Abstract
The Burkholderia pseudomallei rpoS gene was identified, and an rpoS null mutant was constructed. The mutant was shown to have an increased sensitivity to carbon starvation and oxidative stress. By using rpoS-lacZ fusions, transcription of rpoS was shown to be growth phase regulated, reaching a peak upon entry into stationary phase.
Burkholderia pseudomallei is the causative agent of melioidosis, a disease endemic in Southeast Asia and Australia (7). B. pseudomallei can survive inside phagocytes (14, 20) and adapt to many environments (21, 36). Recently, nonpathogenic strains of B. pseudomallei were reassigned as B. thailandensis (5). In gram-negative bacteria, the sigma factor RpoS (σS) activates expression of genes required in response to various stresses including acid, heat shock, UV light, osmotic, oxidative stresses, and carbon starvation (4, 15, 33, 35). σS also controls expression of extracellular virulence factors and is important for pathogenicity in members of the Enterobacteriaceae and Pseudomonadaceae (9, 19, 35).
Expression of virulence factors is also correlated with entry into stationary phase and nutrient limitation in the intracellular pathogen Legionella pneumophila (3). In Escherichia coli, σS is regulated by transcription, translation and proteolysis, and different stress conditions differentially affect these levels of control (16, 24). However, expression of rpoS in pseudomonads is predominantly controlled at the transcriptional level (38). In this study, we isolated the rpoS gene from B. pseudomallei and present evidence that σS is involved in the response to several environmental stresses. Expression experiments with translational and transcriptional fusions were performed to examine the regulation of rpoS in relation to growth phase.
Isolation of the rpoS gene from B. pseudomallei.
Recently, the complete genome sequence of B. pseudomallei has been determined (http://www.sanger.ac.uk/Projects/B_pseudomallei). Using the rpoS sequence from P. aeruginosa PAO1 (34) as the query in a TBLASTX search, an open reading frame of 1,080 bp encoding the predicted σS protein of 359 amino acids was identified. The B. pseudomallei rpoS gene was amplified from PP844 (Table 1) genomic DNA with Vent DNA polymerase (New England Biolabs, Beverly, Mass.) and primers RPOSF and RPOSR (Fig. 1, Table 2). PCR was performed for 30 cycles at 94°C for 1 min., 68°C and 72°C for 45 s each cycle. The amplified rpoS DNA was cloned into the SmaI site of pUC19 to generate pUCBS2 (Table 1) and the nucleotide sequence was determined (GenBank accession number AY183467).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or relevant characteristics | Source or reference |
|---|---|---|
| B. pseudomallei | ||
| PP844 | Prototroph, blood culture isolate from a patient at Khon kaen University Hospital | 37 |
| KN100 | PP844/pKBS1 | This study |
| PP844(pBBS1) | PP844 containing pBBS1 | This study |
| KN100(pBBS1) | PP844/pKBS1 containing pBBS1 | This study |
| Z2BS1 | PP844/pZ2BS1 containing single copy of rpoS-lacZ transcriptional fusion | This study |
| Z3BS1 | PP844/pZ3BS1 containing single copy of rpoS-lacZ translational fusion | This study |
| S. enterica serovar Typhimurium | ||
| BSST100 | Clinical isolate from a patient at Royal Hallamshire Hospital | This study |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK− mK+) supE44 λ−thi-1 relA1 gyrA96 | 13 |
| CC118λpir | Δ(ara, leu)7697 araD139 ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB (Rfr) argE(Am) recA1 λpir+ | 17 |
| JM83 | F−ara Δ(lac-proAB) rpsL(Smr) φ80dlacZΔM15 | 39 |
| SM10λpir | RP4-2-tet::Mu-1 (Kmr) thi-1 thr leu tonA lacY supE recA | 32 |
| S17-1λpir | RP4-2-tet::Mu-1 kan::Tn7 (Tpr Smr) thi proA hsdR recA | 32 |
| Y. enterocolitica | ||
| 08081c | Inv+ | 29 |
| Plasmids | ||
| pUCBS1 | pUC19 containing a 600-bp internal segment of B. pseudomallei rpoS | This study |
| pUCBS2 | pUC19 containing the full-length rpoS | This study |
| pKNOCK-Tc | Mobilizable suicide vector for construction of gene knockouts in Gram-negative bacteria | 2 |
| pKBS1 | pKNOCK-Tc containing a 600-bp internal segment of B. pseudomallei rpoS | This study |
| pBBR1MCS | Broad-host-range cloning vector, Cmr | 23 |
| pBBS1 | pBBR1MCS containing the full-length B. pseudomallei rpoS gene | This study |
| pZINT2 | Mobilizable integrative vector for construction of single-copy lacZ transcriptional fusions | 27 |
| pZINT3 | Mobilizable integrative vector for construction of single-copy lacZ translational fusions | This study |
| pZ2BS1 | pZINT2 containing the 5′ region of nlpD and 3′ region of rpoS fused to lacZ | This study |
| pZ3BS1 | pZINT3 containing the 5′ region of nlpD and 3′ region of rpoS fused to lacZ | This study |
| pFR97 | Plasmid vector for construction of lacZ translational fusions | 31 |
FIG. 1.
