Abstract
In response to low iron availability, Vibrio parahaemolyticus synthesizes and secretes a polyhydroxycarboxylate-type siderophore vibrioferrin which is composed of 1 mol each of 2-ketoglutaric acid, l-alanine, ethanolamine, and citric acid. We have previously reported the cloning and characterization of the pvuA gene, which encodes the 78-kDa outer membrane receptor protein for ferric vibrioferrin. In this study, nine genes involved in the biosynthesis and transport of vibrioferrin have been identified in the genomic regions surrounding the pvuA gene. The genes were sequenced, and gene disruptants were constructed by insertion mutation for phenotype analysis. Five of the genes, named pvsABCDE, constitute an operon that is expressed under iron-limiting conditions. Homology searches of their predicted protein products suggested that the four genes pvsABDE are implicated in the biosynthesis of the siderophore. Another gene in the same operon, pvsC, encodes a putative exporter that is homologous to members of the major facilitator superfamily of multidrug efflux pumps. The remaining four genes, named pvuBCDE, encode proteins strongly homologous to Escherichia coli FecBCDE, respectively, which are components of the ATP-binding cassette transporter system for ferric dicitrate. Reverse transcriptase PCR analysis revealed that these transport genes are transcribed as a single mRNA with the upstream genes, psuA and pvuA. Phenotypic comparison between the wild-type strain and its targeted gene disruptants supported the biological functions for the respective operons that were expected on the basis of the homology search.
Iron is an essential element for virtually all living organisms, and most bacteria require micromolar levels of bioavailable iron for their optimal growth (20). An effective mechanism for assimilating iron necessary for growth under iron-limiting conditions involves the synthesis and secretion of siderophores, which are low-molecular-weight, high-affinity iron-binding compounds that can scavenge extracellular iron and also remove transferrin- and lactoferrin-bound iron and facilitate its uptake by the bacterium (9, 34, 37, 42). The siderophores have shown considerable structural diversity, being classified mainly into hydroxamate and phenol-catecholate types according to their chelating groups (37). Recent discoveries of more hydrophilic siderophores have established a separate type, polyhydroxycarboxylate (complexone) (59). This type of siderophores has been reported to include rhizobactin (51), rhizoferrin (59), staphyloferrin B (13), vibrioferrin (71), and achromobactin (36). In gram-negative bacteria, all siderophore-mediated iron acquisition systems so far described strictly require specific outer membrane receptors as well as ATP-binding cassette (ABC) transport systems (4).
On the other hand, the concentration of free iron must be tightly regulated, since it can catalyze the formation of dangerously reactive radicals (60). The ferric uptake regulation protein Fur is the major repressor of iron uptake systems in gram-negative bacteria (2). Fur complexed with Fe2+ binds to DNA with a 19-bp consensus palindromic sequence, called the Fur box, within the promoter regions of iron uptake genes (2, 12), so that transcription of the genes is shut off, thus limiting the entry of excess iron into the cell. When iron becomes scarce in the cell, Fur is inactivated by the release of the iron cofactor, so that iron uptake genes are transcribed. Regulation of other genes involved in pathogenicity as well as in general metabolism also occurs through the action of Fur, and therefore the restricted availability of iron in the host has been proposed to constitute a major signal which coordinately regulates the expression of such genes (14, 28, 33).
Vibrio parahaemolyticus is a halophilic gram-negative pathogen that naturally inhabits marine and estuarine environments and causes seafood-related gastroenteritis, particularly in Japan and throughout Asia (24). It is capable of acquiring iron through the action of the native siderophore vibrioferrin (70, 71) and of utilizing heme compounds as sole iron sources (69). The chemical structure of vibrioferrin has been confirmed by total synthesis (56). In addition, the Fur protein overexpressed from the fur gene cloned from this species (68) was shown to bind the Fur box for the sodA gene by gel mobility shift assay (16). However, relatively little is known about the genetic basis for the siderophore-mediated iron acquisition system in V. parahaemolyticus, while much is known about those of other Vibrio species, such as V. cholerae (6, 7, 64, 66, 67), V. vulnificus (29, 63), and V. anguillarum (10, 64).
Our previous study identified the V. parahaemolyticus pvuA gene, encoding the ferric vibrioferrin receptor (17). In the present study, we cloned and analyzed the genetic regions surrounding the pvuA gene and identified two iron-regulated operons, containing five genes (named pvsABCDE [pvs stands for V. parahaemolyticus vibrioferrin synthesis]) and four genes (named pvuBCDE [pvu stands for V. parahaemolyticus vibrioferrin utilization]). Homology searches of the respective protein products suggested that these operons are involved in the biosynthesis of vibrioferrin and transport of its ferric complex. Finally, the functions of the operons were confirmed by construction of V. parahaemolyticus mutants with insertional disruptions in some of the genes, coupled with their phenotypic analysis.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise noted, strains were grown at 37°C in Luria-Bertani (LB) medium (46) containing 0.5% (for Escherichia coli) or 3% (for V. parahaemolyticus) NaCl. Cloning and plasmid preparation were performed in E. coli DH5α (21) or E. coli JM109 (72). To impose iron limitation on bacteria, either 2,2′-dipyridyl or ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDA) (Sigma) was added to the medium at the indicated concentrations. All glassware used for preparation of iron-limiting media was washed in 6 M HCl and rinsed in distilled deionized water. When required, appropriate antibiotics were added to the media at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 10 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| V. parahaemolyticus | ||
| WP1 | Clinical isolate | 17 |
| AQ3354 | Clinical isolate | 17 |
| TNB1 | AQ3354, pvsA disrupted; Cmr | This study |
| TNB2 | AQ3354, pvsD disrupted; Cmr | This study |
| TNB3 | AQ3354, pvsE disrupted; Cmr | This study |
| TNB4 | AQ3354, pvuB disrupted; Cmr | This study |
| E. coli | ||
| DH5α | endA1 hsdr17 (rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 deoR [φ80d lac Δ(lacZ)M15)]; general cloning host | Promega |
| JM109 | recA1 endA1 gylA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F′ (traD36 proAB+lacIqlacZΔM15); general cloning host | 72 |
| SY327λpir | Δ(lac pro) argE(Am) recA56 gyrA rpoB λpir, host for π-requiring plasmids | 35 |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu, λpir; Kmr; host for π-requiring plasmids; conjugal donor | 35 |
| Plasmids | ||
| pUC19 | High-copy-number cloning vector; Apr | 61 |
| pBluescript II KS(+) | High-copy-number cloning vector; Apr | Stratagene |
| pMW118 | Low-copy-number cloning vector; Apr | Nippon Gene |
| pACYC184 | Low-copy-number cloning vector; Tcr Cmr | 8 |
| pKTN701 | R6K-ori suicide vector for gene replacement; Cmr | 39 |
| pVP3151 | Initially isolated FURTA-positive clone; pUC19 containing chromosomal 3,151-bp PstI fragment from WP1; Apr | 17 |
| pVS1 | pMW118 containing chromosomal 3.2-kb SalI-SalI fragment from WP1; Apr | This study |
| pVS1-1 | pBluescript II KS(+) containing 373-bp PstI-KpnI fragment from pVS1; Apr | This study |
| pVS2 | pMW118 containing chromosomal 7.5-kb HindIII-HindIII fragment from WP1; Apr | This study |
| pVPV2995 | pBluescript II KS(+) containing the chromosomal 2,995-bp EcoRI-SacI fragment from WP1; Apr | 17 |
| pVFRD1 | pACYC184 containing chromosomal 7.7-kb SalI-HindIII fragment from WP1; Cmr | This study |
| pTNB1 | pKTN701 containing 682-bp XbaI-EcoRI fragment within pvsA from pVS2; Cmr | This study |
| pTNB2 | pKTN701 containing 748-bp SacI-KpnI fragment PCR amplified with primers PVSD-Kpn and PVSD-Sac with pVS2 as a template; Cmr | This study |
| pTNB3 | pKTN701 containing 688-bp KpnI-EcoRI fragment PCR amplified with primers PVSE-Kpn and PVSE-Eco with pVS2 as a template; Cmr | This study |
| pTNB4 | pKTN701 containing 601-bp SacI-KpnI fragment PCR amplified with primers PVUB-Sac and PVUB-Kpn with pVFRD1 as a template; Cmr | This study |
DNA manipulations.
