Abstract
Telomere dysfunction causes genomic instability. However, the mechanism that initiates this instability when telomeres become short is unclear. We measured the mutation rate and loss of heterozygosity along a chromosome arm in diploid yeast that lacked telomerase to distinguish between mechanisms for the initiation of instability. Sequence loss was localized near chromosome ends in the absence of telomerase but not after breakage of a dicentric chromosome. In the absence of telomerase, the increase in mutation rate is dependent on the exonuclease Exo1p. Thus, exonucleolytic end resection, rather than chromosome fusion and breakage, is the primary mechanism that initiates genomic instability when telomeres become short.
Chromosome rearrangements and region-specific amplifications and deletions occur frequently in cancer (41). This genomic instability begins early during tumorigenesis (64), and the mechanisms underlying it are poorly understood (6, 41). The association of telomere dysfunction with genomic instability and increased tumor formation in mice (1, 4, 59) suggests that telomere shortening might contribute to tumorigenesis in humans. Because telomere shortening occurs as a normal consequence of cell division in most human cell types, short telomeres could play a role in the initiation of tumor formation (12). In fact, telomere dysfunction has been linked to terminal deletion and complex chromosomal abnormalities in human tumors (22). Short telomeres can also limit tumor progression (24, 60). Understanding the mechanism by which short telomeres cause genomic instability could provide insight into tumor initiation.
Telomeres consist of repeats of short TG-rich sequences that are elongated by the ribonucleoprotein enzyme telomerase (25). Saccharomyces cerevisiae telomeres contain ∼300 bp of the repeat TG1-3 (75), whereas human telomeres consist of 10 to 15 kb of TTAGGG repeats. In most somatic human cells, telomerase is inactive and telomeres undergo progressive telomere shortening with each cell division (29). This observation has suggested the possibility that telomere shortening might participate in tumor initiation by increasing genome instability (reviewed in references 12, 28, and 50). We previously demonstrated a direct link between telomere shortening and chromosome rearrangements in S. cerevisiae (27).
Although telomere shortening has been associated with increased genomic instability in several systems, little is known about the molecular mechanism that initiates these chromosome rearrangements. The terminal deletions induced by telomere shortening in the absence of telomerase may be initiated by end-to-end chromosome fusion and breakage (51) or by exonucleolytic end resection (27). End-to-end chromosome fusion is the most prominent chromosomal abnormality in telomerase-deficient mice (4). In these mice, the chromosomes with the shortest telomeres are most frequently involved in fusion, suggesting that these fusions could be a primary consequence of telomere shortening (30). Chromosome fusions are also the predominant cytogenetic abnormality in human cells expressing a dominant-negative allele of the telomere-binding protein TRF2 (69). Circular chromosomes form in telomerase-deficient Schizosaccharomyces pombe (54) and chromosome fusion junctions can be isolated from S. cerevisiae (11, 27). In some situations, telomere fusion requires proteins involved in nonhomologous end joining. For example Ku is required for telomere fusions in S. pombe (19) and mice (16), and in some situations DNA ligase IV is required for fusions in S. cerevisiae (43) and human cells (62). However, Ku is not required for telomere fusions in Arabidopsis (57) and is not required for chromosome fusion in cells that survive in the absence of telomerase in S. pombe (3). The chromosome rearrangements in telomerase-deficient S. cerevisiae with short telomeres occur at similar rates in the presence or absence of DNA ligase IV (27), suggesting that end-to-end chromosome fusion may not initiate rearrangements that occur when telomeres shorten in the absence of telomerase.
Even though they are frequently observed, chromosome fusions might not be the primary mechanism that initiates chromosome rearrangements but rather stable by-products. Rapid telomere shortening can be induced in yeast by loss of telomere end-binding proteins. These cells exhibit rapid and extensive degradation of sequence from the chromosome ends (2, 7, 20). If similar end resection occurs in telomerase-deficient cells, it could induce chromosome rearrangements. Distinguishing between end-to-end fusion and end resection would provide insight into the molecular mechanisms responsible for the initial recognition and processing of dysfunctional telomeres.
Here we describe chromosome rearrangements in diploid cells that occur in response to short telomeres. These chromosome rearrangements occur predominantly in the terminal regions of the chromosomes. The exonuclease Exo1p plays a major role in the generation of rearrangements, providing evidence that end resection initiates genomic instability at dysfunctional telomeres. In addition to elucidating the mechanism by which dysfunctional telomeres induce chromosome rearrangements, the present study also provides new insight into the possible role of single-stranded DNA (ssDNA) in the telomere checkpoint.
MATERIALS AND METHODS
Plasmids and strains.
pJHU618 is a HIS3 integrating galactose-inducible EST1 expression vector (27). A constitutive EST1 expression vector (pJHU840) was created by cloning the EST1 open reading frame and the entire surrounding intergenic sequence into pRS412 (ADE2 CEN/ARS). An EST1 deletion vector was created by replacing the BspEI fragment within EST1 with kanMX in pJHU618 (pJHU832). The knockout fragment was excised with BamHI. A galactose-inducible CRE expression vector (pJHU791) was created by cloning GAL1/CRE (26) into pRS411 (MET15 CEN/ARS).
est1Δ/EST1 and est1Δ/est1Δ diploid mutation rate strains.
Strain genotypes are summarized in Table 1. In JHUY669, CAN1 was inserted just centromeric to ADE5, 63,020 bp from the telomere, between YGL232w and YGL231c. URA3 was inserted 11 kb to the right of the centromere of chromosome VII, between MUQ1 and STF2. JHUY669 was an est1Δ spore of a heterozygous EST1/est1Δ diploid strain that was dissected on galactose to maintain EST1 expression from pJHU618. YPH450 and JHUY669 were mated on galactose and transformed with pJHU840 to create JHUY676. The remaining genomic EST1 allele was deleted with the est1Δ::kanMX BamHI fragment of pJHU832 to create JHUY677.
TABLE 1.
S. cerevisiae strains used in this studya
| Strain | Genotype | Source and/or reference |
|---|---|---|
| JHUY669 | MATaest1Δ::LEU2 his3Δ1::pJHU618 ade2Δ::hisG can1Δ::MET15 ura3Δ0 leu2Δ0 lys2Δ0 met15Δ0; VII-L, CAN1; VII-R, URA3 | |
| YPH450 | MATα ade2 can1 ura3 leu2 his7 trp1; VII-L, ade5 lys5 cyh2 trp5 leu1 | Derived from A364A (7) |
| JHUY676 | MATa/MATα est1Δ::LEU2/EST1 his3Δ1::pJHU618/HIS3 ade2Δ::hisGlade2 can1Δ::MET15/can1 ura3Δ0/ura3 leu2Δ0/leu2 lys2Δ0/LYS2 met15Δ0/MET15 HIS7/his7 TRP1/trp1; VII-L, (ADE5 CAN1 LYS5 CYH2 TRP5 LEU1)/(ade5 lys5 cyh2 trp5 leu1); VII-R, URA3/( ); pJHU840 | JHUY669 × YPH450 + pJHU840 |
| JHUY677 | MATa/MATα est1Δ::LEU2/est1Δ::kanMX his3Δ1::pJHU618/HIS3 ade2Δ::hisGlade2 can1Δ::MET15/can1 ura3Δ0/ura3 leu2Δ0/leu2 lys2Δ0/LYS2 met15Δ0/MET15 HIS7/his7 TRP1/trp1 VII-L, (ADE5 CAN1 LYS5 CYH2 TRP5 LEU1)/(ade5 lys5 cyh2 trp5 leu1); VII-R, URA3/(); pJHU840 | est1Δ/est1ΔJHUY676 |
