Abstract
The Ets2 transcription factor is regulated by mitogen-activated protein (MAP) kinase phosphorylation of a single threonine residue. We generated by gene targeting a single codon mutation in Ets2 substituting Ala for the critical Thr-72 phosphorylation site (Ets2A72), to investigate the importance of MAP kinase activation of Ets2 in embryo and tumor development. Ets2A72/A72 mice are viable and develop normally. However, combining the Ets2A72 allele with a deletion mutant of Ets2 results in lethality at E11.5 and shows that Ets2A72 is a hypomorphic allele. Mammary tumors caused by transgenic polyomavirus middle T antigen, activated Neu(Erbb2), or the combination of Neu and transgenic VEGF (Neu; VEGF-25) were all restricted in Ets2A72/A72 females. The Ets2A72/A72 restriction on Neu; VEGF-25 tumor growth was associated with increased p21Cip1 expression. The size of tumors transplanted into fat pads of mice with Ets2 targeted alleles was correlated directly with Ets2 activity and fewer stromal cells expressing matrix metalloproteinase 9 (MMP-9). Decreased MMP-3 and MMP-9 mRNAs were confirmed in Ets2A72/A72 macrophages. Activation of Ets2 at Thr-72 acts in the stroma, downstream of vascular endothelial growth factor production, in part through the regulation of macrophage proteases to support the progression of Neu- and polyomavirus middle-T-initiated mammary tumors.
Most cancers are epithelial in origin. However, a significant and sometimes predominate portion of breast, colon, stomach, and pancreas tumors are host fibroblasts, endothelial cells, inflammatory cells, smooth muscle cells, adipocytes, and extracellular matrix known collectively as stroma (6). Much less is known about the molecular genetics of tumor-stroma interaction than is known about epithelial transformation. Tissue stroma can both suppress and promote tumor progression (16, 57). Among the examples of stroma-neoplasia communication is the role of matrix metalloproteinases produced by fibroblasts and invasive inflammatory cells in the angiogenic switch associated with pancreatic and skin neoplastic progression (5, 10).
Ets2 is one of over 25 transcription factors in the human genome that utilize the Ets winged helix-loop-helix DNA-binding domain (30, 54). Extracellular signal-regulated kinase 2 (Erk2) binds to Ets2 through a docking site in the evolutionarily conserved, N-terminal pointed domain of a subset of Ets transcription factors (44). The signal transduction pathway from growth factor receptors, such as fibroblast growth factor (FGF) receptor and epidermal growth factor receptor (EGFR; Erbb1), through the Ras-mitogen-activated protein kinase pathway to the pointed domain of Ets transcription factors is used for regulating developmental transitions in Drosophila melanogaster and Caenorhabditis elegans and growth factor signaling in humans and mice. In Drosophila the closest relative of Ets2, the Pointed-P2 product of the pointed gene, is key in Ras-pathway-dependent photoreceptor development (40) and in tracheal ductal morphogenesis (42). In mammals, Ets2 has been implicated in growth factor stimulation of macrophages (7, 31, 45, 49); FGF stimulation (59); and oncogenic stimulation by Neu (23), Ras (60), and Raf (36). The expression of either wild-type Ets2 or a dominant-negative form of Ets2 can reverse the transformed characteristics of human or mouse cancer cell lines (17, 18). Inactivation of Ets2 by a gene-targeted deletion of the DNA-binding domain and nuclear localization signal (Ets2db1) results in embryonic lethality associated with deficient expression of matrix metalloproteinase 9 (MMP-9) and CD31 in trophoblastic derivatives (59). Haploinsufficiency of Ets2 is sufficient to restrict mammary tumors arising from polyomavirus middle T (PyMT) oncogene expression driven by the mouse mammary tumor virus (MMTV) long terminal repeat promoter (38).
Here, we show that mutation of Thr-72 creates a hypomorphic allele of Ets2 (Ets2A72), which acts, at least in part, in host stroma to restrict mammary tumors. This tumor restriction is genetically downstream of vascular endothelial growth factor (VEGF) production and is correlated with decreased macrophage MMP-3 and MMP-9 expression.
MATERIALS AND METHODS
Gene targeting.
The Ets2A72 targeting vector was prepared from a 14.4-kb Ets2 genomic clone containing exons 3 to 9 (26). The codon for Thr-72 and a PstI restriction site encoded by codons 73 and 74 were changed using synthetic oligonucleotides and PCR of a 662-bp HindIII fragment spanning exon 4. A phosphoglycerate kinase-Neo gene flanked by LoxP recombination sites was inserted into an FspI site in intron 4, and a thymidine kinase gene was added to the 3′ end of the vector to utilize ganciclovir selection (35). The final vector contained the Neo gene flanked by 2 and 4 kb of Ets2 genomic sequence. DNA was introduced into embryonic stem (ES) cells by electroporation, and G418- and ganciclovir-resistant colonies were isolated by the Burnham Institute Mouse Molecular Genetics service. Two targeted ES cell clones were identified from 86 clones screened by PCR with one primer 5′ of targeting vector sequences (TTTGCTGTGCTCCCTTCTCTCAGT) and a primer, MA-AM11B-loxPneo (AAGAACGAGATCAGCAGCCTCTGT), within the vector, resulting in a 2.28-kb product. One clone was confirmed by Southern blot analysis with probes flanking the targeting vector. This allele was designated Ets2A72-Neo.
Chimeric animals were generated by the injection of Ets2A72-Neo/+ ES cells into C57/BL6 blastocysts by standard methods (28). Subsequent progeny were screened by PCR of DNA isolated from tail biopsy samples with the primer cDNA5BR (CTCTGCAGACTGGGGCTTATTC) and the MA-AM11B-loxPneo primer, generating a 506-bp product. The Neo gene was excised by mating Ets2A72-Neo/+ mice with pCXxNLS-Cre15, Cre-expressing deletion mice, kindly provided by Steve O'Gorman (Salk Institute, La Jolla, Calif.) (39). Mice positive for Cre-mediated excision were screened by PCR with the primers cDNA5BR and MA-AM11-4165 (GCAGGGAGAGCAAGAAAGGACAC), giving a 506-bp product for wild-type Ets2 and a 607-bp product for Ets2A72. To screen for the PyMT transgene, the primers MT-665 (CCAACCCGAGATGTGCTGAA) and MT-1310 (CTGCAAATCCCGAAGAATCA) were used for PCR of tail DNA for both the MMTV-PyMT (Jackson Laboratory; FVB/N-TgN [MMTVPyVT] 634 Mul) and MMTV-PyVMTY315,322FDb-4 (MMTV-PyMT) (56) lines. The MMTV-VEGF-25 (VEGF-25) transgenic mouse line expresses the mouse VEGF164 form of VEGF in mammary epithelium (40a). The transgene was screened by tail PCR with primers GAAAGACCGATTAACCATGTCAC and TCAGCAGTAGCCTCATCATCA.
Tumor transplantation.
The MMTV-PyMTY315,322F-DB7 serially transplanted mouse mammary tumor line was established and carried in the FVB/N mouse strain by serial transplantation of a tumor arising due to the MMTV-driven expression of PyMT antigen containing point mutations of the codons for tyrosines 315 and 322 (8, 56). Adult female mice heterozygous for the Ets2db1 targeted allele (59) on an FVB/N genetic background or wild-type littermates of 78- to 114-day-old mice were used as recipients. Ets2A72/+ or Ets2A72/A72 littermates bred five or more generations into the FVB/N background were also used as hosts. Pieces of PyMTY315,322F-DB7 tumors 1 mm3 in volume were surgically inserted into both no. 4 mammary fat pads of each recipient as described previously (8, 61). The genotypes of the recipients were not known to the surgeon at the time of implantation. After 8 to 9 days (experiment 1) and after 9 days (experiments 2 and 3) animals were sacrificed and the fat pads were removed, mounted on glass slides, fixed in acidic ethanol or Carnoy's fixative, stained with carmine alum, dehydrated through ethanol and toluene, and mounted in Permount. Tumors were photographed with an Olympus Z40 dissection microscope and a Nikon Coolpix 990 digital camera. Tumor size was measured with the use of NIH Image software by identifying the border of the tumors as a gray-scale threshold and converting the area in pixels to square millimeters by reference to a standard grid. The area was converted to volume by assuming a sphere with cross-sectional area equal to the measured value. After photography, the tissue was removed from the slide and embedded in paraffin, and 10-μm sections were stained with hematoxylin and eosin. Lymph nodes were measured by the same method as a control and reference.
The same incisions used for the mammary gland clearing technique were made for the subcutaneous transplantation of PyMTY315,322F tumor segments 1 mm3 in size. The skin was folded back, and the first bifurcation of the blood vessel anterior to the inguinal lymph node in the no. 4 fat pad was located. A tiny pocket was made in the connective tissue fascia, and the tumor tissue was placed between the bifurcation of the vein, beneath the connective tissue layer and the dermis. After 9 days, the tumors were dissected attached to the skin. The skin was mounted on cork and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS). Tumor size was assessed by image analysis of digital images acquired after brief carmine staining and using incident light.
BMMs.
Bone marrow-derived macrophages (BMMs) were prepared by isolating bone marrow from the femurs of 7- to 10-week-old female mice. Bone marrow was cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum-20 ng of colony-stimulating factor 1 (CSF-1) (Chemicon)/ml for 7 days and then dispersed to 500,000 cells per well in six-well trays. BMMs undergoing CSF-1 stimulation were cultured without CSF-1 for 16 h and then treated with 20 ng of CSF-1/ml for 8 h. BMMs undergoing lipopolysaccharide (LPS)-gamma interferon (IFN-γ) treatment were cultivated with 20 ng of CSF-1/ml for 16 h and then stimulated with an additional 100 ng of LPS/ml and 100 U of IFN-γ/ml for 8 h.
Immunohistochemical detection of MMP-9, F4/80, and factor VIII.
