Abstract
The GATA family of transcription factors participates in gastrointestinal (GI) development. Increases in GATA-4 and -5 expression occur in differentiation and GATA-6 expression in proliferation in embryonic and adult settings. We now show that in colorectal cancer (CRC) and gastric cancer promoter hypermethylation and transcriptional silencing are frequent for GATA-4 and -5 but are never seen for GATA-6. Potential antitumor target genes upregulated by GATA-4 and -5, the trefoil factors, inhibinα, and disabled-2 (Dab2) are also silenced, in GI cancers, with associated methylation of the promoters. Drug or genetically induced demethylation simultaneously leads to expression, in CRC cells, of all of the GATA-4, -5, and downstream genes. Expression of exogenous GATA-5 overrides methylation at the downstream promoters to activate the target genes. Selection for silencing of both upstream transcription factors and their target genes in GI cancers could indicate that epigenetic silencing of the involved genes provides a summated contribution to tumor progression.
GATA factors are a family of transcription regulatory proteins containing two conserved zinc finger DNA-binding domains recognizing the sequence WGATAR (28, 39). GATA-1, -2, and -3 are important in the development and differentiation of the hematopoietic cell lineage (26). GATA-4, -5, and -6 guide development and differentiation in endoderm-derived organs (24), including the induction of the differentiation of embryonic stem cells (11), specification of proper gut embryogenesis, and guidance of epithelial cell differentiation in the adult (14, 22, 29, 31). GATA-4, -5, and -6 have been implicated in cancer development. In this regard, GATA-6 might be predicted to have oncogenic effects since it is predominantly expressed in proliferating progenitor cells (14, 22, 29, 31). In contrast, GATA-4 and -5 would be more likely to behave as tumor suppressor genes since increased expression levels correlate with terminal differentiation in intestinal epithelium (14) and terminal differentiation induced in colorectal cancer (CRC) cells by sodium butyrate (14, 20). GATA-6 expression decreases in these latter settings, and GATA-6 may function through a repressive effect on GATA-4 (14). Diminished GATA-4 and/or GATA-5 expression has been reported in serous ovarian cancers (23) and gastric cancer (GC) (3), and the chromosome regions for GATA-4 (8p23.1-p22) (23) and GATA-5 (20q13.2-q13.3) (32), are frequent targets of deletion in cancer (13, 18). Importantly, GATA proteins bind the promoters of, and have been suggested as transcriptional activators for, a number of proposed antitumor genes, as discussed in detail below.
Despite growing evidence linking loss of GATA-4 and -5 and downstream target functions to cancer development, mutations in these genes have not been frequently found. We now show a high incidence for epigenetic silencing of GATA-4 and -5 in both human CRC and GC. Surprisingly, a series of proposed downstream GATA target antitumor genes are also silenced with associated epigenetic silencing marks at their promoters. Both the upstream and the downstream genes are simultaneously reactivated by drug and genetic demethylating strategies. Overexpression of GATA-5 alone can also activate the target genes. We suggest that a hierarchy of related gene silencing events may cooperate to drive the progression of individual tumors.
MATERIALS AND METHODS
Cell lines and tissue samples.
We studied 6 CRC cell lines (RKO, HCT116, DLD-1, HT29, LoVo, and SW480), 1 GC cell line (AZ521), and 45 primary CRC and 27 primary GC samples. All primary normal and neoplastic tissues studied were collected under clinical research guidelines at all participating institutions.
Drug treatment of cells and RNA extraction.
The CRC and AZ521 cell lines were grown in Dulbecco modified Eagle medium, minimal essential medium, or McCoy's supplemented with 10% fetal bovine serum, penicillin, and streptomycin. For demethylation studies, cells were treated daily with 5 μM 5-aza-2′deoxycytidine (DAC; Sigma) for 48 h (41). We also treated AZ521 and HCT116 cells with a histone deacetylase inhibitor, TSA (Wako), alone and in a combination of DAC plus TSA (6, 41). Total RNA was isolated by using the Trizol reagent (Invitrogen).
RT-PCR procedures.
For reverse transcription-PCR (RT-PCR), 2 μg of total RNA was reverse transcribed by using the Superscript kit (Invitrogen), and we amplified all genes with multiple cycle numbers (28 to 35 cycles) to obtain semiquantitative differences in their expression levels. GATA-4, -5, and -6 primer pairs were those previously described (3), and the primer sequences and RT-PCR conditions for all other genes are available upon request.
Methylation analyses.
DNA extraction, bisulfite treatment, DNA sequencing (Johns Hopkins University School of Medicine Biosynthesis and Sequencing Facility, Department of Biological Chemistry), and methylation-specific PCR (MSP) were performed as previously described (7, 17), and the primer sequences utilized for all genes are available upon request.
Recombinant adenovirus generation and infection procedure.
Full-length GATA-5 was amplified from human GC cDNA according to GenBank sequences (NM080473 and AL499627) and subcloned into a pAdTrack-CMV shuttle plasmid (16). The virus titer was determined by plaque assay in low-passage 293 cells, and infection was performed at doses of 0.4 PFU/cell in HCT116 cells, 8 PFU/cell in RKO cells, and 4 PFU/cell in AZ521 cells to give at least 70% green fluorescent protein-reactive cells with minimal to no cytotoxicity.
Immunoblotting.
For examination of GATA-5 protein expression, adenovirus-infected cells were harvested after 48 or 72 h, lysed in sample buffer (LB broth, dithiothreitol, and benzenesulfonyl fluoride), and Western blotting was performed on 5 μg of cell lysate with a goat GATA-5 polyclonal antibody (1:200 dilution; Santa Cruz Biotechnology). For examination of trefoil factor 1 (TFF1/p52) expression, we performed Western blotting with 40 μl of cell culture medium with a mouse anti-pS2 peptide (1:150 dilution; Zymed Laboratories).
RESULTS
Frequent epigenetic silencing of GATA-4 and GATA-5 in CRC and GC.
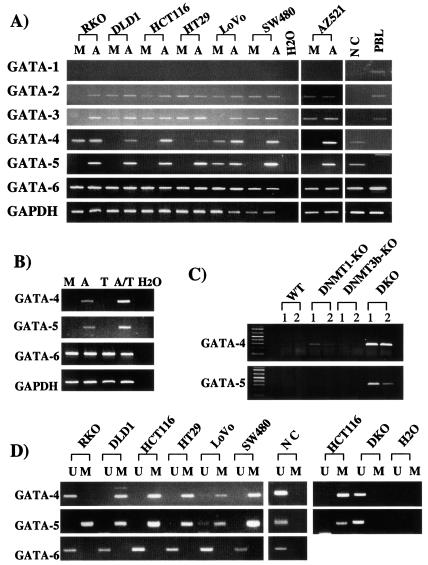
As shown by semiquantitative RT-PCR, GATA-1, -2, and -3 are expressed in lymphocytes but not in normal colon (Fig. 1A). GATA-4, -5, and -6 are all expressed in normal colon, whereas only GATA-6 is also expressed in lymphocytes (Fig. 1A and Table 1). GATA-1 is not expressed in any of the cancer cell lines, whereas GATA-6 is expressed in each. Interestingly, GATA-2 is expressed in all of the cell lines except RKO CRC cells, and GATA-3 is absent from RKO and LoVo CRC cells (Fig. 1A). Most strikingly, four of six CRC cell lines and the GC line do not express GATA-4; all but LoVo CRC cells lack GATA-5, whereas five of seven lines lack both GATA-4 and -5 (Fig. 1A and Table 1).