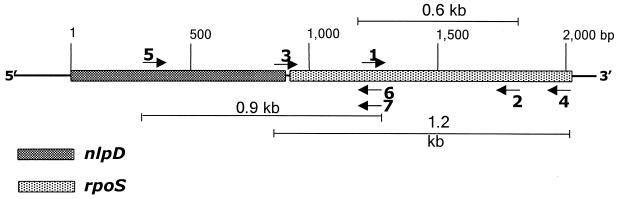
Arrangement of the nlpD-rpoS genes in the genome of B. pseudomallei. Arrows indicate the location and orientation of primers used to construct rpoS plasmids in this study: 1, SBKF; 2, SBKR; 3, RPOSF; 4; RPOSR; 5, PNSF; 6, PNSR; and 7, PNSFR97.
TABLE 2.
PCR primers
| Primer | Sequence | Restriction sitea |
|---|---|---|
| SBKF | 5′-GACGACTTCCGGGCGCTTCT-3′ | |
| SBKR | 5′-ATCGTCGGGCAGCAGATCGAG-3′ | |
| RPOSF | 5′-CCTGTCGATCCGCTGAAGTATT-3′ | |
| RPOSR | 5′-CCAGAATCGGTGTCATTGATGAA-3′ | |
| PNSF | 5′-GCCCGCATGCCGCTTTATCGGATCGCGCTCG-3′ | SphI |
| PNSR | 5′-GCCGTCTAGAAGAAGCGCCCGGAAGTCGTC-3′ | XbaI |
| PNSFR97 | 5′-GCCGTCTAGAAAGCGCCCGGAAGTCGTCCAG-3′ | XbaI |
Underlined in the sequence.
The deduced amino acid sequence exhibits 93% amino acid identity to σs of Burkholderia cepacia, 67% to Ralstonia solanacearum σs, 46% to Pseudomonas aeruginosa σs and 43% to E. coli σs. Similar to the situation in R. solanacearum, P. aeruginosa, and E. coli, rpoS in B. pseudomallei is located downstream of nlpD (Fig. 1). In several gram-negative bacterial species rpoS transcription is directed from promoters located within and upstream of nlpD (12, 22, 25, 30). Only 11 bp separate the nlpD coding sequence from rpoS, suggesting that transcription of rpoS in B. pseudomallei also initiates within or upstream of nlpD. Perusal of the B. pseudomallei nlpD sequence reveals two sequences which could serve as promoters for transcription of rpoS. Nineteen base pairs upstream of the nlpD translation initiation codon is the sequence TTGATC(N17)TAAAAT, which probably serves as the promoter for nlpD and possibly also for rpoS. Located 131 bp upstream of the nlpD stop codon is the sequence TGCACA(N17)TAAAAG, which could also serve as a promoter for rpoS transcription.
Role of RpoS in response to environmental stresses.
An rpoS knockout mutant, KN100 (Table 1), was created with pKBS1 according to a previously described procedure (26). The plasmid pUCBS1 was constructed by blunt-end ligation of a 632-bp internal fragment of rpoS, generated by PCR with primers SBKF and SBKR, into the SmaI site of pUC19. pKBS1 was constructed by transferring the 632-bp KpnI-XbaI fragment from pUCBS1 into the mobilizable suicide vector pKNOCK-Tc (2). The effects of rpoS inactivation on cell survival during prolonged carbon starvation as well as other functions were analyzed with this mutant. To confirm that all changes in phenotypes were caused by the disruption of rpoS and were not due to polar effects on downstream genes, a plasmid (pBBS1) containing the complete rpoS coding sequence under control of the lacZ and cat gene promoters was constructed and transferred into B. pseudomallei wild-type and mutant strains for complementation analysis.
