Abstract
CAK1 encodes an essential protein kinase in Saccharomyces cerevisiae that is required for activation of the Cdc28p Cdk. CAK1 also has several CDC28-independent functions that are unique to meiosis. The earliest of these functions is to induce S phase, which is regulated differently in meiosis than in mitosis. In mitosis, Cdc28p controls its own S-phase-promoting activity by signaling the destruction of its inhibitor, Sic1p. In meiosis, Sic1p destruction is signaled by the meiosis-specific Ime2p protein kinase. Our data show that Cak1p is required to activate Ime2p through a mechanism that requires threonine 242 and tyrosine 244 in Ime2p's activation loop. This activation promotes autophosphorylation and accumulation of multiply phosphorylated forms of Ime2p during meiotic development. Consistent with Cak1p's role in activating Ime2p, cells lacking Cak1p are deficient in degrading Sic1p. Deletion of SIC1 or overexpression of IME2 can partially suppress the S-phase defect in cak1 mutant cells, suggesting that Ime2p is a key target of Cak1p regulation. These data show that Cak1p is required for the destruction of Sic1p in meiosis, as in mitosis, but in meiosis, it functions through a sporulation-specific kinase.
Meiosis is the pathway used by sexually reproducing organisms to produce haploid gametes. In the yeast Saccharomyces cerevisiae, mitotically growing diploid cells undergo sporulation, a highly regulated program of meiotic development, when starved of key nutrients (18). Successful sporulation depends on the ordered execution of landmark events. After induction, cells undergo a single round of DNA synthesis (meiotic S phase) followed by high levels of homologous recombination. Next, cells complete two rounds of chromosomal segregation without an intervening round of DNA synthesis. The resulting haploid products are engulfed by the prospore membrane and subsequently surrounded by a multilayered spore wall that is capable of protecting the cell from a variety of environmental insults. Coordination of the landmark events of sporulation is key to ensuring genetic diversity and proper chromosome number.
One level of regulation of the sporulation program occurs through the transcriptional cascade (20). Genome-wide transcriptional analysis has shown that as many as 1,600 of the 6,000 genes in yeast exhibit transcriptional regulation during sporulation (3, 25). These genes have been temporally classified in up to 12 subgroups. Here, we broadly refer to the timing of gene induction as early, middle, and late. Another way the cell regulates events in sporulation is through networks of protein kinases. Many of the 120 protein kinase homologs in yeast function in the program. Thirty-two of the kinases are transcriptionally induced at specific times during sporulation (25). A subset of these kinases regulate the transcriptional program through feedback loops, while others are required to regulate the execution of specific landmark events.
The progression through mitotic and meiotic cell division pathways in most eukaryotic organisms requires changes in the activities of cyclin-dependent kinases (Cdks) that are controlled by phosphorylation and by binding to inhibitors and to cyclin activators. For activation, many Cdks need to be phosphorylated in their activation loops (T-loops) by Cdk-activating kinases (CAKs) (16, 32). In S. cerevisiae, the sole essential Cdk required to drive the mitotic cell cycle, Cdc28p, is activated by phosphorylation of threonine 169 by the monomeric Cak1p enzyme (7, 14, 36). The only essential role of Cak1p in mitosis is to activate Cdc28p, as shown by the observation that Cak1p is dispensable in cells harboring an allele of CDC28 (CDC28-43244), which contains an acidic substitution of T169 and additional activating point mutations (5). Other Cdk homologs, Bur1p and Kin28p, are also phosphorylated by Cak1p on the conserved threonine (T240 and T162, respectively), but their phosphorylation is not essential for cell division under standard conditions (17, 41). Cak1p appears to be largely unregulated during mitosis (15). In contrast, during meiotic development, CAK1 is transcriptionally induced as a middle sporulation gene. We previously identified CAK1 as a multicopy suppressor of mutants in SMK1, which encodes a mitogen-activated protein kinase (MAPK) required for spore wall morphogenesis (38). Further studies using the CAK1-independent allele of CDC28 (CDC28-43244) revealed that Cak1p functions in a CDC28-independent manner at two or more steps during sporulation (27). Cak1p is first required for timely progression through meiotic S phase and then later required for spore wall formation through the SMK1 MAPK pathway. Cdc28p is required for both execution of meiotic S phase and the meiotic divisions, raising the possibility that Cak1p could also regulate these steps in a CDC28-dependent fashion (1, 29). Thus, Cak1p appears to play a central role in regulating multiple steps in meiosis through multiple substrates.
IME2 encodes a meiosis-specific kinase that promotes meiotic S and M phases (9, 21, 31). Similar to the cak1-Δ phenotype, cells lacking IME2 fail to perform a timely and efficient meiotic S phase. Ime2p is thought to stimulate S phase by promoting the degradation of the Cdk inhibitor Sic1p (6). Ime2p also enhances transcription of the early genes and destabilizes the transcriptional activator of the early genes, Ime1p, after it has been induced (10). Ime2p is later responsible for promoting middle gene expression by enhancing the production and activation of the middle sporulation gene (MSG) transcriptional activator, Ndt80p, which in turn induces the B-type cyclins (Clbs) that are required to activate Cdc28p, thereby promoting the nuclear divisions (1, 2, 11, 34). Ime2p also negatively regulates Sum1p, a transcriptional repressor of middle genes (19, 23, 24, 39).
Although cells use the same machinery to carry out mitotic and meiotic S phases, many organisms have developed meiosis-specific regulation of the machinery. For example, destruction of Sic1p is a key step for entry into both mitotic and meiotic S phase. Sic1p binds to and inhibits the kinase activity of S-phase-promoting Cdc28p-Clb5p and Cdc28p-Clb6p complexes (28). When mitotically growing cells are ready to initiate DNA synthesis, the G1 cyclin-Cdc28p complex phosphorylates Sic1p, targeting it for ubiquitin-mediated proteolysis, thus liberating Cdc28p-Clb5p/6p to trigger S phase (37). Recent data have shown that Cdc28p is required for meiotic S phase, consistent with the dependence of meiotic S phase on Clb5p and Clb6p (1, 35). In meiosis however, Cdc28p is dispensable for Sic1p degradation (1). Instead, Sic1p destruction in meiosis requires Ime2p (6). CDC28-43244 is able to bypass the requirement for CAK1 in mitotic S phase but not in meiotic S phase, suggesting that Cak1p controls the activity of a meiosis-specific regulator of S phase in a CDC28-independent manner (27). Here, we further examined the CDC28-independent roles of Cak1p in promoting meiotic S phase.
We have found that strains lacking CAK1 are deficient in the timing of Sic1p degradation. Removal of SIC1 in this genetic background partially suppresses the meiotic S-phase defect, suggesting that one essential early CDC28-independent role of Cak1p is to promote Sic1p degradation. This role is likely to operate via Ime2p, since overexpression of IME2 can also partially suppress the S-phase delay of cak1-Δ mutants. While Ime2p is phosphorylated and is catalytically active in cells containing CAK1, it is not phosphorylated and is catalytically inactive in cells lacking CAK1. Mutational analyses of IME2 indicate that the Cak1p-dependent activation of Ime2p requires a threonine residue and a tyrosine residue located in the presumed activation loop of Ime2p. These data suggest that Cak1p promotes meiotic S phase by stimulating the phosphorylation and activation of the Ime2p meiosis-specific protein kinase.
MATERIALS AND METHODS
Yeast strains and plasmids.