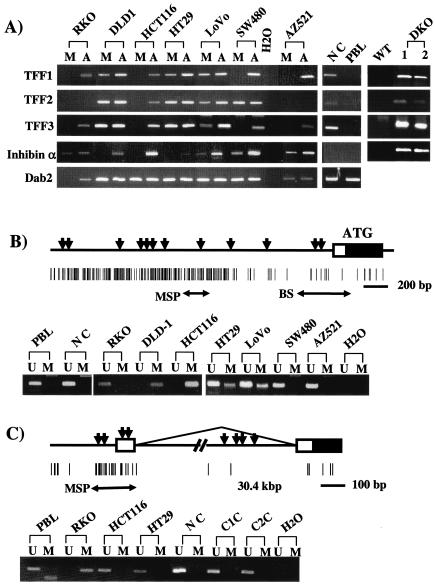
FIG. 1.
GATA-4, -5, and -6 expression in GI cancer cell lines. (A) GATA-4, -5, and -6 expression levels were examined by RT-PCR in seven cancer cell lines (CRC lines RKO, DLD1, HCT116, HT29, LoVo, and SW480 and GC cell line AZ521) with (lanes A) and without (lanes M [mock]) treatment with DAC and in normal colonic mucosa (lane NC) and peripheral blood lymphocytes (lane PBL). GAPDH expression is used as an internal loading control for the RT-PCR, and H2O indicates no RNA added. (B) GC cell line AZ521 was treated with low-dose DAC alone (lane A), TSA (lane T), a combination of these two drugs (lane A/T), or mock treatment (lane M), and examined by RT-PCR as described in panel A. (C) GATA-4 and GATA-5 expression was examined as described in panel A in wild-type (WT) HCT116 colon cancer cells and two clones each of these cells in which both alleles of DNA methyltransferases 1 (DNMT1-KO) and 3b (DNMT3b-KO) or both DNA methyltransferases (DKO) were knocked out (21). (D) MSP analysis of the promoter CpG islands of GATA-4 and -5 (primer regions depicted by black arrows [MSP] with an asterisk in Fig. 2) in six CRC lines and normal colon mucosa. PCR products recognizing unmethylated (lanes U) and methylated (lanes M) CpG sites are analyzed in 2.5% agarose gels stained by ethidium bromide. To the right, this MSP analysis is shown for GATA-4 and -5 in the HCT116 colon cancer wild-type and the DNMT1 plus DNMT3b knockout cells (DKO) used in panel C.
TABLE 1.
Summary of methylation status, expression levels of GATA genes, and candidate target genes in cancer cells before and after DAC treatment
| Gene | RKO
|
DLD1
|
HCT116
|
HT29
|
LoVo
|
SW480
|
AZ521
|
Normal colon
|
PBLc
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mta | Exprb
|
Mt | Expr
|
Mt | Expr
|
Mt | Expr
|
Mt | Expr
|
Mt | Expr
|
Mt | Expr
|
Mt | Expr | Mt | Expr | ||||||||
| M | A | M | A | M | A | M | A | M | A | M | A | M | A | ||||||||||||
| GATA-4 | U | + | + | M | − | + | M | − | + | M | − | + | U/M | + | ++ | M | − | + | M | − | + | U | + | U | − |
| GATA-5 | M | − | + | M | − | + | M | − | + | M | − | + | U/M | + | ++ | M | − | + | M | − | + | U | + | U | − |
| GATA-6 | U | + | + | U | + | + | U | + | + | U | + | + | U | + | + | U | + | + | U | + | + | U | + | U | + |
| TFF1 | M | − | + | U | + | ++ | M | − | + | U | + | ++ | U | + | ++ | M | − | + | M | − | + | U/M | + | M | − |
| TFF2 | NA | − | − | ND | + | + | M | − | + | M | + | + | ND | + | + | ND | + | + | NA | − | − | ND | + | ND | − |
| TFF3 | ND | − | + | ND | + | + | M | − | + | M | + | + | ND | + | ++ | ND | + | + | ND | − | + | ND | + | ND | − |
| inhibinα | U | + | + | M | − | + | M | − | + | U/M | − | +/− | U/M | + | ++ | U | + | ++ | U | + | ++ | U | − | U | − |
| Dab2 | M | − | + | U | + | + | U | + | + | U | + | + | U | + | + | U | + | + | U | + | + | U | + | U | + |
Mt, methylation status. Abbreviations: U, unmethylated; M, methylated; NA; genomic sequences were not dected by either MSP and bisulfite sequencing; ND; not done.
Expr, expression. Abbreviations: M, mock treatment; A, DAC treatment. Key: −, undetectable level; +, detectable level; ++, increased expression level from the “+” basal level. Analyses were done by RT-PCR.
PBL, peripheral blood lymphocytes.
We used the demethylating agent DAC to initially study the epigenetic status of GATA-4, -5, and -6 in each of the cell lines (Fig. 1A and Table 1). Each basally silent GATA gene, except GATA-1, which is not expressed in normal colon, is reexpressed by this treatment. Furthermore, the silenced GATA-4 and -5 genes had characteristics of hypermethylated tumor suppressor genes (41), (6), since treatment with the histone deacetylation inhibitor TSA alone fails to reactivate these genes but is synergistic with a low dose of DAC in doing so (Fig. 1B). Finally, expression of both GATA-4 and -5 is restored in HCT116 CRC DKO cells (Fig. 1C) in which two key DNA methyltransferase genes, DNMT1 and DNMT3b, have been biallelically disrupted with resultant virtual abolition of DNA methyltransferase activity (34), and there is very minor expression of GATA-4 in HCT116 cells in which DNMT1 alone (35) is knocked out (Fig. 1C).
GATA-4 and -5 have aberrant promoter CpG island methylation in cultured and primary CRC and GC.