Chromosomal DNAs of V. parahaemolyticus strains were extracted from overnight cultures with a Wizard genomic DNA purification kit (Promega), and plasmid DNA was routinely prepared with a plasmid miniprep kit (Bio-Rad), according to the manufacturer's protocols. Cloning, restriction endonuclease digestion, and DNA ligation were carried out according to standard protocols (46). Restriction fragments were isolated, as required, from agarose gels by using a Prep-A-Gene purification kit (Bio-Rad), and electroporation was performed in a Gene Pulser apparatus (Bio-Rad), as detailed by the manufacturer. Restriction enzymes and a DNA ligation kit were purchased from Takara Biomedicals (Kyoto, Japan). PCR was performed basically as previously described (17). When PCR fragments required minimal errors, the high-fidelity KOD-plus DNA polymerase (Toyobo, Osaka, Japan) was used.
Southern blotting and colony hybridization.
Southern blotting and colony hybridization were performed according to the DIG system user's guide for filter hybridization (Roche Diagnostics). Restriction enzyme-digested DNA fragments were separated through a 1% agarose gel, transferred to a positively charged nylon membrane (Roche Diagnostics) with a vacuum blotter (Bio-Rad), and fixed to the membrane by baking it for 15 min at 80°C. Also, colonies on a nylon membrane for colony and plaque hybridization (Roche Diagnostics) were denatured and neutralized, and the transferred DNA was fixed to the membrane by baking it for 30 min at 120°C. Blots were incubated overnight at 68°C with an appropriate digoxigenin (DIG)-labeled probe. After treatment of the membrane with alkaline phosphatase-labeled anti-DIG Fab fragments, the hybridized DNA was detected with a CSPD reagent for Southern blot analysis and with colorimetric detection reagents for colony hybridization (Roche Diagnostics).
Cloning of the pvs and pvu genes.
The chromosomal DNA of V. parahaemolyticus WP1 was fully digested with various combinations of restriction enzymes, and the DNA fragments were examined by Southern hybridization with the following DIG-labeled probes (Fig. 1C). DIG-labeled probes A (780 bp), B (533 bp), and C (645 bp) were prepared with the oligonucleotide primer sets FVp 28F (5′-GGGCAGAAGGTTTGGTCTTTATTGT-3′) and FVp 28R (5′-ATGGTTTCGGTTGCCGATGG-3′), VP-VFB-F (5′-CGAACAACCTGATTGCTGGC-3′) and VP-VFB-R (5′-GTTGATAGTACTGCTCCGCC-3′), and VF-F5 (5′-GGGTACTCGTTATGTTAACG-3′) and VF-R4 (5′-GTGTAAGGCAGCTGGTTACC-3′), respectively, under the PCR conditions recommended in the DIG PCR probe synthesis kit (Roche Diagnostics). Plasmids pVP3151 and pVP2995 (17) were used as the templates for preparation of probes A and C, respectively, and plasmid pVS1 (Fig. 1B) was used for preparation of probe B. After electrophoresis, DNA fragments which had been confirmed to hybridize with the respective probes were excised and extracted from the agarose gel. The DNA fragments were ligated into plasmids cut with appropriate restriction enzymes, and the resulting ligates were transformed into competent E. coli DH5α or JM109 cells. Colonies on LB agar (1.5%) containing an appropriate antibiotic were screened by colony blot hybridization with the same probe. The nucleotide sequences of the positive plasmid clones, termed pVS1, pVS2, and pVFRD1 (Fig. 1B), were determined with the subclones derived from them.
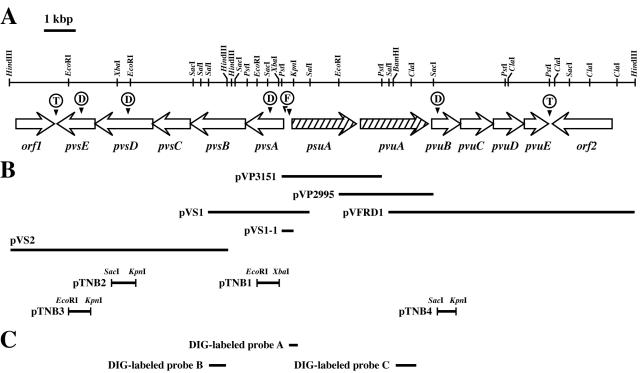
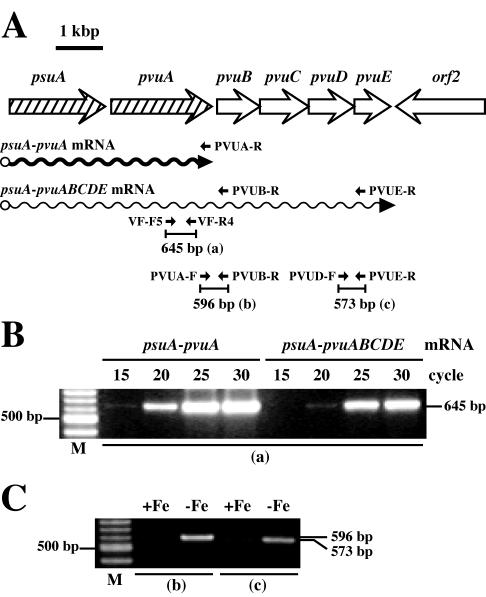
FIG. 1.
(A) Restriction enzyme map and genetic organization of the pvs and pvu loci of V. parahaemolyticus WP1, including the cognate vibrioferrin receptor gene (pvuA) and a receptor gene (psuA) for a siderophore of unknown origin (18). The transcriptional direction for each ORF is shown by an open arrow. D, F, and T indicate positions of disruption, putative Fur boxes, and termination signals, respectively. (B) Inserts of the relevant recombinant plasmids. pVP3151 was initially isolated as a clone conferring a Lac+ phenotype in the FURTA (53). The smaller solid bars with restriction enzyme names represent the insert fragment ligated into the suicide vector pKTN701 to construct the gene disruptants. (C) DIG-labeled probes constructed for hybridization experiments.
Determination and analysis of nucleotide sequences.
Double-stranded DNA sequencing was performed with a DNA sequencer (SQ5500E; Hitachi, Tokyo, Japan) and a Thermo Sequenase premixed or core cycle sequencing kit (Hitachi) by primer walking in two directions. The custom-synthesized primers were labeled with a 5′-oligonucleotide Texas red labeling kit (Amersham Pharmacia Biotech). Sequence analysis was conducted with the Genetyx-Mac 9.0 software package (Genetyx Software Development Co., Tokyo, Japan). The BLASTN and BLASTP programs (1) of the Institute for Chemical Research, Kyoto University, were used to determine homologies of the deduced amino acid sequences with other proteins. Conserved regions within proteins were identified by using the BLOCKS program (22). The hydropathic index profile was calculated by using the Kyte-Doolittle formula (27).
RNA isolation and analysis.
A culture of V. parahaemolyticus WP1 grown in LB medium to an A660 of 0.15 was split into two aliquots; one was left untreated (iron-replete cells), the other was supplemented with 2,2′-dipyridyl at 200 μM (iron-limiting cells). Both were then further incubated for 1 h. Total RNA from each cell sample was prepared by using an RNeasy Protect Bacteria kit (Qiagen) according to the manufacturer's protocol, and the amount of total RNA was quantified by measuring the A260.
(i) Primer extension.
Primer extension was performed with an oligonucleotide primer, PVSA-PE (5′-GAGTGACTGCATCATTAACG-3′), complementary to pvsA, which was 5′ labeled with Texas red as described above. The labeled primer was annealed to 10 μg of total RNA, and the annealed primer was extended by use of avian myeloblastosis virus reverse transcriptase (RT) XL (Takara Biomedicals) at 50°C for 60 min according to the manufacturer's protocol. The extension products were separated by electrophoresis on a sequencing gel together with the DNA sequence ladder of the control region synthesized with the same primer and with pVS1 as a template.
(ii) RT-PCR.
