Abstract
Surface biology aims to observe and control biological processes by combining bio-, surface, and physical chemistry. Self-assembled monolayers (SAM) on gold surfaces have provided excellent methods for nanoscale surface preparation for such studies. However, extension of this work requires the specific immobilization of whole protein domains and the direct incorporation of recombinant proteins into SAM is still problematic. In this study a short random coil peptide has been designed to insert into thioalkane layers by formation of a hydrophobic helix. Surface plasmon resonance (SPR) studies show that specific immobilization via the internal cysteine is achieved. Addition of the peptide sequence to the terminus of a protein at the genetic level enables the production of a range of recombinant fusion-proteins with good yield. SPR shows that the proteins display the same gold-binding behavior as the peptide. It is shown that cell growth control can be achieved by printing the proteins using soft lithography with subsequent infilling with thioalkanes The expression plasmid is constructed so that any stable protein domain can be easily cloned, expressed, purified and immobilized.
Keywords: self-assembling monolayer, protein immobilization, nanotechnology, circular dichroism spectroscopy, surface plasmon resonance, osteoblast
The specific immobilization of proteins upon surfaces has the potential to revolutionize both the study of their natural properties and their utilization in novel, self-assembling nanostructures (Terrettaz et al 2002). Patterned proteins have applications in molecular biosensors and protein arrays (Bertone and Snyder 2005), but can also contribute to cell patterning for research (Liu et al 2005; Sniadecki et al 2006) tissue engineering and cell-based biosensors (Csucs et al 2003; Hasirci and Kenar 2006; Petty et al 2007; Yap and Zhang 2007). Protein two-dimension (2D) arrays are following in the wake of DNA arrays by increasing the rate of biomolecular interaction analysis. The most widespread examples are based upon contact printed antibodies or immobilized protein libraries. Typical formats use glass supports and apply the proteins onto gels, into microwell arrays or directly to glass surfaces (Bertone and Snyder 2005). The latter uses developments of the DNA arraying technology including robotic arrayers and a range of immobilization methods. Nonspecific adsorption of proteins to a solid support or simple chemical coupling are the most popular methods. The latter include noncovalent adsorption to poly-L-lysine, polyvinylidene difluoride (PVDF), and nitrocellulose, cross-linking via aldehyde or epoxy (MacBeath and Schreiber 2000) to surface lysine residues, histidine tag (Klenkar et al 2006), avidin (Delehanty and Ligler 2002), or glutathione-S-transferase (GST) (Jung et al 2005) based immobilization using fusion proteins. Recombinant tags such as oligohistidine or GST improve specificity and orientation but these are reversible and lack long-term stability. Finally the need to array purified protein samples is removed if the proteins are produced using cell-free synthesis in response to locally arrayed DNA (NAPPA approach) (Ramachandran et al 2004). This method can be accompanied by incorporation of an immobilization tag. However, to reduce the scale of arrays whilst still obtaining quantitative data, the quality of the surface needs to be constantly improved (Ekins and Chu 1999). One way to overcome protein denaturation at surfaces and problems of nonspecific binding is to combine proteins with self assembling oriented layers at the solid substrate–protein interface. Work by several groups has demonstrated the wide potential of self assembling monolayers of immobilized amphiphiles incorporating small peptides (Zhang et al 1999; Miura et al 2000; Huang et al 2003). However, a significant obstacle to the further development of such technologies is the lack of methods that enable the anchoring of large soluble protein molecules to these surfaces (Hirano and Mooney 2004). Further requirements include surfaces with low nonspecific binding and a potential for incorporation into detection technologies such as fluorescence, surface acoustic wave (SAW), quartz crystal microbalance (QCM), impedance spectroscopy (IS), or surface plasmon resonance (SPR) methods (Cooper 2003, 2006). A simple and widely applicable immobilization strategy of large proteins with SAM has yet to be developed. Two of the best methods come from the Mrksich group who has developed a fusion protein system with chitninase fusion proteins binding to SAM immobilized chitin analogues (Hodneland et al 2002) or a cysteine-directed immobilization into mixed maleimide and ethylene glycol terminated SAM (Houseman et al 2003) An alternative approach is to fuse the protein of interest to a membrane protein scaffold which self assembles with SAM (Terrettaz et al 2002; Shah et al 2007). It has also been shown that combined surfaces of lipid and protein could provide patterned arrays in which the nonprotein surface had extremely low nonspecific binding due to the properties of lipid layers (Kung et al 2000). To provide a further option we present here the combination of a high-level expression system (Anderluh et al 2003) with a minimal 12 amino-acid peptide to produce proteins capable of oriented co-assembly with SAM. The peptide is termed the “switch-tag” since it can switch from a water soluble coil to a hydrophobic helix. This sequence is easily added at the genetic level to the C-terminus of any protein and expressed recombinantly. The subsequent insertion of the tag into the SAM ensures orientation and low nonspecific binding, plus the option to combine the products with many detection technologies.