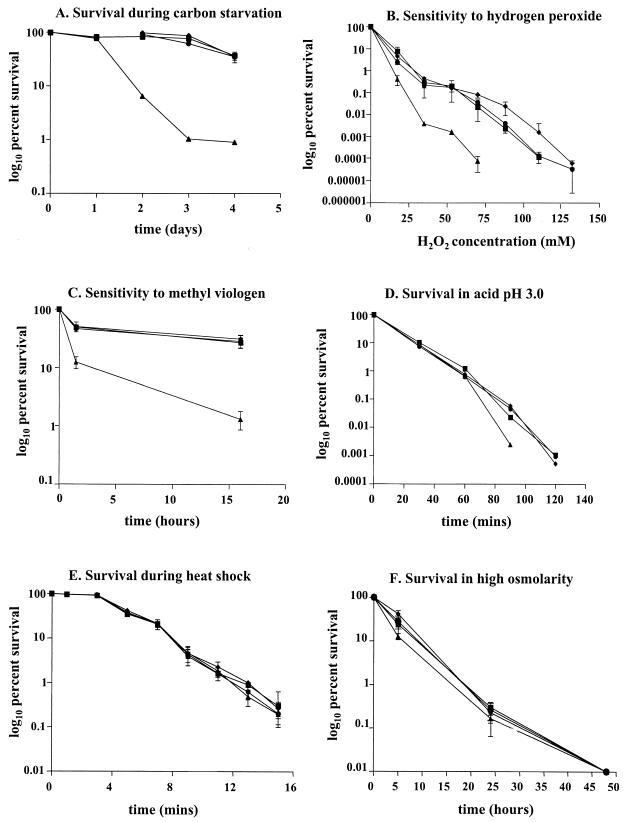
To examine the ability of B. pseudomallei to survive during prolonged carbon starvation, overnight cultures were diluted 100-fold into glucose minimal medium and grown at 37°C with aeration. After the cells entered the stationary phase of growth, incubation was continued for several days, during which time cell viability was determined by measuring CFU. Seventy-two hours after entry into the stationary phase, a 100-fold decrease in the number of viable cells was observed for the rpoS mutant compared to the wild type. This finding suggests that the rpoS mutant is more sensitive to carbon starvation than the wild type. This result is in accordance with previous findings, which have demonstrated a general function for σS as a growth phase-dependent regulator of metabolism under conditions of nutrient limitation (10, 16, 35). Introduction of plasmid pBBS1, containing the full-length rpoS gene, completely restored starvation tolerance to the wild-type level in the mutant, indicating that the loss of viability of the mutant was specifically caused by inactivation of the rpoS gene (Fig. 2A).
FIG. 2.
Effect of rpoS mutation on stress responses. B. pseudomallei parent strain PP844 (▪), B. pseudomallei rpoS mutant (▴), B. pseudomallei parent strain harboring pBBS1 (♦), and B. pseudomallei rpoS mutant harboring pBBS1 for complementation (•). (A) Survival during carbon starvation. (B) Sensitivity to hydrogen peroxide. (C) Sensitivity to methyl viologen. (D) Survival in acid pH (pH 3.0). (E) Survival during heat shock. (F) Survival in high osmolarity.
The effect of rpoS inactivation on sensitivity to H2O2 and the redox-cycling agent methyl viologen was also examined. Sensitivity to hydrogen peroxide was measured on cells grown to mid-logarithmic phase (optical density at 600 nm = 0.4). H2O2 was added to the cells at the indicated concentrations, and the cells were incubated for 20 min at 37°C. Catalase was added (400 units/ml), and cell viability was measured as described above. After a 20-min incubation in 50 mM H2O2, the number of viable cells for the mutant was approximately 80-fold lower than in the wild type (Fig. 2B). The sensitivity of the mutant to methyl viologen was also examined by exposing mid-logarithmic-phase cells to 40 mM methyl viologen. Growth of the wild-type cells was not significantly affected by the presence of 40 mM methyl viologen, whereas the mutant showed a 50-fold reduction in viability after exposure for 16 h (Fig. 2C). The sensitivity of the mutant to H2O2 and methyl viologen suggests a role for σS in oxidative stress resistance. This is consistent with the role of rpoS in resistance to oxidative stress in B. cepacia and P. aeruginosa (1, 35). On the other hand, inactivation of the rpoS promoter in R. solanacearum did not result in increased sensitivity to H2O2 (10).
