Abstract
Saccharomyces cells with a single unrepaired double-strand break adapt after checkpoint-mediated G2/M arrest. We have found that both Rad51 and Rad52 recombination proteins play key roles in adaptation. Cells lacking Rad51p fail to adapt, but deleting RAD52 suppresses rad51Δ. rad52Δ also suppresses adaptation defects of srs2Δ mutants but not those of yku70Δ or tid1Δ mutants. Neither rad54Δ nor rad55Δ affects adaptation. A Rad51 mutant that fails to interact with Rad52p is adaptation defective; conversely, a C-terminal truncation mutant of Rad52p, impaired in interaction with Rad51p, is also adaptation defective. In contrast, rad51-K191A, a mutation that abolishes recombination and results in a protein that does not bind to single-stranded DNA (ssDNA), supports adaptation, as do Rad51 mutants impaired in interaction with Rad54p or Rad55p. An rfa1-t11 mutation in the ssDNA binding complex RPA partially restores adaptation in rad51Δ mutants and fully restores adaptation in yku70Δ and tid1Δ mutants. Surprisingly, although neither rfa1-t11 nor rad52Δ mutants are adaptation defective, the rad52Δ rfa1-t11 double mutant fails to adapt and exhibits the persistent hyperphosphorylation of the DNA damage checkpoint protein Rad53 after HO induction. We suggest that monitoring of the extent of DNA damage depends on independent binding of RPA and Rad52p to ssDNA, with Rad52p's activity modulated by Rad51p whereas RPA's action depends on Tid1p.
Saccharomyces cerevisiae cells with a single unrepaired double-strand break (DSB) arrest prior to mitosis, but after eight or more hours they adapt and resume cell cycle progression even though they still harbor a broken chromosome (24, 25, 40, 49). Arrest in these cells depends on a network of checkpoint proteins that sense DNA damage and trigger a cascade of protein kinases (reviewed in references 7, 18, 27, 36, and 55). These checkpoint proteins include Rad17p, Rad24p, Mec3p, and Ddc1p that appear to act as DNA damage sensors (19, 20, 28), as well as Rad9p, which may be both a sensor and a modulator of protein kinases (5, 12). These proteins activate the ATM/ATR homologue, Mec1p, which phosphorylates both Rad53p and Chk1p, two protein kinases that control different aspects of the DNA damage response (38, 39). Both the Ddc1p-Rad17p-Mec3p and Mec1p-Ddc2p complexes have been shown to bind directly to a HO endonuclease-induced DSB (20, 28, 37). Rad53p is required for the phosphorylation of both Dun1p, a protein kinase that controls several DNA damage-inducible genes (1, 4), and Cdc5p, a polo-like kinase that has been implicated in the regulation of several steps in mitosis (for review, see reference 32). Both Rad53p and Chk1p must be functional to ensure mitotic arrest of DNA-damaged cells (38). An observation relevant to the work described below is that RAD51 mRNA transcripts are up-regulated in response to DNA damage and that this induction is dependent both on the intact homologous recombination machinery (i.e., RAD52) and on the DNA damage signal transduction cascade enforced by Rad53 checkpoint kinase (3).
A recent study has examined the phosphorylation and protein kinase activity of Rad53p after induction of a single unrepaired DSB (34). Following the induction of the DSB, Rad53p kinase activity increases an hour or more after the DNA damage is inflicted, suggesting that cells respond to the damage only after the DNA has been degraded to form 3′-ended single-stranded DNA (ssDNA) tails. Kinase activity remains elevated for 8 to 12 h and decreases at the time cells adapt. In a similar fashion, Chk1p becomes hyperphosphorylated in parallel with Rad53p and these activated forms of Chk1p disappear at the time of adaptation (34). This suggests that adaptation requires the dephosphorylation and/or turnover of the activated kinases. This hypothesis is strengthened by the fact that overexpressing the phosphatase Ptc2 suppresses prolonged G2/M arrest of DNA-damaged cells and suppresses all adaptation-defective mutations tested (26).
Several adaptation-defective mutations, which cause cells to remain permanently arrested when cells have a single, unrepaired DSB, have been identified (24, 25, 49). This prolonged arrest depends on the continued activity of both the Mec1p and Rad53p checkpoint kinases (25, 34). These adaptation-defective mutations appear to affect the process in different ways. Deletion of the genes encoding yKu70p and yKu80p enhances the DNA damage signal by causing a twofold increase in the rate of 5′-to-3′ exonuclease resection, thus apparently increasing the extent of ssDNA and prolonging G2/M arrest (24). Consistent with this idea, cells also become permanently arrested when there are two DSBs, each resected at a normal rate (24). In both cases, a single amino acid substitution mutation, rfa1-t11 (Rfa1-L45E), in the largest subunit of the ssDNA binding complex, RPA, suppresses permanent arrest (24, 50). These results suggest that the cell monitors the extent of DNA damage by measuring the extent of ssDNA (10). This idea has been strongly supported by results of recent experiments by Zou and Elledge (58) showing that the recruitment of Mec1-Ddc2 to sites of DNA damage is defective in an rfa1-t11 mutant.
In contrast, the adaptation-defective cdc5-ad mutation does not alter 5′-to-3′ resection of DNA and is not suppressed by rfa1-t11 (34). Nevertheless, permanent arrest in cdc5-ad cells depends on the continued activity of Rad53p and Mec1p (34). Cdc5p appears therefore to be both a downstream target of Rad53p kinase (directly or indirectly) and its feedback regulator. Deletion of the casein kinase II proteins also causes permanent cell cycle arrest in the face of DNA damage, but these less severe mutations have not been analyzed in much detail (49).
Four other adaptation-defective mutations have recently been described. The recombination protein Tid1p, a homologue of Rad54p, interacts with the Rad51p strand exchange protein (6). In mitotic cells, a tid1 deletion has a relatively minor role in gene conversion repair of a DSB but does play an important role in a RAD51- and RAD54-independent pathway of DNA repair known as break-induced replication (42). Nearly 100% of G2/M-arrested tid1Δ cells permanently arrest, as do yku70Δ cells, in the presence of a single unrepaired DSB (25). Unlike yku mutations, tid1Δ does not affect the rates of resection of DSB ends. Like yku70Δ, tid1Δ is suppressed by rfa1-t11. The three other adaptation-defective mutations are distinct in that they also prevent cells from resuming cell cycle progression when DNA damage has been repaired. A deletion of the Srs2 helicase (53) prevents both adaptation and recovery. Recent evidence has suggested that Srs2p acts to displace Rad51p from ssDNA (23, 54), although in the checkpoint studies we carried out, it appears that Srs2p's checkpoint role is at a later step, when DNA has been repaired in a RAD51-independent process. Interestingly, however, rad51Δ suppressed the recovery defect. The remaining two adaptation- and recovery-defective mutations, ptc2Δ and ptc3Δ, together abolish PP2C phosphatase activity that is apparently required to turn off the checkpoint kinase cascade beginning with Mec1p (26).