RNA samples prepared as described above were pretreated with 30 U of RNase-free DNase I (Qiagen) for 1 h at 37°C to exclude the possibility of contamination with traces of chromosomal DNA, and the amount of total RNA was quantified again by measuring the A260. RT-PCR analysis was carried out with an RNA PCR kit (AMV version 2.1; Takara Biomedicals) according to the manufacturer's protocol. In this analysis, we employed the V. parahaemolyticus fur-specific transcript (68) both as an internal control for total RNA isolation efficiency and as a negative control for iron-regulated expression. cDNA synthesis was performed at 42°C for 1 h with 5 μg of DNase I-treated total RNA and the following primers: VPFUR-R2 (5′-ACCACCGTCACTGCATTTAC-3′), complementary to the V. parahaemolyticus fur gene (68); PVSE-R (5′-GCATTACTCAGTTCGGACTC-3′), complementary to pvsE; PVUA-R (5′-CGGAAGTAATACTCCTCGTC-3′), complementary to pvuA; PVUB-R (5′-GGATGACTCGTATGGCATCG-3′), complementary to pvuB; and PVUE-R (5′-GCTTGTCAGCAAACTCAATC-3′), complementary to pvuE. Subsequent PCR was carried out with 2 μl of RT reaction mixture as a template as follows: after an initial denaturation of 2 min at 94°C, DNA was amplified for 30 cycles (unless otherwise indicated), with each cycle consisting of denaturation at 94°C for 30 s, annealing at appropriate temperatures as indicated for 30 s, and extension at 72°C for 1 min. For PCR amplification, the following primer pairs were used: for fur, VPFUR-F1 (5′-TCTAGAAGTACTTCAGCAGCCAG-3′) and VPFUR-R1 (5′-TAGCTGCTATTTCGCGTTGG-3′); for the pvsAB region, PVSA-F (5′-AAATTAACTACCGCAGCATTGG-3′) and PVSB-R (5′-GCTACTGAACTTGGCGTTCG-3′); for the pvsDE region, PVSD-F (5′-GCCAGAACTAGAAGCATTGC-3′) and PVSE-R; for the pvuA region, VF-F5 and VF-R4; for the pvuAB region, PVUA-F (5′-GATTACGCCAACACCAGTGC-3′) and PVUB-R; and for the pvuDE region, PVUD-F (5′-CACAAGTTGTTGATTCCTGC-3′) and PVUE-R. The RT-PCR products were analyzed by agarose gel electrophoresis and stained with ethidium bromide. The relative amounts of RT-PCR products on the gel were assessed with NIH Image, version 1.2 (http://rsb.info.nih.gov/nih-image/). Under our conditions, RT-PCR was able to specifically detect mRNA, because no band was observed when RT was omitted.
Construction of gene-targeted disruptants of V. parahaemolyticus.
To investigate their specific functions, the pvsA, pvsD, pvsE, and pvuB genes were each inactivated by inserting a suicide vector into the V. parahaemolyticus chromosome by homologous recombination as originally described by Miller and Mekalanos (35). Strain WP1 provided no mutants with disruption in these genes (17), and so we constructed such mutants from another vibrioferrin-producing strain, AQ3354, which appeared to have an arrangement of these genes identical to that of strain WP1. The EcoRI-XbaI fragment internal to pvsA (17) was cut out and ligated to R6K-ori suicide vector pKTN701 (39) digested with the same restriction enzymes. The resulting recombinant plasmid, termed pTNB1 (Fig. 1B), was used to transform competent cells of E. coli SY327λ pir. Chloramphenicol-resistant (Cmr) transformants were selected, and the insert of the plasmid DNA extracted was confirmed by restriction mapping. In the same way, plasmids pTNB2, pTNB3, and pTNB4, containing the DNA fragments internal to pvsD, pvsE, and pvuB, respectively, were constructed and selected (Fig. 1B). In these cases, the inserted DNA fragments in pTNB2, pTNB3, and pTNB4 were generated by PCR with the following respective primer sets and templates: PVSD-Kpn (5′-TAGCGTGCTGGGTACCGAGC-3′), PVSD-Sac (5′-ATGCAAACAGAGCTCCAGCG-3′), and pVS2; PVSE-Kpn (5′-CGGGTACCTGTATGACCTAAAC-3′), PVSE-Eco (5′-ATGAGGAATTCCAACTTGTCGC-3′), and pVS2; and PVUB-Sac (5′-CTTTTGAGCTCGAAGCGATACC-3′), PVUB-Kpn (5′-ACTAGGTACCAATCAG GGTTGG-3′), and pVFRD1 (the nucleotides changed for generation of the restriction enzyme sites are underlined). The PCR products were digested with the respective restriction enzymes to insert the resulting fragments into pKTN701. The plasmids thus constructed were transformed into E. coli SM10λ pir as a donor and transferred to V. parahaemolyticus AQ3354 by membrane filter mating conjugation, followed by incubation for 3 h at 37°C on a nitrocellulose filter laid over LB agar plates with 1.5% NaCl. Transconjugants were selected by overnight incubation at 37°C on LB agar plate containing ampicillin and chloramphenicol. Some of the Cmr colonies were isolated, and single-crossover recombination with respect to the corresponding genes was verified by comparative Southern blot hybridization and PCR amplification for the chromosomal DNAs from the mutant and wild-type strains (data not shown). The mutant strains with disruptions in pvsA, pvsD, pvsE, and pvuB thus obtained were designated TNB1, TNB2, TNB3, and TNB4, respectively.
Assay methods for siderophore production and growth under iron-limiting conditions.
Vibrioferrin production by V. parahaemolyticus AQ3354 and its disruptants was tested by incubating them on chrome azurol S (CAS) agar plates (48). Bacterial growth was monitored by measuring the A660s of cultures initially inoculated with stationary-phase cells at an A660 of 0.005 and then shaken (125 rpm.) at 37°C. When required, vibrioferrin (71) and EDDA were added to LB medium.
Nucleotide sequence accession numbers.
The nucleotide sequence data have been deposited in the DDBJ database under accession numbers AB082122 (pvu) and AB082123 (pvs).
RESULTS
Cloning and nucleotide sequences of the chromosomal regions surrounding the psuA-pvuA locus.
Our previous study has identified the operon containing psuA and pvuA, which encode a receptor for a ferric siderophore of unknown origin and the receptor for ferric vibrioferrin, respectively (17). To identify and characterize the genes involved in the vibrioferrin-mediated iron assimilation system in V. parahaemolyticus, we attempted to clone the chromosomal regions upstream and downstream of this locus by chromosomal walking. As a results, three kinds of successive recombinant plasmids, termed pVS1, pVS2, and pVFRD1, were selected from V. parahaemolyticus WP1 genomic libraries by Southern blot hybridization, and analysis of their DNA sequences disclosed the genetic organization shown in Fig. 1A.
(i) pvs locus.