| JHUY704 | MATacdc13-1 his3Δ1::pJHU618 ade2Δ::hisG can1Δ::hphMX ura3Δ0 lys2Δ0 met15Δ0; VII-L, CAN1; VII-R, kanMX | |
| JHUY671 | MATα cdc13-1 ade2 can1 ura3 leu2 his7 trp1; VII-L, ade5 lys5 cyh2 trp5 leu1 | |
| JHUY705 | MATa/MATα cdc13-1/cdc13-1 his3Δ1::pJHU618/HIS3 ade2Δ::hisG/ade2 can1Δ::hphMX/can1 ura3Δ0/ura3 LEU2/leu2 lys2Δ0/LYS2 met15Δ0/MET15 HIS7/his7 TRP1/trp1; VII-L, (ADE5 CAN1 LYS5 CYH2 TRP5 LEU1)/(ade5 lys5 cyh2 trp5 leu1); VII-R, kanMX/() | JHUY704 × JHUY671 |
| JHUY707 | MATahis3Δ1::pJHU618 ade2Δ::hisG can1Δ::hphMX ura3Δ0 lys2Δ0 met15Δ0; V-L, ARS121 3A-loxP; VII-L, UR-loxP CAN1; VII-R, kanMX | |
| JHUY706 | MATα ade2 can1 ura3 leu2 met15Δ0 his7 trp1; VII-L, ade5 lys5 cyh2 trp5 leu1 | Deletion of MET15 with pAD4 (5) in YPH450 |
| JHUY708 | MATa/MATα his3Δ1::pJHU618/HIS3 ade2Δ::hisGlade2 can1Δ::hphMX1can1 ura3Δ0/ura3 LEU2/leu2 lys2Δ0/LYS2 met15Δ0/met15Δ0 HIS7/his7 TRP1/trp1; V-L, (ARS121 3A-loxP)/(); VII-L, (UR-loxP ADE5 CAN1 LYS5 CYH2 TRP5 LEU1)/(ade5 lys5 cyh2 trp5 leu1); VII-R, kanMX/() | JHUY707 × JHUY706 |
| JHUY709 | MATa/MATα est1Δ::LEU2/EST1 mre11Δ::hphMX/MRE11 yku70Δ::kanMX/YKU70 his3Δ1::pJHU618/his3Δ1::pJHU618 ade2Δ::hisGlade2Δ::hisG can1Δ::MET15/can1Δ::MET15 ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 met15Δ0/met15Δ0; XV-L, (ADE2-CAN1 URA3)/(ADE2-CAN1 URA3) (TELO) | Derived from JHUY635 (27) |
| JHUY710 | MATa/MATα est1Δ::LEU2/EST1 exo1Δ::kanMX/EXO1 his3Δ1::pJHU618/his3Δ1::pJHU618 ade2Δ::hisGlade2Δ::hisG can1Δ::MET15/can1Δ::MET15 ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 met15Δ0/met15Δ0; XV-L, (ADE2-CAN1 URA3)/(ADE2-CAN1 URA3) (TELO) | Derived from JHUY635 (27) |
| JHUY711 | MATa/MATα est1Δ::LEU2/EST1 mre11Δ::hphMX/MRE11 exo1Δ::kanMX/EXO1 his3Δ1::pJHU618/his3Δ1::pJHU618 ade2Δ::hisGlade2Δ::hisG can1Δ::MET15/can1Δ::MET15 ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 met15Δ0/met15Δ0; XV-L, (ADE2-CAN1 URA3)/(ADE2-CAN1 URA3) (TELO) | Derived from JHUY635 (27) |
| BY4741 | MATahis3Δ1 ura3Δ0 leu2Δ0 met15Δ0 | 5 |
| JHUY728 | MATacdc13-1 his3Δ1 ura3Δ0 leu2Δ0 met15Δ0 | |
| JHUY729 | MATa/MATα est1Δ::LEU2/EST1 rad1Δ::kanMX/RAD1 his3Δ1::pJHU618/his3Δ1::pJHU618 ade2Δ::hisG/ade2Δ::hisG can1Δ::MET15/can1Δ::MET15 ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 met15Δ0/met15Δ0; XV-L, (ADE2-CAN1 URA3)/(ADE2-CAN URA3) (TELO) | Derived from JHUY635 (27) |
Genotypes on specific chromosome arms are listed from telomere to centromere. Empty parentheses indicate no gene was inserted on the chromosome arm.
cdc13-1 diploid mutation rate strain.
The cdc13-1 allele was introduced into the endogenous CDC13 locus in appropriate haploids by transformation with XhoI-digested pVL451(obtained from Ted Weinert) and selection on SC lacking uracil (SC−Ura), followed by selection on 5-fluoorotic acid (5-FOA). JHUY671 and JHUY704 were mated to create JHUY705.
Diploid dicentric mutation rate strain.
JHUY707 is an EST1 version of JHUY669 in which URA3 was replaced with kanMX, and can1Δ::MET15 was replaced with can1Δ::hphMX. A 3,070-bp vector fragment containing the 5′ half of URA3 and a loxP site (UR-loxP) was inserted 12,080 bp from the left telomere of chromosome VII, between YGL258w and MNT2. A 1,365-bp vector fragment containing ARS121, the 3′ half of URA3, and a loxP site (3A-loxP) was inserted 10,469 bp from the left telomere of chromosome V, between YEL073c and YEL072w. The 5′ and 3′ halves of URA3 were separated from the loxP sites by the 5′ or 3′ half of the ACT1 intron. These URA3-ACT1 intron sequences were amplified from pAGE1638 (74). The ACT1 intron was split at the MfeI site. YPH450 was transformed with pAD4 (5) to introduce the met15Δ0 allele (JHUY706). JHUY706 and JHUY707 were mated to create JHUY708.
Haploid mutation rate strains.
JHUY709 to JHUY711 and JHUY729 were derived from JHUY635, a strain containing ADE2 and CAN1 located 3,482 bp from the telomere and URA3 between YOL155c and YOL154w on the left arm of chromosome XV (27). We refer to this localization of CAN1, ADE2, and URA3 as TELO. MRE11 was deleted with hphMX that was amplified from pAG32 (23) with the primers 5′-ATGAAATCCATTTCGTTACAAGATGTTCCCCATTTGAGGCAGCTGAAGCTTCGTACGC-3′ and 5′-TTCAATAAGATACTTAGACGTAGCATCTTTATCGTGAGGATAGGCCACTAGTGGATCTG-3′. Deletion was confirmed by Southern blotting. The exo1Δ allele and the rad1Δ allele were crossed into the JHUY635 strain background from strains created in the Saccharomyces Genome Deletion Project (73). JHUY635 had been created from the same strain background used in the Deletion Project. The average white-colony mutation rates (point mutation rates) were 1.4 × 10−7 (wild type), 1.5 × 10−7 (est1Δ), 1.2 × 10−6 (exo1Δ), 7.8 × 10−7 (est1Δ exo1Δ), 2.3 × 10−7 (rad1Δ), and 1.9 × 10−7 (est1Δ rad1Δ).
Mutation rate analysis.
For diploid mutation rate analysis, est1Δ/EST1 (JHUY676) and est1Δ/est1Δ diploids (JHUY677) were plated to single colonies on yeast extract-peptone-dextrose (YPD) and grown for 40 h at 30°C. Ten red colonies that had lost the EST1/pRS412 vector were chosen and grown in log phase for 21 h each day in 5 ml of medium (90% minimal medium-10% YEP medium supplemented with 87 mg of adenine/liter, 30 mg of leucine/liter, 30 mg of lysine/liter, 20 mg of uracil/liter, 20 mg of tryptophan/liter, 200 mg of ornithine/liter, 2% raffinose, and 0.1% dextrose). Cultures were inoculated at appropriate concentrations to grow to ∼3 × 107 cells/ml. The plotted cell densities were adjusted to conform to the convention in which cultures are inoculated at the same concentration each day (104 cells/ml). The plotted cell density is equal to the (final concentration × 104 cells/ml)/initial concentration. Each day 103 cells were plated on 2% galactose SC lacking arginine and lacking histidine (SC−Arg−His) control plates. An appropriate number of cells to form ∼200 canavanine-resistant (CanR) colonies (determined in preliminary experiments) were plated on CAN1 mutant selection plates (2% galactose SC−Arg−His−Ura with 60 mg of canavanine/liter). Chromosome loss rate was measured by plating 1.5 ml on 2% galactose SC−Arg−His with 60 mg of canavanine and 0.5 g of 5-FOA/liter. Mutation rate experiments for haploid strains were performed for 8 to 10 independent spores derived from heterozygous diploids (27). Mutation rates and chromosome loss rates were calculated by using the maximal-likelihood method as previously described (27, 38).
To measure mutation rate and loss of heterozygosity (LOH) frequency for diploid cdc13-1/cdc13-1 strains, JHUY705 cells were plated as single colonies on YPD and grown 3 days at 22°C. Ten independent colonies were inoculated into 5 ml of the liquid medium used for diploid mutation rate experiments and grown in log phase for 21 h at 22°C. Cultures were diluted to 5 × 106 cells/ml in two 5-ml cultures. A total of 5 ml was grown at 22°C for 7 h. A total of 5 ml was grown at 37°C for 3.5 h and then shifted to 22°C for 3.5 h (20). For all cultures, 103 cells were plated on SE lacking arginine (SE−Arg) control plates and 2 × 105 cells were plated on mutant selection plates (SE−Arg supplemented with 60 mg of canavanine/liter and 200 μg of G418/ml) to select CanR colonies that retained kanMX. SE medium contains 1 g of glutamate/liter as a nitrogen source and allows for the use of G418 in dropout media (10).