The yeast strains and plasmids used in this study are described in Tables 1 and 2, respectively. All strains used in this study are in the SK1 genetic background. The galactose-inducible allele of SIC1 was generated by a two-step amplification of overlapping PCR fragments to generate a chimeric construct containing 483 bp 5′ to the SIC1 initiator ATG, a 1-kb URA3 marker cassette, 678 bp of the GAL1 promoter, and 415 bp of the SIC1 open reading frame. In the first step, a URA3::GAL1-SIC1 fragment was generated from PCR amplification of pMDM169 (22). In the second step, the upstream region of SIC1 was amplified from S288C genomic DNA, yielding a product containing 21 bp with 3′ homology to URA3. These two PCR products were then mixed together in a final reaction and amplified by using the outermost primers described above. The final 2.6-kb PCR product was used to transform a cak1-Δ::TRP1/CAK1 CDC28-43244/CDC28-43244 strain. Transformants growing on SD medium lacking uracil were sporulated, tetrads were dissected, and segregants were mated to construct homozygous diploids on galactose-containing media. Galactose-dependent induction and noninduced levels of SIC1 were monitored by immunoblotting.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference | |
|---|---|---|---|
| LNY3 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 | L. Neigeborne | |
| LNY65 | MATa/MATα ime2::LEU2/ime2::LEU2 ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 | L. Neigeborne | |
| JLY3 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 CDC28-43244/CDC28-43244 | 27 | |
| JLY4 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 CDC28-43244/CDC28-43244 cak1-Δ::TRP1/cak1-Δ::TRP1 | 27 | |
| KSY111 | JLY4 + GAL1-SIC1::URA3/GAL1-SIC1::URA3 | This study | |
| KSY112 | JLY3 + GAL1-SlC1::URA3/GAL1-SlC1::URA3 | This study | |
| KSY137 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1-ΔFA::hisG or trp1::hisG/trp1-ΔFA::hisG or trp1::hisG lys2/lys2 ho::hisG or ho::LYS2/ho::hisG or ho::LYS2 IME2-myc::TRP1/IME2-myc::TRP1 CDC28-43244/CDC28-43244 | This study | |
| KSY138 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1-ΔFA::hisG or trp1::hisG/trp1-ΔFA::hisG or trp1::hisG lys2/lys2 ho::hisG or ho::LYS2/ho::hisG or ho::LYS2 IME2-myc::TRP1/IME2-myc::TRP1 CDC28-43244/CDC28-43244 cak1-Δ::TRP1/cak1-Δ::TRP1 | This study | |
| KSY189 | KSY137 + ime2-K97R-myc::TRP1/ime2-K97R-myc::TRP1 | This study | |
| KSY220 | KSY137 + ime2-T242A, Y244F-myc::TRP1/ime2-T242A, Y244F-myc::TRP1 | This study | |
| KSY222 | KSY137 + ime2-Y244F-myc::TRP1/ime2-Y244F-myc::TRP1 | This study | |
| KSY190 | KSY138 + ime2-K97R-myc::TRP1/ime2-K97R-myc::TRP1 | This study | |
| KSY218 | KSY138 + ime2-T242A, Y244F-myc::TRP1/ime2-T242A, Y244F-myc::TRP1 | This study | |
| KSY219 | KSY138 + ime2-Y244F-myc::TRP1/ime2-Y244F-myc::TRP1 | This study | |
| KSY223 | KSY138 + ime2-T242A-myc::TRP1/ime2-T242A-myc::TRP1 | This study | |
| AMY22 | LNY65 + pAM4 | This study | |
| AMY23 | LNY65 + pSM1 | This study | |
| AMY24 | LNY65 + pSM2 | This study | |
| AMY25 | LNY65 + pSM3 | This study | |
| AMY28 | LNY65 + pSM6 | This study | |
| AMY34 | LNY65 + pSM10 | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Markers | Source or reference |
|---|---|---|
| YEp351 | 2μm LEU2 | 12 |
| pS324 | 2μm LEU2 YEp13 + IME2 | Saul Honigberg |
| pMR1 | 2μm URA3 pRS426 + IME2 | 27 |
| pAM4 | pMR1 + 13myc | This study |
| pSM1 | pAM4 + ime2-Y244F-myc | This study |
| pSM2 | pAM4 + ime2-T242A, Y244F-myc | This study |
| pSM3 | pAM4 + ime2-S246A-myc | This study |
| pSM6 | pAM4 + ime2-Y241F-myc | This study |
| pSM10 | pAM4 + ime2-T242A-myc | This study |
The myc-tagged IME2-T-loop mutants were generated by site-directed mutagenesis (QuikChange kit; Stratagene) of the IME2 gene in pAM4. The T242 codon was changed to an A by mutating ACG to GCG, the Y244 codon was changed to an F by mutating TAC to TTC, the Y241 codon was changed to an F by mutating TAT to TTT, and the S246 codon was changed to an A by mutating AGC to GCC. To create the chromosomally integrated strains, PCR products containing the IME2 coding information from the mutant plasmids (from 150 bp upstream of the ATG to 1,910 bp downstream of the ATG) were used to transform a haploid ime2ΔM::URA3Kl-13Myc::TRP1 strain (KBY443) (1). Loss of URA3 was selected on 5-fluoroorotic acid (5-FOA), and integration was confirmed by diagnostic PCR and restriction digestion. Homozygous diploids were generated by backcrossing FOAR haploids to wild-type SK1 haploids or to cak1-Δ::TRP1 CDC28-43244 haploids (ALY1125), followed by sporulation, tetrad dissection, and mating.
An IME2-myc high-copy-number plasmid (pAM4) was generated from pMR1 in several steps. First, an SmaI restriction site was introduced in frame just prior to the stop codon of IME2 (QuikChange kit; Stratagene). Next, the myc tag was PCR amplified from KSY137 genomic DNA with a 3′ primer that introduced a KpnI restriction site. Finally, the myc PCR fragment was ligated to the plasmid cut with SmaI and KpnI. The nucleotide sequences of this and all mutant derivatives of this plasmid were determined.
Growth and sporulation of cells.
Vegetative growth was maintained in YEPD (1% yeast extract, 2% peptone, 2% glucose), SD (0.67% yeast nitrogen base without amino acids and 2% glucose plus nutrients essential for auxotrophic strains), or YEPA (1% yeast extract, 2% peptone, 2% potassium acetate). For synchronous sporulation, cells were grown in YEPA to an optical density at 600 nm (OD600) of 0.3 to 0.5. Cells harboring the galactose-inducible allele of SIC1 were pregrown in the media described above containing galactose instead of glucose, harvested, washed in water, and grown in YEPA for 4 generations. Prior to sporulation, cells were harvested, washed in SM (2% potassium acetate plus 10 μg of adenine, 5 μg of histidine, 30 μg of leucine, 7.5 μg of lysine, 10 μg of tryptophan, 5 μg of uracil per ml) and resuspended in SM at 2 OD units/ml. For the experiments shown in Fig. 3 and 4, cells were grown in YEPA to an OD600 of 1.5 to 3.0 prior to sporulation at 1.5 OD unit/ml in 1% potassium acetate plus 0.02% raffinose.
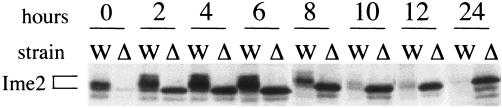
FIG. 3.
Ime2p is unmodified in cak1-Δ strains. Protein was isolated from whole-cell lysates of sporulating cells at the indicated times from wild-type (W) (KSY137) or cak1-Δ (Δ) (KSY138) diploid cells in the CDC28-43244 genetic background. Ime2p-myc protein was assayed by immunoblot analysis with a polyclonal Ime2p antibody.
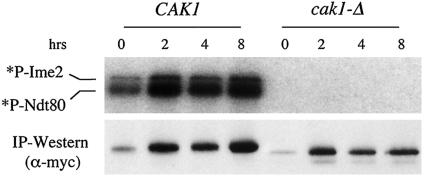
FIG. 4.
Cak1p is required for the catalytic activity of Ime2p. Epitope-tagged Ime2p was immunoprecipitated (IP) from sporulating CAK1 (KSY137) or cak1-Δ (KSY138) diploid cells in the CDC28-43244 background at the indicated times. Protein kinase activity was measured by incubation of Ime2p-myc with purified MBP-Ndt80p substrate and [γ-32P]ATP. *P-Ime2 and *P-Ndt80 correspond to Ime2p autophosphorylation and phosphorylation of Ndt80p, respectively. Immunoblot analysis shows that comparable levels of Ime2p-myc are present in the reaction mixtures.
In vitro kinase assay.