Results and discussion
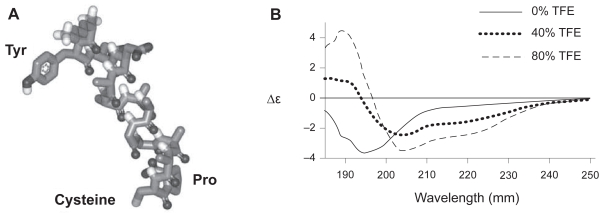
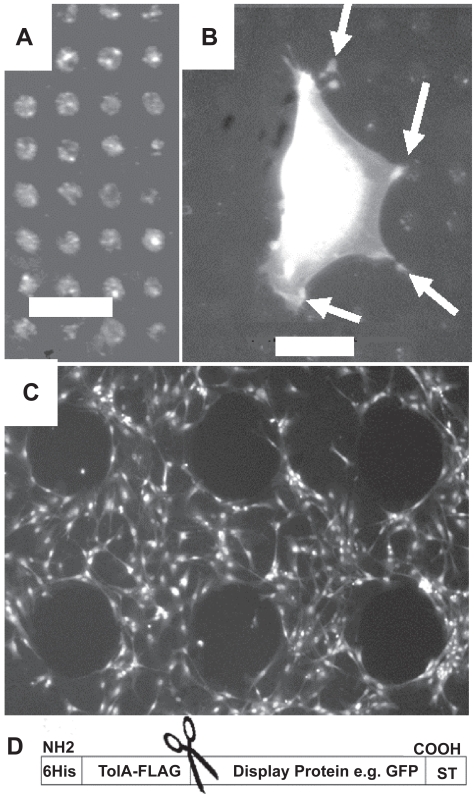
A sequence of NH2-AAAAGAAACP-COOH was used as the switch-tag (ST) (Munoz and Serrano 1994). We have previously observed that C-terminal cysteines, which have thiol and carboxylic acid groups in close proximity, can be self-proteolytic and thus added a protecting C-terminal proline residue which does not hydrogen bond to the helix and, in in silico modeling, orientates the – SH group of the cysteine towards the target binding surface (Figure 1A). A synthetic ST with additional N-terminal residues (KYDD) to aid solubility and concentration measurements showed a coil to helix transition in increasing concentrations of the hydrophobic solvent tri-fluoroethanol (TFE) (Figure 1B). Specific binding of the peptide to gold from aqueous solution was assessed by SPR (Biacore-X) with bare gold (Au-chip) surfaces and a synthetic switch tag peptide incorporating the FLAG antibody-binding motif (DYKDDDDKGG) at its N-terminus (FLAG-ST). To reduce nonspecific adhesion the gold was pretreated with 2-mercaptoethanol (Keegan et al 2005). Specific binding was determined by reference to peptides in which the Cys–SH group was blocked by iodoacetamide. This showed that 75% of surface mass is specifically bound via the Cys after washing. After infilling the remaining surface by injection of PEG-thiol solution, to inhibit nonspecific binding (NSB) (Seigel et al 1997), anti-FLAG antibody was injected. Following a wash with 0.1% sodium dodecyl sulfate (SDS) solution to remove nonspecifically bound antibody, 145 resonance units (R.U. = 0.0001 degree shift in SPR minimum position) remained bound to the iodoacetamide-blocked peptide-treated surface, whereas 1250 R.U. were bound to the nonblocked peptide surface, clearly demonstrating the thiol-directed assembly of FLAG-peptides (Figure 2a). The background is reduced still further if a longer ex situ assembly of PEG-thiol is performed (data not shown). The ST sequence was genetically engineered onto the C-terminus of the bacterial protein TolAIII, fusions to which are known to be well expressed and readily purified from Escherichia coli. (Anderluh et al 2003). The two naturally occurring cysteine residues in the protein were mutated to serine, leaving the ST peptide cysteine as the only thiol group. The complete plasmid allows for insertion of proteins between the TolAIII and ST modules and subsequent proteolytic cleavage to remove the stabilizing TolA fusion partner. This was achieved with a multiple cloning site (MCS) to allow the simple insertion of any chosen protein sequence between the TolA-III and ST coding regions. A thrombin protease cut site