To measure the effect of a dicentric chromosome on LOH frequency, JHUY708 was transformed with GAL1/CRE/pRS411 (pJHU791) or the empty pRS411 vector and plated on SC lacking methionine and cysteine (SC−Met−Cys). Ten colonies from each transformation were chosen and inoculated at 3 × 104 cells/ml in 5 ml of SC−Met−Cys (2% dextrose) and grown for 21 h in log phase. Cells were diluted to 2 × 104 cells/ml in 5 ml of 2% galactose SC−Met−Cys and grown 42 h in log phase. Cells were plated after growth in dextrose and galactose. For all cultures, 103 cells were plated on SE-Arg control plates, 2 × 105 cells were plated on SE−Arg with 60 mg of canavanine/liter, and 200 μg of G418/ml to measure the general mutation rate. To measure the frequency of dicentric chromosome formation (Ura+ cells), 1 ml was plated on SE−Arg−Ura, except that 106 cells were plated from galactose CRE cultures. To measure mutation rate in only Ura+ cells, 3 ml was plated on SE−Arg−Ura with 60 mg/liter canavanine and 200 μg of G418/ml.
LOH calculation.
To measure the extent of LOH, colonies from canavanine plates were replica plated onto plates containing SC−Lys, SC−Trp, SC−Leu, and YEP containing 10 mg of cycloheximide/liter. Plates for est1Δ/est1Δ and est1Δ/EST1Δ diploids contained 2% galactose. Plates for cdc13-1/cdc13-1 and dicentric chromosome strains contained 2% dextrose. The frequency of LOH between specific markers was calculated as the number of white CanR mutant colonies that had lost the relevant distal markers divided by the total number of white CanR mutant colonies.
Pulsed-field gel electrophoresis.
Whole chromosomes were resolved by pulsed-field gel electrophoresis (24 h, 200 V, and 55 to 110 s). The structure of chromosome VII was confirmed in diploid strains by probing whole chromosomes with a URA3 probe. To separate whole chromosomes from the 27-kb chromosome fragment, we used the parameters 11 h, 200 V, and 0.5 to 5 s. The probe to the chromosome fragment was the PCR product of JH109F (5′-ACAATCGATTTCCTTTAGGC-3′ ) and JH109R (5′-GCTTTTACTATGCGATCGCC-3′ ). To confirm break-induced replication (BIR) in haploid TELO strains, whole chromosomes were probed with sequence transferred telomeric to homology region I (HR I) and HR II (probes A and B) (27), and terminal restriction fragment lengths were measured by digestion with PmeI and hybridization with a DCP1 probe using the parameters 18 h, 200 V, and 1 to 15 s (27).
ssDNA measurement.
DNA was isolated from 300-ml log-phase cultures grown in 2% dextrose haploid mutation rate medium according to the 40-ml yeast DNA Miniprep protocol (58). The substitution of dextrose in these cultures slightly alters the shapes of the growth curves, probably by decreasing leaky expression from the galactose-inducible EST1. DNA was also prepared from wild-type (BY4741) and cdc13-1 (JHUY728) haploids grown in YPD at 22°C or shifted to the nonpermissive temperature (37°C) for 4 h (20). Nondenatured (1 μg) and denatured (0.1 μg) DNA samples were dot blotted onto Hybond-N+ membrane as described previously (20). Membranes were probed with a previously described telomeric G-strand oligonucleotide that hybridizes to a 3′ overhang (14, 55). The C-stand telomere probe is the reverse complement of the G-strand probe. To control for telomere length, hybridization of the G-strand telomere probe to nondenatured DNA (Gn) was divided by hybridization of a centromeric gene probe (HSP10) to denatured DNA (Hd). To control for artifactual denaturation of telomeric DNA, the ratio of nondenatured to denatured DNA measured with the C-strand probe was used as a correction factor (Cn/Cd), given the denatured G-strand signal (Gd). Therefore, the amount of ssDNA at the telomeric G-strand per genome is proportional to the following calculated value: [Gn − Gd × (Cn/Cd)]/[Hd × (1 − Cn/Cd)]. Error bars indicate the standard deviation. The percent ssDNA on the C-strand is equal to (100 × Cn/Cd). For pulsed-field gel analysis, agarose-embedded DNA was prepared from cells grown in 300-ml log-phase cultures (21). Agarose-embedded DNA was treated with exonuclease I (NEB) for 24 h at 37°C. Hybridization was quantitated by using a phosphorimager.
RESULTS
Telomere shortening increases mutation rate in diploid yeast.
We previously demonstrated that telomere shortening causes terminal deletions and nonreciprocal translocations in haploid yeast (27). Because loss of essential genes is lethal in haploids, we were unable to determine whether deletions occur primarily at chromosome ends or whether they occur throughout chromosome arms. If terminal deletions arise from breakage of dicentric chromosomes, breaks would likely occur along the length of a chromosome arm. However, if the loss of end protection followed by exonucleolytic end resection creates deletions, sequence loss would be concentrated near chromosome ends.
To examine where sequence loss occurs, we created a diploid strain for mutation rate analysis in which deletion of an arm or even an entire chromosome would not be lethal. We have previously shown that telomere shortening causes terminal deletions but not internal deletions (27). Therefore, we monitored the loss rate of a terminal marker and then measured the extent of sequence loss in the colonies that lost the most terminal marker by assaying the loss of more internal markers. Mutant colonies were selected on the basis of loss of CAN1, which was inserted on one copy of chromosome VII, just centromeric to ADE5 (Fig. 1A). In the red ade2/ade2 strain background, colonies arising from point mutations in CAN1 are red, whereas colonies arising from deletion of both CAN1 and ADE5 are white. The other copy of chromosome VII has recessive mutations in genes distributed along the length of the chromosome arm. Loss of the wild-type alleles of these genes from the CAN1-marked chromosome was determined by growth on plates that select for the presence of each wild-type marker. The absence of growth on specific plates defined the extent of LOH in can1 mutant colonies (Fig. 1A) (7). Selection for URA3 on the opposite arm of chromosome VII ensured that the centromere was retained in the mutant colonies (Fig. 1A). This system allowed us to select for cells that had lost CAN1 through a terminal deletion.
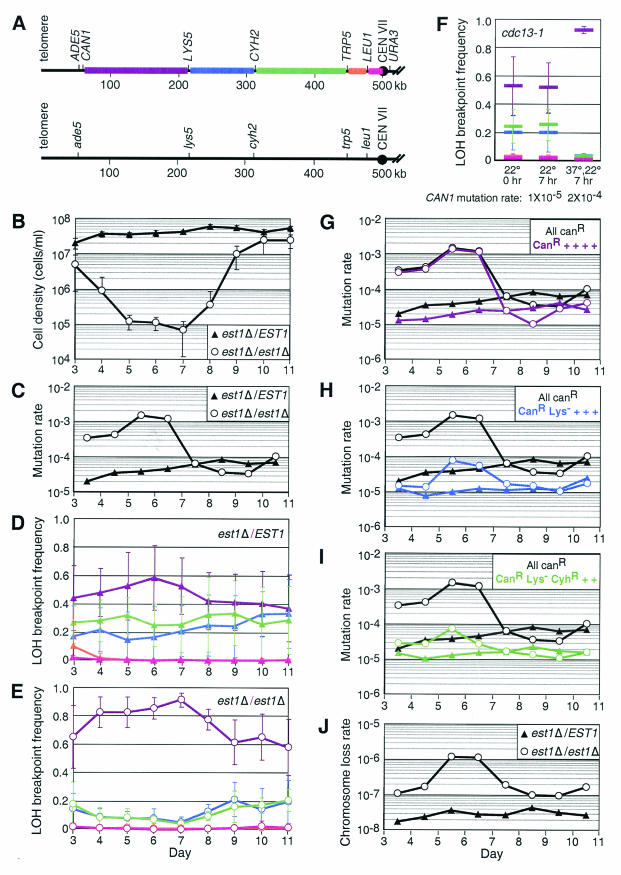
FIG.1.
