene was never seen here. We saw no evidence of transposition, as might have been noted if any subclone was missing the outer regions of the transgene while preserving the center. No indications of chromosomal translocation were seen in the form of a four-band pattern arising with PstI and BamHI codigestion or the appearance of any band in any subclone larger than 6.2 kb. We conclude that in a wild-type-transformed cell, center-break activity does not usually promote aberrations. Further, because Ab-MLV pre-B cells acquire mutations that inactivate p53 (36), we infer that the orderliness of palindrome rearrangement does not require active p53. (Six rearrangements derived directly from a p53 null cell line, M630FL7#1 [Table 2 and data not shown], were all small central deletions as well).
Determination of palindrome rearrangement rates in the germ line and early embryogenesis would be extremely difficult to ascertain, but the germ line rates are likely similar to the results obtained here (1, 39). For example, taking 62 cell divisions for the germ line in male mice (6) and basing the calculation on the rearrangement rate we obtained with somatic tissue culture cells, the fraction of mature sperm bearing palindrome rearrangements should be roughly 27%. This is consistent with the actual fraction of variant mice born to Line 78 fathers, given as 15 to 65% in Akgün et al. (1). It is highly significant that a close correspondence in palindrome modification emerges in two such different experimental systems: in nontransformed germ line cells in intact mice in vivo and in Ab-MLV-transformed pre-B-like cells in tissue culture. One obvious inference is that the ability to modify true palindromes via the center-break mechanism is widespread and is necessary for genome maintenance.
The physiological substrates for center-break activity may be true palindromes.
One might well suppose that true palindromes exist at multiple sites in the human genome, though any examples will, as a consequence of the cloning barrier, have been excluded from the sequence generated by the human genome project. As mentioned in the introduction, sequences on human chromosomes 11, 22, and 15 are possible examples of naturally occurring, long true palindromes in the human genome (7, 9, 17, 18, 33). Further, as demonstrated here, the presence of a long perfect palindrome in the Line 78 strain is not lethal to mammalian cells. In addition, previous breeding experiments indicate that the Line 78 palindrome is inherited in a Mendelian ratio with no overt pathological effects (1). Inherited palindromes and perhaps also palindromes that are generated de novo through replication errors or repair-induced mutation may be the actual physiological targets of the center-break mechanism. Center-break activity is an effective way to limit the cruciformation potential of a structure-prone palindrome and consequently to curb the ability of the sequence to generate disruptive discontinuities in DNA. The palindrome-associated chromosomal translocations and deletions observed in human translocations may thus represent rare examples of center-break activity gone awry.
The center-break mechanism in mammalian cells.
As proposed, the center-break model is initiated by nicking the hairpins of an extruded cruciform. An alternative possibility is that hairpin nicking is secondary to a step involving break and join events at the four-way junction at the cruciform base (23). Evidence for the latter model comes from yeast studies and is based upon analyses of nonpalindromic inverted repeats (23). Possibly both scenarios are correct, with the first step influencing the outcome. It can be imagined that if center-break modification is initiated by a Holliday junction-type resolvase followed by hairpin nicking, there may be somewhat different results than if hairpin nicking is the first step. Conceivably, early involvement of homologous recombination enzymes may support abortive strand invasion-extension events and ultimately give rise to the compound class of rearrangements, whereas more commonly, events that begin with hairpin nicking may simply proceed along a pure NHEJ pathway. Such questions can certainly be addressed in the future with Line 78 tissue culture cells that are either mutant or knocked down for expression of DNA repair enzymes.
Hairpin nicking is key in modification of true palindromes, and there is both biochemical and genetic evidence in support of a role for Mre11 (23 and references cited therein). Another candidate nuclease, Artemis, has recently been described for mammals, but it is less likely to be involved. Although the Artemis nuclease attacks hairpin DNA ends, as formed in V(D)J recombination, in a DNA-protein kinase (DNA-PK)-dependent fashion (24), cells mutant in DNA-PK are still fully competent in center-break-mediated resolution of transiently transfected palindrome circles (20, 21). The Line 78 tissue culture system opens up a straightforward experimental approach to elucidation of these and other functions responsible for successful center-break modification in mammalian cells and should provide the means for exploring the effect on genome stability when center-break modification is blocked.
In summary, we propose that center-break modification in mammals is the main means by which illegitimate recombination events caused by sporadic cruciform extrusion are kept in check. Center-break activity is responsible for virtually all of the palindrome rearrangements observed in wild-type cultured cell lines and gives rise to limited, predictable changes. A provocative implication of the fact that somatic cells are well equipped for palindrome modification is that if left alone, extrusion events may make a hidden but significant contribution to somatically acquired chromosomal aberrations in cancer.