Sections (5 μm) were deparaffinized, hydrated to PBS, incubated in 5% H2O2 in PBS for 5 min, digested with Ficin (Zymed, South San Francisco, Calif.) for 10 min, and blocked for 30 min with 5% normal goat serum (DAKO, Glostrup, Denmark) and 3% bovine serum albumin (BSA) in PBS and with the avidin-biotin blocking kit (Vector Laboratories, Burlingame, Calif.) according to the manufacturer's instructions. Sections were incubated overnight at 4°C with polyclonal rabbit anti-MMP-9 (3) diluted 1:500 in 0.1% BSA-PBS, washed, and incubated for 1 h at room temperature with biotin-conjugated goat anti-rabbit antibody (DAKO) diluted 1:250 in PBS-BSA. After extensive washing with 0.1% BSA-PBS, the sections were incubated with streptavidin-horseradish peroxidase conjugate (Amersham Biosciences, Piscataway, N.J.) diluted 1:100 in 0.1% BSA-PBS and developed by incubating the slides with diaminobenzidine (Zymed) for 10 min. Sections were briefly counterstained with hematoxylin before mounting. The numbers of MMP-9-positive cells in the tumors and the tumor-stromal interface were counted, and the size of the counted area was determined using Image software from the National Institutes of Health (NIH).
F4/80 rat monoclonal antibody (Caltag) diluted 1/60 and rabbit anti-factor VIII-related antigen (DAKO A0082) diluted 1/1,000 were used on Carnoy's fixative-fixed, paraffin-sectioned material. Mouse-absorbed, biotinylated anti-rat secondary antibody (Vector Laboratories) was used at 5 μg/ml. The ABC Delux system (Vector Laboratories) and diaminobenzidine development were used for detection. Rabbit antibody was detected with the DAKO EnVision peroxidase-conjugated dextran complex and diaminobenzidine development. F4/80 staining of transplanted tumor sections was measured by converting digital images to gray scale and then black and white with the use of the threshold command. The total area of staining was measured with the use of NIH Image software.
Digital images of Fig. 2C and D and Fig. 6C to F were captured at 1,026 by 2,060 pixels using an AxioCam digital camera (Zeiss) mounted on a Zeiss microscope. The images were processed using PhotoShop software. Scale bars were inserted before reduction.
FIG. 2.
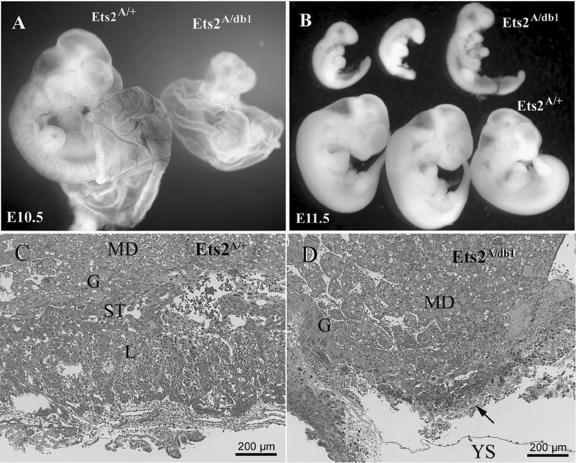
Haploinsufficiency of Ets2A72. (A) Ets2A72/+ and Ets2A72/db1 embryos with partially dissected yolk sacs at E10.5. Ets2A72/db1 embryos are smaller than Ets2A72/+ embryos, with no blood in the yolk sac. (B) Ets2A72/db1 and Ets2A72/+ embryos at E11.5. Ets2A72/db1 embryos are retarded and small. (C and D) Hematoxylin-and-eosin-stained sections of placentas from E11.5 concepti. Maternal decidua (MD), giant cell layer (G), spongiotrophoblast layer (ST), labyrinth layer (L), and yolk sac (YS) are indicated. Note that the spongiotrophoblast layer and labyrinth regions are absent in panel D.
FIG. 6.
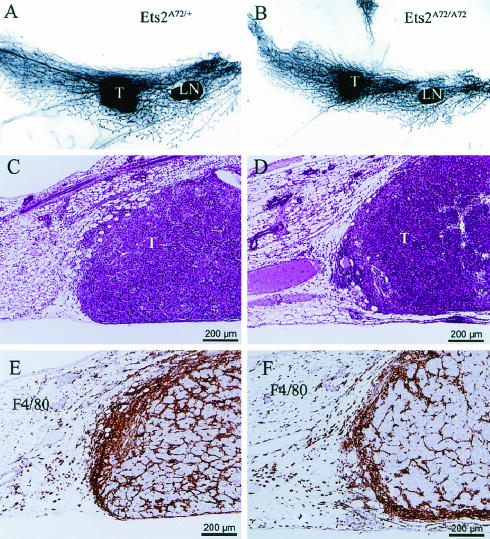
Transplanted PyMTY315,322F-DB7 tumors are smaller in Ets2A72/A72 hosts and are associated with macrophage infiltration. (A, C, and E) Ets2A72/+ hosts; (B, D, and F) EtsA72/A72 hosts. (A and B) Images of whole mounted mammary glands 9 days after tumor transplantation. LN, lymph node; T, tumor. (C and D) Hematoxylin-and-eosin-stained section of typical transplanted tumors. (E and F) Representative tumor fields stained with F4/80 antibody in brown.
RNA analysis.
RNA was prepared with the use of Trizol reagent (Invitrogen) (9). The cDNA was prepared from 1 μg of total RNA using oligo(dT) priming and Superscript II reverse transcriptase (Invitrogen). The cDNA levels were quantified by real-time PCR in a LightCycler instrument using the SYBR Green I PCR kit and the LightCycler software (Roche Diagnostics GmbH). Gene expression was normalized to the level of cyclophilin A (Cph) in the same cDNA sample, and a standard curve generated with Cph primers was used to estimate relative differences in gene expression (27). The two primers for each target were as follows: Cph/(Ppia), AGACCAGCAAGAAGATCACC and GGAAAATATGGAACCCAAAG; Bcl-xL/(Bcl2l set 1), GACGTGATCATTTCCCATC and GAGGTTCTGGTCCTTGTCTC; Bcl-xL/(Bcl2l set 2), TCAGAGCTTTGAGCAGGTAG and AAGGCTCTAGGTGGTCATTC; CycD1/(Ccnd1), AGGTAATTTGCACACCTCTG and ACAAAGCAATGAGAATCTGG; VEGFa/(Vegfa, 3′ noncoding), GAAGAAGAGGCCTGGTAATG and GGAAGATGAGGAAGGGTAAG; VEGF/(Vegfa, coding), CAGGCTGCACCCACGACAGAAG and CTATGTGCTGGCTTTGGTGAGGTTT; p21/(Cdkn1a), GGGTTCTCAGTGACTTCTCC and CCACTAAGTGCTTTGACACC; MMP-3, TTAAAGACAGGCACTTTTGG and CAGGGTGTGAATGCTTTTAG; MMP-9, CGTCTGAGAATTGAATCAGC, and AGTAGGGGCAACTGAATACC; Ets2, CAGAGGCCTAATCCTCAGTC and GGCCAAATTACAAAACCTTC.
Ets2 protein analysis.
Mouse embryo fibroblasts were isolated from 12.5-day Ets2A72/A72 and Ets2+/+ embryos by standard methods (28). Exponentially growing cultures were labeled with 500 μCi of [35S]methionine (Amersham)/ml for 20 min. Cells were harvested either immediately or after incubation in normal complete medium for up to 1 h as indicated. Cells were lysed, and proteins were immunoprecipitated with rabbit anti-Ets2 antiserum and analyzed by acrylamide gel electrophoresis and autoradiography as previously described (28). Fourteen-day film exposures were processed with NIH Image software to measure image intensity.
RESULTS
Generation of an Ets2A72 knock-in allele.
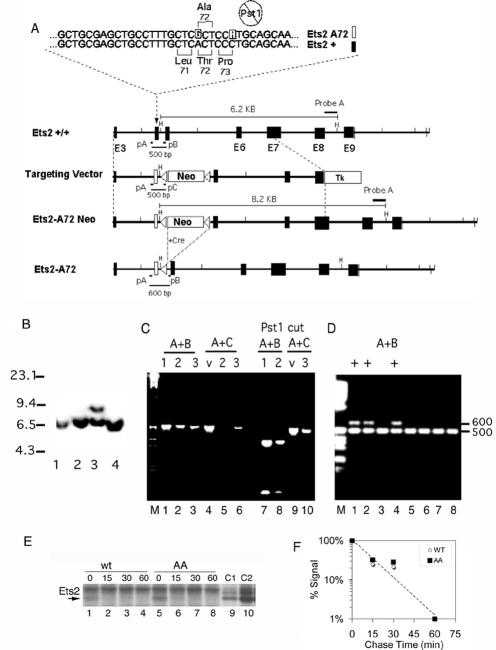
In cultured cells Ets2 basal transcriptional activity is activated by the phosphorylation of threonine-72 by oncogenes or growth factors that utilize the Ras-mitogen-activated protein kinase pathways (60). We introduced a single nucleotide change in codon 72 of mouse Ets2 to change it from Thr to Ala (Ets2A72). In addition, a second, silent, single nucleotide mutation of Pro-73 was introduced to eliminate a diagnostic PstI restriction site (Fig. 1A). The targeting vector containing the mutations and a Neo gene flanked by LoxP recombination sites was introduced into the Ets2 gene of ES cells. Two correctly targeted ES clones were identified. Southern blot analysis confirmed the expected structure of the targeted gene (Fig. 1B). PCR, restriction enzyme digestion (Fig. 1C), and DNA sequencing of amplified genomic DNA (data not shown) confirmed the presence of the two mutant nucleotides. Upon identification and germ line transmission of the targeted Ets2A72-Neo allele, the Neo selectable gene was removed by mating to mice expressing Cre recombinase from the protamine promoter in the male germ line (39) (Fig. 1D). This created the Ets2A72 allele. Ets2A72 RNA was found to be expressed at the same levels as that of wild-type Ets2 (see below). This indicates that the LoxP sequence left in intron 4 in the Ets2A72 allele does not interfere with the expression of the gene at the RNA level. To determine if A72 alteration might alter the degradation of the Ets2A72 protein, the turnover of Ets2 was determined in mouse embryo fibroblasts derived from Ets2A72/A72 and wild-type embryos. Cells were labeled with [35S]methionine for 20 min and then incubated in medium with normal levels of nonradioactive methionine for various times up to 60 min. Ets2 was immunoprecipitated and detected by enhanced autoradiography of a gel used to separate the immunoprecipitated proteins (Fig. 1E). Ets2 was identified by reference to a sample from 3T3 cells, which were transfected with an Ets2 expression vector and then labeled with radioactive methionine (Fig. 1E, lanes 9 and 10). Newly synthesized Ets2 protein was degraded rapidly with an estimated half-life of 10 min (Fig. 1F). The newly synthesized Ets2A72 protein was found at comparable levels and turned over similarly in comparison to wild-type Ets2. This result indicates that the effects of the A72 mutation are not due to increased turnover of the mutant protein. It also confirms the very rapid turnover of Ets2 (20).