Telomere dysfunction causes terminal deletions that are restricted to chromosome ends. (A) Diagram of chromosome VII homologues in diploid strains designed to test mutation rate and LOH. CAN1 is located 63 kb from the telomere on one homologue of chromosome VII that contains wild-type alleles of ADE5, LYS5, CYH2, TRP5, and LEU1. These genes are mutant on the other homologue of chromosome VII. Intervals between CAN1, LYS5, CYH2, TRP5, LEU1, and the centromere are colored so that chromosome breakpoints in these regions can be referenced in panels D to F. (B) Average cell density of 10 est1Δ/EST1 and est1Δ/est1Δ diploid cultures that had lost a covering EST1 plasmid and were grown in continuous log phase for the number of days indicated on the x axis. Error bars indicate the standard deviation of the concentration of each of the 10 independent cultures. (C) Mutation rate (mutations/cell division) calculated by using the maximal-likelihood method (27, 38) from white (ade5) Ura+ CanR colonies that had lost both the CAN1 and the ADE5 genes from the cultures in panel B. (D and E) Frequency of LOH breakpoints between marker genes in white Ura+ CanR colonies from est1Δ/EST1 and est1Δ/est1Δ cultures shown in panel B. Error bars indicate the standard deviation. Colors correspond to recombination in the interval separating specific marker genes in panel A as follows: CAN1-LYS5, purple; LYS5-CYH2, blue; CYH2-TRP5, green; TRP5-LEU1, orange; LEU1-CEN VII, red. (F) The frequency of recombination in the various intervals was measured in cdc13-1 strains. Ten cdc13-1/cdc13-1 cultures grown at the permissive temperature (22°C) were split (0 h) and grown at 22°C for 7 h or at the nonpermissive temperature (37°C) for 3.5 h and then shifted to 22°C for 3.5 h to allow recovery of the culture prior to plating (7, 20). The colors of each bar correspond to recombination in the interval separating specific marker genes as indicated in panel A. (G to I) Mutation rates were calculated separately within each intergenic interval designated in panel A for est1Δ/EST1 and est1Δ/est1Δ cultures. The colors correspond to the relevant LOH breakpoint designated in panel A. The mutation rates from panel C are plotted in black for comparison: triangles, est1Δ/EST1; and circles, est1Δ/est1Δ. (G) Mutation rate in white CanR colonies that had not lost any other marker genes (purple). (H) Mutation rate in white CanR colonies that were Lys− (blue). (I) Mutation rate in white CanR colonies that were Lys− and CyhR (green). (J) Chromosome loss rates calculated from white CanR Lys− CyhR Trp− Leu− colonies that had also lost the URA3 gene and were 5-FOAR.
As telomeres shorten in strains deficient for the telomerase component Est1p, the growth rate declines and eventually recovers due to recombination-mediated telomere elongation (Fig. 1B) (40, 44). Mutant colonies were selected under conditions in which Est1 transcription was reactivated to allow colony growth from cells with declining growth rates (27). White Ura+ CanR colonies that had lost CAN1 and ADE5, but retained URA3, were selected from multiple independent cultures and used to calculate the mutation rate (27). The mutation rate was relatively constant in est1Δ/EST1 heterozygotes, but in the est1Δ/est1Δ strain it increased about 50-fold over the background rate to 10−3 (Fig. 1C).
Telomere dysfunction increases LOH predominantly near chromosome ends.
To gain insight into the mechanism for the increase in mutation rate in est1Δ/est1Δ cultures, the extent of LOH on chromosomes that had lost CAN1 and ADE5 was measured by assessing the presence of LYS5, CYH2, TRP5, and LEU1 through growth on selective media. The frequency of LOH breakpoints between the different loci in est1Δ/EST1Δ colonies was fairly constant throughout the growth of the cultures (Fig. 1D) and was consistent with the pattern of LOH breakpoints observed for random chromosome breakage, where LOH breakpoint frequencies are proportional to the distances between marker genes (7). However, when telomeres were short in est1Δ/est1Δ cultures, the majority of the white CanR colonies lost only the most telomeric marker genes (Fig. 1E). This pattern of LOH is similar to that observed in cdc13-1/cdc13-1 strains in which the loss of end protection and subsequent exonucleolytic end resection occurs at the nonpermissive temperature (7, 20) (Fig. 1F).
To determine whether the increase in mutation rate in est1Δ/est1Δ cultures was restricted to chromosome ends or whether it also extended to more internal loci, we recalculated the mutation rate for white CanR colonies that had lost specific internal marker genes. The mutation rate for loss of CAN1 without loss of internal marker genes overlapped with the previously calculated total mutation rate in est1Δ/est1Δ cultures (Fig. 1G), although moderate increases in mutation rate could be observed for CanR colonies that had also lost internal markers (Fig. 1H and I). Telomere shortening also increases the rate of chromosome loss (Fig. 1J) (32, 45), but this rate is 1,000-fold lower than the rate of terminal deletion. Therefore, the increase in mutation rate when telomeres are short can be explained in large part by a loss of sequence near chromosome ends. This observation is consistent with sequence loss being caused by end resection rather than chromosome fusion and breakage.
Although telomerase deficiency has been associated with complex chromosome rearrangements in haploid cells (27, 53), such complex rearrangements are not common at loci that are predominantly repaired by homologous recombination (27) or in diploids (31, 47, 48). To confirm the structures of chromosomes with terminal deletions in diploid cells, whole chromosomes were separated by pulsed-field gel electrophoresis. Hybridization of a URA3 probe confirmed that the size of the URA3-marked chromosome VII homolog from CanR colonies was the same as that of the wild-type homologue in 20 of 20 est1Δ/EST1Δ and 20 of 20 est1Δ/est1Δ colonies (data not shown). This suggested that the damaged chromosome had been repaired by recombination-mediated copying of sequence from its homolog (BIR), as expected in diploid cells (31, 47).
Elevated LOH in est1Δ/est1Δ cells cannot be explained by breakage of dicentric chromosomes.
We hypothesized that an end-to-end chromosome fusion and subsequent breakage would cause breaks throughout a chromosome arm. However, it was possible that breaks might be near the site of fusion or that those closer to the telomeres might be healed more efficiently than more internal breaks. A bias toward more efficient repair at certain loci would not be expected in the wild-type cells since LOH in these cells predominantly occurs through mitotic recombination (17). To address the possibility that the breakage of dicentric chromosomes might also be biased toward the recovery of terminal deletions, we created a strain in which the formation of a dicentric chromosome with chromosome VII could be induced (Fig. 2A). A loxP site and the 5′ half of URA3 were inserted near the telomere on the left arm of chromosome VII. A loxP site and the 3′ half of URA3 were inserted near the telomere on the left arm of chromosome V. Cre-mediated recombination between the loxP sites creates a dicentric chromosome and releases a 27-kb linear chromosome fragment that expresses URA3 (Fig. 2A). This system for inducing chromosome fusions was introduced into an EST1/EST1 version of the diploid mutation rate strain. Cells were transformed with an empty vector or a galactose-inducible CRE vector (10). No Ura+ colonies were formed in cultures containing the empty vector. The frequency of Ura+ colony formation in cultures containing the CRE vector increased from 2 × 10−8 in uninduced glucose cultures to 4 × 10−4 for induced galactose cultures. The expected 27-kb linear chromosome fragment was observed in Ura+ CRE-expressing colonies only (Fig. 2B), indicating that the CRE-induced recombination worked efficiently. The frequent loss of ADE5 in white and sectored Ura+ colonies suggested that segregation of fused chromosomes at mitosis was causing DNA breaks (Fig. 2C).
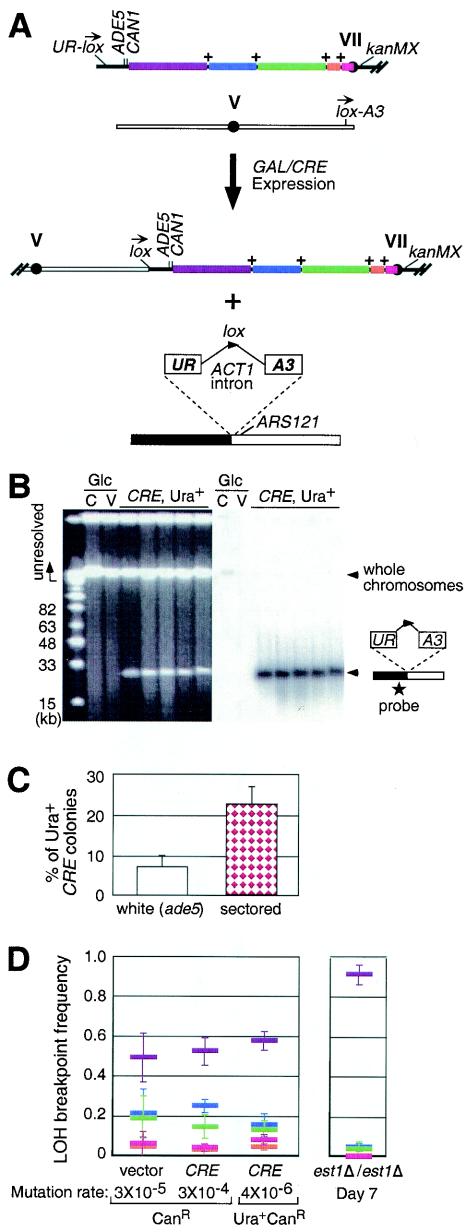
FIG. 2.