Acknowledgments
This study was supported by a grant from the Canadian Institutes of Health Research to S.M.L.
We gratefully acknowledge David F. Andrews, Barbara Thomson, and Homayoun M. Baybourdy of the Statistics Department, University of Toronto, for assistance in the rate analyses. We also thank Yuan Xiao Zhu for technical contributions and Howard Lipshitz and Maria Jasin for comments on the manuscript.
REFERENCES
- 1.Akgün, E., J. Zahn, S. Baumes, G. Brown, F. Liang, T. J. Romanienko, S. Lewis, and M. Jasin. 1997. Palindrome resolution and recombination in the mammalian germline. Mol. Cell. Biol. 17:5559-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benham, C. J., A. G. Savitt, and W. R. Bauer. 2002. Extrusion of an imperfect palindrome to a cruciform in superhelical DNA: complete determination of energetics using a statistical mechanical model. J. Mol. Biol. 316:563-581. [DOI] [PubMed] [Google Scholar]
- 3.Cho, S. K., T. D. Webber, J. R. Carlyle, T. Nakano, S. M. Lewis, and J. C. Zúñiga-Pflücker. 1999. Functional characterization of B lymphocytes generated in vitro from embryonic stem cells. Proc. Natl. Acad. Sci. USA 96:9797-9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collick, A., J. Drew, J. Penberth, P. Bois, J. Luckett, F. Scaerou, A. Jeffreys, and W. Reik. 1996. Instability of long inverted repeats within mouse transgenes. EMBO J. 15:1163-1171. [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham, L. A. 2002. Characterization of palindrome instability in mammalian cell lines. Masters thesis. University of Toronto, Toronto, Canada.
- 6.Drost, J. B., and W. R. Lee. 1998. The developmental basis for germline mosaicism in mouse and Drosophila melanogaster. Genetica 103:421-443. [PubMed] [Google Scholar]
- 7.Edelmann, L., E. Spiteri, K. Koren, V. Pulijaal, M. G. Bialer, A. Shanske, R. Goldberg, and B. E. Morrow. 2001. AT-Rich palindromes mediate the constitutional t(11;22) translocation. Am. J. Hum. Genet. 68:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorbunova, V., and A. A. Levy. 1997. Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 25:4650-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henthorn, P. A., D. L. Mager, T. H. J. Huisman, and O. Smithies. 1986. A gene deletion ending within a complex array of repeated sequences 3′ to the human b-globin gene cluster. Proc. Natl. Acad. Sci. USA 83:5194-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honchel, R., B. A. Rosenzweig, K. L. Thompson, K. T. Blanchard, S. M. Furst, R. E. Stoll, and F. D. Sistare. 2001. Loss of palindromic symmetry in Tg. AC mice with a nonresponder phenotype. Mol Carcinog. 30:99-110. [PubMed] [Google Scholar]
- 11.Hudson, C. R., J. G. Frye, F. D. Quinn, and F. C. Gherardini. 2001. Increased expression of Borrelia burgdorferi vlsE in response to human endothelial cell membranes. Mol. Microbiol. 41:229-239. [DOI] [PubMed] [Google Scholar]
- 12.Hyrien, O., M. Debatisse, G. Buttin, and B. R. de Saint Vincent. 1987. A hotspot for novel amplification joints in a mosaic of Alu-like repeats and palindromic A+T-rich DNA. EMBO J. 6:2401-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacks, T., L. Remington, B. O. Williams, E. M. Schmitt, S. Halachmi, R. T. Bronson, and R. A. Weinberg. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 14.Jin, R., and R. P. Novick. 2001. Role of the double-strand origin cruciform in pT181 replication. Plasmid 46:95-105. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, R. D., and M. Jasin. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19:3398-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, E. L., H. Peng, F. M. Esparza, S. Z. Maltchenko, and M. K. Stachowiak. 1998. Cruciform-extruding regulatory element controls cell-specific activity of the tyrosine hydroxylase gene promoter. Nucleic Acids Res. 26:1793-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurahashi, H., and B. S. Emanuel. 2001. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 10:2605-2617. [DOI] [PubMed] [Google Scholar]