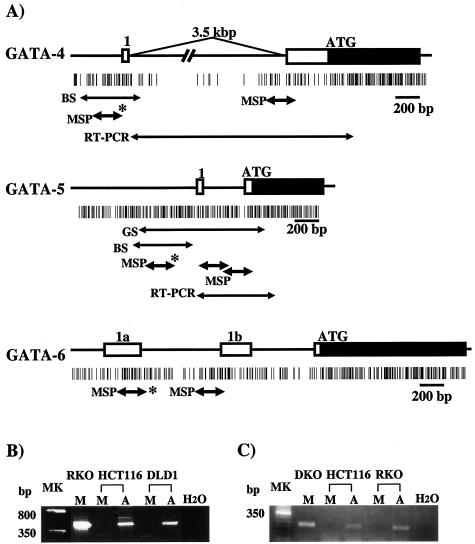
We studied the promoter methylation status of GATA-4, -5, and -6. Through RT-PCR studies combined with new expressed sequence tag (EST) identification in database searches, we have clarified the 5′ structure of each of the genes and identified CpG islands associated with the most 5′ promoter regions of each (Fig. 2). MSP analyses revealed these islands to be typical in having a nonmethylated status regardless of the gene expression state (4) in normal lymphocytes and normal colon from patients without cancer (Fig. 3A and Table 1). GATA-4 has a weak methylation signal in 2 normal colon mucosa samples from patients with cancers in which the gene is hypermethylated, as detailed below, but is not methylated in 12 other normal samples from patients with CRC (Fig. 3A and C). Methylation of GATA-5 is not seen in any normal samples (Fig. 3A and C), and neither gene is methylated in five normal gastric mucosa samples (data not shown).
FIG. 2.
Schematic representation of 5′ regions of GATA-4, -5, and -6 and MSP analyses of promoter methylation status. (A) Schematic representation of the genes. The GATA-4 diagram includes a new exon 1, located 3.5 kb upstream of the previously designated exon 1 (identified from EST BG718444), genomic sequences containing this EST (AC090790 and AC069185), and a confirmatory PCR approach showing the EST to be contained in the single transcript amplified for this gene (see panel B). The newly reported GATA5 cDNA (no. NM080473) includes one 5′-untranslated exon (41 bp). This newly identified exon 1 is located 387 bp upstream of exon 2 that contains the translation start site in the genomic GATA-5 sequence (no. AL499627). The data for the genomic structure of GATA-6 (A and B) was obtained from the newly reported sequence of this gene (GenBank no. AC009669), which reveals two 5′-untranslated exons (1a and 1b). Boxes indicate exons, including coding (black) and noncording (white) regions. Vertical bars show CpG sites. Black arrows below the CpG sites indicate the regions analyzed by MSP, genomic sequencing (GS), and bisulfite sequencing (BS) in the present study. The regions analyzed by MSP for which methylation status corresponded to GATA-4, -5, and -6 expression are indicated by an asterisk. (B) RT-PCR analysis of 5′-untranslated region of GATA-4 by using the primer set (see panel A, RT-PCR) in CRC lines. (C) RT-PCR analysis of 5′-untranslated region of GATA5 with the primers designated as in panel A. Lanes (B and C): MK, 50-bp ladder marker; M, mock treatment of cells; A, treatment with DAC; H2O, no RNA added.
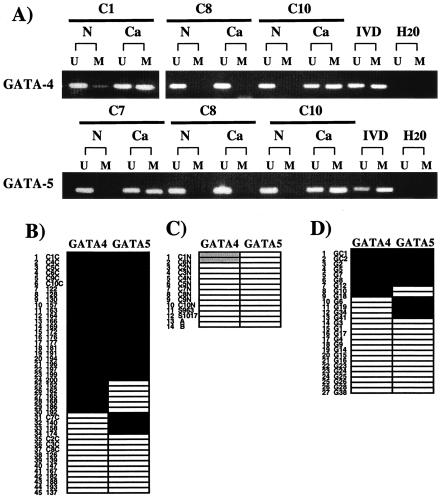
FIG. 3.
Methylation analysis of GATA-4 and -5 in noncultured normal and neoplastic GI samples. (A) Examples of MSP analyses (carried out as described for Fig. 1D) of GATA-4 and -5 in noncultured colon cancer tissues (Ca) and corresponding normal mucosae (N). Lanes: U, unmethylated alleles; M, methylated alleles; IVD, in vitro-methylated control; H2O, no DNA added. (B) Summary of the analyses for GATA-4 and -5 methylation in 45 primary CRCs. Each number in the vertical column represents a single tumor. Key: black, detection of methylated alleles; white, detection of unmethylated alleles only. (C) MSP analyses for normal colon mucosa samples from patients without (n = 2) or with (n = 12) cancer. Shaded boxes indicate weak detection of methylated alleles in two patients with GATA-4 simultaneously hypermethylated in cancer. (D) Summary for 27 primary GCs.
In contrast to the normal patterns described above, the promoters of GATA-4 and -5 were abnormally methylated in cultured gastrointestinal (GI) cancers in which these genes are basally silent, whereas GATA-6 was not methylated in any of these same cultures (Fig. 1D and Table 1). In the CRC HCT116 cells, the wild-type cells contained only signal for methylated alleles of GATA-4 and -5, whereas only unmethylated alleles were found in the DKO cells (Fig. 1D). In addition, GATA-4 and -5 are also frequently hypermethylated in primary tumors, with strong MSP methylation signals in 30 of 45 (66.7%) and 28 of 44 (63.6%) primary CRC tissues, respectively, and both genes are hypermethylated in 24 (53%) of the tumors (see, for example, Fig. 3A and B). Of 27 GC tissues, 9 (33.3%) were found to be GATA-4 methylation positive, 11 (40.7%) were found to be positive for GATA-5, and 7 (26%) had hypermethylation of both genes (Fig. 3D).
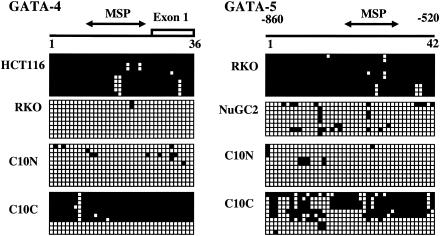
In selected samples, we verified MSP results by bisulfite sequencing (Fig. 4). GATA expression-negative cultured tumor cells (GATA-4 [HCT116] and GATA-5 [RKO]) and methylation-positive primary CRC samples show dense methylation of the promoter CpG islands, but expression-positive cultured tumor cells (GATA-4 [RKO] and GATA-5 [NuGC-2]), a GATA-5-positive GC line (3), and normal colon samples showed only scattered methylation within the examined regions. Even one of the normal colon samples with a weak MSP methylation signal for GATA-4 (C1N) showed hypermethylation only in the 5′ and 3′ borders of the promoter CpG island (data not shown). The primary colon cancer, C10, shown to be hypermethylated for the promoter regions of GATA-4 and -5 by MSP (Fig. 3A), was densely methylated for most alleles, as determined by bisulfite sequencing (Fig. 4).
FIG. 4.