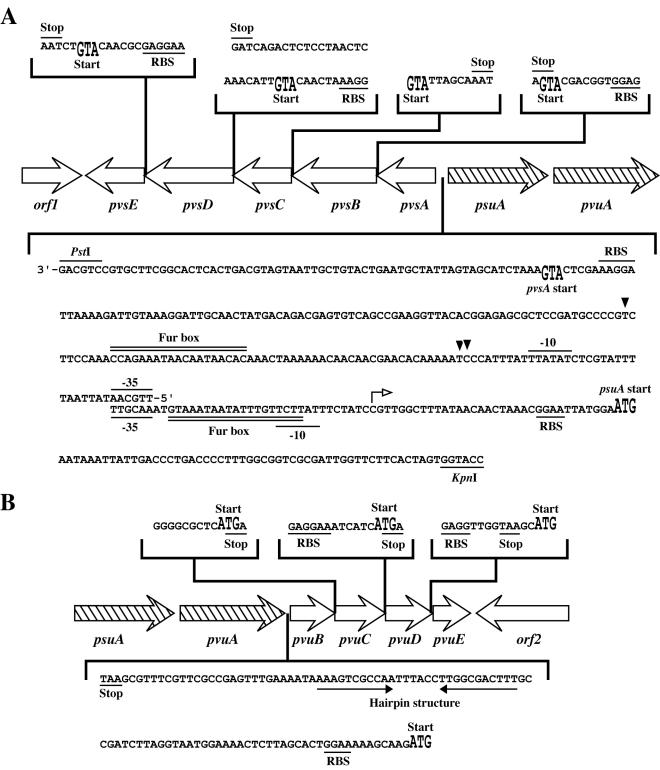
The nucleotide sequences of two overlapping plasmids, pVS1 (3.2 kb) and pVS2 (7.5 kb), were determined for their appropriate subclones. Scanning the sequence led to the detection of five contiguous open reading frames (ORFs) with the same transcription polarity, starting 248 bp upstream of the start codon for the known gene psuA and being transcribed in an opposite direction relative to psuA (Fig. 1A). These genes were designated pvsA, pvsB, pvsC, pvsD, and pvsE. The G+C content of the sequenced DNA including pvsABCDE, is 50.0 mol%, somewhat higher than the average G+C content of V. parahaemolyticus (46 to 47 mol%) (3). However, it is unclear at present whether this difference is statistically significant. The nucleotide sequence of the intergenic region between the psuA and pvsA genes is presented in Fig. 2A. We mapped a putative promoter region upstream of the start codon of pvsA, which shares significant homology (61.5%) to the consensus sequence (TTGACA and TATAAT) of the E. coli promoter. A potential ribosome-binding site (50) for pvsA is located 6 bp upstream of the proposed ATG translation initiation site. A putative Fur box with 11 matches of 19 nucleotides to the E. coli consensus (2, 12) was found upstream of pvsA but without overlapping with the putative promoter sequences. A pBluescript II KS(+) subclone, pVS1-1, containing the PstI-KpnI fragment of pVS1 (Fig. 1B) conferred a Lac+ phenotype to E. coli H1717 in the Fur titration assay (FURTA) (53), confirming the presence of the potential Fur boxes within the intergenic region between the pvsA and psuA genes. Moreover, these genes were suggested to exist in an operon structure, as the start and stop codons for the individual ORFs either overlap or are separated by no more than 6 bp (Fig. 2A). Therefore, transcription of these genes would be regulated by the Fur protein in response to exogenous iron concentrations through the potential Fur box located in the promoter region of the pvsA gene. A potential rho-independent transcription termination signal (45) was found 22 bp downstream of pvsE.
FIG. 2.
Nucleotide sequences in the intergenic regions of the pvs (A) and pvu (B) operons. The putative −35 and −10 promoter sequences as well as putative Fur box sequences in the junction region between pvsA and psuA are indicated. The transcription start sites for the pvs operon are indicated by solid arrowheads. The predicted ribosome-binding site (RBS) and the start and stop codons for each gene are also presented. The transcription start site determined for the psuA-pvuA operon (17) is indicated by a small open arrow above the nucleotide sequence. The hairpin-like structure located between the pvuA and pvuB genes is indicated by converging arrows.
(ii) pvu locus.
Nucleotide sequencing of the 7.7-kb SalI-HindIII insert of pVFRD1 revealed four complete ORFs with the same transcription polarity, which were designated pvuB, pvuC, pvuD, and pvuE (Fig. 1A). The G+C content of the coding region was 49.9 mol%. Although the expected ATG initiation codon of pvuB is 101 bp from the end of the preceding gene pvuA, there was no potential promoter sequence for the pvuB gene in this region (Fig. 2B). The pvuBCDE genes also show either the virtual absence of or 2 bp of intervening sequence (Fig. 2B). In addition, plasmid pVFRD1 conferred a Lac− phenotype in the FURTA (53), indicating that there is no functional Fur box in the promoter region. As described below, transcriptional linkage of the pvuBCDE genes to the upstream genes, psuA and pvuA, in an operon fashion was demonstrated by RT-PCR experiments.
Homology of predicted protein sequences.
The first gene product in the pvs operon, PvsA, shows no homology with any protein of known function but shows 26% identity (40% similarity) to a hypothetical protein annotated as a ligase/carboxylase in Mesorhizobium loti (55). The protein products of pvsB and pvsD bear 24 and 23% identity (38 and 37% similarity) to IucC and IucA, respectively, in the aerobactin biosynthesis pathway of E. coli (32) and 26% identity (41% similarity) to RhbC in the rhizobactin 1021 biosynthesis pathway of Sinorhizobium meliloti (30). IucA, IucC, and RhbC all have been reported to be enzymes catalyzing amide bond formation reactions. No homology, however, was detected between PvsB and PvsD, although they were expected to catalyze formally similar reactions. The products of pvsC and pvsE show 24% identity (38% similarity) to the Streptococcus pneumoniae proton motive force-dependent 12-spanner membrane efflux pump belonging to the major facilitator superfamily (MFS) (19) and 28% identity (45% similarity) to the pyridoxal 5′-phosphate-dependent decarboxylase BtrK in Bacillus circulans (40), respectively. Particularly, PvsC has secondary structures typical of membrane-spanning transport proteins: motif A, a 13-residue sequence (G-[RKPATY]-L-[GAS]-[DN]-[RK]-[FY]-G-R-[RK]-[RKP]-[LIVGST]-[LIM]) between transmembrane spanners 2 and 3, conserved in all 17 families of MFS proteins (41), was found as GNFADRYGRRRSL, and motif D2 (lgx5PvxP), conserved in 12-spanner MFS proteins (44), was detected as LGMPLFLPVVL. Therefore, these findings suggested that the PvsC protein might be an inner membrane exporter involved in the secretion of vibrioferrin from the cell. As described below, since the disruptant in the last gene in the operon, pvsE, showed a phenotype defective in vibrioferrin production, PvsE may be required for decarboxylation of a certain precursor to form, most probably, an ethanolamine moiety.
The amino acid sequences deduced from the pvuBCDE genes showed relatively strong homologies with those of four corresponding components, FecBCDE, of the E. coli ABC transport system for ferric dicitrate (52). The PvuB protein displays 49% identify (66% similarity) to the periplasmic binding protein FecB. SignalP prediction (http://www.cbs.dtu.dk/services/SignalP/) (38) for PvuB identified a putative signal sequence of 31 amino acids at the N terminus, with a most likely signal peptidase cleavage site between 31S and 32Q. The signature sequence 101QPSLEAIAVLKPDLII116, with the three residues (underlined) fully conserved in iron complex-binding proteins (58), also characterizes the PvuB protein. In addition, the relevant motif 129LXXIAP134 for this cluster of periplasmic proteins (31) was also identified in PvuB. PvuC and PvuD were identified as integral cytoplasmic membrane transporters (permeases) with homology to FecC (38% identity and 55% similarity) and FecD (43% identity and 63% similarity), respectively. In addition, PvuC and PvuD are extremely hydrophobic (27), with each containing a distinctive domain known as the EAA motif (47). This motif, which is highly conserved among cytoplasmic membrane transporters, typically occurs in a hydrophilic loop region containing the invariant G residue located approximately 100 amino acid residues from the C terminus of the protein (amino acid positions 102 and 103 for PvuC and PvuD, respectively). Similarly to FecC and FecD in E. coli (52), PvuC and PvuD exhibit homology with each other (38% identity), suggesting that the PvuC and PvuD proteins in V. parahaemolyticus also form a heterodimeric membrane-localized transporter complex. The amino acid sequence deduced from pvuE shows strong homology with a number of ATP-binding proteins associated with the ABC transport systems and is the most homologous to FecE (56% identity and 75% similarity). The multiple alignment of PvuE with these proteins revealed the typical Walker motif A, 34GPNGCGKST42, and motif B, 158VVMLDEPT165(62), and the ABC signature 138LSGG141 (25) in the PvuE sequence. These sites are recognized for their ability to form an ATP-binding pocket and are the distinguishing feature among the family of ABC transporters (23).
The products of orf1 and orf2, with 365 and 628 amino acid residues, respectively, show homologies with a binding protein component of ABC sugar transporter in the Pseudomonas aeruginosa genomic sequence (37% identity in a 353-amino-acid overlap, AE004743) (54) and with a methyl-accepting chemotaxis protein in the V. cholerae genomic sequence (40% identity in a 640-amino-acid overlap, VC1289), respectively. Therefore, at this stage of our investigation, we expected that the pvsABCDE and pvuBCDE genes would direct the biosynthesis of vibrioferrin and the ABC transport system for ferric vibrioferrin, respectively.
Transcription start site for pvsA and analysis of pvsABCDE cotranscription.