The kinase activity of Ime2p-myc was measured as described by Benjamin et al. (1). Briefly, sporulating cells were harvested and frozen in liquid nitrogen at various times postinduction. Next, the cells were lysed by 3 cycles of 1-min bursts of bead beating followed by 2 min on ice (500 μl of 0.5-mm-diameter glass beads; BioSpec Products) in 500 μl of lysis buffer (50 mM HEPES [pH 7.4], 75 mM KCl, 1 mM EGTA, 1 mM MgCl2, 0.1% NP-40, 50 mM NaF, 50 mM β-glycerolphosphate, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 8.8 μg of aprotinin per ml, 4 μg of antipain per ml, 0.1 μg of pefabloc SC per ml, 2 μg of pepstatin A per ml, 1 μg of chymostatin per ml, 1 mM benzamidine, 2 μg of leupeptin per ml). Equal amounts of protein (2 to 6 mg) from clarified supernatants were used for immunoprecipitation of Ime2p-myc with 20 μl of a 1:1 slurry of 9E10 beads (Covance) by mixing for 1.5 h. Immunoprecipitates were collected and washed three times in lysis buffer and once in kinase buffer (20 mM HEPES [pH 7.4], 100 mM KCl, 10 mM MgCl2). A third of the immunoprecipitate was removed for Western blotting, and the remainder was resuspended in 25 μl of kinase buffer plus 5 μCi of [γ-32P]ATP (Amersham), 10 μM ATP, and 10 μg of purified MBP-Ndt80p substrate. The reactions were terminated after a 30-min incubation at 30°C with the addition of sodium dodecyl sulfate-containing gel loading buffer followed by boiling. Samples were resolved on 12% polyacrylamide gels prior to audoradiography.
DNA staining and analysis.
To monitor the meiotic divisions, 2 OD units of sporulating cells was taken at various times, fixed in 90% ethanol (EtOH), and stained with 1 μg of 4′,6-diamidino-2-phenylindole (DAPI) per ml. The completion of the meiotic divisions was scored by counting the proportion of cells containing more than one DAPI-staining body observed by fluorescence microscopy. One hundred cells in at least three independent isolates per strain were analyzed. To measure DNA content, 3 OD units of sporulating cells was fixed in 70% EtOH at various time points. Prior to analysis, 1 OD unit of cells was digested with 1 mg of RNase A per ml and 5 mg of pepsin per ml before staining overnight at 4°C with 0.5 mg of propidium iodide per ml. Cells were then diluted 1:5, lightly sonicated to reduce clumping, and analyzed with a Beckman-Coulter XL cell sorter. Analysis was conducted at least two times per strain.
Immunological analysis of proteins.
Whole-cell lysates were prepared as described by Yaffe and Schatz (40). Briefly, 2 to 4 OD units of sporulating cells was harvested at various times and washed once with and then resuspended in 1 ml of ice-cold water plus 25 mM β-glycerolphosphate, 1 mM Na3VO4, and 100 μg of phenylmethylsulfonyl fluoride per ml. After addition of 150 μl of ice-cold 2 M NaOH and 8% β-mercaptoethanol, samples were stored at −80°C. Samples were thawed on ice, and proteins were precipitated by addition of 150 μl of 50% trichloroacetic acid, incubation for 10 min at 4°C, and centrifugation for 2 min. Pellets were washed twice in cold acetone, briefly dried in a vacuum, resuspended in 300 μl of 3× sample buffer, and boiled for 3 min. Samples were diluted 1:2 in Laemmli sample buffer and loaded on a 10% polyacrylamide gel to analyze Sic1p and Cdc28p or on a 7.5% gel to analyze Ime2p, run at 20 to 25 mV, transferred to Immobilon-P membrane (Millipore), and blocked overnight at 4°C in a mixture containing phosphate-buffered saline (PBS) and 0.1% Tween 20 plus I-Block (Tropix). Antibody staining was performed for 1 h at room temperature with polyclonal antibodies against Sic1p (1:5,000; a gift from M. Tyers), Ime2p (30), Cdc28p (1:1,000) (33), or T-169PO4-Cdc28p (1:100) (26). After washing, the membrane was incubated for 1 h at room temperature with a secondary anti-rabbit antibody (Promega, 1:5,000; or Covance, 1:1,000). The myc epitope on Ime2p was detected with a 9E10 monoclonal antibody (Santa Cruz, 1:1,000; or Covance, 1:2,000) and a secondary anti-mouse antibody (Promega, 1:5,000; or Amersham, 1:2,000). Detection of the secondary antibody was achieved by chemiluminescence (Tropix CDP-Star or the Amersham ECL enhanced chemiluminescence system).
The epitope-tagged Ime2p shown in Fig. 5 was partially purified by freezing and grinding cells in liquid N2 prior to lysis in the buffer described in the “In vitro kinase assay” section (except Tris-Cl was substituted for HEPES) and clarification by centrifugation. The clarified lysate was loaded on a Q-Sepharose column (Amersham), washed with the lysis buffer, and eluted in a step gradient of increasing KCl concentration (0.2 M increments). Ime2p-myc eluted from the beads at 0.4 M KCl.
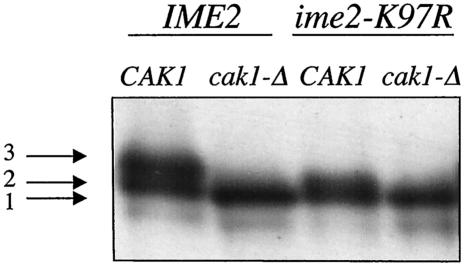
FIG. 5.
Cak1p is required for phosphorylation of Ime2p. The catalytically inactive ime2-K97R allele tagged with the myc epitope was introduced into the CDC28-43244 background in diploid cells either containing (KSY189) or lacking CAK1 (KSY190). Cells were sporulated for 2 h prior to lysis and partial purification of Ime2p by ion-exchange chromatography. Differences in the electrophoretic mobilities of Ime2p were detected by immunoblot analysis with an anti-myc antibody.
RESULTS
CAK1 mutants are deficient in the timing of Sic1p degradation.
During mitosis, the essential function of CAK1 can be bypassed by a CAK1-independent allele of CDC28 (CDC28-43244) that contains a glutamic acid in place of the threonine that is normally phosphorylated by Cak1p as well as additional activating point mutations (5). Unless otherwise indicated, all of the experiments described in this report were conducted in this genetic background. For simplicity, CDC28-43244/CDC28-43244 diploid strains containing and lacking CAK1 will be referred to below as wild type (or CAK1) and cak1-Δ, respectively.
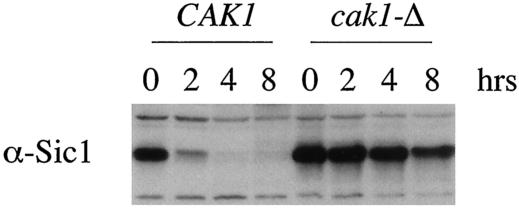
Wild-type SK1 cells complete meiotic S phase around 3 h postinduction under our standard sporulation conditions (see Materials and Methods). We have previously shown that cells lacking CAK1 do not complete meiotic S phase until around 24 h postinduction (27). Furthermore, catalytically inactive cak1 alleles do not complement the S-phase defect, suggesting that Cak1p phosphotransferase activity is required to activate meiotic S phase. To elucidate how Cak1p regulates S phase during meiosis, we compared Sic1p levels in wild-type and cak1-Δ/cak1-Δ strains. Immunoblotting revealed that in wild-type cells, Sic1p levels decreased shortly after meiotic induction and were undetectable at 4 h postinduction (Fig. 1). This interval corresponds to the time in which meiotic S phase occurred in these cells. The timing of the decline in Sic1p levels was similar in CDC28 and CDC28-43244 cells (data not shown). In contrast to the wild type, Sic1p was present at high levels in cak1-Δ/cak1-Δ cells, past the time in which wild-type cells have completed S phase, executed the meiotic divisions, and initiated spore wall formation (Fig. 1). The failure of cak1-Δ/cak1-Δ cells to reduce Sic1p protein levels cannot be due to a defect in Cdc28p activity, since Cdc28p is dispensable for Sic1p destruction in meiosis (1). These results suggest that Cak1p promotes Sic1p destruction in a Cdc28p-independent manner.
FIG. 1.
Cak1p is required for the timely destruction of Sic1p. Whole-cell lysates of CAK1 (JLY3) or cak1-Δ (JLY4) homozygous diploid cells in the CDC28-43244 background were prepared at the indicated times during sporulation and assayed by immunoblot analysis with Sic1p antiserum (α-Sic1). Vegetative growth is represented by the 0-h time point.