Here we report that another protein involved in recombinational repair of DSBs is also adaptation defective. Rad51p is an evolutionarily conserved strand exchange protein that binds to ssDNA in vitro to form filaments that might compete with RPA in the assessment of how much ssDNA has been generated at DSB ends (11, 13, 14, 31, 41, 47). The adaptation defect of rad51Δ is partially suppressed by rfa1-t11, which encodes a mutant protein that has recently been shown to be inefficiently replaced by Rad51p in vitro (17). Quite surprisingly, a rad52Δ rfa1-t11 double mutant is also adaptation defective. Based on these results, we suggest that Rad52p, which is modulated by Rad51p, and RPA, which is modulated by Tid1p, independently monitor the extent of DNA damage and determine whether cells will adapt in the face of a single unrepaired DSB.
MATERIALS AND METHODS
Strains.
All strains are derivatives of JKM179 (hoΔ MATα hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG' ura3-52 ade3::GAL::HO). Construction of the yku70Δ::URA3 strain (JKM181), the rad52Δ::TRP1 strain (JKM168), the rad9Δ::KAN (YSL60) strain, and the rfa1-t11 strain (YSL40) was previously described (24). The tid1Δ::URA3 strain (YSL300) is a segregant from a cross between HK764 (a gift from H. Klein) and JKM179. The rad51Δ::URA3 strain (YSL302) was constructed by transformation using a HindIII fragment of pJH724. The rad54Δ::LEU2 (YSL303), rad55Δ::LEU2 (YSL304), and sgs1Δ::URA3 (YSL305) strains were obtained by transformation with a BglII fragment of pXRAD::LEU2 (a gift from L. Symington), a HindIII fragment of pSTL11 (a gift from S. Lovett), and a XhoI fragment of pJH1340, respectively. The rad59Δ::KAN strain (YSL306) was constructed by crossing the JKM179 derivative CSHYKO (a gift from C. Greider) with the isogenic MATa strain, JKM139. XW652 has been previously described (57). Additional tid1Δ derivatives of the JKM179 background were constructed by transformation with PCR-amplified KAN::MX cassettes from the Research Genetics collection of Saccharomyces strains harboring deletions of different yeast genes. These isogenic tid1Δ strains showed adaptation behavior nearly identical to that of the backcrossed strain.
Plasmids pR51.3, pR51.4, and pR51.5 (kindly provided by P. Sung) express wild-type RAD51, rad51-K191A, and rad51-K191R, respectively, under the PGK promoter (48). pRS413 plasmids carrying Rad51-Y388H, Rad51-L99P, and Rad51-T146A (21) were generously provided by L. Krejci. The promoter and open reading frame of each rad51 mutant was cloned into the URA3-containing centromeric plasmid pRS316.
Analysis of G2/M checkpoint adaptation.
Cells were grown in preinduction medium (yeast extract-peptone [YEP]-lactate) at 30°C overnight and were spread onto agar plates containing COM-galactose medium. G1 (unbudded) cells were micromanipulated onto a grid, and cells were then monitored for growth and division.
Measurement of DNA degradation.
A DSB was induced in YEP-lactate-grown cells by adding 2% galactose. Genomic DNA was isolated at intervals and subjected to slot blot hybridization by using a strand-specific RNA probe as previously described (24). Alternatively, the extent of resection and stability of 3′ ends was examined on Southern blots of DNA separated on denaturing gels, where higher-molecular-weight bands hybridizing to a short probe homologous to sequences near the HO cut site represent partial digestion products caused as restriction sites become single stranded (24, 56).
Fluorescence-activated cell sorter (FACS) analysis.
Flow cytometry analysis was performed with a Becton Dickinson fluorescence-activated cell analyzer, as described by Paulovich and Hartwell (33), by using either propidium iodide or sytox green (Molecular Probes Co.). Each sample was also visualized with a fluorescence microscope to analyze cell morphology and the location of the nuclei in the cells.
Western blotting and in situ autophosphorylation assay.
Crude-extract preparation, the Western blotting procedure, and the in situ autophosphorylation assay have been described by Pellicioli et al. (34).
Chromatin immunoprecipitation.
Cultures were induced with 2% galactose, harvested, and then analyzed by chromatin immunoprecipitation as described previously (8, 45). Polyclonal antibodies directed against Rad51p were kindly provided by P. Sung and D. Bishop. Immunoprecipitated DNA and input DNA were analyzed by PCR using the oligonucleotides 5′ CTTGCTCTTGTTCCCAATGTTTG 3′ and 5′ CCGCATGGGCAGTTTACCT 3′ located distally to MAT. PCR conditions were chosen so that a linear relationship existed between the amount of PCR product and the amount of template DNA, and this relationship was verified with a calibration curve prepared for each set of PCRs. PCR products were visualized on 2% agarose gels stained with ethidium bromide, and images provided by either a Bio-Rad Gel Doc 1000 or Alpha Innotech AlphaImager 1220 were analyzed using Quantity One software (Bio-Rad).
RESULTS
Cells lacking Rad51p fail to adapt.
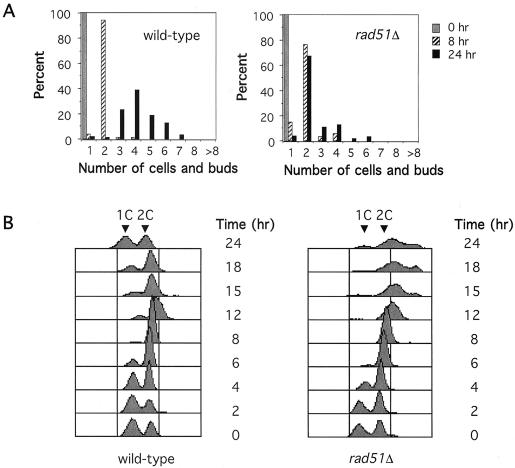
A MATα strain, JKM179, carrying a galactose-inducible HO gene (ade3::GAL::HO) and deletions of the recombination donor cassettes HML and HMR was grown in YEP-glycerol-succinate and placed onto a synthetic complete-galactose plate to induce HO-mediated cleavage of the MAT locus (24). Individual unbudded G1 cells were micromanipulated onto a grid so that their cell cycle progression could be monitored microscopically. By 8 h, nearly all cells had grown into the dumbbell shape characteristic of G2/M-arrested cells. By 24 h, more than 90% of wild-type cells had resumed growth and produced microcolonies with three or more cells and buds, as expected for cells able to adapt to the presence of a single DSB (Table 1 and Fig. 1A).
TABLE 1.
Effect of recombination-defective mutations on adaptation
| Straina | % Adaptation ± SD 24 h post-HO induction |
|---|---|
| Wild type | 93.5 ± 4.0 |
| rad51Δ | 27.6 ± 4.9 |
| rad52Δ | 84.0 ± 0.8 |
| rad54Δ | 74.8 ± 16.0 |
| rad55Δ at 23°C | 84.0 ± 8.5 |
| rad55Δ at 30°C | 89.0 ± 4.8 |
| rad59Δ | 88.4 ± 3.2 |
| rad51Δ rad59Δ | 34.0 ± 6.3 |
| tid1Δ | 1.4 ± 0.1 |
| srs2Δ | 24.8 ± 4.4 |
| sgs1Δ | 70.1 ± 13.8 |
| rad52Δ409-420 | 63.4 ± 10.5 |
All strains are derivatives of JKM179, containing MATα but lacking both HML and HMR. An irreparable DSB was induced from a GAL::HO gene integrated at the ade3 locus.