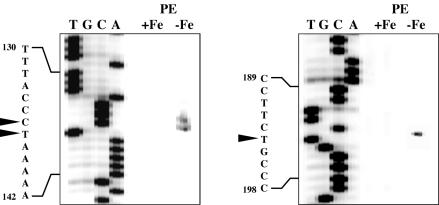
The transcription start site for pvsA was investigated by primer extension with total RNA from V. parahaemolyticus WP1 grown under iron-replete or iron-limiting conditions. Three major extended products were detected only when total RNA extracted from iron-limited cells was used as the template, and the start sites were determined to be at nucleotide positions 136, 137, and 194 near the predicted promoter of pvsA (Fig. 3; see also Fig. 2A). These results are consistent with the presence of a putative Fur box that can account for the iron-regulated expression of the gene via the Fur iron repressor protein. However, it is unclear whether the three major bands are all products of independent transcription initiation at individual sites or whether the smaller band is a specific degradation product of either of the larger bands or both.
FIG. 3.
Transcription start sites of pvsA determined by primer extension (PE). Reverse transcription was performed on total RNA from V. parahaemolyticus WP1 grown either under iron-replete (+Fe) or iron-limiting (−Fe) conditions. The same primer was used to generate a sequence ladder (T, G, C, A) from plasmid pVS1. The relevant sequence is shown to the left of each panel, and the transcription start sites detected are highlighted by arrowheads (see Fig. 2A).
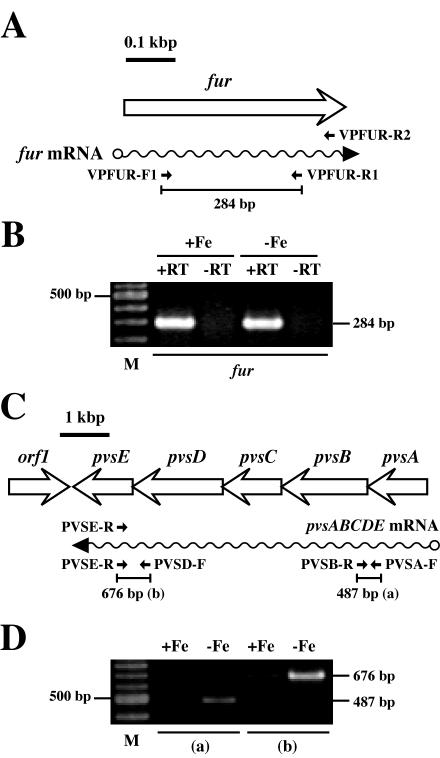
Next, in order to investigate the cotranscription of the pvs genes, RT-PCR analysis was performed with total RNA isolated from strain WP1 grown under iron-replete and iron-limiting conditions. The locations of the primers used for fur and pvs are shown in Fig. 4A and C. Expression of the V. parahaemolyticus fur gene was constitutive (i.e., not influenced by iron availability), and thus this gene was used as a negative control (Fig. 4B). Moreover, without the addition of RT, no products were amplified from the RNA templates isolated from iron-replete or iron-limited cultures (Fig. 4B), indicating that the RT-PCR products detected were attributable exclusively to mRNA. In contract to fur, the pvsABCDE genes were transcribed preferentially under iron-limiting conditions. Indeed, the anticipated 487-and 676-bp fragments were amplified with two primer sites, internal to the pvsA and pvsB genes and to the pvsD and pvsE genes, respectively, from mRNA templates expressed under iron-limiting conditions (Fig. 4D). From these results, we conclude that the pvs genes were cotranscribed as a single polycistronic mRNA and that its expression was controlled by iron availability.
FIG. 4.
Detection of the pvsABCDE operon by RT-PCR. (A and C) Schematic representation of the fur and pvsABCDE genes. The small arrows show the positions and directions of the gene-specific primers used in RT-PCR with lengths of the expected PCR products. The wavy arrows represent the transcript. (B and D) RT-PCR analysis of the constitutive fur gene (internal control) and for detection of the pvsABCDE transcript. The annealing temperatures for panels B and D were 58 and 60°C, respectively. Lanes contain RT-PCR products amplified from total RNAs isolated from V. parahaemolyticus WP1 grown under iron-replete (+Fe) and iron-limiting (−Fe) conditions. The sizes of individual RT-PCR products are indicated at the right of each blot. Lanes +RT and −RT, RT-PCR with and without RT, respectively; lanes M, molecular size standards.
Cotranscription of pvuBCDE with the upstream genes psuA and pvuA.
Their consecutive and close locations suggested that the pvuBCDE genes could be cotranscribed as a single polycistronic transcript. However, we were unable to detect either any extension band by primer extension analysis or any transcript corresponding to the complete pvuBCDE region by Northern blotting, consistent with the fact that no promoter sequence was found in the intergenic region between pvuA and pvuB (Fig. 2B). In addition, we have previously reported that the psuA-pvuA-specific transcript was abundant enough to detect it by Northern blotting (17). These observations gave rise to the possibility that the pvuBCDE genes might be transcribed together with the upstream genes, psuA and pvuA, but that this long mRNA species (psuA-pvuABCDE mRNA) might be present at very low levels. To confirm this hypothesis, relative RT-PCR was performed with total RNA isolated from strain WP1 grown under iron-limiting conditions. The locations of the primers used for detection of the psuA-pvuA and psuA-pvuABCDE transcripts are shown in Fig. 5A. The primers PVUA-R and PVUB-R were used for synthesis of cDNAs specific to the psuA-pvuA and psuA-pvuABCDE mRNAs, respectively, both of which would be expressed under iron-limiting conditions, and the common PCR primer pair, VF-F5 and VF-R4, was then used to amplify two different cDNA templates to yield the anticipated 645-bp amplicons. The intensity of each band at each cycle (20, 25, and 30 cycles) revealed that the psuA-pvuA transcript was more abundant than the psuA-pvuAB(CDE) transcript, with the greatest differential of approximately 14-fold at 20 cycles (Fig. 5B).
FIG. 5.
Demonstration of psuA-pvuA- and psuA-pvuABCDE-specific transcription by RT-PCR. (A) Schematic representation of the psuA and pvuABCDE genes, the primers used for RT-PCR, and differentially transcribed mRNAs. The thick wavy arrow represents the most abundant transcript (psuA-pvuA mRNA), which probably terminates at or adjacent to the hairpin-like structure located in the junction region between the pvuA and pvuB genes (see Fig. 2B), as reported previously (17). The thin wavy arrow represents a low-abundance transcript (psuA-pvuABCDE mRNA) detected in this study. The small arrows show the positions and directions of the gene-specific primers with lengths of the expected RT-PCR products. (B) Demonstration of differential abundance between the psuA-pvuA and psuA-pvuABCDE transcripts. Total RNA isolated from V. parahaemolyticus WP1 grown under iron-limiting conditions was used for cDNA synthesis with primers PVUA-R (for psuA-pvuA) and PVUB-R (for psuA-pvuABCDE), and PCR was performed with various numbers of cycles (15 to 30) with the common primer pair VF-F5 and VF-R4. (C) RT-PCR analysis for detection of the psuA-pvuABCDE transcript. Total RNA isolated from V. parahaemolyticus WP1 grown under iron-replete (+Fe) or iron-limiting (−Fe) conditions was used for reverse transcription with primer PVUE-R, and PCR was performed with two primer pairs, (i) PVUA-F and PVUB-R and (ii) PVUD-F and PVUE-R. The annealing temperatures for panels B and C were 56 and 61°C, respectively. The sizes of individual RT-PCR products are indicated at the right of each blot. Lanes M, molecular size standards.
As a means of confirming the transcriptional linkage of psuA-pvuA to pvuBCDE, we also used RT-PCR to amplify the regions between primers PVUA-F and PVUB-R and between primers PVUD-F and PVUE-R (Fig. 5A). Primer PVUE-R was also used for cDNA synthesis of the psuA-pvuABCDE transcript. The anticipated 596- and 573-bp fragments were preferentially amplified from total RNA isolated from the iron-limiting cultures (Fig. 5C). Detection of the 596-bp fragment containing the pvuA-pvuB intergenic region together with that of the 573-bp fragment containing part of pvuE clearly indicated the transcriptional linkage of psuA-pvuA to pvuBCDE. Moreover, the band sizes of the PCR products obtained in control reactions with the WP1 genomic DNA and the same primer pairs were concordant with those of the RT-PCR products (data not shown). These data demonstrate that the pvuBCDE genes are cotranscribed as a polycistronic mRNA with the psuA and pvuA genes from the Fur box-containing promoter upstream of psuA (17).