Deletion of SIC1 in the cak1-Δ background partially bypasses the S-phase delay.
If the primary defect of cak1-Δ/cak1-Δ cells is in Sic1p degradation, then deletion of SIC1 should promote progression through S phase in this genetic background. We next asked if deletion of SIC1 could bypass the S-phase delay observed in the cak1-Δ mutant strain. It has been reported that sic1-Δ strains are genetically unstable and prone to aneuploidy (22). To avoid this complication, we generated a cak1-Δ/cak1-Δ strain whose sole source of SIC1 was controlled by the GAL1 promoter (referred to here as GAL1-SIC1). GAL1-SIC1 cells grown in galactose expressed Sic1p levels that are comparable to the levels of Sic1p seen in wild-type cells (data not shown). The level of Sic1p rapidly declined when cells were transferred to medium containing acetate as the sole carbon source (presporulation medium), and Sic1p was undetectable during sporulation in this background (Fig. 2C).
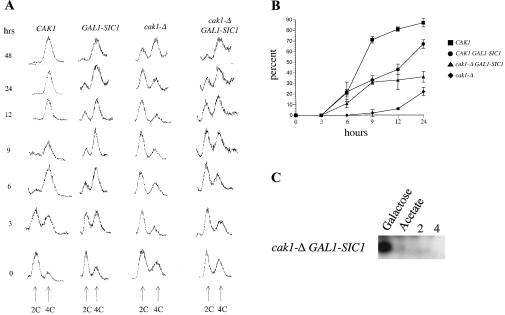
FIG. 2.
Depletion of Sic1p partially rescues the meiotic S-phase delay of cak1-Δ cells. Diploid strains containing a galactose-inducible SIC1 gene allowed depletion of Sic1p specifically during sporulation. (A) Progression through meiotic S phase was monitored by flow cytometry to detect cells with 2C and 4C DNA content as indicated (from left to right: JLY3, KSY112, JLY4, and KSY111). Sporulating cells were fixed at the indicated times postinduction, and DNA was stained with propidium iodide. (B) Completion of the meiotic divisions was monitored by fixing cells of the indicated genotype and staining the DNA with DAPI. Cells were viewed by microscopy under UV light and counted for completion of either MI or MII. All experiments were in the CDC28-43244 genetic background. (C) Protein was isolated from a cak1-Δ strain harboring the GAL1-SIC1 allele (KSY111) grown in inducing medium (galactose) or in noninducing medium (acetate) and at the indicated times after transfer to sporulation medium. Sic1p was monitored by immunoblot analysis.
We used flow cytometry (fluorescence-activated cell sorting [FACS]) to compare the timing of meiotic S phase in the Sic1p-depleted GAL1-SIC1 strains with isogenic strains that expressed SIC1 under native control. The timing of meiotic S phase in wild-type CAK1 cells containing the GAL1-SIC1 allele was similar to that in wild-type cells containing the endogenous SIC1 promoter (compare columns 1 and 2 in Fig. 2A). In contrast, depletion of Sic1p in the cak1-Δ/cak1-Δ background advanced entry into meiotic S phase (compare columns 3 and 4 in Fig. 2A). Approximately 50% of the GAL1-SIC1 cak1-Δ diploid cells accumulated a 4C DNA content by 9 h as compared to 30 h in a cak1-Δ strain expressing SIC1 from its normal promoter (Fig. 2A), demonstrating that the failure of cak1-Δ cells to efficiently degrade Sic1p can account for part of the S-phase delay. However, the depletion of Sic1p did not completely restore the timing of S phase in cak1-Δ cells to that seen in wild-type cells where 50% of cells accumulated a 4C DNA content by 3 h. Thus, in addition to promoting Sic1p destruction, Cak1p might promote S phase by at least one additional mechanism.
We also monitored meiotic divisions in the Sic1p-depleted (GAL1-SIC1) strains by staining cells with DAPI at various times during sporulation. Wild-type cells initiated the meiotic divisions around 5 to 6 h postinduction, and approximately 80% of these cells completed both divisions by 12 h (Fig. 2B). The timing of the meiotic divisions in CDC28-43244 strains was not reproducibly different from that in CDC28 strains (data not shown). Cells lacking CAK1 were stalled in the meiotic program at S phase, and therefore the nuclear divisions began later and occurred in fewer cells. On average, only about 20% of cak1-Δ/cak1-Δ cells completed both divisions even after lengthy incubations (Fig. 2B). This population could be from a subset of cells that slowly degraded Sic1p and therefore completed S phase between 24 and 48 h postinduction (Fig. 2A, column 3). Removal of Sic1p, using the GAL1-SIC1 allele, in wild-type CAK1 cells did not appear to alter the timing of meiotic S phase or initiation of the divisions, but the total percentage of cells successfully completing both meiotic divisions is modestly reduced (Fig. 2A and B). The basis of this decrease has not been investigated further. Importantly, when Sic1p was depleted in the cak1-Δ/cak1-Δ background, the meiotic divisions occurred more often and earlier (Fig. 2B). These results demonstrate that depletion of Sic1p partially suppresses the defects in meiotic S and M phases caused by deletion of CAK1. Therefore, a critical early role of Cak1p is to promote a decrease in Sic1p protein levels and thereby relieve inhibition of the Cdc28p activity required for S phase. Furthermore, these data indicate that a GAL1-SIC1 cak1-Δ diploid mutant cell can carry out the meiotic divisions once meiotic S phase is completed. This suggests that Cak1p may not have any CDC28-independent functions between S phase and the meiotic divisions.
Overexpression of IME2 partially bypasses the S-phase delay in the cak1-Δ mutant.
Sic1p degradation is known to depend on Ime2p in meiosis (6). This result suggests that Cak1p may positively regulate Ime2p. If cak1-Δ/cak1-Δ cells fail to produce active Ime2p, then introduction of additional copies of IME2 on a high-copy-number plasmid might suppress the mutant phenotype. We measured the meiotic divisions in cells containing either an empty 2μm plasmid or a 2μm plasmid containing IME2 after 24 h of sporulation (Table 3). Overexpression of IME2 in wild-type cells did not alter the ability to complete meiosis. In cak1-Δ/cak1-Δ cells, the pS324 high-copy-number IME2 plasmid increased the percentage of cells that completed both nuclear divisions (47% compared with 12%). Moreover, FACS analysis revealed that overexpression of IME2 advanced the timing of meiotic S phase in the cak1-Δ/cak1-Δ mutant (data not shown). In the Sic1p-depleted GAL1-SIC1/GAL-SIC1 cak1-Δ/cak1-Δ cells, the high-copy-number IME2 plasmid only modestly increased the percentage of cells that completed both nuclear divisions from 35% to 46%. These genetic interactions suggest that the cak1-Δ cells may have too little Ime2p activity and thus cannot decrease Sic1p levels. The ability of high-copy-number IME2 plasmids to promote S phase in a cak1-Δ/cak1-Δ strain is consistent with Cak1p and Ime2p functioning in the same pathway. Moreover, the ability of Sic1p depletion to only partially bypass the S-phase defect of a cak1-Δ/cak1-Δ mutant is consistent with Ime2p's requirement for execution of multiple events early in sporulation.
TABLE 3.
Overexpression of IME2 partially suppresses the meiotic defect in cells lacking Cak1pa
| Strain | % Meiosis
|
|
|---|---|---|
| YEp351 | pS324 | |
| CDC28-43244 | 78 ± 12 | 88 ± 1 |
| cak1-Δ CDC28-43244 | 12 ± 8 | 47 ± 10 |
| GAL1-SIC1 CDC28-43244 | 57 ± 6 | 59 ± 3 |
| GAL1-SIC1 cak1-Δ CDC28-43244 | 35 ± 3 | 46 ± 7 |
The indicated homozygous diploid strains harboring a high-copy-number vector containing IME2 (pS324) or a control vector lacking IME2 (Yep351) were sporulated for 24 h before analysis. The completion of both meiotic divisions was determined as described in Materials and Methods.