was also included between the TolA-III and MCS sequences to allow the TolA-III protein to be cleaved from the tagged fusion protein and a FLAG epitope inserted in TolAIII. The TolAIII- FLAG-ST fusion protein was expressed in E. coli BL-21 cells and purified by Ni2+ affinity chromatography as described (Anderluh et al 2003). Far UV-CD analysis showed that it was soluble and correctly folded and SPR revealed that 510 R.U. of fusion protein remained bound after washing versus only 25 R.U. of the iodoacetamide-blocked species. The surfaces were briefly in-filled in situ with PEG-thiol, and probed with anti-FLAG antibody, as described previously; 1580 R.U. of antibody remained bound after washing to the normal surface whereas only 265 R.U. remained on the blocked protein surface (Figure 2b). A longer PEG-thiol incubation would decrease this still further since errors in the SAM are sites for nonspecific binding. Next, green fluorescent protein (GFP) was used as a model for an inserted protein domain as it is easily visualized on surfaces. The coding DNA sequence of GFP was cloned into the plasmid and protein expressed in BL-21 E. coli. Microcontact printing (Xia and Whitesides 1998) was used to pattern arrays of 5-μm diameter spots of a TolAIII-GFP-Switch-Tag fusion protein later in filled with 1 mM PEG-thiol solution in 100% ethanol overnight to assemble a PEG-terminated SAM both in and around the spots. The protein was easily visualized by fluorescence microscopy and thus remains folded (Figure 3a). Rat primary osteoblast cells incubated on the surface adhered to the islands of protein but were unable to bind to the surrounding PEG-thiol coated surface, as shown in Figure 3b. The cells are much larger than the pattern and adjust their contact points to avoid the PEG in-filled areas. GFP has no known specific cell adhesion properties but provides clear anchor points within the PEG-layer. This illustrates that the ST is able to specifically immobilize recombinant proteins as defined patterns within an otherwise homogeneous SAM. The GFP version was also printed using a stamp which contains holes. The stamped surface was backfilled with PEG-thiol and incubated with primary osteoblast cells which avoided the protein-free region (Figure 3c).
Figure 1.
(A) Image of the predicted structure of the switch tag peptide (ST) in its helical conformation. (B) Far UV CD spectra of ST. A 0.2 mg/ml solution of peptide in 50 mM NaH2PO4 buffer, pH 7.0 was used throughout. Increasing the quantity of the hydrophobic solvent TFE promoted the transition of peptide secondary structure from random coil to α-helix.
Abbreviation: TFE, tri-fluoro ethanol.
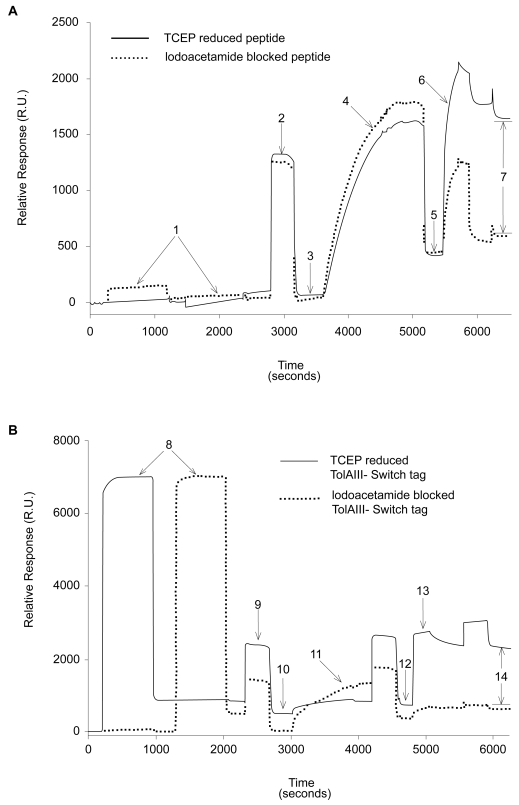
Figure 2.