FIG. 1.
Construction of the Ets2A72 allele. (A) The two nucleotides of the mutant allele are highlighted in the top strand, and the wild-type sequence is highlighted in the lower strand. The second mutation abolishes the PstI site. Maps of the relevant portion of Ets2 are shown with larger blocks and numbers indicating exons and a white block indicating exon 4 containing the Ets2A72 mutations. LoxP recombination sites and orientations are shown as triangles. Recombination of the targeting vector with the Ets2 gene generates the Ets2A72-Neo allele. Mating of mice carrying the Ets2A72-Neo allele with a transgenic mouse, which expresses Cre in the male germ line, results in excision of the Neo gene, leaving one LoxP element. H, HindIII; pA, pB, and pC, PCR primer sites; Tk, thymidine kinase gene used for negative selection in ganciclovir; Neo, phosphoglycerate kinase-Neo gene providing resistance to the drug G418. (B) Southern blot of targeted ES clones. DNA was digested by HindIII. The 6.2-kb wild-type band and the 8.2-kb A72-Neo band were detected by probe A, shown in panel A. Lanes 1, 2, and 4 represent wild-type ES cell clones. Lane 3 is Ets2A72-Neo/+ clone 135. Numbers at left are molecular sizes in kilobases. (C) PCR analysis of ES cells. DNA samples: 1, cloned Ets2+/+ gene fragment; 2, Ets2+/+ genomic DNA; 3, Ets2A72-Neo/+ clone 135; v, targeting vector DNA. Amplification with primers pA and pB or pA and pC is indicated. Samples in lanes 7 to 10 were digested with PstI. Both the A72-Neo targeting vector and clone 135 contain the mutation eliminating the PstI site. (D) PCR products of progeny of breeding Ets2A72-Neo/+ and Cre deletion mice using primers A and B. The wild-type Ets2 gene produces a product of 500 bp. The Ets2A72 generates a 600-bp product. The large Ets2A72-Neo allele does not amplify under these conditions. Lanes 1, 2, and 4 show products generated by the excision of the floxed Neo. (E) Ets2A72 protein turns over at the same rate as does wild-type Ets2. A 7-day exposure of a sodium dodecyl sulfate-acrylamide gel used to separate [35S]methionine-labeled proteins immunoprecipitated from wild-type or Ets2A72/A72 embryo fibroblasts is shown. Cells were labeled for 20 min and then incubated for the indicated time in medium containing normal levels of nonradioactive methionine. (F) Signal intensities were determined by NIH Image software, normalized, and plotted as a function of time of chase.
Developmental haploinsufficiency of Ets2A72.
The previously described Ets2db1 allele represents a deletion of the DNA-binding domain and nuclear localization signal. In a homozygous state Ets2db1 does not support embryonic development (59). After five generations of breeding into the FVB/N genetic background, Ets2A72/+ heterozygotes were inbred to determine viability. Ets2A72/A72 homozygous progeny were found at expected Mendelian ratios (data not shown). Thus, two Ets2A72 alleles are sufficient to prevent extraembryonic tissue deficiency. However, when Ets2A72/A72 mice were bred with Ets2db1/+ heterozygotes, only EtsA72/+ progeny were recovered (Table 1). Thus, while two EtsA72 alleles are permissive, decreasing Ets2 by an additional 50% is not tolerated. Ets2db1/db1 embryos die at about 8.5 days of development (59). Analysis of embryos derived from Ets2A72/A72 and Ets2db1/+ parents revealed that Ets2A72/db1 embryos survived until about E11.5 (Fig. 2B). E10.5 embryos were significantly smaller and retarded in their development (Fig. 2A). Furthermore, the yolk sacs of Ets2A72/db1 embryos failed to develop blood vessels although blood could be seen within the embryos proper (Fig. 2A). The abnormal development of Ets2A72/A72 embryos was associated with smaller, disorganized placentas lacking a labyrinth region where maternal and embryonic vascular systems intermingle (Fig. 2C and D). Ets2A72/db1 embryos survived approximately 3 days longer than Ets2db1/db1 embryos. Without prior knowledge of the complete rescue of Ets2db1/db1 embryos by tetraploid extraembryonic tissues (59), the retarded development of the Ets2A72/db1 embryos might be mistaken for an embryonic growth defect. However, the rescue of Ets2db1/db1 embryos has demonstrated that Ets2 is largely dispensable for embryonic development. Because tetraploid embryo cells are capable of colonizing and complementing both trophoblastic and extraembryonic endoderm deficiencies (14), it remains to be determined if the yolk sac abnormalities of the Ets2A72/db1 embryos reflect an additional specific extraembryonic function for Ets2. However, the later death of Ets2db1/A72 embryos implies a rank order of Ets2 alleles: Ets2+/+ > Ets2A72/+ > Ets2db1/+ ≥ Ets2A72/A72 > Ets2A72/db1 > Ets2db1/db1.
TABLE 1.
Embryonic lethality of Ets2A72/db1
| Age | No. of Ets2A72/A72 × Ets2db1/+ mice
|
|||
|---|---|---|---|---|
| Ets2A72/+ | Ets2A72/db1 | Resorption | Total | |
| E8.5 | 7 | 8 | 0 | 15 |
| E10.5 | 6 | 11 | 1 | 18 |
| E11.5 | 6 | 4 | 5 | 15 |
| E12.5 | 11 | 0 | 12 | 23 |
| Adult | 33 | 0 | 33 | |
Adult Ets2A72/A72 mice appeared normal with respect to fertility and longevity. They also did not develop the hair abnormalities found in rescued Ets2db1/db1 mice. Histological analysis of 50 organs of three Ets2A72/A72 adults did not reveal abnormalities except for a single lung adenoma, which is commonly found in FVB/N mice (data not shown). Mammary gland development of Ets2A72/A72 females was normal and not distinguishable from that of wild-type or heterozygous females (data not shown).
Restriction of PyMT and Neu mammary tumors by limited Ets2 activity.
To test the effect of the Ets2A72 allele on mammary tumors, we generated homozygous and heterozygous Ets2A72 females which also carried the MMTV-PyMT transgene (25). At 90 days, mammary tumors arising in the fat pads of Ets2A72/A72 female mice were less than half the size of those of control animals with one wild-type Ets2 allele (Table 2). This restriction on the development of PyMT tumors in Ets2A72/A72 mice resembled the previous observation of haploinsufficiency of Ets2db1/+;PyMT tumors (38). However, the previous experiment was performed on a mixed, outbred genetic background. On an FVB/N background, the degree of restriction by Ets2A72/A72 was greater than that of Ets2db1/+ (data not shown). However, the cellular composition and organization of tumors arising in different Ets2 genotypes were very similar and characteristic for these tumors (data not shown) (25).
TABLE 2.
Ets2A72/A72 restriction of mammary tumors
| Mouse group and characteristic | Ets2A72/A72 | Ets2A72/+ | Ets2+/+ |
|---|---|---|---|
| MMTV-PyMT at 90 days | |||
| No. of animals | 6 | 12 | |
| Avg tumor wt (g) | 0.62 | 1.6 | |
| SE | 0.12 | 0.2 | |
| MMTV-PyMTY315,322F at 133 days | |||
| No. of animals | 10 | 10 | 6 |
| No. hyperplasia positive | 10 | 10 | 6 |
| No. of tumor-bearing micea | 3 | 10 | 6 |
| No. of tumors/tumor-bearing mouse | 1.3 | 3.1 | 3.3 |
Masses with diameters of 3 mm or greater were counted as tumors.
PyMT signals through both Shc and PI3′K pathways. Mutation of PyMT tyrosines 315 and 322 results in defective activation of phosphatidylinositol 3′-kinase (PI3′K) by PyMT but retention of signaling through the Shc adapter proteins and Src (11, 56). Transgenic expression of a mutant form of PyMT (PyMTY315,322F) results in highly apoptotic, cystic tumors, the phenotype of which can be complemented by forced expression of Akt, a downstream effector of PI3′K (29, 56). As Ets2 might be expected to act primarily downstream of Shc and the Ras pathway, the degree of mammary tumor restriction by limited Ets2 activity might be expected to be even greater in tumors arising from PyMTY315,322F. The Ets2A72 allele was combined with the MMTV-PyMTY315,322F transgene. As expected, tumor development was severely restricted in PyMTY315,322F;Ets2A72/A72 mice compared to PyMTY315,322F mice containing wild-type Ets2 or only one EtsA72 allele (Table 2). At 133 days, only 30% of bigenic PyMTY315,322F;Ets2A72/A72 mice developed tumors (χ2 analysis, P < 0.0001). In addition, fewer tumors were found in those mice that did develop tumors. Thus, limited Ets2 activity restricted the formation of PyMT tumors with deficient PI3′K signaling.