Effect of a dicentric chromosome on patterns of LOH breakpoints. (A) Dicentric chromosome formation in a diploid strain designed to test the mutation rate. loxP sites were inserted near the ends of chromosome VII-left and V-left. Chromosome VII is drawnwith the left side on the left, whereas chromosome V is drawn with the left side on the right in all portions of this panel. The 5′ or 3′ half of URA3 (“UR” and “A3”) were adjacent to the loxP sites and separated by ACT1 intron sequence (74). Arrows indicate the loxP orientation. Cre-mediated recombination at these sites creates a dicentric chromosome and releases a 27-kb chromosome fragment that expresses URA3. ARS121 was inserted telomeric to the 3′ half of URA3 to ensure replication of the released linear fragment. The colored regions on chromosome VII correspond to those in Fig. 1A, with “+” symbols indicating the positions of LYS5, CYH2, TRP5, and LEU1. (B) Ethidium bromide-stained (left) and hybridized (right) pulsed-field gel separating the fragment from unresolved whole chromosomes. The star indicates the probe location. Abbreviations: C, GAL/CRE vector; V, empty vector; Glc, cultures grown in glucose. (C) Percentage of Ura+ induced GAL/CRE colonies that were white (ade5) or red/white sectored. (D) LOH breakpoint frequency for ade5 CanR colonies or Ura+ ade5 CanR colonies from 10 cultures transformed with GAL/CRE or an empty vector and grown in galactose to induce CRE expression (left panel). For comparison, data for the day 7 est1Δ/est1Δ cultures (Fig. 1E) is provided in the right panel. Colors correspond to those in Fig. 1D to F. The mutation rates are indicated under the corresponding culture.
Chromosome fusions induced after CRE expression resulted in an increase in the CAN1 mutation rate from 3 × 10−5 to 3 × 10−4 (Fig. 2D). Although the majority of CanR colonies from cultures with CRE induction resulted from breakage of the dicentric chromosome, the pattern of LOH frequencies at the various loci was similar to those of colonies without CRE induction (Fig. 2D). The pattern of LOH breakpoint frequencies for Ura+ CanR colonies, which arose from cells with dicentric chromosomes, was also similar to those of colonies without CRE induction (Fig. 2D). Therefore, breakage of dicentric chromosomes does not alter the pattern of LOH breakpoint frequencies observed in CanR colonies compared to wild-type colonies. The similarity of these LOH breakpoint patterns (Fig. 2D) confirms that breakage of dicentric chromosomes cannot explain the very different pattern of LOH breakpoints seen in est1Δ/est1Δ diploids (Fig. 2D and 1E).
Deletion of EXO1, but not MRE11, reduces the mutation rate in cells with dysfunctional telomeres.
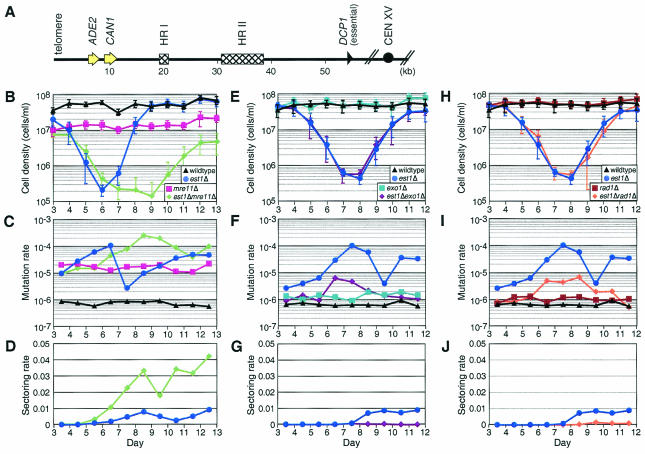
To further test the hypothesis that exonuclease resection at dysfunctional telomeres is the primary mechanism for the initiation of genomic instability, we examined the effect of the exonuclease Exo1p on mutation rate in a haploid est1Δ background. We also examined the role of Mre11p, since it has been implicated in telomere processing (13, 67). We expected that, if a nuclease were responsible for end resection, in the absence of that nuclease we would see a decrease in mutation rate. Mre11p is part of the MRX complex that may contribute to end resection at double-strand breaks (35, 39, 63). Mutation rates were measured in the haploid TELO strain background, where the left end of chromosome XV was marked with ADE2 and CAN1 (27) (Fig. 3A). Terminal deletions result in red CanR colonies. This strain background was chosen because terminal deletions at this locus are preferentially repaired by BIR (47, 52) beginning in regions of shared homology with other chromosomes (HR I and HR II) (27) (Fig. 3A). Knowledge of this repair mechanism allowed us to more easily distinguish between factors affecting initiation of damage versus repair of damage, since the factors that contribute to BIR are well defined (36). Mre11p functions in one BIR pathway but not in the Rad51-dependent BIR pathway that is predicted to be the predominant pathway due to the length of homology at this locus (27, 33). We confirmed that the terminal deletions in red CanR mre11Δ and est1Δ mre11Δ colonies were healed by BIR by pulsed-field gel analysis of the chromosomes (data not shown) (27). The growth rate of the mre11Δ strain was lower than that of the wild type, and both rates were constant over the course of the experiment (Fig. 3B). The est1Δ mre11Δ strains declined in growth rate and generated survivors somewhat more slowly than est1Δ strains, as do rad50Δ strains (37). The rate of terminal deletions as calculated from the frequency of red CanR (ade2 can1) colonies, was ∼10-fold higher for mre11Δ strains than for the wild type (9), and both mutation rates were constant over the course of the experiment (Fig. 3C). The mutation rate of the est1Δ strains increased when telomeres were short, as seen previously (27). The mutation rate of the est1Δ mre11Δ cultures increased to a level slightly higher than that of est1Δ cultures. Since deletion of Mre11p did not decrease the mutation rate, the MRX complex is probably not involved in telomere processing to initiate telomere dysfunction.
FIG. 3.
Mutation rate in haploid double mutant strains. (A) Diagram of chromosome XV in the TELO strain background (27). ADE2 and CAN1 are inserted near the left telomere. DCP1 is the most telomeric essential gene. HR I and HR II are the sites that are substrates for repair by BIR. (B) The average cell density of 10 independent cultures of each strain grown in log phase is plotted: wild type (black triangles), est1Δ (blue circles), mre11Δ (magenta squares), and est1Δ mre11Δ (green diamonds). Error bars indicate the standard deviation. The growth rate for the est1Δ strain decreases somewhat more rapidly than has previously been observed. This may be the effect of a slight telomere phenotype in the parent diploid strain that was heterozygous for EST1 and MRE11. (C) Mutation rates for the strains shown in panel B, calculated from red (ade2) CanR colonies. (D) Rate of red/white sectoring of colonies on nonselective control plates for strains shown in panel B. No wild-type colonies (n = 26,366) and 6 of 36,968 mre11Δ colonies were sectored (data not shown). (E) Average cell density of wild-type (black triangles), est1Δ (blue circles), exo1Δ (turquoise squares), and est1Δ exo1Δ (purple diamonds) TELO strains. Error bars indicate the standard deviation. (F) Mutation rates for strains in panel E, calculated from red CanR colonies. (G) Rate of red/white sectoring of colonies on nonselective control plates for strains in panel E. No wild-type (n = 24388) or exo1Δ colonies (n = 25603) were sectored (data not shown). (H) Average cell density of wild-type (black triangles), est1Δ (blue circles), rad1Δ (brown squares), and est1Δ rad1Δ (orange diamonds) TELO strains. Error bars indicate the standard deviation. Note that data for wild-type and est1Δ cultures are identical to those in panels E to G since these experiments were performed concurrently. (I) Mutation rates for strains in panel H, calculated from red CanR colonies. (J) Rate of red/white sectoring of colonies on nonselective control plates for strains in panel H. No wild-type (n = 24,388) or rad1Δ (n = 21,190) colonies were sectored (data not shown). Sectoring rates were calculated in panels D, G, and J by replacing the number of mutant colonies with the number of sectored colonies in mutation rate calculations (27).