SPR traces showing differential binding of reduced versus blocked FLAG-ST (A) and FLAG-TolAIII-ST (B) protein to gold surfaces. A buffer of 200 mM NaCl, 0.5 mM EDTA and 20 mM Tris pH 8 was used throughout. 25 μl injections of 0.1 mg/ml peptide or protein solution plus 0.5 mM TCEP at 2 μl/min were used (points 1 and 8). A 25 μl wash of running buffer plus 1% SDS at 5 μl/min (points 2 and 9) removed nonspecifically bound material, leaving immobilized molecules on the surface (3 and 10). 25 μl of 0.5 mM PEG-Thiol solution in running buffer was then injected at 2 μl/min and the surface again washed with 1% SDS as before. This led to a mass increase of around 350 R.U. (5 and 12). To ensure that the PEG-thiol had not displaced protein, 20 μl of 0.1 mg/ml anti-FLAG antibody solution was injected at 5 μl/min (6 and 13), followed by a wash of 25 μl 0.1% SDS solution. The amounts of specifically bound antibody (points 7 and 14) was found to be 1105 R.U. on the peptide surface and 1315 R.U. on the protein surface.
Abbreviations: SDS, sodium dodecyl sulfate; SPR, surface plasmon resonance; ST, switch tag peptide; TCEP, tris(2-carboxyethyl)phosphine.
Figure 3.
(A) Fluorescence microscopy images demonstrating patterning of TolAIII-GFP-ST protein, micro-contact printed in arrays on gold with a feature size of 5 μm followed by a PEG-thiol backfill. (B) Rat primary osteoblast cell, visualized by DAPI staining, adhering specifically to the GFP protein spots and avoiding the background of PEG-thiol. This demonstrates that the protein is exposed and providing attachment sites. The cell was incubated on the surface in serum free media for 24 h. (C) Printing with a “negative” stamp with holes instead of pillars provides protein free areas and allows for control of cell adhesion. Cell free features are nonprinted 300 μm diameter areas containing only back filled PEG-thiol. In this case the protein is printed for 10 s with a 4h PEG-thiol backfill. Cells were incubated for 24 h and stained with DAPI. (D) Design of the Tol-fusion-ST system. The fusion protein provides solubility and FLAG tag antibody site which can be later removed by specific proteases indicated by scissors leaving a Protein-ST combination.
The ability of phospholipid layers to limit nonspecific binding has been exploited in combination with patterned arrays of soluble proteins (Kung et al 2000), whilst other approaches have combined the peptide with the lipid itself (Svedhem et al 2003) or printed proteins onto a reactive SAM (Rozkiewicz et al 2006). Incorporation of functional synthetic peptides into SAM is straightforward and patterning has been achieved by micro-contact printing ethylene glycol-terminated SAM in patterns leaving exposed gold for subsequent peptide assembly (Zhang et al 1999). The system described here provides a generic method to co-assemble proteins with classical SAM. We have designed a plasmid which attaches a short peptide sequence which can insert into an 11 carbon chain thick SAM (Figure 3d). It was previously shown that hydrophobic helical peptides can be assembled into dense SAM and retain a helical structure (Kitagawa et al 2004). The current system avoided altering the protein’s hydrophobicity by allowing the peptide to be random coil in solution and helical in the SAM. Although the behavior of the peptide and modified proteins confirmed the first property of water solubility, it was unfortunate that the relatively low immobilization density prevented FTIR confirmation of helix formation in the SAM (Kitagawa et al 2004). Nevertheless the TolAIII-ST expression system is able to produce engineered proteins that show specific adhesion without compromising water solubility or expression levels in E. coli. We have previously shown that by pre-treating the surface with β-mercapto-ethanol surface interactions are reduced and proteins keep their 3D structure (Terrettazet al 2002; Keegan et al 2005) the subsequent assembly with lipids stabilizes the array and further contributes to retaining protein structure (Terrettaz et al 2002; Holt et al 2005; Cisneros et al 2006). This expression system has the potential to increase the range and complexity of proteins that can be specifically immobilized on surfaces designed to modify cellular growth (Shah et al 2007) or the development of new detector elements for SPR, SAW, or QCM.