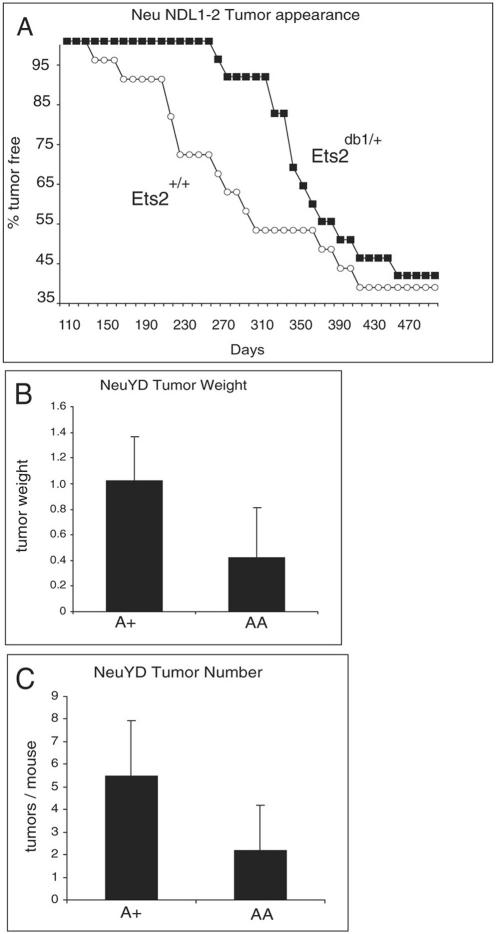
The PyMT oncogene utilizes the same signal transduction pathways as the Neu EGFR family member implicated in human breast cancer. To determine if Ets2 is also important in Neu tumor development, we evaluated the effect of Ets2 on tumor appearance in the MMTV-NeuNDL1-2 mice (Fig. 3A). The MMTV-NeuNDL1-2 line expresses a form of Neu which is activated by a deletion in the extracellular domain (48). Tumor appearance was delayed in animals heterozygous for the Ets2db1 allele. However, restricting Ets2 activity did not alter the penetrance of tumor formation that occurred in approximately 60% of the NeuNDL1-2 animals. Because of the long latency and incomplete penetrance of the NeuNDL1-2 model, the impact of Ets2A72 was evaluated further in the MMTV-NeuYD mouse model (12). In this model signaling by Neu is restricted by mutation of four of five tyrosines implicated in signaling. A single remaining Shc binding site (tyrosine-1222) is sufficient for tumor formation in all transgenic females within about 20 weeks. MMTV-NeuYD females developed fewer and smaller tumors at an age of 20 weeks in combination with Ets2A72 (Fig. 3B and C). These results show that activation of Ets2 through Thr-72 supports the development of mammary tumors caused by oncogenic Neu-Erbb2. The restriction of mammary tumors by limiting Ets2 activity is not confined only to tumors arising as the consequence of PyMT expression and implies a similar function downstream of common signal transduction pathways utilized by PyMT and Neu (13).
FIG. 3.
An Ets2 restriction of Neu mammary tumors. (A) Female NeuNDL1-2 animals with the indicated Ets2 genotype were examined for tumors three times weekly by palpation. (B and C) Six female NeuYD mice either heterozygous (A+) or homozygous (AA) for Ets2A72 were sacrificed at 20 weeks of age. The average weight (grams) of the largest tumor of each animals is shown in panel B, and the average number of tumors per mouse is shown in panel C. Error bars indicate standard deviations. Two-tailed t test, P = 0.034 for panel B and 0.027 for panel C.
Ets2 acts downstream of VEGF.
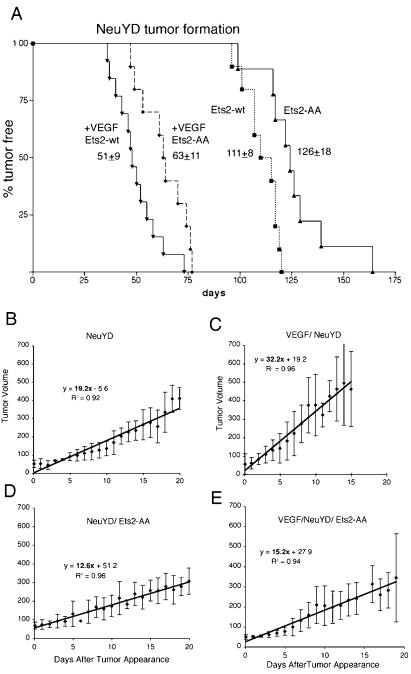
To test whether Ets2 regulates mammary tumors by an angiogenic mechanism, we utilized transgenic mice which overexpress mouse VEGF164 in mammary epithelial cells. The MMTV-VEGF-25 (VEGF-25) line expresses approximately 25-fold-higher VEGF levels in virgin mammary gland without altering mammary gland development. However, tumor formation in NeuYD;VEGF-25 bigenic animals was accelerated and was associated with greatly increased vascularity of hyperplastic and tumor tissues (40a). The Ets2A72 gene was combined with the NeuYD and VEGF-25 transgenes to evaluate tumor appearance. Ets2A72/A72 restricted the time of appearance of NeuYD tumors as expected from the measurements of tumor number and size at a single time point (Fig. 3B and C and 4A; log rank test, P ≤ 0.004). The average time of tumor appearance was delayed 15 days in Ets2A72/A72;NeuYD bigenic females. However, once the tumors appeared, the growth rates of the two types of tumors were similar (Fig. 4B and D; note similar tumor volumes at 15 days for both genotypes) for the first 15 days.
FIG. 4.
Ets2-dependent restriction of VEGF-25-accelerated NeuYD tumor appearance. A comparison of the time of tumor detection in NeuYD female mice (Ets2-wt, squares) and NeuYD female mice homozygous for Ets2A72 (Ets2-AA, triangles) with bigenic NeuYD;VEGF-25 female mice (+VEGF Ets2-wt, inverted triangles) or NeuYD;VEGF-25 mice homozygous for Ets2A72 (+VEGF Ets2-AA, diamonds, broken line) is shown as a function of age. The averages and standard deviations of the time of first detection are indicated. Data from NeuYD animals and bigenic NeuYD;VEGF-25 animals have been submitted for publication elsewhere (40a) and are presented here for comparison to the Ets2A72/A72;VEGF-25 animals. Log rank tests of survival plots indicated a statistically significant difference between Ets2+/+;NeuYD;VEGF-25 and Ets2A72/A72;NeuYD;VEGF-25 tumor appearance (P ≤ 0.03) and between Ets2+/+;NeuYD and Ets2A72/A72;NeuYD tumor appearance (P ≤ 0.025). (B to E) Tumor growth. Tumor size was measured with calipers, and the volume was estimated (length × weight2/2). Average tumor volumes are plotted as a function of elapsed time after first detection. Error bars indicate the standard deviations. Linear curve fit equations with correlation coefficients are shown for ease of comparison of growth rates. Linear functions fit all sets of data better than exponential curve fit except for panel B.
If the Ets2 restriction of mammary tumors was due to a function of Ets2 in regulating VEGF production, transgenic VEGF expression might be expected to bypass an Ets2-sensitive restriction. NeuYD tumors are restricted by angiogenesis because transgenic expression of VEGF in the mammary epithelium dramatically accelerates tumor formation coincident with increased vascularization (40a) (Fig. 4A). However, Ets2A72/A72;NeuYD;VEGF-25 trigenic females developed tumors more slowly (63 days) than did control Ets2+/+;NeuYD;VEGF-25 animals (Fig. 4A) (log rank test, P ≤ 0.008). In addition, the growth of NeuYD;VEGF tumors was dramatically diminished by limited Ets2 activity (Fig. 4C and E). However, the CD31-positive blood vessel density of Ets2-restricted NeuYD;VEGF-25 tumors was not distinguishable from that of control NeuYD;VEGF-25 tumors (data not shown). Furthermore, the Ets2A72/A72 genotype did not have lower levels of endothelial markers CD31, Flk-1, and VE-cadherin and T-cadherin RNAs in these tumors, although all of the endothelial markers were elevated in comparison to NeuYD tumor without transgenic VEGF (data not shown) (40a). Thus, limiting Ets2 activity restricts the appearance of both NeuYD tumors (Fig. 4A; log rank test, P ≤ 0.004) and highly vascularized, NeuYD;VEGF-25 tumors. Ets2A72/A72 did not restrict the increased vascularity caused by forced VEGF expression but still restricted tumor appearance and growth. Western blots of NeuYD;VEGF-25 tumors with differing Ets2 genotypes revealed similar levels of Neu, Akt, and Grb2 proteins (data not shown). Thus, the Ets2 restriction on VEGF-accelerated tumors was not due to modulation of Neu expression or the parallel stimulation of the Akt signaling pathway.
Increased p21Cip1 in Ets2-restricted tumors.
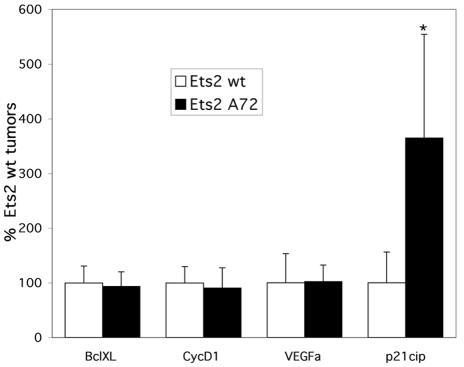
Expression of several putative Ets2 target genes whose products could influence tumor growth was assayed in the NeuYD;VEGF tumors containing either wild-type Ets2 or Ets2A72. These target genes included cyclin D1 and Bcl-xL, both reported to be Ets2 responsive (1, 46); p21Cip1, a reported target of the E1AF Ets factor (21); and VEGF, whose expression in human breast tumors has been correlated with Ets1 expression levels (50). Figure 5 shows the results of real-time PCR quantitation of the relative gene expression. While there was not a significant effect of the tumor Ets2 genotype on the expression of CycD1, Bcl-xL, or endogenous VEGF, there was a 3.6-fold-higher level of p21Cip1 expression in the Ets2A72/A72 tumors, which was highly significant (P = 0.008). The increased p21Cip1 expression is consistent with the decreased growth rate of the Ets2-limited tumors.
FIG. 5.
Increased p21Cip1 expression in Ets2-restricted bigenic tumors. Expression of the indicated genes in NeuYD;VEGF tumors containing either wild-type Ets2 or Ets2A72/A72 was analyzed by quantitative reverse transcription-PCR. The cDNA levels for each sample were normalized to Cph (Ppia) expression. The percent expression shown for each gene is relative to the level of the average expression of the same gene in the wild-type Ets2 tumors. Results shown are the averages and standard deviations from duplicate experiments from at least two different tumors of each genotype.
A stromal function for Ets2 in mammary tumors.