The ability of cells to survive in pH 3.0 was measured following an adaptation step at pH 4.0 (11). Bacterial cells were grown in Luria-Bertani (LB) (pH 7.0) to stationary phase overnight, washed, and suspended in LB adjusted to pH 4.0. Incubation was continued for 4 h before the cells were resuspended in medium adjusted to pH 3.0. Cell viability was then determined at different time intervals. Figure 2D shows that the sensitivity of the rpoS mutant was not significantly different from that of the wild type after exposure to acid pH 3.0 for up to 60 min. However, at 90 min the mutant demonstrated a 10-fold decrease in survival compared to the wild type. Acid tolerance in several gram-negative bacteria has also been shown to be mediated by σS (10, 11, 30).
To measure the ability of cells to survive extreme heat shock, cells were grown to stationary phase, whereupon they were washed and diluted in minimal medium to a density of approximately 7,000 CFU/ml. One milliliter of the diluted culture was placed in a prewarmed tube at 53°C, and viability was determined after different time intervals. To measure sensitivity to osmotic stress, cells were grown to stationary phase, washed, and resuspended in minimal medium containing 4.0 M NaCl. The resuspended cells were incubated at 37°C with aeration, during which time cell viability was determined. Our results showed that there is no significant difference between the wild type and the mutant in their sensitivity to heat shock or osmotic stress (Fig. 2E and 2F). In contrast, a B. cepacia rpoS knockout mutant exhibited hypersensitivity to high temperature but, like B. pseudomallei, showed a wild-type hyperosmotic response (1).
Role of rpoS in invasion of epithelial and macrophage cell lines.
Invasion assays were performed as previously described with slight modifications (8). HEp-2 cells and the Abelson murine leukemia virus-induced tumor cell line RAW264.7 were seeded in 24-well plates at 5 × 105 cells per well. Cells were grown overnight at 37°C with 5% CO2 in Eagle's minimal essential medium (EMEM) and Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (Gibco BRL) for HEp-2 and RAW264.7, respectively. The cell monolayers were washed and incubated with 1 ml of the same medium containing 4% bovine serum albumin for 30 min at 37°C with 5% CO2. After incubation, the monolayers were washed with fresh medium. A log-phase culture of bacteria was resuspended in 1 ml of EMEM medium for HEp-2 cells and DMEM medium for RAW264.7 cells, and 25 μl of the suspension was added to cell monolayers and incubated for 2 h at 37°C with 5% CO2. Cells were washed with phosphate-buffered saline and medium and then incubated with 1.5 ml of medium containing kanamycin for an additional 2 h to eliminate extracellular bacteria. Cell monolayers were then lysed with 1% saponin (Sigma) to release the intracellular bacteria.
The inocula and intracellular bacteria were quantified by plating serial dilutions. For overnight invasion assays, the monolayers were washed with phosphate-buffered saline and incubated overnight with 1.5 ml of medium containing kanamycin. Table 3 shows that both the wild type and rpoS mutant demonstrated an equal ability to invade RAW2643 cells and HEp-2 cells, although at an order of magnitude less efficiently than Salmonella enterica serovar Typhimurium and Yersinia enterocolitica. This indicates that rpoS is not required for survival within the intracellular compartment and suggests that B. pseudomallei is not exposed to or is able to circumvent carbon starvation and oxidative stress inside the host cell. Similar observations were made with Y. enterocolitica, in which inactivation of rpoS had no significant effects on virulence in mice or on the expression of the inv or ail genes, which are involved in cell invasion (4). In contrast, expression of the rpoS gene in S. enterica serovar Typhimurium increased rapidly after phagocytosis (6), and inactivation of rpoS reduced virulence in mice (18). The use of culture cell lines instead of human phagocytes could explain the discrepancy between our study and that of Jones et al. (20), who showed that B. pseudomallei was able to replicate within human phagocytes. Furthermore, bacterial growth within cells could ultimately result in host cell lysis that would release the bacteria into the extracellular environment, where they would be killed by the antibiotic.