Materials and methods
The ST sequence was developed using Agadir (Munoz and Serrano 1994) (http://www.embl-heidelberg.de/Services/serrano/agadir/agadir-start.html). The helix-disrupting glycine residue was added to a poly-alanine sequence at different positions and the resulting helix propensities calculated. A sequence with a central glycine retained sufficient helical propensity that an increase in environmental hydrophobicity would cause this sequence to rearrange from random coil to α-helix. Molecules based on this sequence, with the addition of a C-terminal cysteine and proline residues and N-terminal sequences of either KYDD to aid solubility and concentration measurements or DYKDDDDKGG, a FLAG epitope, were synthesised by f-MOC chemistry and purified to >95% as assessed by high-pressure liquid chromatography and MALDI-TOF mass spectrometry (Invitrogen, EvoTrack, Paisley, UK). Helical structure and thio alkane monolayer were modeled and energy minimized using the Quanta v4.0 package (Accelrys, Orsay Cedex, France).
Circular dichroism spectroscopy
Solutions of 0.2 mg/ml peptide in 50 mM NaH2PO4, pH 7.0 were made with increasing concentrations of the hydrophobic solvent TFE (Sigma-Aldrich, Gillingham, UK) up to 80% TFE, the limit of peptide solubility. All measurements were made with 0.2 mm path length quartz Suprasil cuvettes (Hellma UK, Southend on Sea, UK) using a JASCO J-810 spectropolarimeter, over a wavelength range of 250–195 nm at a scan speed of 20 nm min−1. The mean spectrum derived from 10 scans was corrected by subtraction of a background buffer blank.
Surface plasmon resonance measurements
Surface plasmon resonance measurements of SAM formation used a Biacore X system with “Au” gold sensor chips. The gold surface was cleaned with Piranha solution (Kitagawa et al 2004) for 30 minutes, rinsed with H2O, 1% SDS and finally extensively with H2O before drying with air and immediate docking into the Biacore system. A buffer of 200 mM NaCl, 0.5 mM EDTA, and 20 mM Tris pH 8.0 was used for all solutions and also as the running buffer. Peptide and protein solutions at a concentration of 0.1 mg/ml were used in all experiments. The PEG-thiol (11-mercapto-1-triethyleneglycol-undecane, HSC11-EG3) (ProChimia Surfaces, Sopot, Poland) was used at a concentration of 0.5 mM. The solutions used for thiol binding steps were supplemented with 0.1 mM TCEP (Tris(2-carboxyethyl)phosphine hydrochloride) for 1 h before use. Anti-FLAG monoclonal antibodies were obtained from Sigma-Aldrich and used at a dilution concentration of 0.1 mg/ml prepared daily in running buffer.
TolAIII-Switch tag fusion protein expression constructs were based on the pTol-T vector (Anderluh et al 2003). They were expressed in E. coli BL-21 cells and purified as detailed previously (Anderluh et al 2003).
Micro-contact printing
Micro-contact printing using PDMS stamps was used to transfer patterns of ST protein onto gold substrates (Xia and Whitesides 1998). The gold was dried before printing. The stamps were inked using the relevant alkanethiol or protein and dried in a stream of compressed air, placed on the gold and left for 15 seconds. The stamps were removed and the surface air dried for 10 minutes. If required the surface of the gold was then immersed in the relevant infill solution and left for a minimum of 4 hours to allow formation of a SAM.