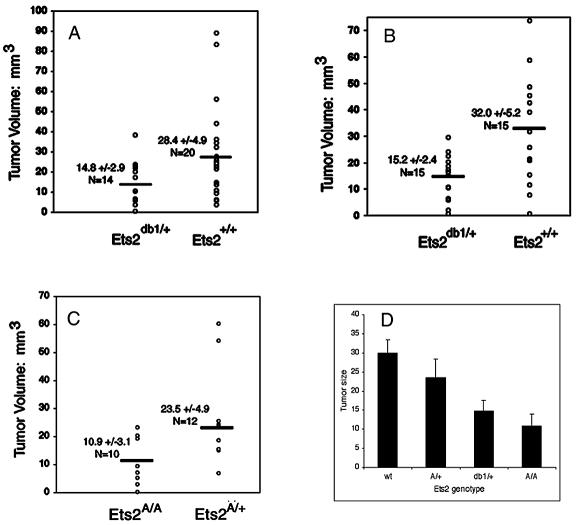
Ets2 may influence tumors in either epithelial or stromal supporting cells. The stromal contribution of the Ets2-sensitive restriction of PyMTY315,322F tumors was tested by tumor transplantation. A well-characterized, serially transplantable tumor line from a PyMTY315,322F tumor (PyMTY315,322F-DB7) (8, 56) was surgically inoculated bilaterally into mammary fat pads of adult female Ets2db1/+ and Ets2+/+ FVB/N strain mice. After 8 or 9 days, the fat pads were excised and the tumor size was measured. Figure 6A and B shows average examples of tumor growth in the two Ets2 genotypes. All 34 transplants (20 in Ets2+/+ mice and 14 in Ets2db1/+ mice) resulted in tumors. No correlation was found between tumor size and age or weight of the hosts (data not shown). Tumors that arose in wild-type animals were frequently larger than those in Ets2db1/+ mice (Table 3; Fig. 7A). The average tumor arising in Ets2db1/+ mice was only 52% of the volume of those in wild-type animals. The difference between the two populations was statistically significant (Student t test, P = 0.02).
TABLE 3.
Ets2 dependence of transplanted PyMTY315,322F-DB7 tumor size in Ets2+/+ and Ets2db1/+ mice
| Tumor type |
Ets2+/+
|
Ets2db1/+
|
|||
|---|---|---|---|---|---|
| Size (mm3)a | n | Size (mm3)a | n | Pb | |
| Mammary pad (expt 1) | 28.4 ± 4.9 | 14 | 14.8 ± 2.8 | 20 | 0.021 |
| Mammary pad (expt 2) | 32.0 ± 45.2 | 15 | 15.2 ± 2.4 | 15 | 0.008 |
| Combined | 29.9 ± 3.5 | 29 | 15.0 ± 3.5 | 37 | 0.0004 |
| Subcutaneous | 4.8 ± 0.9 | 9 | 5.3 ± 0.9 | 7 | 0.67 |
| Lymph node | 9.7 ± 0.9 | 16 | 10.0 ± 0.8 | 16 | 0.85 |
Values are means ± standard errors of the means.
t test, two sample, assuming unequal variance. The difference between means of the compared genotypes is not significantly different for subcutaneous transplants or for lymph nodes.
FIG. 7.
PyMTY315,322F tumor size depends on the Ets2 genotype of the host. Tumors were measured in digital images of whole-mount mammary glands prepared 9 days after transplant of 1-mm3 pieces of the PyMTY315,322F-DB7 serially transplanted tumor line. (A and B) Individual values of two replicate experiments using Ets2+/+ and Ets2db1/+ female hosts. (C) Results of transplantation into Ets2A72/+ and Ets2A72/A72 hosts. (D) Average sizes and variation in different Ets2 genotype hosts.
A second trial was performed on eight animals of each genotype in which the animals were all sacrificed at 9 days (Table 3; Fig. 7B). In order to test the importance of the mammary gland environment, six wild-type animals and four Ets2db1/+ animals of the second transplant series also received bilateral subcutaneous transplants. Only one mammary transplant of each series failed to generate a tumor. Both the volumes and variability of the mammary tumors were very similar to those in the first trial. The average tumor volume of Ets2db1/+ mice was only 47.5% of that for Ets2+/+ animals (P = 0.008). Combined, the average volume of PyMTY315,322F tumors arising in Ets2db1/+ mice was 50% of that of tumors growing in animals with two wild-type Ets2 alleles (P = 0.0004). The volume of the lymph node was not significantly different between genotypes.
To determine if activation of Ets2 through Thr-72 phosphorylation is involved in the stromal support of transplanted tumors, we performed the transplant experiment in Ets2A72/A72 and Ets2A72/+ mice. We found that Ets2A72/A72 hosts had an average tumor volume 54% smaller than that of Ets2A72/+ mice (P = 0.04) (Table 4; Fig. 7C). Previously, on the basis of the duration of embryonic development, we deduced a rank order of the Ets2db1 and Ets2A72 alleles. Transplanted tumor size in all trials decreased with decreasing Ets2 activity of the host (Fig. 7D).
TABLE 4.
Ets2 dependence of transplanted PyMTY315,322F-DB7 tumor size in Ets2A72/+ and Ets2A72/A72 mice
| Tumor type |
Ets2A72/+
|
Ets2A72/A72
|
|||
|---|---|---|---|---|---|
| Size (mm3)a | n | Size (mm3)a | n | Pb | |
| Mammary pad | 23.5 ± 4.9 | 12 | 10.9 ± 3.0 | 8 | 0.043 |
| Lymph node | 7.7 ± 0.7 | 12 | 7.3 ± 1.0 | 8 | 0.73 |
Values are means ± standard errors of the means.
t test, two sample, assuming unequal variance. The difference between means of the compared genotypes is not significantly different for lymph nodes.
Limiting Ets2 does not restrict subcutaneous tumor growth.
Subcutaneous transplants of PyMTY315,322F-DB7 tumors grew significantly slower than did identically sized transplants placed in the mammary fat pad (Table 3). The Ets2 genotype had no significant impact upon subcutaneous tumor growth after 9 days. This result is consistent with previous observations that mammary tumor growth can be profoundly influenced by the site of implantation (37). The Ets2-sensitive restriction on mammary tumor growth is site specific and may involve a stimulatory effect of the mammary stroma.
MMP-9 and factor VIII expression in transplanted tumors depends on Ets2.
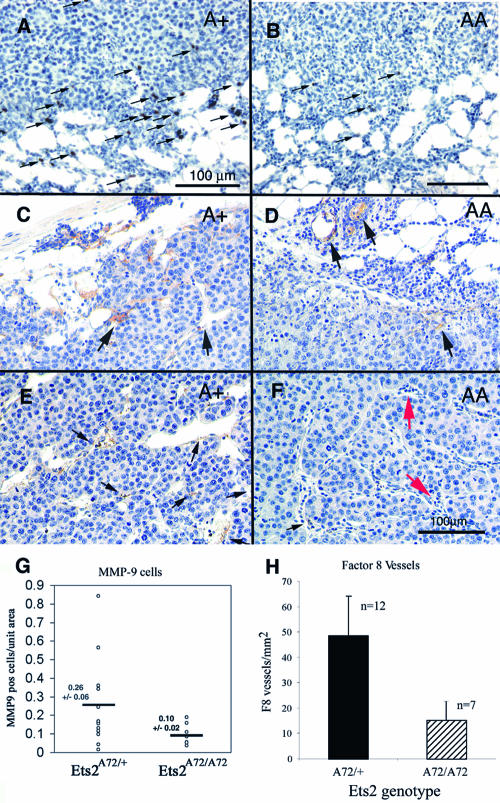
Histological analysis of transplants into Ets2A72/A72 and Ets2A72/+ mice revealed that transplanted PyMTY315,322F tumors resembled those previously described (Fig. 6C and D) (8). Tumors were poorly differentiated carcinomas with a solid growth pattern. Mitotic cells were visible, as were areas of necrosis. Tumors had a significant host inflammatory reaction to the transplants (Fig. 6C and D). Inflammatory cells and fibroblastic proliferation were evident in the mammary fat pads of all genotypes. Antibody staining of macrophages with the F4/80 antibody revealed significant macrophage presence within the tumor and in the tumor periphery (Fig. 6E and F). However, quantitation of total F4/80 staining did not reveal a significant difference between Ets2 genotypes (data not shown).
While the total infiltration of the transplanted tumors by macrophages was not distinguishable by F4/80 staining, a subset of cells that expressed elevated MMP-9 concentrated near the tumor-host interface (Fig. 8A and B). More MMP-9-positive cells were found in Ets2A72/+ hosts than in Ets2A72/A72 hosts (Fig. 8A, B, and G). These MMP-9-positive cells may represent a subset of macrophages, other inflammatory cells, or reactive fibroblasts. As stromal expression of MMP-9 has been implicated in making VEGF available during the angiogenic switch of transgenic pancreatic cancer, we evaluated the vasculature of the transplanted tumors by staining tumor sections with antibody to the endothelial marker factor VIII-related antigen. Neither CD31 nor Flk-1 antibody reactions were compatible with the preparation of these specimens. Mature vessels within the fat pads and at the tumor-stromal interface of the tumors of both Ets2 genotypes were clearly reactive with factor VIII antibody (Fig. 8C and D). However, reactivity was greatly reduced within the body of the tumors of Ets2A72/A72 hosts (Fig. 8E and F) in spite of typical vessel-like structures and the general absence of necrotic areas. Quantitation of factor VIII antibody-reactive vessels within the body of the tumors revealed a greater-than-threefold difference between Ets2A72 homozygous and heterozygous hosts (Fig. 8H). These results suggest a difference in vascular density or maturation.
FIG. 8.
MMP-9 and factor VIII-reactive vasculature is Ets2 dependent. (A and B) Representative fields of transplanted PyMTY315,322F-DB7 tumor sections stained with MMP-9 antibody and hematoxylin. The Ets2 genotype of the host is indicated. Arrows indicate representative positive reactive cells near the tumor-stromal interface. (C to F) Immunohistochemical detection of vessels expressing factor VIII-related antigen. (C and D) Areas at the interface between the tumor and stroma. (E and F) Representative fields within the tumor. Black arrows indicate representative positive areas. Red arrows in panel F show putative vessels not stained with factor VIII antibody. The bar in panel F represents 100 μm and is applicable to panels C to F. (G) The numbers of MMP-9-positive cells were counted. Each value represents the average of one tumor transplant section. The averages and standard errors are shown. (H) Factor VIII-positive vessel density was determined by counting independent fields away from the tumor edge. The averages and standard deviations are shown.
Ets2 regulates macrophage MMP-9 and MMP-3.