TABLE 3.
Effect of rpoS mutation on invasion of cell linesa
| Strain | RAW2643 cells
|
HEp-2 cells
|
||||
|---|---|---|---|---|---|---|
| Inoculum (CFU) | Mean no. of cells recovered at 4 h | Mean no. of cells recovered at 24 h | Inoculum (CFU) | Mean no. of cells recovered at 4 h | Mean no. of cells recovered at 24 h | |
| PP844 (wild type) | 2.6 × 108 | 8.6 × 105 ± 5.1 × 105 | 2.0 × 105 ± 1.0 × 105 | 8.0 × 108 | 1.3 × 104 ± 3.2 × 103 | 3.0 × 104 ± 1.3 × 104 |
| KN100 (rpoS knockout mutant) | 8.0 × 108 | 6.8 × 105 ± 1.5 × 105 | 2.2 × 105 ± 3.0 × 104 | 5.0 × 108 | 3.6 × 104 ± 2.2 × 104 | 2.9 × 104 ± 1.4 × 104 |
| PP844 with pBBS1 (wild type + rpoS plasmid) | 2.0 × 108 | 2.9 × 105 ± 5.7 × 103 | 1.7 × 105 ± 6.0 × 104 | 5.0 × 108 | 1.8 × 104 ± 4.3 × 103 | 2.3 × 104 ± 1.1 × 104 |
| KN100 with pBBS1 (rpoS knockout mutant + rpoS plasmid) | 8.0 × 108 | 3.7 × 105 ± 3.2 × 104 | 7.0 × 105 ± 2.0 × 104 | 3.0 × 108 | 4.1 × 104 ± 3.4 × 104 | 1.0 × 104 ± 1.5 × 103 |
| E. coli JM83 | 2.2 × 107 | <1.0 × 10° ± 0.0 | ND | 8.0 × 108 | <1.0 × 10° ± 0.0 | ND |
| S. enterica serovar Typhimurium | 2.1 × 107 | 2.2 × 105 ± 7.2 × 104 | ND | ND | ND | ND |
| Y. enterocolitica | ND | ND | ND | 3.0 × 107 | 4.8 × 105 ± 1.0 × 105 | ND |
ND, not done. Values are means ± standard deviations.
Growth phase-dependent regulation of the B. pseudomallei rpoS gene.
To study the regulation of the rpoS gene in B. pseudomallei, rpoS-lacZ transcriptional and translational fusions were constructed and integrated into the chromosome in single copy. The mobilizable suicide plasmid pZINT2 (27) was used for the construction of an rpoS-lacZ transcriptional fusion. The resultant plasmid, pZ2BS1, was generated by amplification of 221 codons of the 3′ region of the nlpD gene and the first 71 codons of rpoS with primers PNSF and PNSR, followed by ligation of the SphI- and XbaI-digested PCR products with pZINT2. pZINT3 was used as the vector for construction of a single-copy rpoS-lacZ translational fusion and was constructed from pZINT2 by replacing a 5.2-kb SalI-StuI fragment containing the lacZYA cassette with a 3.0-kb XhoI-DraI restriction fragment of pFR97 (31), which contains the lacZ gene devoid of the first six codons of lacZ and the Shine-Dalgarno sequence.
To construct the rpoS-lacZ translational fusion plasmid pZ3BS1, a 0.92-kb PCR product obtained with primers PNSF and PNSR97 was ligated as an SphI-XbaI fragment into pZINT3. pZ2BS1 and pZ3BS1 were integrated in single copy at the rpoS locus of B. pseudomallei PP844 by homologous recombination, resulting in strains Z2BS1 and Z3BS1, respectively. Cultures were grown in LB broth, and β-galactosidase assays (28), normalized to culture density (optical density at 600 nm), were performed at various time points.