We previously reported that overexpression of IME2 using pRSB733 did not effect the completion of the meiotic divisions in a cak1-Δ/cak1-Δ mutant (27). pRSB733 contains 270 bp of promoter sequence, whereas the plasmid used here (pS324) contains 1,500 bp of the promoter. Using IME2 2μm plasmids containing various amounts of the IME2 promoter sequence, we found a correlation between the length of the regulatory region and its ability to suppress the S-phase defect in cells lacking CAK1 (data not shown).
Ime2p is not phosphorylated and is catalytically inactive in the cak1-Δ strain.
We next wanted to determine if Cak1p was required for the normal production and phosphorylation of Ime2p. IME2 mRNA is expressed specifically during meiosis starting early in the program and continuing until after the completion of the nuclear divisions. Ime2p can be detected shortly after meiotic induction in a basally phosphorylated form and becomes hyperphosphorylated after DNA replication (1, 34). To determine if Cak1p is required for the phosphorylation of Ime2p, we compared Ime2p in cells containing and lacking CAK1 by immunoblot analysis.
As previously reported, Ime2p was present at low levels in wild-type cells during mitotic growth in acetate. The expression level of Ime2p at 0 h is enhanced as these cells were washed and resuspended in sporulation media prior to processing (see Materials and Methods). By 2 h postinduction, when Sic1p destruction and S phase were occurring, the level of Ime2p increased substantially. During the course of sporulation, Ime2p migrated as a series of electrophoretically distinct isoforms (Fig. 3). Recent data have shown that the more slowly migrating forms of Ime2p are a consequence of phosphorylation of Ime2p (1). We have previously shown that cells lacking CAK1 have a modest delay in inducing the transcriptional program and reduction in early sporulation transcript levels. The differences in Ime2p levels at 0 h between wild-type and cak1-Δ cells are indicative of this difference (27). In the strain lacking CAK1, Ime2p levels increased substantially between 0 and 2 h. Strikingly however, Ime2p migrated as a single unmodified form at all time points tested (Fig. 3). These data indicate that the modification of Ime2p requires Cak1p. The S-phase defect of mutant cak1-Δ/cak1-Δ cells is therefore not due to a failure to make Ime2p but may be due to inadequate posttranslational modification of Ime2p.
We next wanted to test if the CAK1-dependent modification of Ime2p was correlated with the catalytic activity of Ime2p. Ime2p has previously been shown to phosphorylate itself and the Ndt80p transcription factor in vitro (1, 34). Using these reactions, we compared the catalytic activity of Ime2p purified from cells containing and lacking CAK1. Ime2p immunopurified from wild-type cells phosphorylated itself and Ndt80p (Fig. 4). In contrast, Ime2p immunopurified from cak1-Δ/cak1-Δ cells was unable to phosphorylate itself or Ndt80p at any time point tested. These observations demonstrate that Cak1p is required to activate Ime2p.
Cak1p is required for phosphorylation of Ime2p.
To further characterize the role of Cak1p in activating Ime2p, we monitored the electrophoretic mobility of wild-type Ime2p-myc and catalytically inactive Ime2p-K97R-myc in wild-type and in cak1-Δ/cak1-Δ mutant cells. For these experiments, we partially purified Ime2p by ion-exchange chromatography (see Materials and Methods), because this allowed better electrophoretic resolution of the different phosphorylated forms of Ime2p (data not shown).
After 2 h in sporulation medium, Ime2p from wild-type cells migrated as a doublet (bands 2 and 3 in Fig. 5). Even the most rapidly migrating member of the doublet (band 2) appeared to be modified, since it migrated more slowly than the Ime2p from cells lacking CAK1. Ime2p from a cak1-Δ/cak1-Δ strain migrated as a single band (band 1 in Fig. 5). We have been unable to detect heterogeneity in the Ime2p from cak1-Δ/cak1-Δ cells even at late time points after meiotic induction, regardless of whether the Ime2p is wild type or mutant K97R (Fig. 3) (unpublished results). Importantly, in a CAK1 strain, the catalytically inactive Ime2p-K97R-myc protein migrated as a compressed doublet comprised of bands 1 and 2. These data suggest that the most slowly migrating form of Ime2p (band 3) is produced by CAK1-dependent phosphorylation followed by autophosphorylation. These data show that Cak1p can cause modification of Ime2p or catalytically inactive mutant Ime2p. These data raise the possibility that Ime2p is inactive (even to phosphorylate itself) unless it has been previously modified by a CAK1-dependent reaction, consistent with the in vitro kinase assay result in Fig. 4.
Phosphorylation of the T-loop in Ime2p is dependent on Cak1p.
Of the 10 yeast protein kinases that are most similar to Ime2p, 6 are MAP kinases, 2 are Cdks, and 2 are glycogen synthase kinases (13). This family of enzymes is referred to as the CMGC (cyclin/MAPK/glycogen synthase/cyclin-like kinase) family. Many CMGC kinases are activated by phosphorylation of residues in their T-loops. MAPKs are activated by dual phosphorylation of a T-loop TXY motif located 10 residues upstream of the catalytic APE motif in subdomain VIII by upstream dual-specificity MAPK kinases. IME2 contains a TAY that aligns with the TXY of MAPKs and starts at residue 242. T242 also aligns with the T in CDC28p (as well as Kin28p and Bur1p), which are known sites of Cak1p activating phosphorylation.
To investigate if phosphorylation of residues in the T-loop of Ime2p is needed for Ime2p activity, we changed the codons encoding the T or Y in the TAY motif as well as other nearby phosphorylatable amino acid codons (S/T/Y) in the predicted activation loop of IME2 to encode nonphosphorylatable amino acids. High-copy-number plasmids containing mutant ime2 alleles were tested for their abilities to complement the sporulation defect of an ime2-Δ/ime2-Δ mutant. As previously reported, an ime2-T242A mutant completed meiosis and formed spores at wild-type levels (Table 4), however, it did so slightly more slowly than the wild-type control (data not shown). In contrast, the ime2-Y244F mutant completed low levels of meiosis more slowly than the wild-type control and never formed spores (Table 4). Mutation of both T242 and Y244 resulted in the most severe meiotic phenotype observed. The other ime2 T-loop mutants tested behaved indistinguishably from the wild type.
TABLE 4.
Terminal phenotype of cells overexpressing IME2 T-loop mutantsa
| IME2 allele | Spore formation |
|---|---|
| IME2 | + |
| IME2-Y241F | + |
| IME2-T242A | + |
| IME2-Y244F | − |
| IME2-T242A, Y244F | − |
| IME2-S246A | + |
Mutant IME2 alleles were introduced on high-copy-number plasmids into an IME2-Δ/IME2-Δ strain. Spore formation was examined after 48 h in sporulation medium.