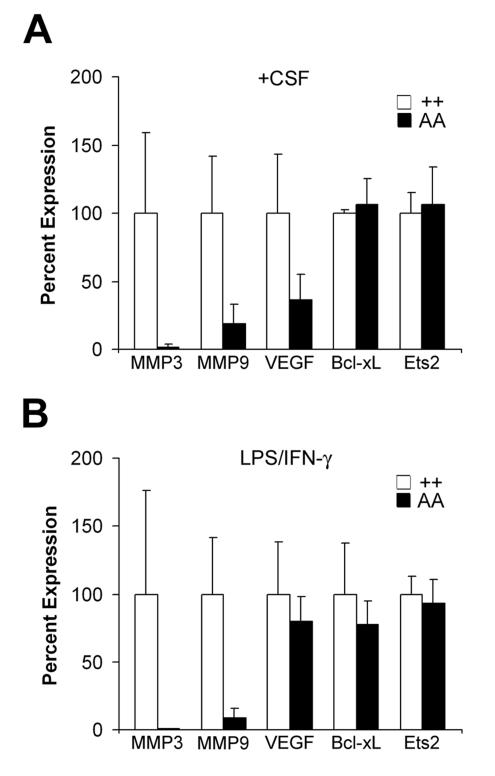
The stromal action of Ets2 and the invasion of transplanted tumors by macrophages stimulated consideration of Ets2-dependent gene expression in isolated macrophages. Macrophages were isolated from bone marrow and activated by exposure to either CSF-1 alone or the combination of IFN-γ and LPS. Ets2A72/A72 macrophages contained dramatically lower levels of MMP-9 and MMP-3 RNAs than did controls after stimulation with either condition (Fig. 9). However, Ets2 and Bcl-xL RNA levels did not differ between Ets2 genotypes (Fig. 9). VEGF RNA levels were lower in Ets2A72/A72 macrophages stimulated with CSF-1 but did not differ significantly after activation with LPS and IFN-γ (Fig. 9). These results show that the induction of MMP-9 and MMP-3 expression is highly dependent on the kinase activation of Ets2.
FIG. 9.
Ets2 regulates MMP-3 and MMP-9 in macrophages. The indicated RNAs derived from BMMs of Ets2+/+ and Ets2A72/A72 animals were measured by quantitative PCR. (A) Seven-day primary macrophage cultures were treated with CSF-1; (B) combination of LPS and IFN-γ. Values represent the averages of three different cell isolates normalized to Cph expression. Target genes were assayed in triplicate for each condition. Error bars represent standard deviations of the averages of three samples.
DISCUSSION
Ets2 activity is modulated by phosphorylation of Thr-72 in vivo.
The developmental defects caused by Ets2db1/db1 and Ets2A72/db1 but not Ets2db1/+ or Ets2A72/A72 provided a basis for deducing the rank order of the Ets2 alleles: Ets2+/+ > Ets2A72/+ > Ets2db1/+ > Ets2A72/A72 > Ets2A72/db1 > Ets2db1/db1. As the A72 targeted mutation does not alter the expression of Ets2A72 RNA or the rapid turnover of Ets2A72 protein, the decreased Ets2 activity of Ets2A72 is due to the absence of the activating phosphorylation site. This confirms in vivo the importance of the Thr-72 residue of Ets2 as deduced in cell culture transfection analysis (60) but also demonstrates that the basal activity of Ets2 is functionally important. The developmental deficiencies caused by Ets2A72/db1 and Ets2db1/db1 are due to trophoblast malfunction because tetraploid embryo aggregation can rescue Ets2db1/db1 embryo development (59). The basal activity of the Ets2A72 allele permits development to proceed 3 days longer than Ets2db1/db1 to E11.5. The absence of embryonic blood cells in the Ets2A72/db1 placenta reflects the absence of the labyrinth layer due to an apparent failure of chorioallantoic fusion. This could be considered a vascular defect as the allantois is the source of the embryonic blood vessels which normally interdigitate with the maternal circulation in the placenta. However, the failure to form the labyrinth region may be secondary to a proliferation defect of trophoblast cells. A deficiency of trophoblast cell growth in response to factors such as FGF is consistent with the formation of giant cells, which form in the absence of continued FGF stimulation (52), and the absence of spongiotrophoblast and labyrinth layers.
Limited Ets2 restricts PyMT and Neu mammary tumors.
Haploinsufficiency of Ets2 restricts the development of PyMT (38) and NeuNDL1-2 mammary tumors. The similar restriction of PyMT, PyMTY315,322F, and NeuYD tumors by Ets2A72/A72 indicates that kinase activation of Ets2 through the Thr-72 residue is important in the development of these mammary tumors. One trivial explanation for the Ets2 restriction of tumor development in these models might be a direct or indirect Ets2 dependence of the MMTV promoter. There are four observations that make this unlikely. First, PyMTY315,322F RNA and Neu protein levels were similar in Ets2A72/+ and Ets2A72/A72 tumors and matched hyperplastic tissues. Second, subcutaneous tumor transplants grew at the same rate in Ets2A72 heterozygous and homozygous hosts. This suggests that circulating levels of tumor growth factors including hormones are likely similar in the two Ets2 genotypes. This conclusion is further reinforced by the normal hormone-dependent development of the mammary epithelium in Ets2A72/A72 females. Finally, transient-transfection analysis of MMTV promoter constructions did not reveal significant activation by Ets2 (data not shown).
NeuNDL and NeuYD both activate Shc-dependent signaling that leads to Ras and downstream kinase activation. Both also activate the PI3′K pathway by increasing Erbb3 protein levels and downstream signaling targets including Akt. While PyMT activates both Shc and PI3′K pathways directly, PyMTY315,322F requires a secondary event in order to progress to a tumor. EGFR signaling complements the defective PI3′K signaling impaired by the Y315,322F mutations (56). Ets2 has been implicated in mediating EGFR signaling because Ets2-deficient mice, rescued from early placental insufficiency, have the same hair phenotype as the hypomorphic EGFR mutation waved-2 (34), and in cultured fibroblasts, Ets2 mediates EGF transcriptional activation of MMP-3 (59). While it is clear that Ets2 can mediate the downstream signaling of activated Neu and EGFR, identification of the specific target cell(s) responsible for restricting mammary tumors is essential to understanding the molecular role of Ets2.
Ets2-dependent, stromal support of PyMTY315,322F tumors.
Ets2 deficiency in the stroma of the mammary fat pad restricts tumor growth. This Ets2 stromal restriction is specific to the mammary fat pad and was not manifest in subcutaneous positions. This restriction was revealed with the use of a transplantable tumor which generates lower vessel density and decreased vessel tortuosity as a consequence of the mutation of the PI3′K binding sites of PyMT (8). This vascular deficiency of the PyMTY315,322F-DB7 tumor cells may make them more dependent upon a stromal source of angiogenic activity. The decreased factor VIII-positive vessel density and MMP-9-positive cells associated with the Ets2A72/A72 hosts are consistent with a role of Ets2 in promoting host vascularization of the transplanted tumors. However, the absence of significant necrosis in transplanted Ets2A72/A72 host tumors and the presence of factor VIII-negative vessels suggest that Ets2 might be more important to the function of endothelial cells.
Ets2 regulates MMP activity in macrophages.
One of the stromal cell types which is clearly impacted by Ets2A72 is the macrophage. Again, Ets2 deficiency appears more important for the function than for the development of the target cells because Ets2 is not necessary for the development of macrophages (26, 59) or their invasion of transplanted tumors. However, isolated macrophages had dramatic deficiencies in the expression of both MMP-3 and MMP-9. MMP-3 has been implicated previously in mouse mammary tumorigenesis and the epithelial-to-mesenchymal transition of tumor cells (33, 51). Ets2-deficient fibroblasts are also defective in the induction of MMP-3 in response to either FGF or EGF (59). MMP-9 is expressed by fibroblasts, endothelial cells, and cells of hematopoietic origin, but generally not carcinoma cells (15). Ets2-deficient trophoblast cells express little MMP-9 (59). Ets transcription factors have been implicated in the EGF and Ras regulation of MMP-9 (24, 55). The MMP-9-positive cells in our tumor transplantation studies may correspond to a subpopulation of macrophages, eosinophils, neutrophils, or reactive fibroblasts.
The phosphorylation of Ets2 has been implicated previously in CSF-1 stimulation of macrophages (19, 49). This is consistent with an important role of Ets2 in macrophage function, although a specific role for Ets2 in the regulation of Bcl-xL, another reported Ets2 target gene, was not found. Macrophages have a significant effect on PyMT tumor progression (32). However, the Ets2A72/A72 restriction of PyMT tumors appears more severe than that imposed by the deficiency of macrophages in the Csf1op/op mice. Thus, it is difficult to ascribe all of the Ets2 restriction of mammary tumors to defective macrophage function.
Ets2 acts downstream of increased vascular density.
Several observations suggest a possible role for Ets2 in tumor angiogenesis or endothelial function. First, VEGF RNA expression was lower in Ets2A72/A72 macrophages stimulated by CSF-1 (but not LPS and IFN-γ). Second, MMP-9 from inflammatory cells has been previously implicated in making VEGF available for the progression of pancreatic cancer (5). Third, factor VIII staining of the tumor blood vessels of transplanted PyMTY315,322F mice distinguished Ets2-deficient and normal hosts. Finally, the developmental defect of Ets2A72/db1 concepti could be considered a vascular defect. Clearly, the transgenic expression of VEGF164 overcomes a major restriction on NeuYD tumor progression. However, Ets2 deficiency still restricted VEGF-accelerated NeuYD tumors. Thus, Ets2 does not limit tumors by regulating VEGF production by epithelial cells. In addition, Ets2 does not likely regulate the mitogenic response of endothelial cells to VEGF because Ets2A72/A72;NeuYD;VEGF-25 tumors had similar vessel density as those with wild-type Ets2. Despite the similar tumor vessel density, Ets2A72/A72;NeuYD;VEGF-25 tumors grew significantly more slowly. This Ets2 dependence in tumor growth rate was revealed only after accelerating the appearance and growth of tumors by forced VEGF expression. The growth rates of both PyMT and NeuYD tumors without transgenic VEGF are not dependent on Ets2 genotype even though the time of tumor onset is Ets2 sensitive (38). The elevated level of the cell cycle progression inhibitor p21Cip1 in Ets2A72/A72;NeuYD;VEGF-25 tumors is consistent with the observed lower tumor growth rate. However, p21Cip1 is activated by strong stimulation of the Ras-Raf signaling pathway (58) and is increased in response to overexpression of the E1AF Ets transcription factor (21). The elevated p21Cip1 expression in the Ets2A72/A72 tumors would not be expected for an Ets2 loss-of-function mutation, unless Ets2 acts by a negative mechanism (for example, see reference 2). Alternatively, elevated p21Cip1 expression may reflect a secondary effect of slower growth of the tumors caused by limited Ets2 activity. The Ets2 restriction on VEGF-accelerated tumors is likely distinct from the effects seen on vascularly restricted PyMT and NeuYD tumors. The role of Ets2 in endothelial cell function will be of great future interest.