Sodium bisulfite DNA sequencing of GATA-4 and -5 in colorectal (RKO and HCT116) and gastric (NuGC2) cancer cell lines and in various noncultured GI tissue samples. Each horizontal row of squares represents analysis, in a single clone of bisulfite-treated DNA, of 36 (GATA-4) or 42 (GATA-5) CpG sites contained in the region shown. Solid and open squares represent methylated and unmethylated CpG sites, respectively. GATA expression-negative cell lines (GATA-4 in HCT116 and GATA-5 in RKO) show densely methylated clones, but expression-positive cells (GATA-4 in RKO and GATA-5 in NuGC2 GC cells) have predominantly unmethylated clones. A primary colon cancer (case C10C) has predominantly methylated clones of GATA-4 and -5, and normal colon mucosa from the same patient (C10N) has unmethylated clones.
GATA genes and their candidate downstream targets have independent epigenetic silencing in GI cancers.
We next sought to determine how GATA gene silencing might correlate with the expression of candidate downstream genes that have been reported to be upregulated by these transcription factors and some of which are speculated to act as putative antitumor genes. One such group, the TFF genes, are predominantly expressed in gastric (44) and colonic epithelium (40) and encode for secreted proteins that help guide epithelial cells properly during repair of damaged GI epithelium (25) (9). TFF1 (also known as pS2) is known to be a tumor suppressor gene. Approximately 30% of TFF1 knockout mice develop gastric adenomas and carcinomas (25), and occasional mutations, allelic deletions, and reduced expression occur in human primary GCs (12, 33). Importantly, TFF1 and TFF2 (1) are upregulated by the CRC prevention agents, nonsteroidal anti-inflammatory drugs, indomethacin, and aspirin (2). We found that TFF-1 to -3, while not expressed in normal lymphocytes, were all expressed in normal colon but that TFF1 expression was absent from four, TFF2 was absent from three, and TFF3 was absent from three of seven cancer cell lines (Table 1 and Fig. 5A).
FIG. 5.
Expression and methylation status of GATA downstream target genes in cultured colon cancer and GC cells and normal tissues. (A) RT-PCR analyses of the expression of each gene in the same colon cancer and GC cell lines and normal tissues as given for GATA analyses in Fig. 1A. Also, to the far right, is shown an analysis of expression, carried out as described above, of the TFF genes and inhibinα in the HCT116 colon cancer wild-type (WT) and DNMT1−/− plus DNMT3b−/− cells (DKO) shown in Fig. 1C. Lanes: M, mock treatment of cells; A, treatment with DAC; NC, normal colon; PBL, normal lymphocytes; H2O, no RNA added. (B) Methylation status of the inhibinα gene. A schematic of the 5′ region of the gene is shown above in which the rectangle depicts the first exon, and the blackened area denotes the coding region within this exon. The black triangles represent positions of consensus GATA-binding sites, and the vertical lines each represent a CpG site. The large arrow (BS) denotes a region of bisulfite sequencing for the CpG poor region previously thought to be the only promoter region (see the text), and the smaller arrows (MSP) represent the positions of primers used for the MSP analysis in all of the cancer cell lines of the newly defined CpG island discussed in the text and shown in the lower part of panel B. Lanes: U, unmethylated alleles; M, methylated alleles; PBL, normal lymphocytes; NC, normal colon; H2O, no DNA added. (C) Methylation status of Dab2. A schematic of Dab2 is shown above in which a 5′ untranslated exon 1 (open box) is located upstream from exon 2 which contains the ATG for start of the coding region (black area within the square for exon 2). Arching line, mRNA splicing which joins exon 1 to exon 2; black triangles, positions of GATA-binding sites; vertical lines, CpG sites and the island around exon 1; arrows at the bottom (MSP), position of MSP primers used to analyze the methylation status of the CpG island as shown in the panel below. The lower part of panel C shows examples of MSP results for the methylation status of the Dab2 5′ CpG island. Lanes: U, unmethylated alleles; M, methylated alleles; PBL, normal lymphocytes; NC, normal colon; C1C and C2C, colon cancers.
Both GATA-1 and -4 are reported to bind to and upregulate the promoter of the inhibinα gene, a member of the transforming growth factor β superfamily (10, 21), which induces gonadal sex cord-stromal tumors when disrupted in mice (27). We could not detect expression of inhibinα in either normal lymphocytes or colon. However, a serial analysis for gene expression tag and EST (GenBank no. BM987739) have been identified for normal colon mucosa, and this gene was clearly expressed in some of the tumor cell lines (Fig. 5A and Table 1). However, expression of this gene was absent in two of the seven cancer lines studied, and the gene was barely to poorly expressed in two other cancer lines (Fig. 5A and Table 1).
Disabled-2 (Dab2) is an important candidate tumor suppressor gene reported to be directly activated by GATA-6 (30). Studies of this activation, however, have been confusing because a more recent study (37) suggested that the true promoter region and exon 1 for this gene are at least 14 kb farther upstream from the promoter region described in the earlier report. Our findings, presented below, clarify the role of this new region even further and link it to expression of the gene. Dab2 is not expressed at a very high frequency in breast and ovarian cancers, and its absence has been correlated to the ability of epithelial cancer cells to grow independently of the basement membrane (38). We found that Dab2 is expressed in normal colon and lymphocytes. However, using both RT-PCR primers for the internal coding region and primers linking the newly reported upstream first exon to this coding area, we found that expression was absent in one of the CRC lines (i.e., RKO cells) tested here (Table 1 and Fig. 5A).
In considering the pattern for basal expression of all of the candidate genes for GATA regulation (Table 1), line RKO lacks five of the six genes, line HCT116 lacks four, and line AZ521 lacks three. However, despite evidence that GATA factors can upregulate the expression for all of the downstream genes studied, the simple absence of GATA-4 or GATA-5 expression does not seem to account for the loss of downstream gene expression. For example, cell lines HCT116, DLD1, HT29, SW480, and AZ521 all demonstrated a decrease in GATA-4 and -5 expression. However, TFF1 was not expressed in HCT116, SW480, and AZ251 but was expressed in DLD1 and HT29 (Table 1). TFF2 was not expressed in HCT116 and AZ251 but was expressed in DLD1, HT29, and SW480. Similar discrepancies were apparent for other genes (Table 1).
Importantly, however, silencing of all of the downstream genes, despite this lack of coordination between expression of upstream and downstream genes, appeared to be, as for GATA-4 and -5, under epigenetic regulation in many of the GI cancer cell lines. When the cells were treated with DAC, each downstream gene is activated in virtually every cell line in which the gene lacks basal expression, and in many instances (TFF1, TFF3, and inhibinα) a low basal expression was further increased (Fig. 5A and Table 1). Two exceptions occurred in cell lines RKO and AZ521, wherein TFF2 was not reexpressed after DAC treatment (Fig. 5A and Table 1). As described below, the promoter region for TFF2 appeared to be either homozygously mutated and/or deleted from the genome of these above two cell lines. In the CRC cell line HCT116, in which the TFF2 promoter is present, the gene was readily activated by DAC (Fig. 5A and Table 1). Also, all four silent genes in wild-type HCT116 cells, TFF1 to -3 and inhibinα, were reexpressed in the HCT116-DKO cells (Fig. 5A).