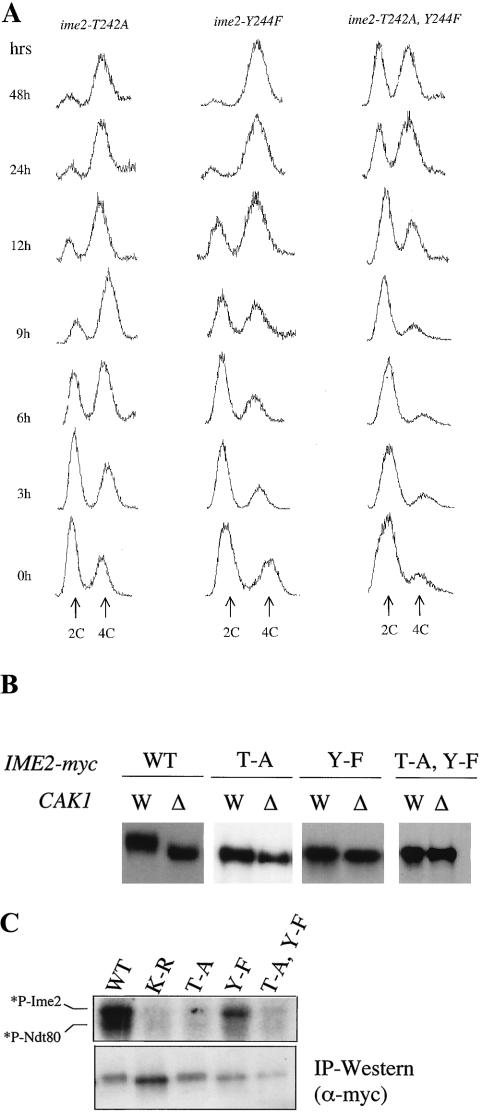
In order to assess the phenotypes of the ime2-T242A and Y244F single and double mutants in more detail, we replaced the wild-type IME2 coding sequences with mutant sequences in a CDC28-43244 diploid. We then assayed the abilities of these ime2 mutants to undergo S phase, complete the nuclear divisions, and form spores. As in the high-copy-number plasmid assay, the ime2-T242A mutant exhibited wild-type levels of meiosis and sporulation, but the initiation and completion of S phase were delayed by 3 h, thus delaying completion of subsequent events (Fig. 6A and Table 5). The ime2-Y244F mutant had more severe defects. In this background, initiation of S phase was delayed by about 9 h, and only a subset of cells accumulated with 4C DNA content by 48 h. About half of the Y244F cells completed the nuclear divisions, and none formed normal spores. The double-mutant ime2-T242A, Y244F strain was more defective than the single mutants. These cells initiated meiotic S phase even later than the Y244F mutant, and only 17% of the cells completed meiosis. This mutant was also deficient in Sic1p degradation (data not shown). Similar to the Y244F mutant, the double mutant also failed to form spores. Thus, the ime2-T242A, Y244F/ime2-T242A, Y244F strain exhibited the same meiotic defects as the cak1-Δ/cak1-Δ strain. These results suggest that both T242 and Y244 may function as Ime2p T-loop phospho-acceptors whose phosphorylation stimulates Ime2p activity, similar to the regulation of MAPKs.
FIG. 6.
Ime2p T-loop residues T242 and Y244 are required for timely S phase. (A) The progression through meiotic S phase in diploid strains harboring IME2 T-loop point mutations (from left to right: KSY221, KSY222, and KSY220) was monitored by flow cytometry. Sporulating cells were fixed at the indicated times, and nuclear DNA was stained with propidium iodide to detect 2C and 4C populations. In the wild type (WT), 50% of cells complete S phase by around 3 h (see Fig. 2B for wild-type control). (B) Analysis of the electrophoretic mobilities of the IME2 T-loop mutants was conducted by comparing the epitope-tagged proteins in diploids containing CAK1 (W; from left to right: KSY137, KSY221, KSY222, and KSY220) and lacking CAK1(Δ; from left to right: KSY138, KSY223, KSY219, and KSY218). Whole-cell lysates from sporulating cells were prepared after 4 h and probed with an antibody against the myc epitope. Each pair represents the most clearly resolved image obtained, because the Cak1p-dependent modification of the single mutants resulted in a small, but reproducible, change in electrophoretic mobility. (C) The protein kinase activity of epitope-tagged Ime2p-myc mutants was assayed by mixing immunopurified Ime2p-myc with purified Ndt80p (upper panel). *P-Ime2 and *P-Ndt80 correspond to Ime2p autophosphorylation and phosphorylation of Ndt80p, respectively. The relative amounts of protein used in the reaction mixtures are shown in the lower panel by Western blot analysis of immunoprecipitated material.
TABLE 5.
Meiotic phenotypes of IME2 T-loop mutantsa
| Strain | % Meiosis | Spore formation |
|---|---|---|
| IME2 CDC28-43244 | 85 ± 1 | + |
| ime2-T242A CDC28-43244 | 86 ± 3 | + |
| ime2-Y244F CDC28-43244 | 53 ± 6 | − |
| ime2-T242A, Y244F CDC28-43244 | 17 ± 1 | − |
The wild-type IME2 chromosomal locus was replaced with mutant ime2 alleles, and the indicated homozygous diploid strains were generated. Cells were assayed after 48 h in sporulation medium. The completion of both meiotic division and the formation of spores were scored as described in Materials and Methods.
We also tested the T-loop mutant alleles of IME2 in a wild-type CDC28 background. Similar to the phenotypes in the CDC28-43244 background, we found that the single Y244F mutant was partially impaired in completing the landmark events and that the double mutant was more defective. However, the ime2-Y244F and ime2-T242A, Y244F substitutions showed slightly more severe defects in the CDC28-43244 background than in the wild-type CDC28 background. For example, the double ime2-T242A, Y244F/ime2-T242A, Y244F mutant allowed completion of both meiotic divisions in 35% of CDC28/CDC28 cells but only 12% of CDC28-43244 cells. As in the CDC28-43244 genetic background, no spores were formed in CDC28 cells with doubly mutant ime2-T242A, Y244F. The influence of CDC28 on the abilities of the ime2 mutants to complete S phase and the nuclear divisions is consistent with Cdc28p and Ime2p functioning in the same pathway to promote these events (1).
Our evidence shows clearly that T242 and Y244 are important for Ime2p function in vivo. We examined CAK1-dependent changes in the electrophoretic mobility of mutant Ime2p proteins isolated from sporulating cells. Wild-type Ime2p-myc exhibited a significant shift in mobility in the presence of CAK1 compared to its absence (Fig. 6B, [also see Fig. 3 and 5]). Importantly, the double-mutant Ime2p-T242A, Y244F-myc protein migrated as a single band, whose mobility was indistinguishable in cells containing or lacking CAK1. The effect of the single substitutions on the CAK1-dependent electrophoretic mobility shift is similar to the effect of the double substitution. We consistently observed a minor difference in mobility of the single T-loop mutant proteins between wild-type and cak1-Δ strains. However, these differences are at the limits of resolution of the electrophoretic systems tested. These data indicate that the threonine and tyrosine are required for the modification to Ime2p seen in wild-type cells and suggest that both residues are modified in a CAK1-dependent manner.
Since mutation of the T-loop residues reduced the in vivo activity of Ime2p, we asked whether it altered the catalytic activity of the kinase. We mixed the various immunopurified epitope-tagged Ime2 proteins isolated from 2-h sporulating cultures with purified Ndt80p and radiolabeled ATP. As also shown in Fig. 4, wild-type Ime2p-myc had autocatalytic activity and phosphorylated Ndt80p (Fig. 6C). In contrast, Ime2p-T242A, Y244F-myc did not have phosphotransferase activity in vitro. The Ime2p-T242-myc protein behaved like the double mutant in vitro. Surprisingly, mutation of Y244 to F abolished the ability of Ime2p to phosphorylate Ndt80p under the in vitro conditions tested, but only partially reduced Ime2p's ability to phosphorylate itself. These in vitro data raise the possibility that phosphorylation of each T and Y in Ime2p's T-loop residues plays unique roles in regulating the activity of the enzyme.
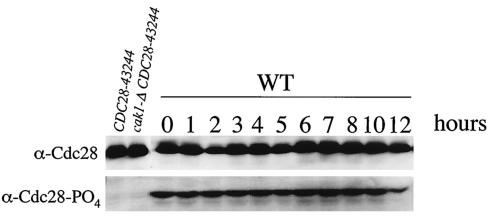
Our data have demonstrated that Cak1p functions in a CDC28-independent manner to activate Ime2p early in sporulation. We have previously shown that Cak1p functions later in the pathway to promote spore formation through the activation of the SMK1 MAPK pathway (27). Although Cak1p has multiple CDC28-independent functions during meiosis, Cak1p could also modulate phosphorylation of Cdc28p during meiosis. To examine whether CAK1-dependent phosphorylation of Cdc28p changes during sporulation, we probed an immunoblot from a sporulation time course with a phospho-specific antibody against T169 of Cdc28p. Just as Ross et al. have shown that this phosphorylation is not regulated in mitotic cells (26), we found that phosphorylation of Cdc28p on T169 is uniform throughout meiosis (Fig. 7). These data suggest that the phosphorylation of Cdc28p by Cak1p in meiosis is constitutive and thus not subject to regulatory control.
FIG. 7.
Cdc28p-T169 phosphorylation levels do not change during sporulation. Whole-cell lysates taken from sporulating wild-type (WT) cells (LNY3) at the indicated times were probed with an antibody against Cdc28p (upper panel) and a phosphospecific antibody recognizing the T169 phosphorylated form of Cdc28p (lower panel). Strains containing and lacking CAK1 in the CDC28-43244 genetic background (JLY3 and JLY4, respectively) were used as controls.