Ets2 acts downstream of signaling pathways stimulated by FGF and EGF (59), Erbb2 (23), Ras (22), Raf (36), and Erk (19, 44). However, the functional importance of Ets2 in mediating signaling through the Ras pathway in vivo is dependent upon the specific cell type. The Ets2-dependent stromal support of tumors does not rule out additional effects of Ets2 in mammary epithelial cells. Three other Ets transcription factors, PEA3, ESX/Elf-3, and Elf-1, have been implicated in mammary tumors (4, 47, 53), normal mammary differentiation, and Erbb2 regulation (43), respectively. Furthermore, increased phosphorylation of Ets2 on Thr-72 is found in ovarian cancer cell lines (41). Given the similarities in DNA-binding specificities of multiple Ets members, selective inactivation of individual Ets members in vivo provides the advantage of dissecting the specific role of individual Ets factors. Gene targeting has now implicated Ets2 in placenta development, hair morphogenesis, and mammary tumor progression. Ets2 may mediate growth factor signaling in all these cases.
Ets2 is necessary for embryonic development and for optimal growth of tumors, but in both cases it may act primarily in the support of the embryo or tumor rather than in the embryo or tumor itself. It will be necessary to compare the relative contributions of epithelial and endothelial cells, macrophages, and other supporting cell types and appropriate growth factors to understand fully the multiple molecular roles that Ets2 may play during mammary tumorigenesis. The sensitivity of mammary tumors to Ets2 activity stimulates consideration of whether genetic variability of Ets2 activity may modify human breast cancer disease.
Acknowledgments
This work was supported by grants from the National Cancer Institute (CA74547 and CA98778 to R.G.O.; CA57621 and CA72006 to Z.W.) and a Cancer Center support grant (CA30199) to the Burnham Institute; Department of Defense Breast Cancer Research Program grants (DAMD17-01-1-0170 and DAMD17-00-1-0175 to R.G.O.), the Canadian Breast Cancer Initiative (W.J.M.), an NIH Predoctoral Training Grant (T32-CA77109) in the support of A.K.M. and J.T.; and a California Breast Cancer Fellowship to M.E.
We thank Grace Cecena and Robbin Newlin for excellent technical assistance, Robert J. Munn for his excellent digital photoimaging, and Abraham Gomez for care and attention to mouse husbandry.
REFERENCES
- 1.Albanese, C., J. Johnson, G. Watanabe, N. Eklund, D. Vu, A. Arnold, and R. G. Pestell. 1995. Transforming p21ras mutants and c-ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 270:23589-23597. [DOI] [PubMed] [Google Scholar]
- 2.Baker, K. M., G. Wei, A. E. Schaffner, and M. C. Ostrowski. 2003. Ets-2 and components of mammalian SWI/SNF form a repressor complex that negatively regulates the BRCA1 promoter. J. Biol. Chem. 278:17876-17884. [DOI] [PubMed] [Google Scholar]
- 3.Behrendtsen, O., C. M. Alexander, and Z. Werb. 1992. Metalloproteinases mediate extracellular matrix degradation by cells from mouse blastocyst outgrowths. Development 114:447-456. [DOI] [PubMed] [Google Scholar]
- 4.Benz, C. C., R. C. O'Hagan, B. Richter, G. K. Scott, C.-H. Chang, X. Xiong, K. Chew, B.-M. Ljung, S. Edgerton, A. Thor, and J. A. Hassell. 1997. HER2/Neu and the ets transcription activator PEA3 are coordinately upregulated in human breast cancer. Oncogene 15:1513-1525. [DOI] [PubMed] [Google Scholar]
- 5.Bergers, G., R. Brekken, G. McMahon, T. H. Vu, T. Itoh, K. Tamaki, K. Tanzawa, P. Thorpe, S. Itohara, Z. Werb, and D. Hanahan. 2000. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat. Cell Biol. 2:737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bissell, M. J., and D. Radisky. 2001. Putting tumours in context. Nat. Rev. Cancer 1:46-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulukos, K. E., P. Pognonec, E. Sariban, M. Bailly, C. Lagrou, and J. Ghysdael. 1990. Rapid and transient expression of Ets2 in mature macrophages following stimulation with CMGF, LPS, and PKC activators. Genes Dev. 4:401-409. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, A. T. W., L. J. T. Young, P. C. Y. Chen, C. Y. Chao, A. Ndoye, P. A. Barry, W. J. Muller, and R. D. Cardiff. 1997. Microcirculation and metastasis in a new mouse mammary tumor model system. Int. J. Oncol. 11:69-77. [PubMed] [Google Scholar]
- 9.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 10.Coussens, L. M., C. L. Tinkle, D. Hanahan, and Z. Werb. 2000. MMP-9 supplied by bone marrow-derived cells contributes to skin carcinogenesis. Cell 103:481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahl, J., R. Freund, J. Blenis, and T. L. Benjamin. 1996. Studies of partially transforming polyomavirus mutants establish a role for phosphatidylinositol 3-kinase in activation of pp70 S6 kinase. Mol. Cell. Biol. 16:2728-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dankort, D., B. Maslikowski, N. Warner, N. Kanno, H. Kim, Z. Wang, M. F. Moran, R. G. Oshima, R. D. Cardiff, and W. J. Muller. 2001. Grb2 and Shc adapter proteins play distinct roles in Neu (ErbB-2)-induced mammary tumorigenesis: implications for human breast cancer. Mol. Cell. Biol. 21:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dankort, D. L., and W. J. Muller. 2000. Signal transduction in mammary tumorigenesis: a transgenic perspective. Oncogene 19:1038-1044. [DOI] [PubMed] [Google Scholar]
- 14.Duncan, S. A., A. Nagy, and W. Chan. 1997. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of Hnf-4−/− embryos. Development 124:279-287. [DOI] [PubMed] [Google Scholar]
- 15.Egeblad, M., and Z. Werb. 2002. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2:161-174. [DOI] [PubMed] [Google Scholar]
- 16.Elenbaas, B., and R. A. Weinberg. 2001. Heterotypic signaling between epithelial tumor cells and fibroblasts in carcinoma formation. Exp. Cell Res. 264:169-184. [DOI] [PubMed] [Google Scholar]
- 17.Foos, G., J. J. Garcia-Ramirez, C. K. Galang, and C. A. Hauser. 1998. Elevated expression of Ets2 or distinct portions of Ets2 can reverse Ras-mediated cellular transformation. J. Biol. Chem. 273:18871-18880. [DOI] [PubMed] [Google Scholar]
- 18.Foos, G., and C. A. Hauser. 2000. Altered Ets transcription factor activity in prostate tumor cells inhibits anchorage-independent growth, survival, and invasiveness. Oncogene 19:5507-5516. [DOI] [PubMed] [Google Scholar]
- 19.Fowles, L. F., M. L. Martin, L. Nelsen, K. J. Stacey, D. Redd, Y. M. Clark, Y. Nagamine, M. McMahon, D. A. Hume, and M. C. Ostrowski. 1998. Persistent activation of mitogen-activated protein kinases p42 and p44 and ets-2 phosphorylation in response to colony-stimulating factor 1/c-fms signaling. Mol. Cell. Biol. 18:5148-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujiwara, S., R. J. Fisher, N. K. Bhat, S. M. D. Espina, and T. S. Papas. 1988. A short-lived nuclear phosphoprotein encoded by the human ets-2 proto-oncogene is stabilized by activation of protein kinase C. Mol. Cell. Biol. 8:4700-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Funaoka, K., M. Shindoh, K. Yoshida, M. Hanzawa, K. Hida, S. Nishikata, Y. Totsuka, and K. Fujinaga. 1997. Activation of the p21Waf1/Cip1 promoter by the ets oncogene family transcription factor E1AF. Biochem. Biophys. Res. Commun. 236:79-82. [DOI] [PubMed] [Google Scholar]
- 22.Galang, C. K., C. J. Der, and C. A. Hauser. 1994. Oncogenic Ras can induce transcriptional activation through a variety of promoter elements, including tandem c-ets-2 binding sites. Oncogene 9:2913-2921. [PubMed] [Google Scholar]
- 23.Galang, C. K., J. J. Garcia-Ramirez, P. A. Solski, C. J. Der, N. N. Neznanov, R. G. Oshima, and C. A. Hauser. 1996. Oncogenic neu/ErbB-2 increases ets, AP-1, and NF-kB-dependent gene expression, and inhibiting ets activation blocks Neu-mediated cellular transformation. J. Biol. Chem. 271:7992-7998. [DOI] [PubMed] [Google Scholar]
- 24.Gum, R., E. Lengyel, J. Juarez, J. H. Chen, H. Sato, M. Seiki, and D. Boyd. 1996. Stimulation of 92-kDa gelatinase B promoter activity by ras is mitogen-activated protein kinase kinase 1-independent and requires multiple transcription factor binding sites including closely spaced PEA3/ets and AP-1 sequences. J. Biol. Chem. 271:10672-10680. [DOI] [PubMed] [Google Scholar]
- 25.Guy, C. T., R. D. Cardiff, and W. J. Muller. 1992. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12:954-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henkel, G. W., S. R. McKercher, H. Yamamoto, K. L. Anderson, R. G. Oshima, and R. A. Maki. 1996. PU.1 but not ets-2 is essential for macrophage development from ES cells. Blood 88:2917-2926. [PubMed] [Google Scholar]
- 27.Hever, A., R. G. Oshima, and C. A. Hauser. 2003. Ets2 is not required for Ras or Neu/ErbB-2 mediated cellular transformation in vitro. Exp. Cell Res. 290:132-143. [DOI] [PubMed] [Google Scholar]