FIG. 1.
Arrest of cell cycle progression by a single unrepaired DSB in wild-type and mutant cells. (A) Arrest and adaptation of cells experiencing an irreparable DSB in G1. At least 300 G1 unbudded cells were initially plated (gray bar). The numbers of cells and buds at 8 h (hatched bar) and 24 h (black) are shown. After 24 h, wild-type cells initially plated as G1 unbudded cells onto galactose-containing medium to induce HO endonuclease have mostly adapted and resumed cell division, whereas adaptation-defective tid1Δ and rad51Δ strains remain arrested prior to anaphase. (B) Results of FACS analysis of wild-type and mutant cells induced for HO expression at 0 h. The apparent increase in DNA content of checkpoint-arrested cells above the 2C level is caused by light scattering of G2/M-arrested cells because of their greatly enlarged size and is not an indication of continued DNA replication (53).
We examined the effects of deleting a series of genes in the RAD52 epistasis group, important in homologous recombination. Neither rad52Δ, rad54Δ, rad55Δ, rad57Δ, nor rad59Δ mutants exhibited any significant defect in arrest or adaptation; however, a rad51Δ deletion mutant had a clear defect in adaptation. Approximately 75% of rad51Δ cells remained arrested at the first cell division (Fig. 1A). Another 20% of the cells completed the first division but became permanently arrested at the next G2/M boundary, even at 48 h. This differs from the more severely adaptation-defective phenotype of yku70Δ or tid1Δ mutant cells, nearly all of which arrest at or prior to the first cell division. As with yku70Δ and tid1Δ cells, permanently arrested, DNA-damaged rad51Δ cells exhibit a terminal morphology with an especially enlarged mother cell (25 and data not shown). As expected, FACS analysis of rad51Δ cells with a single, unrepaired DSB shows that they become arrested with the fully replicated (2C) DNA content expected for cells that had replicated their DNA but failed to progress through the cell cycle (Fig. 1B). We note that rad51Δ has no effect on the establishment of DNA damage-induced arrest but affects the cell's exit from arrest.
Permanent arrest of rad51Δ cells is bypassed by the rad9Δ checkpoint mutation and partially suppressed by rfa1-t11.
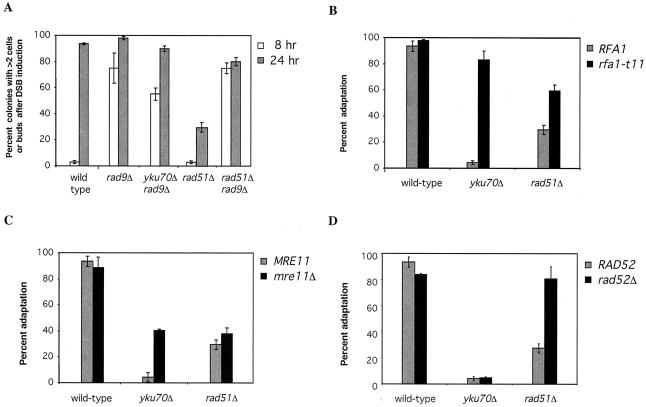
G2/M arrest of rad51Δ cells depends on a functional checkpoint system. Hence, a rad9Δ rad51Δ double mutant did not show significant cell cycle arrest after induction of HO cleavage (Fig. 2A). Again, this result is similar to what was previously observed with yku70Δ (24) and tid1Δ (25). Thus, the permanent arrest phenotype of this new adaptation-defective mutation is dependent on a functional DNA damage arrest checkpoint system.
FIG. 2.
Adaptation of wild-type and mutant cells. Cells progressing beyond the two-cell-plus-bud stage at 24 h after HO induction were scored as having adapted. (A) Effect of rad9Δ on the arrest and adaptation of wild-type and mutant cells at 8 and 24 h. Also shown are the effects of rfa1-t11 (B), mre11Δ (C), and rad52Δ (D) on the adaptation of wild-type and mutant cells at 24 h.
A key finding concerning both yku70Δ and tid1Δ was that the rfa1-t11 mutation restores adaptation in a strain with a single unrepaired DSB (24, 25). cdc5-ad is not suppressed by rfa1-t11 (34). Here we found that rad51Δ is only partially suppressed by rfa1-t11 (Fig. 2B). We also found that rfa1-t11 partially suppresses rad51Δ tid1Δ, but the double mutation is suppressed to a lesser degree than is either single mutation, suggesting that the defects caused by rad51Δ and tid1Δ mutations may not be in the same pathway.
Previously it was shown that the permanent arrest of yku70Δ cells at the G2/M checkpoint could be partially suppressed by the absence of functional Mre11p, although tid1Δ cells were not affected (24, 25). The absence of Mre11p did not significantly affect the failure of adaptation of rad51Δ cells (Fig. 2C). Thus, the arrest phenotype of rad51Δ cells is checkpoint dependent but different from those of yku70Δ, tid1Δ, and cdc5-ad cells.
rad51Δ does not alter the rate of formation of ssDNA by 5′-to-3′ exonucleases.
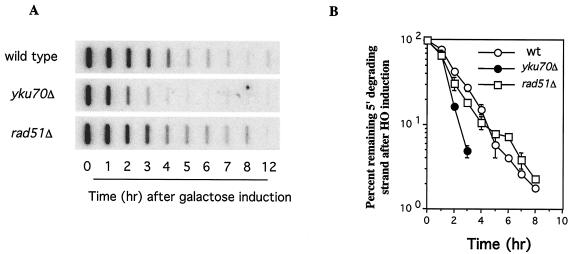
The failure of yku70Δ strains to adapt was explained by the observation that they exhibited a twofold higher rate of 5′-to-3′ resection of DNA ends, thus creating a larger amount of ssDNA (24). The defect in rad51Δ cells must affect adaptation in a different way, as it does not affect the rate of 5-to-3′ resection as measured by slot blot hybridization (Fig. 3). In addition, rad51Δ does not significantly alter the stability of the 3′-ended ssDNA (data not shown). These results are similar to those found for tid1Δ (25).
FIG. 3.
5′-to-3′ resection of an HO-induced DSB in adaptation-defective mutants. (A) Slot blot hybridization was performed with a probe specific for the strand ending 5′ distal to the HO-cleaved MAT locus. (B) Percentages of the sequences remaining at different times. wt, wild type.
Epistatic relationships among adaptation-defective mutants.
We also tested double adaptation-defective mutants to understand their epistatic relationships. tid1Δ rad51Δ cells arrest and fail to adapt, similar to tid1Δ cells, but unlike tid1Δ alone, the double mutation cannot be suppressed by rfa1-t11 (Table 2). The more severe arrest phenotype of yku70Δ cells is also epistatic to rad51Δ (that is, the double mutant resembles the yku70Δ strain rather than the rad51Δ strain). As expected, the double mutant yku70Δ tid1Δ is indistinguishable from either single mutant (data not shown).
TABLE 2.