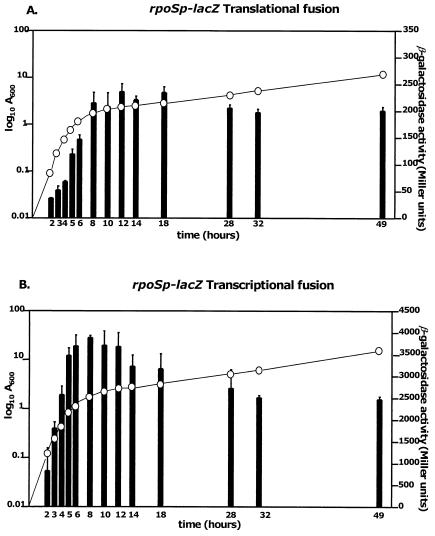
The strain harboring the translational fusion (Z3BS1) showed an increase in β-galactosidase activity starting from the exponential phase and reaching a peak on entry into stationary phase (Fig. 3A). This result showed that RpoS production is growth phase regulated. As the β-galactosidase activities for the translational fusion represent the sum of transcriptional and translational control, the transcriptional fusion was used to see whether this regulation is exerted at the transcriptional or translational level. The β-galactosidase activity of the strain harboring the transcriptional fusion followed a similar pattern (Fig. 3B). In both fusion strains, the activity on entry into stationary phase was approximately fivefold higher than in the early logarithmic phase. The results indicate that B. pseudomallei rpoS is regulated according to growth phase at the transcriptional level. However, the possibility that rpoS is subject to cell density-dependent regulation cannot be ruled out.
FIG. 3.
Growth phase-dependent regulation of the B. pseudomallei rpoS gene. The bars represent the β-galactosidase activity in Miller units, while the circles represent the cell density (optical density at 600 nm). (A) Expression of B. pseudomallei rpoS-lacZ translational fusion according to growth phase. (B) Expression of B. pseudomallei rpoS-lacZ transcriptional fusion according to growth phase.
In conclusion, we have shown that RpoS is likely to play an important role in the response of B. pseudomallei to carbon starvation and oxidative stress. Furthermore, our results show that the rpoS gene in this organism is likely to be regulated according to the growth phase.
Acknowledgments
This work was supported by research grants from the Faculty of Medicine, Ramathibodi Hospital. B.S. was supported by a Royal Golden Jubilee Ph.D. Scholarship from the Thailand Research Fund.
We thank P. Sonthayanon and S. Butrapet for providing laboratory facilities, S. Loprasert and S. Mongkolsuk for technical support, S. Sirisinha for providing B. pseudomallei PP844, and M. F. Alexeyev for providing the pKNOCK-Tc plasmid used in this study.
REFERENCES
- 1.Aguilar, C., I. Bertani, and V. Venturi. 2003. Quorum-sensing system and stationary-phase sigma factor (RpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 69:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26:824-826, 828. [DOI] [PubMed] [Google Scholar]
- 3.Bachman, M. A., and M. S. Swanson. 2001. RpoS cooperates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol Microbiol. 40:1201-1214. [DOI] [PubMed] [Google Scholar]
- 4.Badger, J. L., and V. L. Miller. 1995. Role of RpoS in survival of Yersinia enterocolitica to a variety of environmental stresses. J. Bacteriol. 177:5370-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaiyaroj, S. C., K. Kotrnon, S. Koonpaew, N. Anantagool, N. J. White, and S. Sirisinha. 1999. Differences in genomic macrorestriction patterns of arabinose-positive (Burkholderia thailandensis) and arabinose-negative Burkholderia pseudomallei. Microbiol. Immunol. 43:625-630. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C. Y., L. Eckmann, S. J. Libby, F. C. Fang, S. Okamoto, M. F. Kagnoff, J. Fierer, and D. G. Guiney. 1996. Expression of Salmonella typhimurium rpoS and rpoS-dependent genes in the intracellular environment of eukaryotic cells. Infect. Immun. 64:4739-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dharakul, T., and S. Songsivilai. 1999. The many facets of melioidosis. Trends Microbiol. 7:138-140. [DOI] [PubMed] [Google Scholar]
- 8.Elsinghorst, E. A. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol. 236:405-420. [DOI] [PubMed] [Google Scholar]
- 9.Fang, F. C., S. J. Libby, N. A. Buchmeier, P. C. Loewen, J. Switala, J. Harwood, and D. G. Guiney. 1992. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. 89:11978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flavier, A. B., M. A. Schell, and T. P. Denny. 1998. An RpoS (sigmaS) homologue regulates acylhomoserine lactone-dependent autoinduction in Ralstonia solanacearum. Mol. Microbiol. 28:475-486. [DOI] [PubMed] [Google Scholar]
- 11.Foster, J. W. 1995. Low pH adaptation and the acid tolerance response of Salmonella typhimurium. Crit. Rev. Microbiol. 21:215-237. [DOI] [PubMed] [Google Scholar]