- 18.Kurahashi, H., T. Shaikh, M. Takata, T. Toda, and B. S. Emanuel. 2003. The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am. J. Hum. Genet. 72:733-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leach, D. R. F. 1994. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. BioEssays 16:893-900. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, S. M. 1994. P nucleotide insertions and the resolution of hairpin DNA structures in mammalian cells. Proc. Natl. Acad. Sci. USA 91:1332-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, S. M. 1999. Palindromy is eliminated through a structure-specific recombination process in rodent cells. Nucleic Acids Res. 27:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis, S. M., E. Akgün, and M. Jasin. 1999. Palindromic DNA and genomic stability: further studies. Ann. N. Y. Acad. Sci. 870:45-57. [DOI] [PubMed] [Google Scholar]
- 23.Lobachev, K. S., D. A. Gordenin, and M. A. Resnick. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108:183-193. [DOI] [PubMed] [Google Scholar]
- 24.Ma, Y., U. Pannicke, K. Schwarz, and M. R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781-794. [DOI] [PubMed] [Google Scholar]
- 25.Malagon, F., and A. Aguilera. 1998. Genetic stability and DNA rearrangements associated with a 2 x 1.1-Kb perfect palindrome in Escherichia coli. Mol. Gen. Genet. 259:639-644. [DOI] [PubMed] [Google Scholar]
- 26.Mizuuchi, K., M. Mizuuchi, and M. Gellert. 1982. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J. Mol. Biol. 156:229-243. [DOI] [PubMed] [Google Scholar]
- 27.Mostecki, J., A. Halgren, A. Radfar, Z. Sachs, J. Ravitz, K. C. Thome, and N. Rosenberg. 2000. Loss of heterozygosity at the Ink4a/Arf locus facilitates Abelson virus transformation of pre-B cells. J. Virol. 74:9479-9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Repping, S., H. Skaletsky, J. Lange, S. Silber, F. Van Der Veen, R. D. Oates, D. C. Page, and S. Rozen. 2002. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am. J. Hum. Genet. 71:906-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenberg, N. 1994. abl-mediated transformation, immunoglobulin gene rearrangements and arrest of B lymphocyte differentiation. Semin. Cancer Biol. 5:95-102. [PubMed] [Google Scholar]
- 30.Rosenberg, N., and D. Baltimore. 1976. A quantitative assay for transformation of bone marrow cells by Abelson murine leukemia virus. J. Exp. Med. 143:1453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberg, N., D. Baltimore, and C. D. Scher. 1975. In vitro transformation of lymphoid cells by Abelson murine leukemia virus. Proc. Natl. Acad. Sci. USA 72:1932-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka, H., S. J. Tapscott, B. J. Trask, and M. C. Yao. 2002. Short inverted repeats initiate gene amplification through the formation of a large DNA palindrome in mammalian cells. Proc. Natl. Acad. Sci. USA 99:8772-8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tapia-Paez, I., M. Kost-Alimova, P. Hu, B. A. Roe, E. Blennow, L. Fedorova, S. Imreh, and J. P. Dumanski. 2001. The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum. Genet. 109:167-177. [DOI] [PubMed] [Google Scholar]
- 34.Thome, K. C., A. Radfar, and N. Rosenberg. 1997. Mutation of Tp53 contributes to the malignant phenotype of Abelson virus-transformed lymphoid cells. J. Virol. 71:8149-8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unnikrishnan, I., A. Radfar, J. Jenab-Wolcott, and N. Rosenberg. 1999. p53 mediates apoptotic crisis in primary Abelson virus-transformed pre-B cells. Mol. Cell. Biol. 19:4825-4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unnikrishnan, I., and N. Rosenberg. 2003. Absence of p53 complements defects in Abelson murine leukemia virus signaling. J. Virol. 77:6208-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weckert, H. A., J. A. Hughes, E. M. Benson, and I. S. Dunn. 2000. Quantifiable analysis of human immunoglobulin heavy chain class-switch recombination to all isotypes. J. Immunol. Methods 233:141-158. [DOI] [PubMed] [Google Scholar]
- 38.Willwand, K., A. Moroianu, R. Horlein, W. Stremmel, and J. Rommelaere. 2002. Specific interaction of the nonstructural protein NS1 of minute virus of mice (MVM) with [ACCA](2) motifs in the centre of the right-end MVM DNA palindrome induces hairpin-primed viral DNA replication. J. Gen. Virol. 83:1659-1664. [DOI] [PubMed] [Google Scholar]
- 39.Zhou, Z. H., E. Akgun, and M. Jasin. 2001. Repeat expansion by homologous recombination in the mouse germ line at palindromic sequences. Proc. Natl. Acad. Sci. USA 98:8326-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]