Suppression of adaptation defects caused by rad51Δ and other mutations
| Strain description | % Adaptation ± SD at 24 h after galactose induction |
|---|---|
| Strains with epistatic relationships | |
| rad51Δ | 27.6 ± 4.9 |
| rad51Δ srs2Δ | 29.5 ± 3.5 |
| rad51Δ tid1Δ | 0.3 ± 0.1 |
| srs2Δ tid1Δ | 0.0 ± 0.0 |
| srs2Δ rad52Δ | 85.3 ± 3.6 |
| rfa1-t11 | 95.1 ± 3.5 |
| tid1Δ rfa1-t11 | 86.0 ± 8.4 |
| rad51Δ rfa1-t11 | 64.0 ± 15.2 |
| srs2Δ rfa1-t11 | 18.8 ± 7.6 |
| rad51Δ tid1Δ rfa1-t11 | 28.0 ± 4.1 |
| rad52Δ tid1Δ rfa1-t11 | 17.8 ± 9.9 |
| rad51Δ rad52Δ tid1Δ | 14.3 ± 12.1 |
| rad52Δ rfa1-t11 | 25.0 ± 2.8 |
| Strains with suppression by Rad51 mutants | |
| rad51Δ pPGK::RAD51, 2μm | 68.3 ± 8.3 |
| rad51Δ pPGK::rad51-K191A, 2μm | 71.9 ± 3.7 |
| rad51Δ pPGK (empty vector) CEN ARS | 25.7 |
| rad51Δ pPGK::rad51-K191A CEN ARS | 71.7 |
| srs2Δ pPGK (empty vector) CEN ARS | 20.8 |
| srs2Δ pPGK::rad51-K191A CEN ARS | 33.0 |
| rad51Δ pPGK::rad51-K191R, 2μm | 82.3 ± 2.1 |
| rad51Δ pADH::rad51-L99P, 2μm | 77.3 ± 10.3 |
| rad51Δ pADH::rad51-S146A, 2μm | 62.8 ± 6.1 |
| rad51Δ pADH::rad51-Y388H, 2μm | 27.9 ± 7.2 |
A rad52Δ mutation suppresses rad51Δ.
Although a rad52Δ mutation had no discernible effect on adaptation, we were surprised to discover that the absence of RAD52 suppressed the adaptation defect of the rad51Δ strain (Fig. 2D). This is especially intriguing because our recent studies have shown that the absence of Rad52p abolishes Rad51p binding to HO-cleaved DNA, as detected by chromatin immunoprecipitation (45); hence we might have expected rad52Δ and rad51Δ strains to have identical phenotypes. Instead, rad52Δ suppresses rad51Δ; however, rad52Δ had no effect on either yku70Δ or tid1Δ (24, 25) or on cdc5-ad (49). This result raised the question of whether some aspect of checkpoint monitoring of DNA damage was integrated with one or more processes of RAD52-dependent but RAD51-independent recombination. Although the DSB we study is created in a haploid strain with no extensive homology elsewhere in the genome (i.e., HML and HMR have been deleted), it is possible that, as the ends are resected for many kilobases, recombination is attempted between dispersed repeated DNA sequences such as Ty elements. Alternatively, recombinational interactions may occur between the two unequally resected broken sister chromatid strands after replication of the broken DNA molecules.
Recent studies have revealed a RAD52-dependent, RAD51-independent process that requires the RAD59 gene, so that recombination in a rad51Δ rad59Δ double mutant is reduced almost to the very low level seen in a rad52Δ strain (2, 15, 42). We therefore examined whether the permanent checkpoint arrest of rad51Δ cells could be suppressed by rad59Δ, but an isogenic rad59Δ rad51Δ strain was not different from the rad51Δ mutant (Table 1). This indicates that the role of RAD52 in the checkpoint response is revealed only in the absence of Rad51p.
To examine how rad52Δ may suppress rad51Δ, we created a rad51Δ rad52Δ tid1Δ strain. This triple mutant adapted more than the rad51Δ tid1Δ strain but less than the rad51Δ strain (Table 2). This result indicates again that Rad52p affects adaptation by a pathway different from that used by Tid1p or that Tid1p acts upstream of these other proteins.
The adaptation-defective phenotypes of rad51Δ mutants are in many respects similar to those of srs2Δ mutants (53). About 20% of srs2Δ cells also arrest in the second cell cycle; moreover, srs2Δ is also suppressed by rad52Δ. A rad51Δ srs2Δ double mutant has the same adaptation defect as either single mutant (Table 2). However, the srs2Δ strain also has a defect in recovery when a DSB is repaired after a long checkpoint-mediated delay. rad51Δ cells are slow to recover, but do so, and rad51Δ suppresses the recovery defect of the srs2Δ mutant (53).
Separation-of-function mutations in Rad51p implicate its interaction with Rad52p during adaptation.
Further support for the importance of the Rad51p-Rad52p interaction in the checkpoint response has come from analysis of three DNA repair-defective mutants of Rad51p. The Rad51-Y388H mutant protein is defective in its interaction with Rad52p, as shown by two-hybrid tests (21). We transformed a rad51Δ strain with a centromeric plasmid expressing Rad51-Y388H transcribed from its own promoter and found that the cells remained adaptation defective (Table 2). In contrast, when rad51Δ was complemented with plasmids expressing Rad51-L99P and Rad51-T146A, which are defective in interactions with Rad54p and/or Rad55p (21), adaptation was restored (Table 2).
The repair-defective Rad51p-K191A protein that fails to bind ssDNA is still active in regulating adaptation.
A K191A mutation in Rad51p prevents in vitro DNA binding and strand exchange activity and renders cells radiation sensitive (29, 52). We transformed cells with both a high-copy-number plasmid, pR51.4, and a centromeric plasmid derivative, each expressing Rad51-K191A, and found that these cells could still adapt. These results suggest that it is not the normal recombination activity of Rad51p—such as DNA binding and strand exchange—that regulates adaptation but rather its interaction with some other proteins; this interpretation is again consistent with the idea that adaptation is dependent on an interaction between Rad51p and Rad52p.
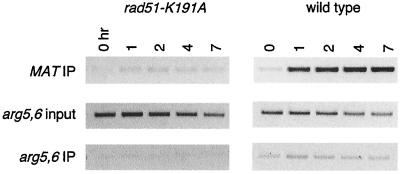
To confirm that Rad51-K191A protein is indeed defective in binding to ssDNA, we used chromatin immunoprecipitation techniques to examine how Rad51-K191A protein binds to DNA adjacent to an unrepaired DSB (45). The rad51Δ strain YSL306 (GAL::HO MATα hmlΔ hmrΔ) was transformed with centromeric plasmids expressing either wild-type Rad51p or Rad51-K191A. DNA and proteins were cross-linked with formaldehyde at intervals after the creation of an HO-induced DSB at MAT. The results of this experiment showed that Rad51-K191A binds to ssDNA ends with 50- to 100-fold less efficiency than the wild type during the first 2 h following HO cleavage (Fig. 4). The very weak binding seen in a strain expressing Rad51-K191A (from plasmid pR51.4) may be a reflection of the fact that the protein is overexpressed from a high-copy vector.
FIG. 4.
Lack of binding of Rad51-K191A protein to ssDNA in vivo. Rad51p binding to ssDNA adjacent to a HO endonuclease-induced DSB was monitored by chromatin immunoprecipitation (IP) in a strain that cannot repair the break by homologous recombination. The amount of input DNA was monitored by PCR amplification of the arg5,6 gene, and the same primer pairs were used to show the low background of arg5,6 DNA in the samples immunoprecipitated by antibody against Rad51.