Characterization of the pvsA, -D, and -E and pvuB disruptants.
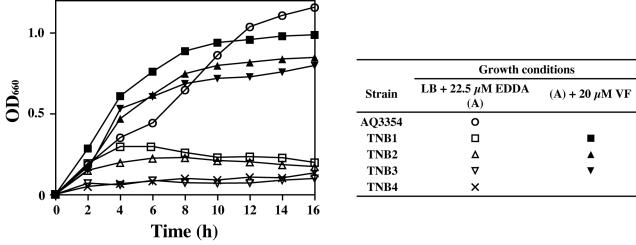
To demonstrate the dependence of vibrioferrin biosynthesis on the aforementioned pvs genes, pvsA, -D, and -E gene disruptants were constructed by homologous recombination with the pKTN701 suicide vector (39). Disruption of the respective genes was confirmed by Southern blot hybridization and PCR (data not shown). Vibrioferrin production was first examined for each disruptant by incubation on CAS agar plates (48). As shown in Fig. 6, the disruptants TNB1, TNB2, and TNB3 all showed a negative vibrioferrin production phenotype. In agreement with this, the disruptants failed to grow under iron-limiting conditions imposed by EDDA at a concentration of 22.5 μM, although the wild-type strain AQ3354 grew well under the same conditions (Fig. 7). However, growth of each disruptant was restored to the level of the wild-type strain in medium supplemented with vibrioferrin at 20 μM, confirming that the disruptants are in fact defective in vibrioferrin biosynthesis. On the other hand, although the pvuB disruptant TNB4, which had been constructed in a way similar to that described above, showed positive vibrioferrin production on the CAS agar plate (Fig. 6), it exhibited almost no growth under iron-limiting conditions compared to that of the wild-type (Fig. 7), Thus, the phenotype exhibited by each disruptant correlated well with the functions expected for the respective operons on the basis of the homology searches.
FIG. 6.
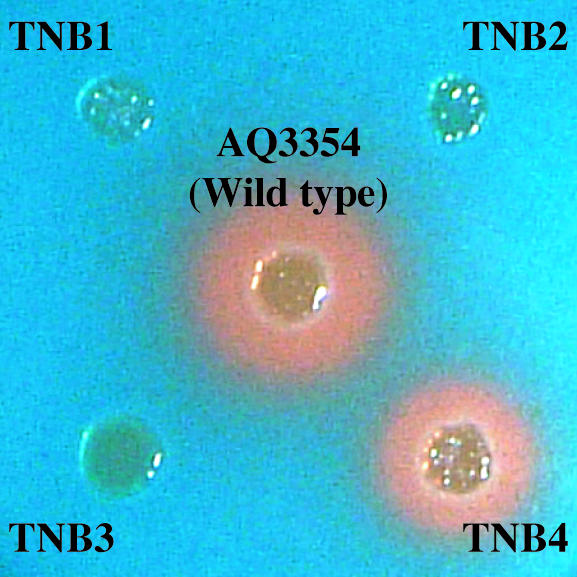
Vibrioferrin production assay of V. parahaemolyticus AQ3354 and its gene disruptants on a CAS agar plate. Each strain was grown in LB medium for 8 h, and after centrifugation the pellet was washed three times with 3% NaCl containing 10 mM MgCl2. The washed cell suspension (2 μl), adjusted to an optical density at 660 nm of 0.5, was placed on a CAS agar plate and incubated for 48 h at 37°C. The distinctive orange halos (desferrated CAS) observed for the wide-type strain AQ3354 and TNB4 are indicative of vibrioferrin production. The gene disruptants TNB1, TNB2, and TNB3 showed no vibrioferrin production.
FIG. 7.
Restoration of growth of the gene disruptants TNB1, TNB2, and TNB3 by the addition of vibrioferrin and impaired growth of TNB4 under iron-limiting conditions. Overnight cultures of V. parahaemolyticus AQ3354 and its gene disruptants TNB1, TNB2, TNB3, and TNB4 grown in LB medium were inoculated at an optical density of 660 nm (OD660) of 0.005 into the same medium containing 22.5 μM EDDA with or without 20 μM vibrioferrin (VF), and the growth was monitored by measuring the OD660. All of the strains except AQ3354 were cultured in the presence of chloramphenicol (10 μg/ml). Results from a representative experiment of three independent experiments are shown.
DISCUSSION
We previously reported that V. parahaemolyticus manganese-resistant mutants with impaired Fur function produced appreciable amounts of vibrioferrin even under iron-sufficient conditions that normally repress vibrioferrin synthesis in the wild-type strain (16). This study identified nine ORFs within two iron-regulated operons and demonstrated that one operon is involved in the biosynthesis of the cognate siderophore vibrioferrin and that the other is involved in the transport of its ferric complex into the cytosol. These results are consistent with the notion that genes required for the bacterial iron acquisition systems frequently form operons or are closely linked, with expression being negatively regulated by the Fur protein in conjunction with iron status. To the best of our knowledge, this is the first report of a functionally characterized gene cluster which is involved in the biosynthesis of a polyhydroxycarboxylate-type siderophore.
We assumed that three enzymes would be required to complete the vibrioferrin biosynthesis pathway from the four components, because they may be readily available from the cell. PvsB and PvsD show homology with E. coli IucC and IucA, respectively, catalyzing the amide bond formation reactions in the aerobactin biosynthesis pathway, the same arrangement as encountered in vibrioferrin, which contains two amide bonds. Thus, owing to its location within the same operon, the remaining pvsA gene seemed a likely candidate to encode an enzyme catalyzing the ester bond formation between citrate and ethanolamine or its derivatives. Its participation in the biosynthetic pathway for vibrioferrin is supported by the disruption in the pvsA gene, which resulted in impaired vibrioferrin production, although we should take into consideration that disruption of this gene has a polar effect on transcription of downstream genes within the pvs operon. On the other hand, PvsE shows homology with many bacterial pyridoxal 5′-phosphate-dependent decarboxylases, particularly with diaminopimelate decarboxylases, suggesting that it catalyzes the decarboxylation of a new kind of precursor amino acid, possibly containing a serine moiety, to produce an ethanolamine moiety. Further experiments will be required for determination of an indigenous substrate of PvsE.
The products of four pvs genes, not including pvsA, also are 23 to 42% identical to hypothetical proteins (SA0114, -0116, -0117, and -0119) in the Staphylococcus aureus genomic sequence (26) and 26 to 43% identical to the proteins (AcsA, AcsD, and LysA) annotated to be the achromobactin biosynthesis enzymes in Erwinia chrysanthemi (accession number AF416739) (36). These hypothetical proteins in S. aureus may be responsible for the biosynthesis of staphyloferrin B, the siderophore produced by this species (13, 49). It should be noted that these three siderophores contain a chiral citrate moiety and 2-ketoglutarate (in the ring form of pyrrolidinone) (Fig. 8). Interestingly, the hypothetical proteins more homologous (42 to 54% identity) to the five Pvs proteins, including PvsA, have recently been found in the Xanthomonas campestris pv. Campestris ATCC 33913 genomic sequence (11). Although there is so far no report on the production of a siderophore by this strain, a putative siderophore, if any, must be structurally very similar to vibrioferrin. Thus, the occurrence in these species of genes encoding proteins functionally homologous to those in V. parahaemolyticus suggests that biosynthetic pathways for the respective siderophores are genetically closely related and thus that the genes involved are clustered to encode a family of proteins specific to this type of siderophore.
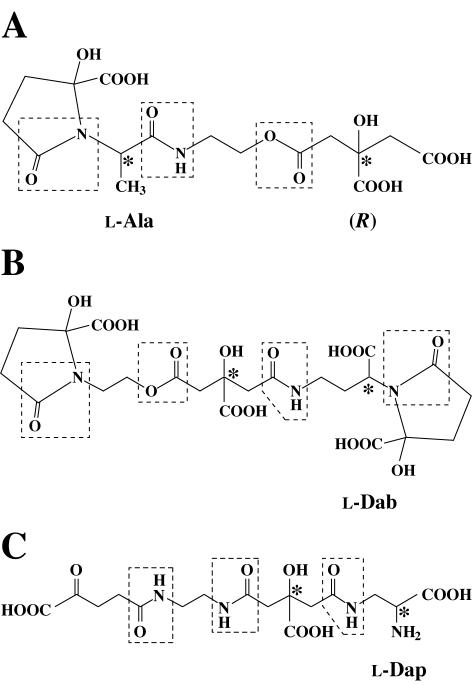
FIG. 8.