The mismatch repair exonuclease Exo1p is also active at double-strand breaks and plays a role in end resection at telomeres in cdc13-1 strains (42, 49, 65, 66). However, the role of Exo1p in telomere processing in the absence of telomerase is unclear. In an assay for the ability of a telomere to fuse to a double-strand break, deletion of EXO1 reduced the ability of tlc1Δ telomeres to fuse by only ∼2-fold, suggesting that EXO1 may not be required for telomere dysfunction to occur in the absence of telomerase (8). Deletion of EXO1 in the TELO strain background did not alter the growth of wild-type or est1Δ cultures (Fig. 3E). The mutation rate for exo1Δ cultures was similar to that of the wild type (Fig. 3F). The mutation rate for exo1Δ est1Δ cultures was reduced ∼10-fold compared to est1Δ cultures. This suggests that Exo1p contributes to the end resection that generates terminal deletions in cells with dysfunctional telomeres and strengthens the argument that end resection is the primary cause of terminal deletions in est1Δ cells. Pulsed-field gel analysis of chromosomes from exo1Δ and est1Δ exo1Δ colonies indicated that the terminal deletions were repaired by BIR (27; data not shown).
In the TELO strain background, colonies in which ADE2 is unstable and is being lost due to terminal deletion showed red/white sectoring on control, non-canavanine-containing plates (27). Sectoring provides a measure of ongoing chromosomal instability in the culture. The rate of colony sectoring was higher in est1Δ mre11Δ cultures than in est1Δ cultures (Fig. 3D), providing further evidence that Mre11p does not significantly participate in end resection at dysfunctional telomeres. In contrast, the rate of sectoring in est1Δ exo1Δ control colonies was very low, further supporting a role for Exo1p-mediated end resection of short dysfunctional telomeres (Fig. 3G).
Deletion of RAD1 reduces the mutation rate in cells with dysfunctional telomeres.
Because Exo1p is a 5′-to-3′ exonuclease, it creates ssDNA on the 3′ strand. This is the strand that primes DNA replication during the BIR reaction that repairs the terminal deletions at the end of chromosome XV (Fig. 3A) (27). However, any 3′ nonhomologous sequence must be removed from the single strand before it can prime BIR. A likely candidate for an endonuclease that can remove the nonhomologous sequence is Rad1p, a 3′ flap endonuclease that functions in nucleotide excision repair and single-strand annealing (34, 61). If Rad1p plays a role in chromosome rearrangements, deletion of RAD1 should reduce the est1Δ mutation rate by decreasing BIR repair of ends with terminal deletions. Deletion of RAD1 in the TELO strain background did not alter the growth of wild-type or est1Δ cultures (Fig. 3H). The mutation rate for rad1Δ cultures was similar to that of the wild type (Fig. 3I). As in exo1Δ est1Δ cultures, the mutation rate for rad1Δ est1Δ cultures was reduced ∼10-fold compared to est1Δ cultures (Fig. 3I). Pulsed-field gel analysis of chromosome XV end structure showed that the ends were repaired by BIR in rad1Δ and rad1Δ est1Δ cultures (data not shown), indicating that deletion of RAD1 reduces, but does not prevent, repair by BIR. There is also a reduced rate of colony sectoring in rad1Δ est1Δ cultures (Fig. 3J). The role of Rad1p in generating the chromosome rearrangements in est1Δ cultures is consistent with its role as a flap endonuclease in single-strand annealing (34). These data further support a model in which terminal deletions are initiated by a 5′-to-3′ exonuclease activity in est1Δ cells.
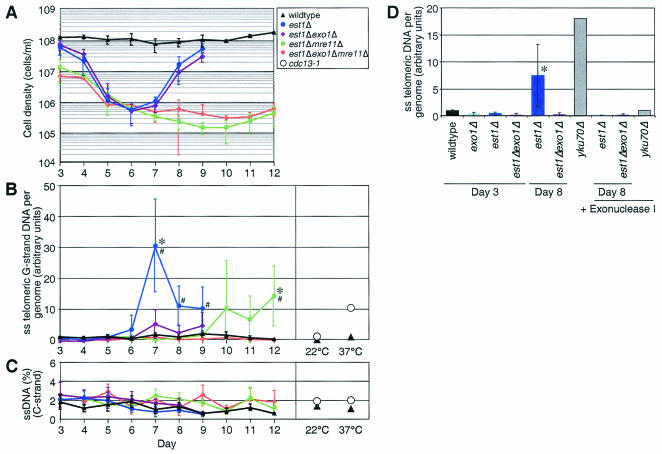
Accumulation of ssDNA in est1Δ cells is dependent on Exo1p.
If Exo1p functions at dysfunctional telomeres, it should increase the amount of ssDNA on the G-rich strand of the telomere in est1Δ cells. Such increased telomeric ssDNA is observed in cdc13-1 cells (20, 49). We measured the fraction of ssDNA in haploid cultures derived from the sporulation of a diploid heterozygous for EST1, EXO1, and MRE11 by using dot blot analysis. There was a significant increase in 3′ G-strand ssDNA at telomeres in est1Δ strains late in the growth curve (Fig. 4A), when telomeres were short, compared to wild-type cells (Fig. 4B, days 7 to 9). The amount of ssDNA in the est1Δ strains was significantly higher than in est1Δ exo1Δ strains (Fig. 4B, day 7). In addition, the amount of ssDNA in est1Δ exo1Δ mre11Δ strains was less than or equal to the amount in wild-type strains throughout the course of the experiment (Fig. 4B). To determine the strand specificity, we measured the ssDNA on the telomeric 5′ C strand and found no increase (Fig. 4C).
FIG. 4.
Accumulation of ssDNA in est1Δ strains is Exo1p dependent. (A) Genomic DNA was prepared from TELO cultures derived from independent spores of a diploid strain heterozygous for EST1, EXO1, and MRE11. Three cultures were grown for each EST1 strain, and five cultures were grown for each est1Δ strain. Average cell density of cultures grown in continuous log phase are shown. Error bars indicate the standard deviation. Wild-type (black triangles), est1Δ (blue circles), est1Δ exo1Δ (purple diamonds), est1Δ mre11Δ (green squares), and est1Δ exo1Δ mre11Δ (orange diamonds) strains are plotted as indicated. The slight variation in the growth of these cultures compared to those in Fig. 3 reflects the fact that these cultures were grown in 2% glucose rather than 2% raffinose-0.1% glucose. Glucose allows the cells to grow faster, compressing the growth curves. It also more completely represses expression from the galactose-inducible EST1 vector, preventing the same recovery of est1Δ mre11Δ cultures that is seen in Fig. 3B. (B) Telomeric ssDNA per genome measured on the 3′ G strand by dot blot analysis for strains in panel A (left side). Symbols correspond to those in panel A. DNA was not prepared from mre11Δ cultures on day 3 due to insufficient cell numbers. As a control, the amount of ssDNA was also measured in a cdc13-1 strain (○) grown at the permissive (22°C) or nonpermissive temperature (37°C) for 4 h (20) (right side). The asterisks indicate that the values for the est1Δ and est1Δ mre11Δ cultures are significantly different from those for the est1Δ exo1Δ and est1Δ mre11Δ exo1Δ cultures, respectively (P < 0.05). The “#” symbol indicates that the value is significantly different from that of the wild type (P < 0.05). Statistical comparisons were performed by using a two-sample Student t test. Averages for exo1Δ (0.23, day 3), mre11Δ (−2.07, day 4), and exo1Δ mre11Δ (−2.42, day 4) cultures (data not shown) were slightly less than for the wild type (0.94, day 3; 0.66, day 4; or 1.04, average of all days). These differences were statistically significant (P < 0.05), but only three cultures were tested. (C) Dot blots were probed for the presence of single-stranded C-strand DNA. The percentages of single-stranded telomeric 5′ C strand for strains in panel A and cdc13-1 controls are shown. Error bars indicate the standard deviation. Averages for exo1Δ, mre11Δ, and exo1Δ mre11Δ cultures were similar to the other genotypes (data not shown). Symbols correspond to those in panels A and B. (D) Telomeric ssDNA per genome. Agarose-embedded DNA was prepared from three to four spores per genotype derived from a TELO diploid heterozygous for EST1 and EXO1. Whole chromosomes were resolved by pulsed-field gel electrophoresis, and a native gel was hybridized with a poly-CA probe or a denatured gel was hybridized with a probe to an essential gene on chromosome V, PCM1. The telomeric signal at chromosome V was divided by the PCM1 signal. The asterisk indicates that the average for est1Δ cultures is significantly different than that of est1Δexo1Δ cultures. Day 8 represents the low point in the growth of est1Δ cultures, as in Fig. 3E. Error bars indicate the standard deviation. There was no PCM1 hybridization to a native gel (data not shown).