As Western blotting confirmed that the level of Rad51-K191A expressed from the centromeric plasmid is about 10-fold higher than that of the wild type (data not shown), we also entertained the hypothesis that when the mutant protein is expressed at this level, Rad51-K191A may act by sequestering Rad52p, making the cells phenotypically Rad52− as rad52Δ suppresses rad51Δ (Table 2). However, this is unlikely to be the explanation. Whereas overexpressing Rad51-K191A suppresses rad51Δ alone, it does not suppress the adaptation defect of an srs2Δ mutant, although rad52Δ does suppress the adaptation defect of an srs2Δ mutant (Table 2). Hence, overexpressing Rad51-K191A does not create a phenocopy of rad52Δ.
Changes in the phosphorylation and kinase activity of Rad53p in different genetic backgrounds.
The presence of damaged DNA stimulates the phosphorylation of Rad53p by the Mec1 protein kinase, activating Rad53p as a protein kinase (39). This activated state is evident in Western blots as slower-migrating phosphorylated forms of the protein. It is also possible to measure Rad53 kinase activity by autophosphorylation by using an in-gel assay (34). When wild-type cells carry an unrepaired DSB, Rad53p phosphorylation and kinase activity increase dramatically approximately 1 h after HO-mediated DNA cleavage and peak at 6 to 8 h after HO induction (Fig. 5) at the time that all cells in the population are arrested. Rad53p activation persists until 8 to 15 h, when most cells adapt and resume cell cycle progression. Thus, cells turn off the Rad53p checkpoint kinase, even though DNA damage continues to be present.
FIG. 5.
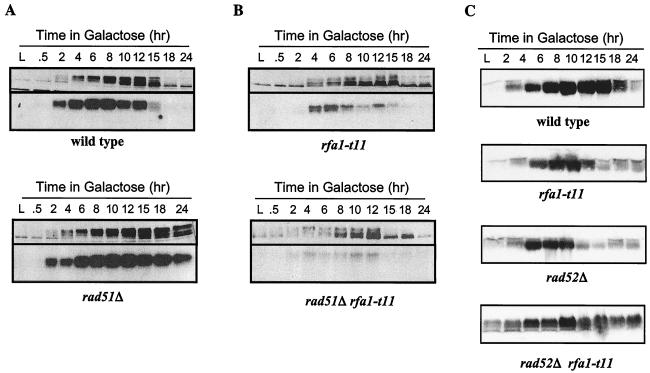
Phosphorylation and kinase activity of Rad53p. The activation of Rad53p kinase is shown both by Western blot analysis (top of each set), in which phosphorylated forms of Rad53p exhibit slower migration, and by an in-gel activity autophosphorylation assay. (A) Activation of Rad53p by a single HO-induced DSB in logarithmically growing wild-type and adaptation-defective mutant cells. L represents cells in logarithmic growth prior to HO induction. (B) Suppression of the adaptation defect of rad51Δ cells by the rfa1-t11 mutation. (C) Persistent hyperphosphorylation of Rad53p in rad52Δ rfa1-t11 cells after induction of an irreparable DSB. This is not seen for either rad52Δ or rfa1-t11 alone. No autophosphorylation assay was performed.
In adaptation-defective cells, however, Rad53p is maintained in a kinase-activated state even after 24 h. As shown in Fig. 5, Rad53p is hyperphosphorylated and activated in rad51Δ cells at 24 h (Fig. 5A) (25). We then examined Rad53p phosphorylation and kinase activity in strains carrying rfa1-t11 (Fig. 5B). rfa1-t11 suppresses the activated state of Rad53p in rad51Δ cells (Fig. 5B), similar to what is seen in rfa1-t11 tid1Δ cells (data not shown). The effect of rfa1-t11 on kinase activity in rad51Δ cells appears to be more complete than the partial suppression of the cells' permanent cell cycle arrest.
Overexpression of Rad51p does not alter the DNA damage checkpoint response.
In a recent study of Rad51p binding to HO-cleaved DNA in these same strains, it was found that the abundance of Rad51p is quite limited. As continued 5′-to-3′ resection of DNA continues, Rad51p fails to “fill up” the newly generated ssDNA after about 1 to 2 h (i.e., beyond a total of about 5 kb on either side of the DSB); however, the ssDNA binding protein RPA does occupy all the ssDNA (45). However, overexpression of Rad51p by about 10-fold allowed Rad51p to bind to newly generated ssDNA over at least 12 kb on one side of the DSB that was monitored (45). These observations raise the question of whether the activation of the checkpoint signal or its intensity may be influenced by the amount of ssDNA bound by RPA but not by Rad51p. However, this does not seem to be the case, as an isogenic strain overexpressing RAD51 showed a similar extent and duration of G2/M arrest after HO induction of a single DSB (data not shown).
A rad52Δ rfa1-t11 double mutant fails to adapt.
Both rfa1-t11 and rad52Δ cells are adaptation proficient; in fact, rfa1-t11 cells adapt somewhat faster than wild-type cells (34). However, a double mutant rfa1-t11 rad52Δ fails to adapt, exhibiting an arrest pattern similar to that seen for rad51Δ cells (Table 2). This arrest appears to be checkpoint mediated because rad9Δ rad52Δ rfa1-t11 cells do not permanently arrest. Moreover, Rad53p remains hyperphosphorylated at 24 h in rad52Δ rfa1-t11 cells (Fig. 5C). This persistent activation of the checkpoint is notably different from what occurs in cells with rad52Δ or rfa1-t11 alone. We note that rad52Δ and especially rad52Δ rfa1-t11 cause persistent, but low-level, activation of Rad53p kinase even before HO expression, but the response after induction of the unrepaired DSB is evident.
These results indicate that the ability to adapt depends on the presence of either Rad52p or wild-type RPA. Moreover, whereas an rfa1-t11 tid1Δ strain adapts, a rfa1-t11 tid1Δ rad52Δ strain does not. A rad52Δ rad51Δ strain adapts, but a rad52Δ rad51Δ rfa1-t11 strain does not (Table 2). A rad52Δ rad51Δ tid1Δ strain also does not adapt.
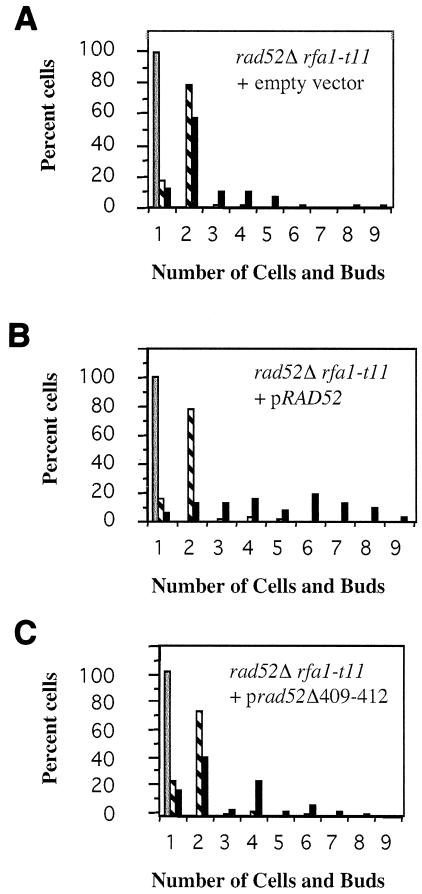
If the interaction between Rad51p and Rad52p is essential for regulation of adaptation, then a rad52 mutant that is impaired in interaction with Rad51p may also be adaptation defective. We examined the C-terminal deletion mutant rad52Δ409-420, which lacks Rad51p binding and is recombination defective (22). Cells with the rad52Δ409-420 mutation alone are slightly impaired in adaptation (Table 1), but those with the mutation in combination with rfa1-t11 are about as adaptation defective as the rad52Δ rfa1-t11 double mutant (Fig. 6).