What might account for the independent epigenetic regulation of upstream GATA and downstream target genes? We found that, as for GATA-4 and -5, the expression of the downstream genes was associated, for the most part, with methylation of their promoter regions, although several different types of methylation patterns were involved. Basal silencing in cancer cells of the candidate GATA target genes, inhibinα and Dab2, appeared to involve classic tumor specific hypermethylation of promoter CpG islands. We found that the initial region defined as the promoter for inhibinα (Fig. 5B) was CpG poor, contained only two GATA-binding motifs (36), and was methylated in normal and tumor cell lines regardless of expression status (data not shown). However, we found a typical CpG island located ca. 700 bp upstream that contained multiple additional GATA-binding sites (Fig. 5B). The methylation status of this CpG island correlated exactly with the expression status being unmethylated in normal colon or lymphocytes, unmethylated or only partially methylated in cancer lines that basally express inhibin α, and fully methylated in the three lines in which the gene is basally silent (Table 1 and Fig. 5B). The upstream promoter region and untranslated exon 1 of Dab2 (37) has an associated CpG island, which in current databases lies 30 kb from exon 2 (Fig. 5C). By MSP analysis, this island was determined to be unmethylated in normal colon and lymphocytes and in the six cell lines that express the gene but fully methylated in RKO cells in which this gene is basally silent and reactivated after DAC treatment (Table 1 and Fig. 5C).
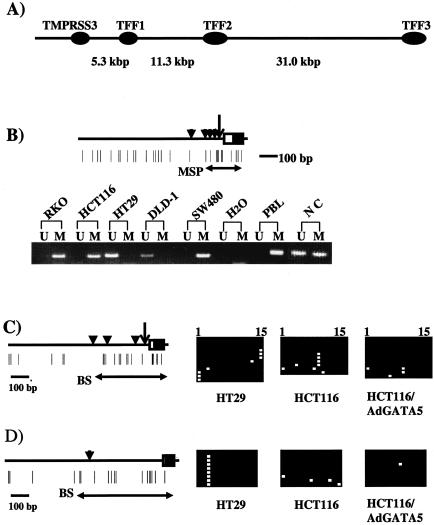
The epigenetic silencing of the TFF genes appears to be much different than for all of the above genes. No CpG islands are found between these three genes, which cluster together in order within 45 kb on chromosome 21q22.3, nor between TFF1 and a separate gene located 5 kb upstream (Fig. 6A). The CpG-poor promoters of these genes are thus more typical for those of tissue-specific genes, which can normally be differentially methylated in correlation to gene expression status (4). Indeed, for each of the cell lines and normal tissues in which TFF1 was basally silent, only methylated alleles for this gene were detected by MSP, whereas the promoter is only partially methylated in normal colon and unmethylated in the three cell lines in which TFF1 was basally expressed (Fig. 6B and Table 1). The promoter regions for TFF2 and TFF3 were examined by bisulfite sequencing. As noted above, no promoter region sequences could be amplified for TFF2 in lines RKO and AZ521, wherein this gene was not basally expressed or reactivated by the demethylating maneuvers. Otherwise, both genes were methylated in cell lines in which the genes were basally expressed or silent (Fig. 6C and D). Thus, these were the only two downstream genes in our study for which promoter methylation did not correlate with expression status.
FIG. 6.
Methylation status of TFF1 to TFF3 in cultured colon cancer cells. (A) Schematic of the alignment of the three TFF genes on chromosome 21q22.3. The location of a separate gene, TMPRSS3, upstream from TFF1 is also shown. No CpG islands could be located anywhere along the depicted stretch of genomic sequence. (B) Methylation status of TFF1. A schematic of the 5′ region of the gene depicts the transcription start site (large vertical arrow) and exon 1 is shown in the rectangle, with the coding region portion shown in solid black. Vertical black triangles, GATA binding sites; vertical lines, CpG sites. Horizontal arrows (MSP) show the primer sites for the MSP analysis in the panel below for selected cancer cell lines and normal tissues (NC, normal colon; PBL, peripheral blood lymphocytes). (C) Schematic of the 5′ region of TFF2. All symbols are exactly as for those in Fig. 6A except that the horizontal arrow (BS) show the area represented in the bisulfite sequencing shown directly beside the schematic. For the sequencing all horizontal squares represent CpG sites in individual sequenced clones (white, unmethylated; black, methylated). The sequencing is shown for (i) HT29 cells in which the gene is expressed, (ii) HCT116 cells in which it is not, and (iii) these same cells which express the gene after adenoviral expression of GATA-5 (see Fig. 7). (D) Schematic of the 5′ region of TFF3. All symbols are as described for the other panels and, again, the horizontal arrow (BS) shows the area represented in the bisulfite sequencing shown directly beside the schematic.
GATA-5 overexpression partially overrides epigenetic silencing of downstream target genes.
The epigenetic profile derived above suggests a complicated scenario in GI cancers wherein there is a potentially redundant epigenetic silencing of both upstream transcription factors and genes that have been implicated as potential downstream targets for activation by these factors. We thus questioned whether the changes at the downstream genes might be insufficient to completely prevent their activation by GATA genes. Using an adenovirus system (16), we transiently expressed exogenous GATA-5 protein (Fig. 7A and B) in HCT116, RKO, and AZ521 cells, in which this gene is basally silent. This resulted in reactivation, at the transcript level (Fig. 7C and Table 1) of inhibinα and TFF1 in all cell lines in which these genes are basally silent. When examined at the protein level, the GATA-5 overexpression resulted in expression of the secreted TFF1 protein (15) in the media of the cell cultures (Fig. 7D). TFF2 is not activated in RKO and AZ521 lines, in which the promoter is mutated or deleted, as noted previously, but is reexpressed in HCT116 cells, in which the GATA-binding sites for this gene are present (Fig. 7C). Dab2 is also reactivated at the transcript level in the one cell line, RKO, in which it is silent (Fig. 7C). Only one of the downstream genes, TFF3, is not reactivated in the lines in which the gene is basally silenced (Fig. 7C) and thus may not be a target for GATA-5.
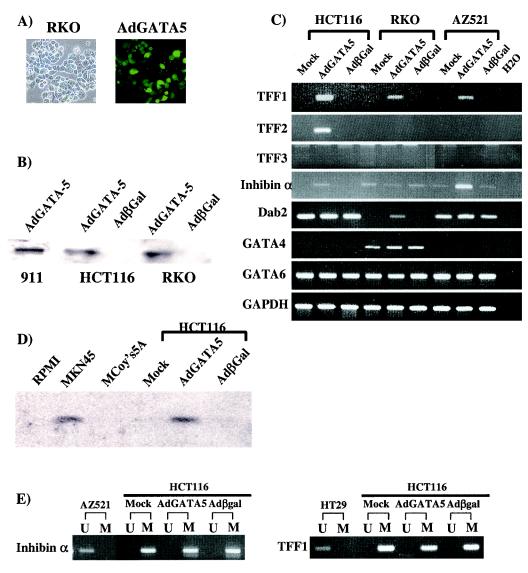
FIG. 7.