To further examine the Exo1p-dependent generation of ssDNA at telomeres in est1Δ cells, whole chromosomes prepared in agarose plugs were run on pulsed-field gels. The ssDNA in the est1Δ cultures was significantly greater than that of est1Δ exo1Δ cultures (P < 0.05) (Fig. 4D). As an additional control, we treated the chromosomal DNA with bacterial 3′-to-5′ exonuclease I and found that the ssDNA signal was significantly decreased (P < 0.05) (Fig. 4D), indicating the presence of 3′ ssDNA in the est1Δ cells. We therefore conclude that Exo1p plays a role in generating ssDNA at the telomere in est1Δ cells, although other exonucleases may also contribute to this process.
DISCUSSION
Telomere shortening primarily induces terminal deletions.
Loss of telomere function can initiate chromosome instability. Here we show that end resection, rather than end-to-end fusion, is the predominant mechanism for the initiation of genomic instability at dysfunctional telomeres. This is demonstrated by the restriction of terminal deletion breakpoints to chromosome ends in est1Δ/est1Δ diploid cells and by the reduction in the est1Δ mutation rate by deletion of the gene for the exonuclease Exo1p. The extensive sequence loss (63 kb) necessary for deletion of CAN1 (Fig. 1A) indicates that many kilobases of sequence can be deleted before repair occurs, even in diploid cells where homology is shared throughout chromosome arms.
Telomere dysfunction is a powerful mutagen that can induce DNA damage at a very high rate. We estimate that the probability of deletion of CAN1 is about 0.001 in each cell cycle because the mutation rate in est1Δ/est1Δ cells reaches 10−3 mutations per cell division (Fig. 1C). Thus, 6.2% of diploid cells will suffer sequence loss of at least one of their 64 chromosome ends in each cell cycle when telomeres are short. This rate may be even higher at sites closer to the telomeres than our CAN1 marker. In addition to loss of genes and local damage, this end degradation has the potential to induce more global genomic changes.
Exo1p-dependent end resection is not required for cell cycle arrest in est1Δ cells.
Telomere shortening in the absence of telomerase leads to loss of end protection and to chromosome rearrangements. The evidence presented here indicates that the initial event in this telomere dysfunction is the generation of ssDNA. ssDNA is known to induce cell cycle arrest in S. cerevisiae (56, 70). Surprisingly, although the amount of ssDNA in est1Δ exo1Δ cells is decreased compared to est1Δ cells, there is no difference in the growth kinetics of these cells. In contrast, ssDNA does play a role in the cell cycle arrest in response to the loss of end protection in cdc13-1 cells (46). The amount of ssDNA in est1Δ cells may be significantly less; ssDNA is not detectable until cultures begin to recover. Thus, ssDNA accumulation does not appear to be essential for the cell cycle arrest that occurs in est1Δ cultures (15, 32). This observation is consistent with the hypothesis that the cell cycle checkpoint in telomerase-deficient cells is different from the Rad9p-dependent checkpoint that is activated in cdc13-1 cells or in response to DNA breaks (15, 32, 71, 72). The lack of ssDNA in est1Δ cells as cells arrest suggests that the checkpoint in telomerase-deficient cells may not respond to ssDNA. The cell cycle arrest might function to prevent sequence degradation at dysfunctional telomeres, although ssDNA may still play a role in the checkpoint at a single cell level that cannot be observed in the bulk population.
The accumulation of ssDNA in est1Δ cells is somewhat delayed in the time course compared to the point where the mutation rate begins to increase. Because a very small fraction of cells in the population may have dysfunctional telomeres, it is likely that ssDNA cannot be measured in a bulk population at early time points. ssDNA is detected only as the culture begins to recover, at which point many cells in the population have dysfunctional telomeres. In fact the largest fraction of colonies are sectored just at the point where survivors are generated (27) (Fig. 3D, G, and J). The cells that have terminal deletions might even be those that have bypassed the checkpoint that induces the cell cycle arrest in telomerase-deficient cultures (15, 32).
Global chromosomal instability is a secondary consequence of end resection at dysfunctional telomeres.
Evidence for global chromosomal instability in the absence of telomerase comes from the elevated rate of chromosome loss (32, 45) (Fig. 1J) and the increased sectoring frequency when no selection for loss of any specific end is applied (27) (Fig. 3D and G). This sectoring may represent secondary processes such as breakage-fusion-bridge cycles that are initiated by telomere dysfunction (27). A moderate increase in LOH initiated at internal sites also suggests ongoing global chromosome instability. In diploid cells, the rate of loss of both CAN1 and LYS5 increased ∼8-fold when telomeres were short (Fig. 1H). There was also a slight increase in the rate of loss of CAN1, LYS5, and CYH2 (Fig. 1I). Therefore, telomere dysfunction can cause loss of internal sequences at a low rate. Interestingly, in contrast to terminal loci, there is little difference in mutation rate between est1Δ/est1Δ and est1Δ/EST1 strains at internal loci at early time points, suggesting that the increase at later time points may result from a secondary process.
End resection can directly initiate global chromosome instability. In a diploid cell or at a haploid locus where repair of a resected end can initiate repair at a region of sequence homology, end resection would be predicted to primarily result in chromosome rearrangements that are restricted to chromosome ends, as we observe in diploid est1Δ/est1Δ yeast (Fig. 1E). However, at a haploid locus without homologous sequence or at a diploid locus that contains highly repetitive sequence, repair of the resected end might result in the formation of a dicentric chromosome that can induce subsequent global genome instability through breakage fusion-bridge cycles. In fact, when loss of CAN1 from its native locus on chromosome V is monitored in haploid cells, short telomeres in telomerase-deficient cells frequently cause chromosome circularization (4 of 14 CanR colonies) (27). Although DNA breaks in diploid cells most commonly are repaired by copying sequence from the homologous chromosome (31, 47, 48), breaks that occur in regions containing the repetitive Ty1 retrotransposons can cause rearrangements involving two chromosomes that are not homologs (68). Recombination between such repetitive elements can create dicentric chromosomes. This situation may be even more relevant in mammalian cells that contain much more repetitive sequence than yeast.
Additional evidence from both yeast and mammals supports the idea that global chromosome instability might be a secondary effect of end resection at short telomeres. Chromosome fusions that result from telomere dysfunction have extensive sequence loss at the junctions in yeast and mouse (27, 30), indicating that end resection is required before chromosome fusion occurs. Therefore, although end-to-end chromosome fusion may occur in both yeast and mammals, it may not be the initial cause of genome instability. Nonreciprocal translocations in tumors could also be secondary effects of the initial genome instability created by end resection at dysfunctional telomeres. In human tumors with telomere dysfunction, deletions in terminal regions of chromosomes precede more global instability (22). This bias toward terminal deletions in early tumorigenesis could be explained if end resection is the initial cause of instability (18). It will be interesting to determine whether mitotic recombination near chromosome ends also increases in mammalian cells with dysfunctional telomeres.
Acknowledgments
We thank Jef Boeke, Forrest Spencer, David Feldser, Chris Frank, and Tom Cunningham for critical reading of the manuscript and Joe Lawler for useful discussions. We thank Jef Boeke, Abram Gabriel, Marc Gartenberg, and Ted Weinert for providing yeast strains and vectors. We thank David Feldser for deletion of MRE11.
This work was supported by the Predoctoral Training Program in Human Genetics and Molecular Biology and the National Science Foundation (J.A.H.) and by NIH grant GM43080 (C.W.G).