FIG. 6.
A C-terminal truncation of Rad52 (Δ409-420) is adaptation defective. A rad52Δ rfa1-t11 strain was transformed with a centromeric plasmid carrying either RAD52 (B) or rad52Δ409-420 (C) or with an empty vector (A). G1 cells were plated onto YEP-galactose as described above. Cells were scored at 8 h (hatched bars) and 24 h (black bars) after induction of the DSB.
DISCUSSION
Previous work has suggested that both the establishment of a checkpoint response and its maintenance depends on the extent of ssDNA created by 5′-to-3′ resection at the ends of DSBs (10, 24, 58). Our finding that two additional ssDNA binding proteins, Rad51p and Rad52p, play a role in adaptation from the G2/M checkpoint further emphasizes the role of ssDNA in checkpoint control. Our results also indicate that Rad52p and RPA function in parallel to regulate adaptation; whereas neither rad52Δ nor rfa1-t11 strains are adaptation defective, the double mutant is unable to adapt after DNA damage. Rad51p and Rad52p play different roles in adaptation. From results for the various mutation combinations summarized in Table 2, we propose that the regulation of adaptation consists of two separate monitors that determine the nature and extent of DSB damage, one represented by Rad52p and the other by RPA. RPA and Rad52p, independently and possibly also through their mutual interactions (11, 14, 41), serve as monitors of the presence of unrepaired ssDNA and thus control the cell's decision to adapt. It is also important to emphasize that none of the proteins discussed here prevent the imposition of the checkpoint, which must be initially sensed in a Rad51-, Rad52-, Tid1-, and Rfa1-t11-independent fashion; rather, these recombination proteins affect the termination of the checkpoint.
Several checkpoint proteins have been shown to bind to HO-induced DSB ends (20, 28, 37). The observed increase in the intensity of Ddc1-green fluorescent protein (GFP) and Ddc2-GFP foci over time (28) suggests that the level of Ddc1p and Ddc2p binding to ssDNA is proportional to the extent of 5′-to-3′ resection of DNA adjacent to the DSB. When cells adapt, however, the intensity of Ddc2-GFP foci decreases (28), suggesting either that the checkpoint protein is displaced from ssDNA or that the ssDNA substrate has changed. It is possible that both Rad52p and RPA complexes may interact with one or both of the checkpoint complexes (Mec1p-Ddc2p and Rad17p-Mec3p-Ddc1p) to modulate the signals during checkpoint-mediated G2/M arrest. So long as one of these DNA damage sensors remains bound to DNA, cells will remain checkpoint arrested.
There appear to be two pathways to remove or inactivate the sensor protein(s), one involving RPA and the other involving Rad52p. These proteins themselves may not displace or inactivate the sensors, but each may facilitate the binding of other proteins that can remove or inactivate them. Given that rad52Δ prevents Rad51p filament formation but that the mutant is adaptation proficient, it is unlikely that Rad51p filament formation itself regulates checkpoint signaling. Moreover, overexpression of Rad51p did not modulate adaptation, even though normally Rad51p is unable to bind to more than about 10 kb of ssDNA. By the time cells adapt, as much as 50 kb or more has apparently been resected, although over time the 3′ ends of ssDNA do also become degraded. Rad52p itself may form multiple rings on ssDNA (51), and this activity may be modulated by the interaction of Rad52p with Rad51p. Similarly, once RPA is bound, it will facilitate the binding of some other proteins, a process that apparently involves Tid1p. Either when Rad52p is absent or when RPA carries the t11(L45E) mutation, the regulation of adaptation is normal, but when either of these two ssDNA binding pathways is blocked at a downstream step (rad51Δ affecting Rad52p or tid1Δ affecting RPA), the checkpoint is maintained. This suggests that there are several steps in creating protein complexes that result in inactivation of the checkpoint.
Recently, Zou and Elledge (58) reported that the rfa1-t11 mutation reduces the binding of Mec1p at a HO nuclease-induced DSB. We note, however, that there is significant Mec1p-dependent hyperphosphorylation of Rad53p in an rfa1-t11 strain after induction of a DSB, albeit the persistence of this state is shorter (34). Even more striking, in a rad52 rfa1-t11 mutant the checkpoint remains permanently activated, suggesting that Rfa1-t11 mutant protein is entirely competent in recruiting sufficient DNA damage checkpoint proteins to impose permanent cell cycle arrest. Thus, it is possible that the establishment of the DNA damage checkpoint does not involve an interaction between Mec1p and RPA but that the maintenance of the checkpoint depends on this interaction.
Link between homologous recombination and G2/M checkpoint control.
Rad52p plays a central role in recombination. It binds to 3′-ended ssDNA created at DSBs, both at the end and along the strand (51). In vitro, Rad52p plays an important role in the formation of the Rad51p filament and in strand exchange (22, 30, 31, 41, 46, 47). Rad52 has been shown genetically and physically to be necessary for RAD51-dependent recombination. In fact, in vivo, the absence of Rad52p prevents the association of Rad51p with ssDNA, as assessed by chromatin immunoprecipitation (45). Hence, one may have expected that rad52Δ and rad51Δ strains should have similar adaptation phenotypes, but this is not the case. Here we show that Rad52p plays a role in DNA damage sensing in which Rad51p apparently needs to make contact with Rad52p but does not necessarily form a filament. We imagine that the Rad52p complex, modulated by Rad51p, monitors whether the DSBs ends have engaged in homologous recombination.
Currently, it is unclear how Rad52p performs its signaling role during adaptation to the DNA damage checkpoint. One can imagine that Rad52p, through its ability to bind to ssDNA and to promote homologous recombination, is used to assess the progress of the DNA repair and to convey this information to the checkpoint complex that may also reside at the site of a DSB (20, 28, 37). However, the role of Rad51p is clearly different from its normal function in recombination, because the Rad51-K191A mutant that exhibits markedly reduced ssDNA binding and is devoid of recombination activity is nevertheless competent for adaptation. It is possible, however, that the mutant protein makes transient contacts with DNA that are sufficient for some role in checkpoint response but not for recombination. Moreover, a partially active Rad51-Y388H mutant, defective in its interaction with Rad52p (21), is adaptation defective, whereas two other Rad51p mutants that are similarly sensitive to methylmethane sulfonate but are defective in interactions with Rad54p and/or Rad55p are adaptation competent. We supported this conclusion by showing that a Rad52Δ409-429 protein, defective in its interaction with Rad51p, is also adaptation defective, especially as measured in combination with rfa1-t11.