Overexpression of GATA-5 in GI cancer cells. (A) Morphological analysis in RKO cells after a GATA-5 construct was overexpressed by using an adenovirus system (20). The left subpanel shows the phase-contrast morphological appearance, and right subpanel shows green fluorescent protein expression in the same fields. (B) Immunoblotting with anti-GATA-5 antibody in cancer cells. The 911 cell line (16) used to package the viral construct served as a positive control for production of the protein. GATA-5 protein is basally undetectable in HCT116 and RKO colon cancer cells, which exhibit GATA-5 promoter methylation. Adenovirus (AdGATA-5) overexpression of GATA-5 results in a strong expression of the expected Mr 45,000 form in both cancer cell lines. The virus expressing the Escherichia coli β-galactosidase gene was used as negative control (Adβgal). (C) RT-PCR analyses of expression for GATA-5 target genes. RNA was extracted from adenovirus-infected cells for PCR analyses for expression of each gene. Note that GATA-5 overexpression induces reexpression of each candidate target gene, except for TTF3 in all cell lines and TFF2 in RKO and AZ521 cells, where the gene is homozygously mutated and/or deleted, in each cell line where basal expression is absent. (D) Immunoblotting with anti-TFF1 antibody in culture medium collected after 48 h from a positive control GC cell line MKN45, which has an unmethylated and expressed TFF1 gene, and colon cancer HCT116 cells in which the gene is hypermethylated and silenced. Note the TFF1 protein in the MKN45 cell media but only in the HCT116 media when cells are infected with adeno-GATA-5 but not when cells are infected with adeno-β-Gal. The RPMI culture medium used for MKN45 and the McCoy's 5A medium used for HCT116 cells are shown as additional negative controls. (E) MSP analyses of 5′ region methylation status of inhibinα and TFF1 before and after overexpression of GATA-5.
In each case described above in which gene reactivation by GATA-5 occurred, the involved genes remain fully methylated (for examples for TFF2, see Fig. 6C and D; for examples for inhibinα and TFF1, see Fig. 7E) even though our analyses indicated robust activation of the genes in short-term studies.
DISCUSSION
Despite growing evidence linking loss of function for GATA transcription factor genes and several of their candidate downstream target genes to cancer development, mutations in such genes have not generally been found. Since epigenetic gene silencing, as well as mutations, can account for tumor suppressor gene loss in cancer (19), our present data suggest that GATA-4, GATA -5, inhibinα, Dab2, and TFF genes could play tumor suppressor gene roles in CRC and GC. In addition to growing evidence that loss of function for the GATA-4 and -5 genes could be important for cancer, inhibinα and TFF1 both induce tumors when knocked out in mice (25, 27), although this is true for the former gene only in the setting of the testes. Interestingly, with respect to the TFF genes, we have also now found homozygous genetic disruption of TFF2 sequences in two cell lines, as well as epigenetic silencing of this gene, findings that also attest to the potential importance of its antitumor effects.
The potential of our findings for the GATA-4, -5, and -6 genes to be important in GI tumor development are particularly compelling. Many cancers involve various degrees of failure to complete cell differentiation and therefore manifest a phenotype of maturation arrest. Our patterns of silencing of the GATA genes would foster this situation. GATA-4, -5, and -6 are known to play distinct roles in embryonic GI development and also appear to do so for differentiation of mature GI epithelium (14, 20, 24). Loss of the differentiation stimuli of GATA-4 and -5, with concomitant retention of the proliferative stimulus of GATA-6, would predictably impede differentiation and could thus play a distinct role in the progression of cancers with this gene expression profile.
An intriguing aspect of our findings concerns why tumors might select for simultaneous epigenetic silencing of both upstream activating transcription factors and multiple downstream candidate target antitumor genes. Multiple possible explanations exist. First, it is certainly possible that, in the GI tract, a tight physiologic linkage between the transcription factors and the candidate downstream genes we have studied does not exist and, therefore, the epigenetic events we highlight here might also not be linked. Each epigenetic gene silencing event could then arise stochastically, and some may not play any true role in the progression of the GI tumors under study. However, as discussed previously, there is certainly experimental evidence pointing to the possible regulatory significance of GATAs for activating the genes we have studied. Further, the potential for all of the genes to play a role in cancer suggests that any coordination between the transcriptional factors and activation of the downstream genes would have great ramifications for cancer progression. Our current findings, given the growing recognition of the importance of epigenetic gene silencing for cancer, emphasizes the need for continuing exploration of the suggested relationships in detail.
A second possible scenario may explain some aspects of our findings. Some of the gene silencing events we observed could reflect normal states that transiently precede differentiation in proliferating and renewing GI cell epithelial compartments. This might be true especially in light of the methylation data for the TFF genes that have no promoter CpG islands. Differential promoter methylation is not unusual in normal tissues for nonhousekeeping genes with CpG-poor rich promoters (4). Thus, the presence or absence of promoter methylation in such genes can accompany their different transcriptional status during differentiation (4). However, promoter CpG islands for most genes, like those found in GATA-4, GATA-5, inhibinα, and Dab2, are generally not methylated in normal cells regardless of expression status (4). We could not find evidence, by sensitive techniques, for such methylation in the normal tissues examined. Perhaps expansion of a normally rare population of adult cells, such as stem cells, with the methylation profiles we observed could occur during tumorigenesis. Our data could provide useful markers to explore this possibility in future studies.
We suggest that one final hypothesis is appealing to consider and might well constitute a common paradigm for events that help fuel tumor progression. For the genes we studied, perhaps the most powerful antitumor effect may come from a summation of their epigenetic inactivation during tumor progression. Finding simultaneous mutations for each involved gene in the same tumor might be quite rare. However, individual tumors epigenetically silence multiple genes (8), and this mode of gene inactivation, where protein function is less permanently, and perhaps less completely, disrupted than is the case for mutational events might be an excellent candidate mechanism for inactivating complex antitumor gene networks. Selection of cells might involve any combination of sporadic epigenetic inactivation events, which facilitate evolution of a cancer. For the downstream gene events, such silencing may only partially blunt transcriptional response to upstream activating factors. Thus, continued expression of GATA factors, and probably also other transcription factors with which these proteins are known to partner (5, 42), could activate downstream genes despite their local promoter methylation. Thus, selection during tumorigenesis for inactivation of one or both of the upstream GATA-4 and GATA-5 genes would be additive to the downstream silencing events to ensure the most powerful selection for loss of function of a group of GATA regulated antitumor genes.