- 28.Hogan, B., F. Costantini, and E. Lacy. 1986. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Hutchinson, J., J. Jin, R. D. Cardiff, J. R. Woodgett, and W. J. Muller. 2001. Activation of Akt (protein kinase B) in mammary epithelium provides a critical cell survival signal required for tumor progression. Mol. Cell. Biol. 21:2203-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karim, F. D., L. D. Urness, C. S. Thummel, M. J. Klemsz, S. R. McKercher, A. Celada, C. VanBeveren, and R. A. Maki. 1990. The ets domain: a new DNA binding motif that recognizes a purine-rich core DNA sequence. Genes Dev. 4:1451-1453. [DOI] [PubMed] [Google Scholar]
- 31.Langer, S. J., D. M. Bortner, M. F. Roussel, C. J. Sherr, and M. C. Ostrowski. 1992. Mitogenic signaling by colony-stimulating factor 1 and ras is suppressed by the ets-2 DNA-binding domain and restored by myc overexpression. Mol. Cell. Biol. 12:5355-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, E. Y., A. V. Nguyen, R. G. Russell, and J. W. Pollard. 2001. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 193:727-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lochter, A., S. Galosy, J. Muschler, N. Freedman, Z. Werb, and M. J. Bissell. 1997. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J. Cell Biol. 139:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luettke, N. C., H. K. Phillips, T. H. Qiu, N. G. Copeland, H. S. Earp, N. A. Jenkins, and D. C. Lee. 1994. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev. 8:399-413. [DOI] [PubMed] [Google Scholar]
- 35.Mansour, S. L., K. R. Thomas, and M. R. Capecchi. 1988. Disruption of the proto-oncogene Int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature 336:348-352. [DOI] [PubMed] [Google Scholar]
- 36.McCarthy, S. A., D. Chen, B.-S. Yang, J. J. Garcia Ramirez, H. Cherwinski, X.-R. Chen, M. Klagsbrun, C. A. Hauser, M. C. Ostrowski, and M. McMachon. 1997. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol. Cell. Biol. 17:2401-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, F. R., D. Median, and G. H. Heppner. 1981. Preferential growth of mammary tumors in intact mammary fatpads. Cancer Res. 41:3863-3867. [PubMed] [Google Scholar]
- 38.Neznanov, N., A. K. Man, H. Yamamoto, C. A. Hauser, R. D. Cardiff, and R. G. Oshima. 1999. A single targeted Ets2 allele restricts development of mammary tumors in transgenic mice. Cancer Res. 59:4242-4246. [PubMed] [Google Scholar]
- 39.O'Gorman, S., N. A. Dagenais, M. Qian, and Y. Marchuk. 1997. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. USA 94:14602-14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Neill, E. M., I. Rebay, R. Tijan, and G. M. Rubin. 1994. The activities of two ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78:137-147. [DOI] [PubMed] [Google Scholar]
- 40a.Oshima, R. G., J. Lesperance, V. Munoz, L. Hebbard, B. Ranscht, N. Sharan, W. J. Muller, C. A. Hauser, and R. D. Cardiff. Angiogenic acceleration of Neu induced mammary tumor progression and metastasis. Cancer Res., in press. [DOI] [PubMed]
- 41.Patton, S. E., M. L. Martin, L. L. Nelsen, X. Fang, G. B. Mills, and M. C. Ostrowski. 1998. Activation of the ras-mitogen-activated protein kinase pathway and phosphorylation of ets-2 at position threonine 72 in human ovarian cancer cell lines. Cancer Res. 58:2253-2259. [PubMed] [Google Scholar]
- 42.Samakovlis, C., G. Manning, P. Steneberg, N. Hacohen, R. Cantera, and M. A. Krasnow. 1996. Genetic control of epithelial tube fusion during Drosophila tracheal development. Development 122:3531-3536. [DOI] [PubMed] [Google Scholar]
- 43.Scott, G. K., C. H. Chang, K. M. Erny, F. Xu, W. J. Fredericks, F. J. Rauscher, A. D. Thor, and C. C. Benz. 2000. Ets regulation of the erbB2 promoter. Oncogene 19:6490-6502. [DOI] [PubMed] [Google Scholar]
- 44.Seidel, J. J., and B. J. Graves. 2002. An ERK2 docking site in the Pointed domain distinguishes a subset of ETS transcription factors. Genes Dev. 16:127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sevilla, L., C. Aperlo, V. Dulic, J. C. Chambard, C. Boutonnet, O. Pasquier, P. Pognonec, and K. E. Boulukos. 1999. The Ets2 transcription factor inhibits apoptosis induced by colony-stimulating factor 1 deprivation of macrophages through a Bcl-xL-dependent mechanism. Mol. Cell. Biol. 19:2624-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sevilla, L., A. Zaldumbide, F. Carlotti, M. A. Dayem, P. Pognonec, and K. E. Boulukos. 2001. Bcl-XL expression correlates with primary macrophage differentiation, activation of functional competence, and survival and results from synergistic transcriptional activation by Ets2 and PU.1. J. Biol. Chem. 276:17800-17807. [DOI] [PubMed] [Google Scholar]
- 47.Shepherd, T. G., L. Kockeritz, M. R. Szrajber, W. J. Muller, and J. A. Hassell. 2001. The pea3 subfamily ets genes are required for HER2/Neu-mediated mammary oncogenesis. Curr. Biol. 11:1739-1748. [DOI] [PubMed] [Google Scholar]
- 48.Siegel, P. M., D. L. Dankort, W. R. Hardy, and W. J. Muller. 1994. Novel activating mutations in the neu proto-oncogene involved in induction of mammary tumors. Mol. Cell. Biol. 14:7068-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, J. L., A. E. Schaffner, J. K. Hofmeister, M. Hartman, G. Wei, D. Forsthoefel, D. A. Hume, and M. C. Ostrowski. 2000. ets-2 is a target for an Akt (protein kinase B)/Jun N-terminal kinase signaling pathway in macrophages of motheaten-viable mutant mice. Mol. Cell. Biol. 20:8026-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Span, P. N., P. Manders, J. J. Heuvel, C. M. Thomas, R. R. Bosch, L. V. Beex, and C. G. Sweep. 2002. Expression of the transcription factor Ets-1 is an independent prognostic marker for relapse-free survival in breast cancer. Oncogene 21:8506-8509. [DOI] [PubMed] [Google Scholar]
- 51.Sternlicht, M. D., A. Lochter, C. J. Sympson, B. Huey, J. P. Rougier, J. W. Gray, D. Pinkel, M. J. Bissell, and Z. Werb. 1999. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell 98:137-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka, S., T. Kunath, A. K. Hadjantonakis, A. Nagy, and J. Rossant. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282:2072-2075. [DOI] [PubMed] [Google Scholar]
- 53.Trimble, M. S., J.-H. Xin, C. T. Guy, W. J. Muller, and J. A. Hassell. 1993. PEA3 is overexpressed in mouse metastatic mammary adenocarcinomas. Oncogene 8:3037-3042. [PubMed] [Google Scholar]
- 54.Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, J. D. Gocayne, P. Amanatides, R. M. Ballew, D. H. Huson, J. R. Wortman, Q. Zhang, C. D. Kodira, X. H. Zheng, L. Chen, M. Skupski, G. Subramanian, P. D. Thomas, J. Zhang, G. L. Gabor Miklos, C. Nelson, S. Broder, A. G. Clark, J. Nadeau, V. A. McKusick, N. Zinder, A. J. Levine, R. J. Roberts, M. Simon, C. Slayman, M. Hunkapiller, R. Bolanos, A. Delcher, I. Dew, D. Fasulo, M. Flanigan, L. Florea, A. Halpern, S. Hannenhalli, S. Kravitz, S. Levy, C. Mobarry, K. Reinert, K. Remington, J. Abu-Threideh, E. Beasley, K. Biddick, V. Bonazzi, R. Brandon, M. Cargill, I. Chandramouliswaran, R. Charlab, K. Chaturvedi, Z. Deng, V. Di Francesco, P. Dunn, K. Eilbeck, C. Evangelista, A. E. Gabrielian, W. Gan, W. Ge, F. Gong, Z. Gu, P. Guan, T. J. Heiman, M. E. Higgins, R. R. Ji, Z. Ke, K. A. Ketchum, Z. Lai, Y. Lei, Z. Li, J. Li, Y. Liang, X. Lin, F. Lu, G. V. Merkulov, N. Milshina, H. M. Moore, A. K. Naik, V. A. Narayan, B. Neelam, D. Nusskern, D. B. Rusch, S. Salzberg, W. Shao, B. Shue, J. Sun, Z. Wang, A. Wang, X. Wang, J. Wang, M. Wei, R. Wides, C. Xiao, C. Yan, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 55.Watabe, T., K. Yoshida, M. Shindoh, M. Kaya, K. Fujikawa, H. Sato, M. Seiki, S. Ishi, and K. Fujinaga. 1998. The ets-1 and ets-2 transcription factors activate the promoters for invasion-associated urokinase and collagenase genes in response to epidermal growth factor. Int. J. Cancer 77:128-137. [DOI] [PubMed] [Google Scholar]
- 56.Webster, M. A., J. N. Hutchinson, M. J. Rauh, S. K. Muthuswamy, M. Anton, C. G. Tortorice, R. D. Cardiff, F. L. Graham, J. A. Hassell, and W. J. Muller. 1998. Requirement for both Shc and phosphatidylinositol 3′ kinase signaling pathways in polyomavirus middle T-mediated mammary tumorigenesis. Mol. Cell. Biol. 18:2344-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiseman, B. S., and Z. Werb. 2002. Stromal effects on mammary gland development and breast cancer. Science 296:1046-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woods, D., D. Parry, E. Cherwinski, E. Bosch, E. Lees, and M. McMahon. 1997. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol. Cell. Biol. 17:5598-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamamoto, H., M. L. Flannery, S. Kupriyanov, J. Pearce, S. R. McKercher, G. W. Henkel, R. A. Maki, Z. Werb, and R. G. Oshima. 1998. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 12:1315-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang, B.-S., C. A. Hauser, G. Henkel, M. S. Colman, C. Van Beveren, K. J. Stacey, D. A. Hume, R. A. Maki, and M. C. Ostrowski. 1996. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activity of c-Ets1 and c-Ets2. Mol. Cell. Biol. 16:538-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young, L. J. T. 2000. The cleared mammary fat pad and the transplantation of mammary gland morphological structures and cells, p. 67-74. In M. Ip and B. Asch (ed.), Methods in mammary gland biology and breast cancer research. Kluwer Academic/Plenum Publishers, New York, N.Y.