DISCUSSION
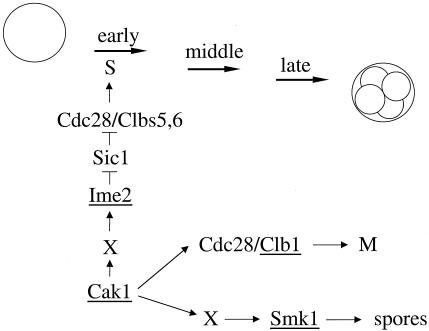
In mitosis, Cak1p's only essential role is to activate Cdc28p. In meiosis, Cak1p is also required in a Cdc28p-independent fashion at multiple steps. We have previously shown that Cak1p is required at or prior to the induction of S phase, while a later step occurs during spore wall assembly when it is required to activate the Smk1p MAPK (27). Here, we have shown that Cak1p functions early in meiosis to promote DNA synthesis through the sporulation-specific Ime2p protein kinase.
The regulation of the S-phase inhibitor Sic1p is different in meiosis than it is in mitosis. In mitosis, Cdc28p complexed to the G1 cyclins is required to phosphorylate Sic1p, targeting it for proteolysis (28). G1 cyclins inhibit meiotic induction and thus are not present in meiotic cells (4). Dirick et al. have proposed that Ime2p is the kinase responsible for targeting Sic1p for proteolysis in meiotic cells (6). Consistent with this proposal, Cdc28p is dispensable for Sic1p degradation in meiosis (1). We found that Sic1p protein is not destroyed on schedule when cak1-Δ/cak1-Δ cells are transferred to sporulation media. Ime2p failed to be modified in its activation loop and was catalytically inactive in cells lacking Cak1p. Moreover, we found that deletion of SIC1 or overproduction of IME2 partially suppressed the meiotic S-phase defect in mutant cak1-Δ cells. These results indicate that Cak1p is required early in meiosis to promote the activation of Ime2p, which in turn stimulates meiotic S phase by targeting Sic1p for destruction (see the model in Fig. 8). Ime2p also positively regulates the transcription of early sporulation genes and is required for timely induction of middle sporulation genes. Thus, a deficit in Ime2p activity in cells lacking CAK1 is also sufficient to explain the defects in the transcriptional program that we previously documented in the cak1-Δ/cak1-Δ background (27). The role of Ime2p in positively regulating early gene expression may be one reason why only a partial bypass of the S-phase defect was observed when Sic1p was depleted. Alternatively, there may be low levels of Sic1p in these experiments (below the level of detection in our immunoblots) that can partially inhibit Cdc28p-Clb5p/Clb6p S-phase-promoting ability. We have shown that Ime2p from cak1-Δ cells is catalytically inactive in vitro. The ability of overexpressed IME2 to partially suppress the S-phase defect in this mutant suggests either that the nonphosphorylatable form of Ime2p has a low level of catalytic activity in vivo or that Ime2p can positively regulate S phase in a noncatalytic fashion. This information provides a glimpse into how the Ime2p meiosis-specific kinase is regulated after the transcriptional program of sporulation is induced.
FIG. 8.
Model for Cak1p's roles in meiosis. Cak1p functions through Ime2p to regulate S phase via Sic1p destruction and through Smk1p to regulate spore formation. Cak1p is also required to activate Cdc28p to initiate meiotic S phase and to execute the meiotic divisions. Not shown is that Cak1p activation of Ime2p also promotes middle gene induction, and thus M phase and spore formation, by inducing expression of CLBs and SMK1, respectively. Genes directly regulated by the transcriptional program of sporulation are underlined. X in the Ime2p and Smk1p pathways could be unique or shared and could represent Cak1p-activated kinases or noncatalytic regulatory components that promote the direct phosphorylation of Ime2p or Smk1p by Cak1p.
Our data suggest that Cak1p is required for phosphorylation of T242 and Y244 in Ime2p's activation loop and that phosphorylation of these residues is necessary for its full catalytic activity in vivo. The CAK1-dependent modification of Ime2p could occur by several possible mechanisms. For instance, Cak1p may directly phosphorylate Ime2p, or it could activate a protein kinase that is in turn required to activate Ime2p. Our laboratory has been unable to detect the phosphorylation of purified Ime2p by purified Cak1p in vitro (27). It is possible, however, that a noncatalytic component may be missing from our reactions. It is worth noting that the dominant activating residue in Ime2p in vivo appears to be Y244, located in the T-loop. While Cak1p has only been shown to phosphorylate threonines located 10 residues amino terminal to the APE motif in subdomain VIII in yeast Cdks, one group has reported in a proteomics study that a Cak1p-GST fusion protein can phosphorylate tyrosine residues in a synthetic peptide (42). It is also possible that Cak1p activates a Cdk, which in turn is required for Ime2p activation. We have ruled out Cdc28p as being uniquely required since the allele of CDC28 used in this study lacks the Cak1p-activatable threonine in its T-loop. The Bur1p and Kin28p Cdks have been shown to be phosphorylated by Cak1p in their T-loops (17, 41). The Ctk1p Cdk has been shown to be phosphorylated on an activation loop threonine in a proteomics study (8). We have tested mutant alleles of BUR1, CTK1, and KIN28 that encode an alanine substitution for the activating threonine in their T-loops. Diploid strains homozygous for these mutant Cdks complete meiotic S phase with wild-type kinetics, suggesting that none of these Cdks are uniquely required to regulate Ime2p (unpublished results).
These issues may also be relevant to our studies involving Cak1p's role in activating the Smk1p MAPK pathway (see model in Fig. 8). Similar to our analysis with Ime2p, we have shown that Cak1p functions upstream of Smk1p (27). The activation loops of Ime2p and Smk1p contain a TXY motif. Dual phosphorylation of the T and Y in Smk1p appears to be required for full catalytic activity (27). Our data suggest that dual phosphorylation is also required for full Ime2p activation. Whether there is a common Cak1p-activated kinase that regulates both Ime2p and Smk1p remains to be determined.
In addition to its roles in activating Ime2p and Smk1p, Cak1p is required to activate Cdc28p during meiosis (see model in Fig. 8). In mitosis, the activation of Cdc28p by Cak1p appears to be largely unregulated, because large pools of Cdc28p phosphorylated on T169 are present throughout the cell cycle (26). We found that the level of phosphorylated Cdc28p is relatively constant throughout meiosis (Fig. 7). These data suggest that while Cak1p-activated Cdc28p is required for meiosis (active Cdc28p is required at both the S and M phases), the modification is constitutive and does not play a direct role in regulating meiotic progression.
Based upon sequence homology, Ime2p has been classified as a Cdk-like kinase, yet no cyclin binding partner has been identified to date. Similar to the roles Cdks play in the cell cycle, Ime2p regulates multiple steps throughout meiosis. These steps include initiating meiotic S phase as well as the nuclear divisions. We and others have shown that Ime2p can be electrophoretically resolved into a series of distinct phospho-forms whose relative abundance changes during meiotic progression (1, 34). We speculate that the sequential phosphorylation of Ime2p represents a modification pathway that is critical for the orderly progression through and successful completion of sporulation. These modifications could alter Ime2p's catalytic potency, substrate specificity, protein-protein interactions, or cellular localization. We have shown here that mutation of the tyrosine to a nonphosphorylatable phenylalanine modestly reduces Ime2p's potency for autophosphorylation and dramatically reduces its ability to phosphorylate Ndt80p in vitro. It is less clear why Ime2p-T242A-myc behaves like a catalytically inactive protein in vitro but is only modestly affected in vivo. These data suggest that there may be complex interactions between T and Y phosphorylation events in activating the Ime2p kinase. Remarkably, Ime2p is completely unmodified, even at late time points, in cells lacking CAK1. These data suggest that Cak1p-dependent modification of Ime2p's activation loop is required for all subsequent phosphorylation events of Ime2p. Catalytically inactive Ime2p appears to be modified in a Cak1p-dependent fashion in its activation loop, yet it fails to get modified further. These data suggest that Cak1p initiates the Ime2p phospho-modification pathway and that autophosphorylation plays an essential role in its progression.