Whatever the final full biological explanation for our data, our results have a significant translational implication. Genetic changes are not reversible but, as we demonstrated here, multiple epigenetically silenced candidate antitumor genes can be simultaneously reactivated in single tumors. This concept should receive careful attention with respect to cancer therapeutic strategies. Also, epigenetic silencing of important genes can occur early in the progression of cancers (19, 43). Upregulation of several of the genes we have studied, especially the TFF factors, has been suggested as important for CRC chemoprevention approaches. Thus, gene reactivation approaches might constitute cancer prevention, as well as therapeutic strategies.
Acknowledgments
This work was supported by grant ESO11858 from the National Institute of Environmental Health Sciences (NIEHS), National Institutes of Health (NIH). Y.A. was supported by a postdoctoral fellowship from JSPS Postdoctoral Fellowships for Research Abroad.
The findings reported here are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH.
We thank Oliver Galm, Kornel Schuebel, Michael House, and Jong-In Park for many helpful suggestions and discussions and Kathy Wieman, Ray-Whay Yen, and Kathy Bender for technical assistance.
J.G.H. and S.B.B. are consultants to Oncogenome Sciences. Under a licensing agreement between the Johns Hopkins University and Oncogenome Sciences, MSP was licensed to Oncogenome Sciences, which is entitled to a share of the royalties received by the university from the sales of the licensed technology. The terms of these arrangements are managed by the university in accordance with its conflict-of-interest policies.
REFERENCES
- 1.Al-Azzeh, E. D., P. Fegert, N. Blin, and P. Gott. 2000. Transcription factor GATA-6 activates expression of gastroprotective trefoil genes TFF1 and TFF2. Biochim. Biophys. Acta 1490:324-332. [DOI] [PubMed] [Google Scholar]
- 2.Azarschab, P., E. Al-Azzeh, W. Kornberger, and P. Gott. 2001. Aspirin promotes TFF2 gene activation in human gastric cancer cell lines. FEBS Lett. 488:206-210. [DOI] [PubMed] [Google Scholar]
- 3.Bai, Y., Y. Akiyama, H. Nagasaki, O. K. Yagi, Y. Kikuchi, N. Saito, K. Takeshita, T. Iwai, and Y. Yuasa. 2000. Distinct expression of CDX2 and GATA4/5, development-related genes, in human gastric cancer cell lines. Mol. Carcinog. 28:184-188. [DOI] [PubMed] [Google Scholar]
- 4.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 5.Boudreau, F., E. H. Rings, H. M. van Wering, R. K. Kim, G. P. Swain, S. D. Krasinski, J. Moffett, R. J. Grand, E. R. Suh, and P. G. Traber. 2002. Hepatocyte nuclear factor-1α, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J. Biol. Chem. 277:31909-31917. [DOI] [PubMed] [Google Scholar]
- 6.Cameron, E. E., K. E. Bachman, S. Myohanen, J. G. Herman, and S. B. Baylin. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21:103-107. [DOI] [PubMed] [Google Scholar]
- 7.Cameron, E. E., S. B. Baylin, and J. G. Herman. 1999. p15INK4B CpG island methylation in primary acute leukemia is heterogeneous and suggests density as a critical factor for transcriptional silencing. Blood 94:2445-2451. [PubMed] [Google Scholar]
- 8.Esteller, M., P. G. Corn, S. B. Baylin, and J. G. Herman. 2001. A gene hypermethylation profile of human cancer. Cancer Res. 61:3225-3229. [PubMed] [Google Scholar]
- 9.Farrell, J. J., D. Taupin, T. J. Koh, D. Chen, C. M. Zhao, D. K. Podolsky, and T. C. Wang. 2002. TFF2/SP-deficient mice show decreased gastric proliferation, increased acid secretion, and increased susceptibility to NSAID injury. J. Clin. Investig. 109:193-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng, Z. M., A. Z. Wu, and C. L. Chen. 1998. Testicular GATA-1 factor up-regulates the promoter activity of rat inhibin alpha-subunit gene in MA-10 Leydig tumor cells. Mol. Endocrinol. 12:378-390. [DOI] [PubMed] [Google Scholar]
- 11.Fujikura, J., E. Yamato, S. Yonemura, K. Hosoda, S. Masui, K. Nakao, J. Miyazaki Ji, and H. Niwa. 2002. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 16:784-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujimoto, J., W. Yasui, H. Tahara, E. Tahara, Y. Kudo, and H. Yokozaki. 2000. DNA hypermethylation at the pS2 promoter region is associated with early stage of stomach carcinogenesis. Cancer Lett. 149:125-134. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara, Y., M. Emi, H. Ohata, Y. Kato, T. Nakajima, T. Mori, and Y. Nakamura. 1993. Evidence for the presence of two tumor suppressor genes on chromosome 8p for colorectal carcinoma. Cancer Res. 53:1172-1174. [PubMed] [Google Scholar]
- 14.Gao, X., T. Sedgwick, Y. B. Shi, and T. Evans. 1998. Distinct functions are implicated for the GATA-4, -5, and -6 transcription factors in the regulation of intestine epithelial cell differentiation. Mol. Cell. Biol. 18:2901-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gouyer, V., A. Wiede, M. P. Buisine, S. Dekeyser, O. Moreau, T. Lesuffleur, W. Hoffmann, and G. Huet. 2001. Specific secretion of gel-forming mucins and TFF peptides in HT-29 cells of mucin-secreting phenotype. Biochim. Biophys. Acta 1539:71-84. [DOI] [PubMed] [Google Scholar]
- 16.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herman, J. G., J. R. Graff, S. Myohanen, B. D. Nelkin, and S. B. Baylin. 1996. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 93:9821-9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, J. W., J. R. Raval, T. F. Beals, M. J. Worsham, D. L. Van Dyke, R. M. Esclamado, G. T. Wolf, C. R. Bradford, T. Miller, and T. E. Carey. 1997. Frequent loss of heterozygosity on chromosome arm 18q in squamous cell carcinomas: identification of 2 regions of loss—18q11.1-q12.3 and 18q21.1-q23. Arch. Otolaryngol. Head Neck Surg. 123:610-614. [DOI] [PubMed] [Google Scholar]
- 19.Jones, P. A., and S. B. Baylin. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3:415-428. [DOI] [PubMed] [Google Scholar]