We have considered whether rad52Δ may suppress rad51Δ by altering the extent of ssDNA, for example by allowing degradation of long 3′-ended molecules or decreasing the rate of resection. This seems unlikely for several reasons. First, rad52Δ does not suppress yku70Δ even though a reduction in the extent of resection by mre11Δ does so (24), and second, cells with the rad52Δ mutation alone are not defective in adaptation. We examined directly the extent of resection and the stability of 3′ ends on denaturing gels, where higher-molecular-weight bands represent partial digestion products caused when restriction sites become single stranded (24, 44, 56). If 3′ ends were extensively degraded, these bands would disappear, as the probe is within 500 bp of the HO cleavage site; conversely, if 5′-to-3′ resection was faster, higher-molecular-weight fragments would appear more rapidly. In fact, DNA from rad52Δ rad51Δ cells at intervals after HO induction was indistinguishable from that from rad51Δ or rad52Δ single mutants (data not shown).
One of the most interesting aspects of our results is that they reveal different functional groupings of recombination proteins than would be expected from the roles of these proteins in homologous recombination. Whereas Rad54p and Rad55p/Rad57p play important roles in DSB-mediated homologous recombination (42), they do not influence adaptation. There is also a RAD52-dependent, RAD51-independent homologous recombination pathway in mitotic cells that depends on RAD50, RAD59, and TID1 (2, 42), but that pathway also does not seem to be involved in regulating adaptation, as rad59Δ did not affect this process. However, tid1Δ strains are profoundly adaptation defective (25). Tid1p exhibits only very weak two-hybrid interaction with Rad51p (6), but there is evidence that Tid1p is capable of improving Rad51p's in vitro strand exchange activity (35). Yet in checkpoint responses, Tid1p and Rad51p are not epistatic (e.g., the rad51Δ rad52Δ strain adapts but the tid1Δ rad52Δ and rad51Δ tid1Δ rad52Δ strains do not; similarly, the tid1Δ rfa1-t11 strain adapts but the tid1Δ rad51Δ rfa1-t11 strain does not).
One may also ask whether rad51Δ and rad52Δ may play a role in the related, but distinct, S phase checkpoint that is seen when cells are exposed to agents such as hydroxyurea (HU) that stall DNA replication. A previous study found that the rad51Δ strain is HU sensitive (1), and the rad51Δ mutation may therefore represent an adaptation defect rather than a problem in DNA repair. We think that this is unlikely, because rad51Δ, rad52Δ, and mre11Δ deletion mutants all have similar HU sensitivities and are similar in their abilities to recover after removal from HU exposure (K. Patterson and J. E. Haber, unpublished data). That three mutants with very different checkpoint phenotypes have similar HU sensitivities is most consistent with the presence of a defect in recombinational repair of lesions generated during HU arrest and subsequent growth in the absence of the inhibitor.
We suggest that adaptation depends on two interacting conditions, the extent of ssDNA and the state of homologous recombination. The amount of ssDNA is monitored through RPA, while the homologous recombinational repair depends on Rad52p. However, these two parts of the checkpoint system likely interact, given that RAD52 physically and genetically interacts with RPA (9, 17, 43) and given the results of the genetic studies we discussed here. We further suggest that RPA and Rad52 proteins each interact with one or more of the checkpoint proteins that continue signaling in adaptation-defective mutants. Once RPA is engaged with the checkpoint, it requires Tid1p to turn off that part of the checkpoint that interacts with RPA. This may involve the ATP-dependent chromatin-remodeling activity assigned to Tid1p, Rad54p, and other members of the Swi2/Snf2 class of proteins (16), but it may also be carried out through allosteric interactions. In any case, in a tid1Δ mutant, the RPA-associated checkpoint proteins apparently continue to signal. The checkpoint system also interacts with Rad52p. Once Rad52p is engaged with the checkpoint, it requires Rad51p to disengage. In rad51Δ cells, the Rad52p-associated checkpoint continues to signal. However, the way in which Rad51p interacts with Rad52p appears to be distinct from the normal recombinational role of Rad51p, as Rad51-K191A that cannot bind DNA is still adaptation proficient but Rad51-Y338P that does not interact with Rad51p is defective. Presumably Rad51-K191A can still interact with Rad52p.
In summary, we have found that a number of proteins involved in homologous recombination play important roles in cellular responses to DNA damage. We suggest that RPA primarily signals the presence of the damaged DNA itself whereas Rad52p signals whether the damage has been successfully taken up into recombination structures. This would explain why cells that are capable of carrying out normal recombination—even though it is a slow process in which there is extensive ssDNA created—are not signaled to arrest (34). In the special case in which a DSB cannot be repaired—the case we study here—both Rad52p and RPA act to assess the extent of the damage itself.
Acknowledgments
We thank all members of J.E.H. and M.F.'s laboratory for enthusiastic discussions. We are grateful to F. Fabre, C. Greider, H. Klein, S. Lovett, and L. Symington for the plasmids and yeast strains. P. Sung and D. Bishop kindly provided anti-Rad51p antibodies.
M.F. was supported by grants from Associazione Italiana per la Ricerca sul Cancro and from Telethon-Italy (grant no. E.1108). S.E.L. is a postdoctoral fellow of The Leukemia and Lymphoma Society. J.E.H. was supported by NIH grants GM61766 and GM20056.