Chemical structures of vibrioferrin (A) and its related siderophores achromobactin (B) and staphyloferrin B (C). Asymmetric carbons are marked with asterisks. The absolute configuration of the chiral carbon in the citrate moiety of vibrioferrin has been established to be R (57), but those in two other siderophores remain to be determined. Complete existence in their cyclized forms has been proposed for vibrioferrin and achromobactin, but not for staphyloferrin B. Abbreviations: Ala, alanine; Dab, diaminobutyric acid; Dap, diaminopropionic acid.
It is very interesting to know how newly synthesized siderophores are exported and how the siderophores are secreted after delivery of iron. However, compared with the extensive work on bacterial iron transport systems, little has been reported on the membrane machinery to export these siderophore molecules into the environment. A 50-kDa iron-regulated outer membrane protein in Pseudomonas aeruginosa was first proposed to function as an exporter for the siderophore pyoverdine (43). The exiT gene, encoding a potential ABC-like exporter in Mycobacterium smegmatis, was linked to exochelin export with siderophore specificity (73). Genes encoding MFS efflux pump homologues were reported for Yersinia pestis (ybtX) (15) and Bordetella pertussis (orfX) (5), both of which are located within the iron-regulated operons. More recently, a 43-kDa MFS efflux pump encoded by entS in the ent-fep cluster of E. coli has been demonstrated to function as an exporter of enterobactin (18). The existence of pvsC within the vibrioferrin biosynthesis operon as well as the homology of its protein product with bacterial MFS efflux pumps implies that PvsC may function as an exporter of vibrioferrin. Very interestingly, genes encoding putative MFS exporters homologous to PvsC were also found in the S. aureus genomic sequence (26) and in the predicted achromobactin biosynthesis operon of E. chrysanthemi (accession number AF416739), respectively. Thus, siderophore expulsion, mediated by a membrane-associated siderophore efflux transporter, may be one of the self-protecting mechanisms devised by siderophore-producing bacteria. Studies under way in our laboratory indicate that the pvsC deletion mutant strain without polar effects over pvsDE exhibited a severe decline in vibrioferrin excretion into the culture medium, and this mutation was complemented with the pvsC gene, as judged by high-performance liquid chromatography (T. Tanabe and S. Yamamoto, unpublished data).
The receptor protein PvuA for ferric vibrioferrin has been found to be homologous to FecA (31% identity and 47% similarity), the E. coli outer membrane receptor for ferric dicitrate (17). The strong homology between the ABC transporter component proteins PvuBCDE and FecBCDE further supports the idea that their structures are very closely related at the iron coordination site. Like the Pvs proteins, the PvuBCDE proteins also share significant homology with the CbrABCD proteins in E. chrysanthemi (31) and with hypothetical proteins (SA1979, SA1978, and SA1977) in the S. aureus genomic sequence (26).
We showed that the pvuBCDE genes were transcribed with the upstream genes psuA and pvuA as a single transcript. However, the amount of the psuA-pvuABCDE transcript was considerably less than that of the psuA-pvuA transcript. We thus propose that a hairpin-like structure between pvuA and pvuB (Fig. 2B) causes transcriptional termination in high abundance for the psuA-pvuA-specific mRNA, while a low level of transcriptional events can pass through this structure to transcribe the genes pvuBCDE, following the same iron regulation pattern as the psuA-pvuA-specific mRNA. Similar transcriptional modulation mediated by the hairpin has been reported for the V. anguillarum fatDCBA and fatDCBA-angRT transcripts (65).
The complete sequences and functional characterization of the two operons presented in this study afford an overall view of the organizational structures and regulation of the gene complex responsible for the biosynthesis of the siderophore vibrioferrin and transport of its ferric complex in V. parahaemolyticus. Enzymological studies are needed for the assignment of biological functions to the genes involved in vibrioferrin biosynthesis and their linkage to specific pathways. It is of special interest to determine whether PvsC functions as an exporter specific to vibrioferrin.
Acknowledgments
We acknowledge T. Kuroda for providing E. coli strains with λpir and for his helpful comments on our work.
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Altschul, S. F., T. I. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagg, A., and J. B. Neilands. 1987. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol. Rev. 51:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann, P., A. L. Furniss, and J. V. Lee. 1984. Genus I. Vibrio Pacini 411AL, p. 518-538. In N. K. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 4.Braun, V., K. Hantke, and W. Köster. 1998. Bacterial iron transport: mechanisms, genetics, and regulation. Metal Ions Biol. Syst. 35:67-145. [PubMed] [Google Scholar]
- 5.Brickman, T. J., and S. K. Armstrong. 1999. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. J. Bacteriol. 181:5958-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butterton, J. R., M. H. Chol, P. I. Watnick, P. A. Carroll, and S. B. Calderwood. 2000. Vibrio cholerae VibF is required for vibriobactin synthesis and is a member of the family of nonribosomal peptide synthetases. J. Bacteriol. 182:1731-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butterton, J. R., J. A. Stoebner, S. M. Payne, and S. B. Calderwood. 1992. Cloning, sequencing, and transcriptional regulation of viuA, the gene encoding the ferric vibriobactin receptor of Vibrio cholerae. J. Bacteriol. 174:3729-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosa, J. H. 1989. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol. Rev. 53:517-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosa, J. H. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61:319-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Silva, A. C. R., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, Jr., L. M. C. Alves, A. M. do Amaral, M. C. Bertolini, L. E. A. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. B. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. S. Ferreira, R. C. C. Ferreira, M. I. T. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, Jr., E. G. M. Lemos, M. V. F. Lemos, E. C. Locali, M. A. Machado, A. M. B. N. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. M. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. M. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira Jr., A. Rossi, J. A. D. Sena, C. Silva, R. F. de Souza, L. A. F. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. D. Tezza, M. Trindade dos Santos, D. Truffe, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 12.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drechsel, H., S. Freund, G. Nicholson, H. Haag, O. Jung, H. Zäjmer, and G. Jung. 1993. Purification and chemical characterization of staphyloferrin B, a hydrophilic siderophore from staphylococci. BioMetals 6:185-192. [DOI] [PubMed] [Google Scholar]
- 14.Escolar, L., J. Pérez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fetherston, J. D., V. J. Bertolino, and R. D. Perry. 1999. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol. 32:289-299. [DOI] [PubMed] [Google Scholar]
- 16.Funahashi, T., C. Fujiwara, M. Okada, S. Miyoshi, S. Shinoda, S. Narimatsu, and S. Yamamoto. 2000. Characterization of Vibrio parahaemolyticus manganese-resistant mutants in reference to the function of the ferric uptake regulatory protein. Microbiol. Immunol. 44:963-970. [DOI] [PubMed] [Google Scholar]
- 17.Funahashi, T., K. Moriya, S. Uemura, S. Miyoshi, S. Shinoda, S. Narimatsu, and S. Yamamoto. 2002. Identification and characterization of pvuA, a gene encoding the ferric vibrioferrin receptor protein in Vibrio parahaemolyticus. J. Bacteriol. 184:936-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furrer, J. L., D. N. Sanders, I. G. Hook-Barnard, and M. A. McIntosh. 2002. Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol. Microbiol. 44:1225-1234. [DOI] [PubMed] [Google Scholar]
- 19.Gill, M. J., N. P. Brenwald, and R. Wise. 1999. Identification of an efflux pump gene, pmrA, associated with fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:187-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerinot, M. L. 1994. Microbial iron transport. Annu. Rev. Microbiol. 48:743-772. [DOI] [PubMed] [Google Scholar]