Eukaryotic cell culture
Primary osteoblastic cells were isolated from the calvariae of five-day old rats (Bokhari et al 2005) They were maintained in a culture medium of Dulbecco’s modified Eagle’s Medium (DMEM, Sigma Aldrich) supplemented with 1% penicillin/streptomycin solution, 1% L-glutamine solution, and 10% fetal calf serum. Cells were passaged at confluency and used within the first four passages.
Acknowledgments
This work was supported by the BBSRC Grant 13/E19051. The authors report no conflicts of interest in this work.
References
- Anderluh G, Gokce I, Lakey JH. Expression of proteins using the third domain of the Escherichia coli periplasmic-protein TolA as a fusion partner. Protein Expr Purif. 2003;28:173–81. doi: 10.1016/s1046-5928(02)00681-2. [DOI] [PubMed] [Google Scholar]
- Bertone P, Snyder M. Advances in functional protein microarray technology. FEBS J. 2005;272:5400–11. doi: 10.1111/j.1742-4658.2005.04970.x. [DOI] [PubMed] [Google Scholar]
- Bokhari MA, Akay G, Zhang SG, et al. Enhancement of osteoblast growth and differentiation in vitro on a peptide hydrogel – polyHIPE polymer hybrid material. Biomaterials. 2005;26:5198–208. doi: 10.1016/j.biomaterials.2005.01.040. [DOI] [PubMed] [Google Scholar]
- Cisneros DA, Muller DJ, Daud SM, et al. An approach to prepare membrane proteins for single-molecule imaging. Angew Chem Int Ed Engl. 2006;45:3252–6. doi: 10.1002/anie.200504506. [DOI] [PubMed] [Google Scholar]
- Cooper MA. Label-free screening of bio-molecular interactions. Anal Bioanal Chem. 2003;377:834–42. doi: 10.1007/s00216-003-2111-y. [DOI] [PubMed] [Google Scholar]
- Cooper MA. Non-optical screening platforms: the next wave in label-free screening? Drug Discov Today. 2006;11:1068–74. doi: 10.1016/j.drudis.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Csucs G, Michel R, Lussi JW, et al. Microcontact printing of novel co-polymers in combination with proteins for cell-biological applications. Biomaterials. 2003;24:1713–20. doi: 10.1016/s0142-9612(02)00568-9. [DOI] [PubMed] [Google Scholar]
- Delehanty JB, Ligler FS. A microarray immunoassay for simultaneous detection of proteins and bacteria. Anal Chem. 2002;74:5681–7. doi: 10.1021/ac025631l. [DOI] [PubMed] [Google Scholar]
- Ekins R, Chu FW. Microarrays: their origins and applications. Trends Biotechnol. 1999;17:217–8. doi: 10.1016/s0167-7799(99)01329-3. [DOI] [PubMed] [Google Scholar]
- Hasirci V, Kenar H. Novel surface patterning approaches for tissue engineering and their effect on cell behavior. Nanomedicine. 2006;1:73–89. doi: 10.2217/17435889.1.1.73. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Mooney DJ. Peptide and protein presenting materials for tissue engineering. Adv Mater. 2004;16:17–25. [Google Scholar]
- Hodneland CD, Lee YS, Min DH, et al. Selective immobilization of proteins to self-assembled monolayers presenting active site-directed capture ligands. Proc Natl Acad Sci U S A. 2002;99:5048–52. doi: 10.1073/pnas.072685299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SA, Lakey JH, Daud SM, et al. Neutron reflectometry of membrane protein assemblies at the solid/liquid interface. Aust J Chem. 2005;58:674–7. [Google Scholar]
- Houseman BT, Gawalt ES, Mrksich M. Maleimide-functionalized self-assembled monolayers for the preparation of peptide and carbohydrate biochips. Langmuir. 2003;19:1522–31. [Google Scholar]
- Huang H, Zhao Y, Liu Z, et al. Enhanced osteoblast functions on RGD immobilized surface. J Oral Implantol. 2003;29:73–9. doi: 10.1563/1548-1336(2003)029<0073:EOFORI>2.3.CO;2. [DOI] [PubMed] [Google Scholar]
- Jung SO, Ro HS, Kho BH, et al. Surface plasmon resonance imaging-based protein arrays for high-throughput screening of protein-protein interaction inhibitors. Proteomics. 2005;5:4427–31. doi: 10.1002/pmic.200500001. [DOI] [PubMed] [Google Scholar]