REFERENCES
- 1.Artandi, S. E., S. Chang, S. L. Lee, S. Alson, G. J. Gottlieb, L. Chin, and R. A. DePinho. 2000. Telomere dysfunction promotes nonreciprocal translocations and epithelial cancers in mice. Nature 406:641-645. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, P., and T. R. Cech. 2000. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell 11:3265-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco, M. A., H.-W. Lee, P. M. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:25-34. [DOI] [PubMed] [Google Scholar]
- 5.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 6.Cahill, D. P., C. Lengauer, J. Yu, G. J. Riggins, J. K. Willson, S. D. Markowitz, K. W. Kinzler, and B. Vogelstein. 1998. Mutations of mitotic checkpoint genes in human cancers. Nature 392:300-303. [DOI] [PubMed] [Google Scholar]
- 7.Carson, M., and L. Hartwell. 1985. CDC17: an essential gene that prevents telomere elongation in yeast. Cell 42:249-257. [DOI] [PubMed] [Google Scholar]
- 8.Chan, S. W.-L., and E. H. Blackburn. 2003. Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol. Cell 11:1379-1387. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C., and R. D. Kolodner. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23:81-85. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, T. H., C. R. Chang, P. Joy, S. Yablok, and M. R. Gartenberg. 2000. Controlling gene expression in yeast by inducible site-specific recombination. Nucleic Acids Res. 28:E108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craven, R. J., P. W. Greenwell, M. Dominska, and T. D. Petes. 2002. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics 161:493-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePinho, R. A. 2000. The age of cancer. Nature 408:248-254. [DOI] [PubMed] [Google Scholar]
- 13.Diede, S. J., and D. E. Gottschling. 2001. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 11:1336-1340. [DOI] [PubMed] [Google Scholar]
- 14.Dionne, I., and R. J. Wellinger. 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA 93:13902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enomoto, S., L. Glowczewski, and J. Berman. 2002. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2626-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espejel, S., S. Franco, S. Rodriguez-Perales, S. D. Bouffler, J. C. Cigudosa, and M. A. Blasco. 2002. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 21:2207-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esposito, M. S., and J. E. Wagstaff. 1981. Mechanisms of mitotic recombination, p. 341-370. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Feldser, D. M., J. A. Hackett, and C. W. Greider. 2003. Telomere dysfunction and the initiation of genome instability. Nat. Rev. Cancer. 3:623-627. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira, M. G., and J. P. Cooper. 2001. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell 7:55-63. [DOI] [PubMed] [Google Scholar]
- 20.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerring, S. L., C. Connelly, and P. Hieter. 1991. Positional mapping of genes by chromosome blotting and chromosome fragmentation. Methods Enzymol. 194:57-77. [DOI] [PubMed] [Google Scholar]
- 22.Gisselsson, D., T. Jonson, A. Petersen, B. Strombeck, P. Dal Cin, M. Hoglund, F. Mitelman, F. Mertens, and N. Mandahl. 2001. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc. Natl. Acad. Sci. USA 98:12683-12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 24.Greenberg, R., L. Chin, A. Femino, K.-H. Lee, G. Gottlieb, R. Singer, C. W. Greider, and R. A. DePinho. 1999. Short dysfunctional telomeres impair tumorigenesis in the INK4aΔ2/3−/− cancer prone mouse. Cell 97:515-525. [DOI] [PubMed] [Google Scholar]
- 25.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 26.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hackett, J. A., D. M. Feldser, and C. W. Greider. 2001. Telomere dysfunction increases mutation rate and genomic instability. Cell 106:275-286. [DOI] [PubMed] [Google Scholar]
- 28.Hackett, J. A., and C. W. Greider. 2002. Balancing instability: dual roles for telomerase and telomere dysfunction in tumorigenesis. Oncogene 21:619-626. [DOI] [PubMed] [Google Scholar]
- 29.Harley, C. B. 1990. Aging in cultured human fibroblasts. Methods Mol. Biol. 5:25-32. [DOI] [PubMed] [Google Scholar]
- 30.Hemann, M. T., M. A. Strong, L. Y. Hao, and C. W. Greider. 2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107:67-77. [DOI] [PubMed] [Google Scholar]
- 31.Hiraoka, M., K. Watanabe, K. Umezu, and H. Maki. 2000. Spontaneous loss of heterozygosity in diploid Saccharomyces cerevisiae cells. Genetics 156:1531-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.IJpma, A., and C. W. Greider. 2003. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14:987-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ira, G., and J. E. Haber. 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 18:6384-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanov, E. L., and J. E. Haber. 1995. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:2245-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanov, E. L., N. Sugawara, C. I. White, F. Fabre, and J. E. Haber. 1994. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3414-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraus, E., W. Y. Leung, and J. E. Haber. 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD51 and RAD50 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lea, D. E., and C. A. Coulson. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 39.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, Mre11/Rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 40.Lendvay, T. S., D. K. Morris, J. Sah, B. Balasubramanian, and V. Lundblad. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144:1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 42.Lewis, L. K., G. Karthikeyan, J. W. Westmoreland, and M. A. Resnick. 2002. Differential suppression of DNA repair deficiencies of yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase). Genetics 160:49-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liti, G., and E. J. Louis. 2003. NEJ1 prevents NHEJ-dependent telomere fusions in yeast without telomerase. Mol. Cell 11:1373-1378. [DOI] [PubMed] [Google Scholar]
- 44.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 45.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 46.Lydall, D., and T. Weinert. 1995. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270:1488-1491. [DOI] [PubMed] [Google Scholar]
- 47.Malkova, A., E. L. Ivanov, and J. E. Haber. 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93:7131-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mangahas, J. L., M. K. Alexander, L. L. Sandell, and V. A. Zakian. 2001. Repair of chromosome ends after telomere loss in Saccharomyces. Mol. Biol. Cell 12:4078-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maringele, L., and D. Lydall. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 16:1919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maser, R. S., and R. A. DePinho. 2002. Connecting chromosomes, crisis, and cancer. Science 297:565-569. [DOI] [PubMed] [Google Scholar]
- 51.McClintock, B. 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 26:234-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morrow, D. M., C. Connelly, and P. Hieter. 1997. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Myung, K., C. Chen, and R. D. Kolodner. 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411:1073-1076. [DOI] [PubMed] [Google Scholar]
- 54.Nakamura, T. M., J. P. Cooper, and T. R. Cech. 1998. Two modes of survival of fission yeast without telomerase. Science 282:493-496. [DOI] [PubMed] [Google Scholar]
- 55.Parenteau, J., and R. J. Wellinger. 1999. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19:4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pellicioli, A., S. E. Lee, C. Lucca, M. Foiani, and J. E. Haber. 2001. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell 7:293-300. [DOI] [PubMed] [Google Scholar]
- 57.Riha, K., and D. E. Shippen. 2003. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc. Natl. Acad. Sci. USA 100:611-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rose, M. D., W. F., and P. Hieter. 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press.
- 59.Rudolph, K. L., S. Chang, H.-W. Lee, M. Blasco, G. Gottlieb, C. W. Greider, and R. A. DePinho. 1999. Longevity, stress response, and cancer in aging telomerase deficient mice. Cell 96:701-712. [DOI] [PubMed] [Google Scholar]
- 60.Rudolph, K. L., M. Millard, M. W. Bosenberg, and R. A. DePinho. 2001. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat. Genet. 28:155-159. [DOI] [PubMed] [Google Scholar]
- 61.Siede, W., A. S. Friedberg, and E. C. and Friedberg. 1993. Evidence that the Rad1 and Rad10 proteins of Saccharomyces cerevisiae participate as a complex in nucleotide excision repair of UV radiation damage. J. Bacteriol. 175:6345-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smogorzewska, A., J. Karlseder, H. Holtgreve-Grez, A. Jauch, and T. de Lange. 2002. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 12:1635-1644. [DOI] [PubMed] [Google Scholar]
- 63.Sugawara, N., and J. E. Haber. 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12:563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thiagalingam, S., S. Laken, J. K. Willson, S. D. Markowitz, K. W. Kinzler, B. Vogelstein, and C. Lengauer. 2001. Mechanisms underlying losses of heterozygosity in human colorectal cancers. Proc. Natl. Acad. Sci. USA 98:2698-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tishkoff, D. X., A. L. Boerger, P. Bertrand, N. Filosi, G. M. Gaida, M. F. Kane, and R. D. Kolodner. 1997. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. USA 94:7487-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsubouchi, H., and H. Ogawa. 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell 11:2221-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsukamoto, Y., A. K. Taggart, and V. A. Zakian. 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11:1328-1335. [DOI] [PubMed] [Google Scholar]
- 68.Umezu, K., M. Hiraoka, M. Mori, and H. Maki. 2002. Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics 160:97-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 70.Vaze, M. B., A. Pellicioli, S. E. Lee, G. Ira, G. Liberi, A. Arbel-Eden, M. Foiani, and J. E. Haber. 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10:373-385. [DOI] [PubMed] [Google Scholar]
- 71.Weinert, T. A., and L. H. Hartwell. 1993. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics 134:63-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weinert, T. A., and L. H. Hartwell. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241:317-322. [DOI] [PubMed] [Google Scholar]
- 73.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 74.Yu, X., and A. Gabriel. 1999. Patching broken chromosomes with extranuclear cellular DNA. Mol. Cell 4:873-881. [DOI] [PubMed] [Google Scholar]
- 75.Zakian, V. A. 1989. Structure and function of telomeres. Annu. Rev. Genet. 23:579-604. [DOI] [PubMed] [Google Scholar]