- 21.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 22.Henikoff, S., and J. G. Henikoff. 1994. Protein family classification based on searching a database of blocks. Genomics 19:97-107. [DOI] [PubMed] [Google Scholar]
- 23.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 24.Honda, T., and T. Iida. 1993. The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct haemolysin and related haemolysins. Rev. Med. Microbiol. 4:106-113. [Google Scholar]
- 25.Hyde, S. C., P. Emsley, M. J. Hartshorn, M. M. Mimmack, U. Gileady, S. R. Pearce, M. P. Gallagher, D. R. Gill, R. E. Hubbard, and C. F. Higgins. 1990. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature 346:362-365. [DOI] [PubMed] [Google Scholar]
- 26.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. Kobayashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 27.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 28.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Litwin, C. M., T. W. Rayback, and J. Skinner. 1996. Role of catechol siderophore synthesis in Vibrio vulnificus virulence. Infect. Immun. 64:2834-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch, D., J. O'Brien, T. Welch, P. Clarke, P. Ó. Cuív, J. H. Crosa, and M. O'Connell. 2001. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183:2576-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahé, B., C. Masclaux, L. Rauscher, C. Enard, and D. Expert. 1995. Differential expression of two siderophore-dependent iron-acquisition pathways in Erwinia chrysanthemi 3937: characterization of a novel ferrisiderophore permease of the ABC transporter family. Mol. Microbiol. 18:33-43. [DOI] [PubMed] [Google Scholar]
- 32.Martínez, J. L., M. Herrero, and V. de Lorenzo. 1994. The organization of intercistronic regions of the aerobactin operon of pColV-K30 may account for the differential expression of the iucABCD iutA genes. J. Mol. Biol. 238:288-293. [DOI] [PubMed] [Google Scholar]
- 33.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mietzner, T. A., and S. A. Morse. 1994. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu. Rev. Nutr. 14:471-493. [DOI] [PubMed] [Google Scholar]
- 35.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Münzinger, M., H. Budzikiewicz, D. Expert, C. Enard, and J. M. Meyer. 2000. Achromobactin, a new citrate siderophore of Erwinia chrysanthemi. Z. Naturforsch. 55C:28-332. [DOI] [PubMed]
- 37.Neilands, J. B. 1981. Microbial iron compounds. Annu. Rev. Biochem. 50:715-731. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 39.Nishibuchi, M., K. Kumagai, and J. B. Kaper. 1991. Contribution of the tdh1 gene of Kanagawa phenomenon-positive Vibrio parahaemolyticus to production of extracellular thermostable direct hemolysin. Microb. Pathog. 11:453-460. [DOI] [PubMed] [Google Scholar]
- 40.Ota, Y., H. Tamegai, F. Kudo, H. Kuriki, A. Koike-Takeshita, T. Eguchi, and K. Kakinuma. 2000. Butirosin-biosynthetic gene cluster from Bacillus circulans. J. Antibiotics. 53:1158-1167. [DOI] [PubMed] [Google Scholar]
- 41.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Payne, S. M. 1988. Iron and virulence in the family Enterobacteriaceae. Crit. Rev. Microbiol. 16:81-111. [DOI] [PubMed] [Google Scholar]
- 43.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenberg, M., and D. Court. 1979. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu. Rev. Genet. 13:319-353. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Saurin, W., W. Köster, and E. Dassa. 1994. Bacterial binding protein-dependent permeases: characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol. Microbiol. 12:993-1004. [DOI] [PubMed] [Google Scholar]
- 48.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 49.Sebulsky, M. T., D. Hohnstein, M. D. Hunter, and D. E. Heinrichs. 2000. Identification and characterization of a membrane permease involved in iron-hydroxamate transport in Staphylococcus aureus. J. Bacteriol. 182:4394-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shine, J., and L. Dalgarno. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to non-sense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, M. J., J. N. Shoolery, B. Schwyn, I. Holden, and J. B. Neilands. 1985. Rhizobactin, a structurally novel siderophore from Rhizobium meliloti. J. Am. Chem. Soc. 107:1739-1743. [Google Scholar]
- 52.Staudenmaier, H., B. Van Hove, Z. Yarachi, and V. Braun. 1989. Nucleotide sequences of the fecBCDE genes and locations of the proteins suggest a periplasmic-binding-protein-dependent transport mechanism for iron(III) dicitrate in Escherichia coli. J. Bacteriol. 171:2626-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stojiljkovic, I., A. Bäumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria: identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 54.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 55.Sullivan, J. T., J. R. Trzebiatowski, R. W. Cruickshank, J. Gouzy, S. D. Brown, R. M. Elliot, D. J. Fleetwood, N. G. McCallum, U. Rossbach, G. S. Stuart, J. E. Weaver, R. J. Webby, F. J. de Bruijn, and C. W. Ronsom. 2002. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184:3086-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takeuchi, Y., T. Akiyama, and T. Harayama. 1999. Total synthesis of the siderophore vibrioferrin. Chem. Pharm. Bull. 47:459-460. [Google Scholar]
- 57.Takeuchi, Y., Y. Nagao, K. Toma, Y. Yoshikawa, T. Akiyama, H. Nishioka, H. Abe, T. Harayama, and S. Yamamoto. 1999. Synthesis and siderophore activity of vibrioferrin and one of its diastereomeric isomers. Chem. Pharm. Bull. 47:1284-1287. [Google Scholar]
- 58.Tam, R., and M. H. Saier. 1993. Structural, functional and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57:320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thieken, A., and G. Winkelmann. 1992. Rhizoferrin: a complexone type siderophore of the Mucorales and Entomophthorales (Zygomycetes). FEMS Microbiol. Lett. 94:37-42. [DOI] [PubMed] [Google Scholar]
- 60.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 61.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 62.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the α and β-subunits of ATP synthase, myosin, kinase and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Webster, A. C. D., and C. M. Litwin. 2000. Cloning and characterization of vuuA, a gene encoding the Vibrio vulnificus ferric vulnibactin receptor. Infect. Immun. 68:526-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Welch, T. J., S. Chai, and J. H. Crosa. 2000. The overlapping angB and angG genes are encoded within the trans-acting factor region of the virulence plasmid in Vibrio anguillarum: essential role in siderophore biosynthesis. J. Bacteriol. 182:6762-6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wertheimer, A. M., W. Verweij, Q. Chen, L. M. Crosa, M. Nagasawa, M. E. Tolmasky, L. A. Actis, and J. H. Crosa. 1999. Characterization of the angR gene of Vibrio anguillarum: essential role in virulence. Infect. Immun. 67:6496-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wyckoff, E. E., S. L. Smith, and S. M. Payne. 2001. VibD and VibH are required for late steps in vibriobactin biosynthesis in Vibrio cholerae. J. Bacteriol. 183:1830-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wyckoff, E. E., J. A. Stoebner, K. E. Reed, and S. M. Payne. 1997. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J. Bacteriol. 179:7055-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamamoto, S., T. Funahashi, H. Ikai, and S. Shinoda. 1997. Cloning and sequencing of the Vibrio parahaemolyticus fur gene. Microbiol. Immunol. 41:737-740. [DOI] [PubMed] [Google Scholar]
- 69.Yamamoto, S., Y. Hara, K. Tomochika, and S. Shinoda. 1995. Utilization of hemin and hemoglobin as iron sources by Vibrio parahaemolyticus and identification of an iron-repressible hemin-binding protein. FEMS Microbiol. Lett. 128:195-200. [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto, S., N. Okujo, S. Matsuura, I. Fujiwara, Y. Fujita, and S. Shinoda. 1994. Siderophore-mediated utilization of iron bound to transferrin by Vibrio parahaemolyticus. Microbiol. Immunol. 38:687-693. [DOI] [PubMed] [Google Scholar]
- 71.Yamamoto, S., N. Okujo, T. Yoshida, S. Matsuura, and S. Shinoda. 1994. Structure and iron transport activity of vibrioferrin, a new siderophore of Vibrio parahaemolyticus. J. Biochem. 115:868-874. [DOI] [PubMed] [Google Scholar]
- 72.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 73.Zhu, W., J. E. L. Arceneaux, M. L. Beggs, B. R. Byers, K. D. Eisenach, and M. D. Lundrigan. 1998. Exochelin genes in Mycobacterium smegmatis: identification of an ABC transporter and two non-ribosomal peptide synthesis genes. Mol. Microbiol. 29:629-639. [DOI] [PubMed] [Google Scholar]