- Keegan N, Wright NG, Lakey JH. Circular dichroism spectroscopy of folding in a protein monolayer. Angew Chem Int Ed Engl. 2005;44:4801–4. doi: 10.1002/anie.200462977. [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Morita T, Kimura S. Observation of single helical peptide molecule incorporated into alkanethiol self-assembled monolayer on gold by scanning tunneling microscopy. J Phys Chem B. 2004;108:15090–5. [Google Scholar]
- Klenkar G, Valiokas R, Lundstrom I, et al. Piezo dispensed microarray of multivalent chelating thiols for dissecting complex protein-protein interactions. Anal Chem. 2006;78:3643–50. doi: 10.1021/ac060024+. [DOI] [PubMed] [Google Scholar]
- Kung LA, Kam L, Hovis JS, et al. Patterning hybrid surfaces of proteins and supported lipid bilayers. Langmuir. 2000;16:6773–6. [Google Scholar]
- Liu LY, Chen SF, Giachelli CM, et al. Controlling osteopontin orientation on surfaces to modulate endothelial cell adhesion. J Biomed Mater Res A. 2005;74A:23–31. doi: 10.1002/jbm.a.30221. [DOI] [PubMed] [Google Scholar]
- Macbeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–3. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- Miura Y, Kimura S, Kobayashi S, et al. Cation recognition by self-assembled monolayers of oriented helical peptides having a crown ether unit. Biopolymers. 2000;55:391–8. doi: 10.1002/1097-0282(2000)55:5<391::AID-BIP1013>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Munoz V, Serrano L. Elucidating the folding problem of helical peptides using empirical parameters. Nat Struct Biol. 1994;1:399–409. doi: 10.1038/nsb0694-399. [DOI] [PubMed] [Google Scholar]
- Petty RT, Li HW, Maduram JH, et al. Attachment of cells to islands presenting gradients of adhesion ligands. J Am Chem Soc. 2007;129:8966–7. doi: 10.1021/ja0735709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran N, Hainsworth E, Bhullar B, et al. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- Rozkiewicz DI, Kraan Y, Werten MWT, et al. Covalent microcontact printing of proteins for cell patterning. Chemistry. 2006;12:6290–7. doi: 10.1002/chem.200501554. [DOI] [PubMed] [Google Scholar]
- Seigel RR, Harder P, Dahint R, et al. On-line detection of nonspecific protein adsorption at artificial surfaces. Anal Chem. 1997;69:3321–8. doi: 10.1021/ac970047b. [DOI] [PubMed] [Google Scholar]
- Shah DS, Thomas MB, Phillips S, et al. Self-assembling layers created by membrane proteins on gold. Biochem Soc Trans. 2007;35:522–6. doi: 10.1042/BST0350522. [DOI] [PubMed] [Google Scholar]
- Sniadecki N, Desai RA, Ruiz SA, et al. Nanotechnology for cell-substrate interactions. Ann Biomed Eng. 2006;34:59–74. doi: 10.1007/s10439-005-9006-3. [DOI] [PubMed] [Google Scholar]
- Svedhem S, Dahlborg D, Ekeroth J, et al. In situ peptide-modified supported lipid bilayers for controlled cell attachment. Langmuir. 2003;19:6730–6. [Google Scholar]
- Terrettaz S, Ulrich W-P, Vogel H, et al. Stable self-assembly of a protein engineering scaffold on gold surfaces. Protein Sci. 2002;11:1917–25. doi: 10.1110/ps.0206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia YN, Whitesides GM. Soft lithography. Angew Chem Int Ed Engl. 1998;37:551–75. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Yap FL, Zhang Y. Protein and cell micropatterning and its integration with micro/nanoparticles assembly. Biosens Bioelectron. 2007;22:775–88. doi: 10.1016/j.bios.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Zhang S, Yan L, Altman M, et al. Biological surface engineering: a simple system for cell pattern formation. Biomaterials. 1999;20:1213–20. doi: 10.1016/s0142-9612(99)00014-9. [DOI] [PubMed] [Google Scholar]