- 12.Fujita, M., K. Tanaka, H. Takahashi, and A. Amemura. 1994. Transcription of the principal sigma-factor genes, rpoD and rpoS, in Pseudomonas aeruginosa is controlled according to the growth phase. Mol. Microbiol. 13:1071-1077. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 14.Harley, V. S., D. A. Dance, G. Tovey, M. V. McCrossan, and B. S. Drasar. 1998. An ultrastructural study of the phagocytosis of Burkholderia pseudomallei. Microbios 94:35-45. [PubMed]
- 15.Hengge-Aronis, R. 1993. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72:165-168. [DOI] [PubMed] [Google Scholar]
- 16.Hengge-Aronis, R. 2002. Signal Transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibanez-Ruiz, M., V. Robbe-Saule, D. Hermant, S. Labrude, and F. Norel. 2000. Identification of RpoS (σS)-regulated genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:5749-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iriarte, M., I. Stainier, and G. R. Cornelis. 1995. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect. Immun. 63:1840-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, A. L., T. J. Beveridge, and D. E. Woods. 1996. Intracellular survival of Burkholderia pseudomallei. Infect. Immun. 64:782-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanai, K., E. Kondo, S. Dejsirilert, and P. Naigowit. 1994. Growth and survival of Pseudomonas pseudomallei in acidic environment with possible relation to the ecology and epidemiology of melioidosis, p. 26-38. In S. D. Puthucheary and M. A. Yasmin (ed.), Melioidosis: revealing problems and future directions. SP-Muda, Kuala Lumpur.
- 22.Kojic, M., C. Aguilar, and V. Venturi. 2002. TetR family member psrA directly binds the Pseudomonas rpoS and psrA promoters. J. Bacteriol. 184:2324-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. 2nd Roop, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 24.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the σS subunit of RNA-polymerase in Escherichia coli is controlled at the levels of transcription, translation and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 25.Lange, R., and R. Hengge-Aronis. 1994. The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol. Microbiol. 13:733-743. [DOI] [PubMed] [Google Scholar]
- 26.Low, K. B. 1991. Conjugational methods for mapping with Hfr and F-prime strains. Methods Enzymol. 204:43-62. [DOI] [PubMed] [Google Scholar]
- 27.Lowe, C. 2001. Ph.D. thesis. University of Sheffield, Sheffield, United Kingdom.
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Portnoy, D. A., H. F. Blank, D. T. Kingsbury, and S. Falkow. 1983. Genetic analysis of essential plasmid determinants of pathogenicity in Yersinia pestis. J. Infect. Dis. 148:297-304. [DOI] [PubMed] [Google Scholar]
- 30.Ramos-Gonzalez, M. I., and S. Molin. 1998. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440. J. Bacteriol. 180:3421-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapira, S. K., J. Chou, F. V. Richaud, and M. J. Casadaban. 1983. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene 25:71-82. [DOI] [PubMed] [Google Scholar]
- 32.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 33.Small, P., D. Blankenhorn, D. Welty, E. Zinser, and J. L. Slonczewski. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stover, C. K., X.-Q. T. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrook-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. M. Lim, K. A. Smith, D. H. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 35.Suh, S. J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas, A. D., J. Forbes-Faulkner, and M. Parker. 1979. Isolation of Pseudomonas pseudomallei from clay layers in defined depths. Am. J. Epidemiol. 110:515-521. [DOI] [PubMed] [Google Scholar]
- 37.Utaisincharoen, P., N. Tangthawornchaikul, W. Kespichayawattana, P. Chaisuriya, and S. Sirisinha. 2001. Burkholderia pseudomallei interferes with inducible nitric oxide synthase (iNOS) production: a possible mechanism of evading macrophage killing. Microbiol. Immunol. 45:307-313. [DOI] [PubMed] [Google Scholar]
- 38.Whistler, C. A., N. A. Corbell, A. Sarniguet, W. Ream, and J. E. Loper. 1998. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor σS and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriol. 180:6635-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]