- 20.Kamitani, H., H. Kameda, U. P. Kelavkar, and T. E. Eling. 2000. A GATA binding site is involved in the regulation of 15-lipoxygenase-1 expression in human colorectal carcinoma cell line, Caco-2. FEBS Lett. 467:341-347. [DOI] [PubMed] [Google Scholar]
- 21.Ketola, I., N. Rahman, J. Toppari, M. Bielinska, S. B. Porter-Tinge, J. S. Tapanainen, I. T. Huhtaniemi, D. B. Wilson, and M. Heikinheimo. 1999. Expression and regulation of transcription factors GATA-4 and GATA-6 in developing mouse testis. Endocrinology 140:1470-1480. [DOI] [PubMed] [Google Scholar]
- 22.Kuo, C. T., E. E. Morrisey, R. Anandappa, K. Sigrist, M. M. Lu, M. S. Parmacek, C. Soudais, and J. M. Leiden. 1997. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 11:1048-1060. [DOI] [PubMed] [Google Scholar]
- 23.Lassus, H., M. P. Laitinen, M. Anttonen, M. Heikinheimo, L. A. Aaltonen, O. Ritvos, and R. Butzow. 2001. Comparison of serous and mucinous ovarian carcinomas: distinct pattern of allelic loss at distal 8p and expression of transcription factor GATA-4. Lab. Investig. 81:517-526. [DOI] [PubMed] [Google Scholar]
- 24.Laverriere, A. C., C. MacNeill, C. Mueller, R. E. Poelmann, J. B. Burch, and T. Evans. 1994. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J. Biol. Chem. 269:23177-23184. [PubMed] [Google Scholar]
- 25.Lefebvre, O., M. P. Chenard, R. Masson, J. Linares, A. Dierich, M. LeMeur, C. Wendling, C. Tomasetto, P. Chambon, and M. C. Rio. 1996. Gastric mucosa abnormalities and tumorigenesis in mice lacking the pS2 trefoil protein. Science 274:259-262. [DOI] [PubMed] [Google Scholar]
- 26.Leonard, M., M. Brice, J. D. Engel, and T. Papayannopoulou. 1993. Dynamics of GATA transcription factor expression during erythroid differentiation. Blood 82:1071-1079. [PubMed] [Google Scholar]
- 27.Matzuk, M. M., M. J. Finegold, J. G. Su, A. J. Hsueh, and A. Bradley. 1992. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature 360:313-319. [DOI] [PubMed] [Google Scholar]
- 28.Molkentin, J. D. 2000. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 275:38949-38952. [DOI] [PubMed] [Google Scholar]
- 29.Molkentin, J. D., Q. Lin, S. A. Duncan, and E. N. Olson. 1997. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 11:1061-1072. [DOI] [PubMed] [Google Scholar]
- 30.Morrisey, E. E., S. Musco, M. Y. Z. Chen, M. M. Lu, J. M. Leiden, and M. S. Parmack. 2000. The gene encoding the mitogen-responsive phosphoprotein Dab2 is differentially regulated by GATA-6 and GATA-4 in the visceral endoderm. J. Biol. Chem. 275:19949-19954. [DOI] [PubMed] [Google Scholar]
- 31.Morrisey, E. E., Z. Tang, K. Sigrist, M. M. Lu, F. Jiang, H. S. Ip, and M. S. Parmacek. 1998. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 12:3579-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemer, G., S. T. Qureshi, D. Malo, and M. Nemer. 1999. Functional analysis and chromosomal mapping of Gata5, a gene encoding a zinc finger DNA-binding protein. Mamm. Genome 10:993-999. [DOI] [PubMed] [Google Scholar]
- 33.Park, W. S., R. R. Oh, J. Y. Park, J. H. Lee, M. S. Shin, H. S. Kim, H. K. Lee, Y. S. Kim, S. Y. Kim, S. H. Lee, N. J. Yoo, and J. Y. Lee. 2000. Somatic mutations of the trefoil factor family 1 gene in gastric cancer. Gastroenterology 119:691-698. [DOI] [PubMed] [Google Scholar]
- 34.Rhee, I., K. E. Bachman, B. H. Park, K. W. Jair, R. W. Yen, K. E. Schuebel, H. Cui, A. P. Feinberg, C. Lengauer, K. W. Kinzler, S. B. Baylin, and B. Vogelstein. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416:552-556. [DOI] [PubMed] [Google Scholar]
- 35.Rhee, I., K. W. Jair, R. W. Yen, C. Lengauer, J. G. Herman, K. W. Kinzler, B. Vogelstein, S. B. Baylin, and K. E. Schuebel. 2000. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature 404:1003-1007. [DOI] [PubMed] [Google Scholar]
- 36.Schmitt, J. F., D. S. Millar, J. S. Pedersen, S. L. Clark, D. J. Venter, M. Frydenberg, P. L. Molloy, and G. P. Risbridger. 2002. Hypermethylation of the inhibin alpha-subunit gene in prostate carcinoma. Mol. Endocrinol. 16:213-220. [DOI] [PubMed] [Google Scholar]
- 37.Sheng, Z., E. R. Smith, J. He, J. A. Tuppen, W. D. Martin, F. B. Dong, and X. X. Xu. 2001. Chromosomal location of murine disabled-2 gene and structural comparison with its human ortholog. Gene 268:31-39. [DOI] [PubMed] [Google Scholar]
- 38.Sheng, Z., W. Sun, E. Smith, C. Cohen, and X. X. Xu. 2000. Restoration of positioning control following Disabled-2 expression in ovarian and breast tumor cells. Oncogene 19:4847-4854. [DOI] [PubMed] [Google Scholar]
- 39.Simon, M. C. 1995. Gotta have GATA. Nat. Genet. 11:9-11. [DOI] [PubMed] [Google Scholar]
- 40.Singh, S., R. Poulsom, A. M. Hanby, L. A. Rogers, N. A. Wright, M. C. Sheppard, and M. J. Langman. 1998. Expression of estrogen receptor and estrogen-inducible genes pS2 and ERD5 in large bowel mucosa and cancer. J. Pathol. 184:153-160. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki, H., E. Gabrielson, W. Chen, R. Anbazhagan, M. van Engeland, M. P. Weijenberg, J. G. Herman, and S. B. Baylin. 2002. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat. Genet. 31:141-149. [DOI] [PubMed] [Google Scholar]
- 42.van Wering, H. M., I. L. Huibregtse, S. M. van der Zwan, M. S. de Bie, L. N. Dowling, F. Boudreau, E. H. Rings, R. J. Grand, and S. D. Krasinski. 2002. Physical interaction between GATA-5 and hepatocyte nuclear factor-1α results in synergistic activation of the human lactase-phlorizin hydrolase promoter. J. Biol. Chem. 277:27659-27667. [DOI] [PubMed] [Google Scholar]
- 43.Widschwendter, M., and P. A. Jones. 2002. DNA methylation and breast carcinogenesis. Oncogene 21:5462-5482. [DOI] [PubMed] [Google Scholar]
- 44.Wright, N. A., W. Hoffmann, W. R. Otto, M. C. Rio, and L. Thim. 1997. Rolling in the clover: trefoil factor family (TFF)-domain peptides, cell migration and cancer. FEBS Lett. 408:121-123. [DOI] [PubMed] [Google Scholar]