REFERENCES
- 1.Allen, J. B., Z. Zhou, W. Siede, E. C. Friedberg, and S. J. Elledge. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2401-2415. [DOI] [PubMed] [Google Scholar]
- 2.Bai, Y., and L. S. Symington. 1996. A RAD52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10:2025-2037. [DOI] [PubMed] [Google Scholar]
- 3.Cohen, Y., M. Dardalhon, and D. Averbeck. 2002. Homologous recombination is essential for RAD51 up-regulation in Saccharomyces cerevisiae following DNA crosslinking damage. Nucleic Acids Res. 30:1224-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de la Torre-Ruiz, M., and N. F. Lowndes. 2000. The Saccharomyces cerevisiae DNA damage checkpoint is required for efficient repair of double strand breaks by non-homologous end joining. FEBS Lett. 467:311-315. [DOI] [PubMed] [Google Scholar]
- 5.de la Torre-Ruiz, M. A., C. M. Green, and N. F. Lowndes. 1998. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 17:2687-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dresser, M. E., D. J. Ewing, M. N. Conrad, A. M. Dominguez, R. Barstead, H. Jiang, and T. Kodadek. 1997. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147:533-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elledge, S. J. 1996. Cell cycle checkpoints: preventing an identity crisis. Science 274:1664-1672. [DOI] [PubMed] [Google Scholar]
- 8.Evans, E., N. Sugawara, J. E. Haber, and E. Alani. 2000. The Saccharomyces cerevisiae Msh2 mismatch repair protein localizes to recombination intermediates in vivo. Mol. Cell 5:789-799. [DOI] [PubMed] [Google Scholar]
- 9.Firmenich, A. A., A. M. Elias, and P. Berg. 1995. A novel allele of Saccharomyces cerevisiae RFA1 that is deficient in recombination and repair and suppressible by RAD52. Mol. Cell. Biol. 15:1620-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasior, S. L., A. K. Wong, Y. Kora, A. Shinohara, and D. K. Bishop. 1998. Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev. 12:2208-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 13.Hays, S. L., A. A. Firmenich, and P. Berg. 1995. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. USA 92:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hays, S. L., A. A. Firmenich, P. Massey, R. Banerjee, and P. Berg. 1998. Studies of the interaction between Rad52 protein and the yeast single-stranded DNA binding protein RPA. Mol. Cell. Biol. 18:4400-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ira, G., and J. E. Haber. 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22:6384-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaskelioff, M., S. Van Komen, J. E. Krebs, P. Sung, and C. L. Peterson. 2003. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 278:9212-9218. [DOI] [PubMed] [Google Scholar]
- 17.Kantake, N., T. Sugiyama, R. D. Kolodner, and S. C. Kowalczykowski. 2003. The recombination-deficient mutant RPA (rfa1-t11) is displaced slowly from single-stranded DNA by Rad51 protein. J. Biol. Chem. 278:23410-23417. [DOI] [PubMed] [Google Scholar]
- 18.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 19.Kondo, T., K. Matsumoto, and K. Sugimoto. 1999. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol. Cell. Biol. 19:1136-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo, T., T. Wakayama, T. Naiki, K. Matsumoto, and K. Sugimoto. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294:867-870. [DOI] [PubMed] [Google Scholar]
- 21.Krejci, L., J. Damborsky, B. Thomsen, M. Duno, and C. Bendixen. 2001. Molecular dissection of interactions between Rad51 and members of the recombination-repair group. Mol. Cell. Biol. 21:966-976. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Krejci, L., B. Song, W. Bussen, R. Rothstein, U. H. Mortensen, and P. Sung. 2002. Interaction with Rad51 is indispensable for recombination mediator function of Rad52. J. Biol. Chem. 277:40132-40141. [DOI] [PubMed] [Google Scholar]
- 23.Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy, H. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305-309. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 25.Lee, S. E., A. Pellicioli, A. Malkova, M. Foiani, and J. E. Haber. 2001. The Saccharomyces recombination protein Tid1p is required for adaptation from G2/M arrest induced by a double-strand break. Curr. Biol. 11:1053-1057. [DOI] [PubMed] [Google Scholar]
- 26.Leroy, C., S. E. Lee, M. B. Vaze, F. Ochsenbein, R. Guerois, J. E. Haber, and M. C. Marsolier-Kergoat. 2003. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol. Cell 11:827-835. (Erratum, 11:1119.) [DOI] [PubMed] [Google Scholar]
- 27.Lowndes, N. F., and J. R. Murguia. 2000. Sensing and responding to DNA damage. Curr. Opin. Genet. Dev. 10:17-25. [DOI] [PubMed] [Google Scholar]
- 28.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15:2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan, E. A., N. Shah, and L. S. Symington. 2002. The requirement for ATP hydrolysis by Saccharomyces cerevisiae Rad51 is bypassed by mating-type heterozygosity or RAD54 in high copy. Mol. Cell. Biol. 22:6336-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.New, J. H., and S. C. Kowalczykowski. 2002. Rad52 protein has a second stimulatory role in DNA strand exchange that complements replication protein-a function. J. Biol. Chem. 277:26171-26176. [DOI] [PubMed] [Google Scholar]
- 31.New, J. H., T. Sugiyama, E. Zaitseva, and S. C. Kowalczykowski. 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391:407-410. [DOI] [PubMed] [Google Scholar]
- 32.Nigg, E. A. 1998. Polo-like kinases: positive regulators of cell division from start to finish. Curr. Opin. Cell Biol. 10:776-783. [DOI] [PubMed] [Google Scholar]
- 33.Paulovich, A. G., and L. H. Hartwell. 1995. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82:841-847. [DOI] [PubMed] [Google Scholar]
- 34.Pellicioli, A., S. E. Lee, C. Lucca, M. Foiani, and J. E. Haber. 2001. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from G2/M arrest. Mol. Cell 7:293-300. [DOI] [PubMed] [Google Scholar]
- 35.Petukhova, G., P. Sung, and H. Klein. 2000. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 14:2206-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhind, N., and P. Russell. 1998. Mitotic DNA damage and replication checkpoints in yeast. Curr. Opin. Cell Biol. 10:749-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rouse, J., and S. P. Jackson. 2002. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9:857-869. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez, Y., J. Bachant, H. Wang, F. Hu, D. Liu, M. Tetzlaff, and S. J. Elledge. 1999. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286:1166-1171. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 40.Sandell, L. L., and V. A. Zakian. 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75:729-739. [DOI] [PubMed] [Google Scholar]
- 41.Shinohara, A., and T. Ogawa. 1998. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391:404-407. [DOI] [PubMed] [Google Scholar]
- 42.Signon, L., A. Malkova, M. Naylor, and J. E. Haber. 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, J., and R. Rothstein. 1995. A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA-binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol. Cell. Biol. 15:1632-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugawara, N., E. L. Ivanov, L. J. Fishman, B. L. Ray, X. Wu, and J. E. Haber. 1995. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature 373:84-86. [DOI] [PubMed] [Google Scholar]
- 45.Sugawara, N., X. Wang, and J. E. Haber. 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12:209-219. [DOI] [PubMed] [Google Scholar]
- 46.Sugiyama, T., and S. C. Kowalczykowski. 2002. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 277:31663-31672. [DOI] [PubMed] [Google Scholar]
- 47.Sung, P. 1997. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272:28194-28197. [DOI] [PubMed] [Google Scholar]
- 48.Sung, P., and S. A. Stratton. 1996. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 271:27983-27986. [DOI] [PubMed] [Google Scholar]
- 49.Toczyski, D. P., D. J. Galgoczy, and L. H. Hartwell. 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90:1097-1106. [DOI] [PubMed] [Google Scholar]
- 50.Umezu, K., N. Sugawara, C. Chen, J. E. Haber, and R. D. Kolodner. 1998. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics 148:989-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Dyck, E., A. Z. Stasiak, A. Stasiak, and S. C. West. 1999. Binding of double-strand breaks in DNA by human Rad52 protein. Nature 398:728-731. [DOI] [PubMed] [Google Scholar]
- 52.Van Komen, S., G. Petukhova, S. Sigurdsson, S. Stratton, and P. Sung. 2000. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell 6:563-572. [DOI] [PubMed] [Google Scholar]
- 53.Vaze, M., A. Pellicioli, S. Lee, G. Ira, G. Liberi, A. Arbel-Eden, M. Foiani, and J. Haber. 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires srs2 helicase. Mol. Cell 10:373-385. [DOI] [PubMed] [Google Scholar]
- 54.Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam, and F. Fabre. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309-312. [DOI] [PubMed] [Google Scholar]
- 55.Weinert, T. 1998. DNA damage checkpoints update: getting molecular. Curr. Opin. Genet. Dev. 8:185-193. [DOI] [PubMed] [Google Scholar]
- 56.White, C. I., and J. E. Haber. 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, X., and J. E. Haber. 1996. A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell 87:277-285. [DOI] [PubMed] [Google Scholar]
- 58.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]