Acknowledgments
We thank Saul Honigberg, Philip Kaldis, Michael Mendenhall, Aaron Mitchell, and Mike Tyers for plasmids and antibodies. We thank Michelle Reed for generating BUR1 and CTK1 mutant yeast strains. We also thank Erica Johnson, David Bungard, Kristy Shuda, and Brendan Maher for helpful discussions and comments on the manuscript.
This work was supported by grants from the NIH (I.H. and E.W.). K.B. was supported by the Cancer Research Fund of the Danyon Runyon-Walter Winchell Foundation and the California Division of the American Cancer Society.
Footnotes
This paper is dedicated to the memory of Ira Herskowitz (deceased 28 April 2003).
REFERENCES
- 1.Benjamin, K. R., C. Zhang, K. M. Shokat, and I. Herskowitz. 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17:1524-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu, S., and I. Herskowitz. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685-696. [DOI] [PubMed] [Google Scholar]
- 3.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 4.Colomina, N., E. Gari, C. Gallego, E. Herrero, and M. Aldea. 1999. G1 cyclins block the Ime1 pathway to make mitosis and meiosis incompatible in budding yeast. EMBO J. 18:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross, F. R., and K. Levine. 1998. Molecular evolution allows bypass of the requirement for activation loop phosphorylation of the Cdc28 cyclin-dependent kinase. Mol. Cell. Biol. 18:2923-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dirick, L., L. Goetsch, G. Ammerer, and B. Byers. 1998. Regulation of meiotic S phase by Ime2 and a Clb5, 6-associated kinase in Saccharomyces cerevisiae. Science 281:1854-1857. [DOI] [PubMed] [Google Scholar]
- 7.Espinoza, F. H., A. Farrell, H. Erdjumentbromage, P. Tempst, and D. O. Morgan. 1996. A cyclin-dependent kinase-activating kinase (Cak) in budding yeast unrelated to vertebrate Cak. Science 273:1714-1717. [DOI] [PubMed] [Google Scholar]
- 8.Ficarro, S., M. McCleland, P. Stukenberg, D. Burke, M. Ross, J. Shabanowitz, D. Hunt, and F. White. 2002. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20:301-305. [DOI] [PubMed] [Google Scholar]
- 9.Foiani, M., E. Nadjar-Boger, R. Capone, S. Sagee, T. Hashimshoni, and Y. Kassir. 1996. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol. Gen. Genet. 253:278-288. [DOI] [PubMed] [Google Scholar]
- 10.Guttmann-Raviv, N., S. Martin, and Y. Kassir. 2002. Ime2, a meiosis-specific kinase in yeast, is required for destabilization of its transcriptional activator, Ime1. Mol. Cell. Biol. 22:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hepworth, S. R., H. Friesen, and J. Segall. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5750-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 13.Hunter, T., and G. D. Plowman. 1997. The protein kinases of budding yeast: six score and more. Trends Biochem. Sci. 22:18-22. [DOI] [PubMed] [Google Scholar]
- 14.Kaldis, P., A. Sutton, and M. J. Solomon. 1996. The cdk-activating kinase (CAK) from budding yeast. Cell 86:553-564. [DOI] [PubMed] [Google Scholar]
- 15.Kaldis, P., P. Zachary, I. Bany, D. Enke, M. Wagner, E. Winter, and M. Solomon. 1998. Localization and regulation of the cdk-activating kinase (Cak1p) from budding yeast. J. Cell Sci. 111:3585-3596. [DOI] [PubMed] [Google Scholar]
- 16.Kaldis, P., V. Tsakraklides, K. E. Ross, E. Winter, and A. Cheng. 2001. Activating phosphorylation of cyclin-dependent kinases in budding yeast p. 13-30. In P. Kaldis (ed.), Cdk-activating kinase (CAK). Landes Bioscience, Georgetown, Tex.
- 17.Kimmelman, J., P. Kaldis, C. J. Hengartner, G. M. Laff, S. S. Koh, R. A. Young, and M. J. Solomon. 1999. Activating phosphorylation of the Kin28p subunit of yeast TFIIH by Cak1p. Mol. Cell. Biol. 19:4774-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kupiec, M., B. Byers, R. Esposito, and A. Mitchell 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In J. Pringle, J. Broach, and E. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Lindgren, A., D. Bungard, M. Pierce, J. Xie, A. Vershon, and E. Winter. 2000. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 19:6489-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell, A. P. 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev. 58:56-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell, A. P., S. E. Driscoll, and H. E. Smith. 1990. Positive control of sporulation-specific genes by the IME1 and IME2 gene products in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2104-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nugroho, T. T., and M. D. Mendenhall. 1994. An inhibitor of yeast cyclin-dependent protein kinase plays an important role in ensuring the genomic integrity of daughter cells. Mol. Cell. Biol. 14:3320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pak, J., and J. Segall. 2002. Role of Ndt80, Sum1, and Swe1 as targets of the meiotic recombination checkpoint that control exit from pachytene and spore formation in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:6430-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierce, M., K. R. Benjamin, S. P. Montano, M. M. Georgiadis, E. Winter, and A. K. Vershon. 2003. Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol. Cell. Biol. 23:4814-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Primig, M., R. Williams, E. Winzeler, G. Tevzadze, A. Conway, S. Hwang, R. Davis, and R. Esposito. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26:415-423. [DOI] [PubMed] [Google Scholar]
- 26.Ross, K., P. Kaldis, and M. Solomon. 2000. Activating phosphorylation of the Saccharomyces cerevisiae cyclin-dependent kinase, Cdc28p, precedes cyclin binding. Mol. Biol. Cell 11:1597-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaber, M., A. Lindgren, K. Schindler, D. Bungard, P. Kaldis, and E. Winter. 2002. CAK1 promotes meiosis and spore formation in Saccharomyces cerevisiae in a CDC28-independent fashion. Mol. Cell. Biol. 22:57-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwob, E., T. Bohm, M. Mendenhall, and K. Nasmyth. 1994. The B-type cyclin kinase inhibitor p40Sic1 controls the G1 to S transition in S. cerevisiae. Cell 79:233-244. [DOI] [PubMed] [Google Scholar]
- 29.Shuster, E. O., and B. Byers. 1989. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics 123:29-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sia, R. A. L., and A. P. Mitchell. 1995. Stimulation of later functions of the yeast meiotic protein kinase Ime2p by the IDS2 gene product. Mol. Cell. Biol. 15:5279-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, H. E., and A. P. Mitchell. 1989. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solomon, M. J., T. Lee, and M. W. Kirschner. 1992. Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol. Biol. Cell 3:13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solomon, M. J., M. Glotzer, T. H. Lee, M. Philippe, and M. W. Kirschner. 1990. Cyclin activation of p34cdc2. Cell 63:1013-1024. [DOI] [PubMed] [Google Scholar]
- 34.Sopko, R., S. Raithatha, and D. Stuart. 2002. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol. Cell. Biol. 22:7024-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuart, D., and C. Wittenberg. 1998. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 12:2698-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thuret, J.-V., J.-G. Valay, G. Faye, and C. Mann. 1996. Civ1 (CAK in vivo), a novel cdk-activating kinase. Cell 86:565-576. [DOI] [PubMed] [Google Scholar]
- 37.Verma, R., R. Annan, M. Huddleston, S. Carr, G. Reynard, and R. Deshaies. 1997. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278:455-460. [DOI] [PubMed] [Google Scholar]
- 38.Wagner, M., M. Pierce, and E. Winter. 1997. The CDK-activating kinase CAK1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J. 16:1305-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter, and A. K. Vershon. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yaffe, M. P., and G. Schatz. 1984. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA 81:4819-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao, S., and G. Prelich. 2002. Activation of the Bur1-Bur2 cyclin-dependent kinase complex by Cak1. Mol. Cell. Biol. 22:6750-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu, H., J. F. Klemic, S. Chang, P. Bertone, A. Casamayor, K. G. Klemic, D. Smith, M. Gerstein, M. A. Reed, and M. Snyder. 2000. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26:283-289. [DOI] [PubMed] [Google Scholar]