Abstract
A total of 12 VanA-type vancomycin-resistant enterococci, consisting of 10 Enterococcus faecium isolates and two Enterococcus avium isolates, were examined in detail. The vancomycin resistance conjugative plasmids pHTα (65.9 kbp), pHTβ (63.7 kbp), and pHTγ (66.5 kbp) were isolated from each of three different E. faecium strains. The plasmids transferred highly efficiently between enterococcus strains during broth mating and were homologous with pMG1 (Gmr; 65.1 kb).
Gene transfer systems are an essential requirement for the spread of drug resistance in microorganisms. In general, the systems of efficient plasmid transfer have not been well characterized for the gram-positive bacteria. However, enterococci possess potent and unique capabilities of transferring plasmids among themselves and to other genera (4, 5, 21, 35). One type of enterococcal plasmid consists of the group of narrow-host-range and pheromone-responsive plasmids (4, 5, 9). The other type consists of the broad-host-range pAMβ1 and pIP501 plasmids, which were originally isolated from Enterococcus faecalis (8, 24) and Streptococcus agalactiae (13, 18), respectively, and transfer on a solid surface at low frequency (8, 13, 18, 24, 27, 40).
We have described the isolation of the pheromone-independent gentamicin resistance conjugative plasmid pMG1 (Gmr; 65.1 kb) from an Enterococcus faecium clinical isolate in Japan (20). pMG1 transfers efficiently among enterococcus strains during broth mating. pMG1-like plasmids are widely disseminated in vancomycin-resistant E. faecium clinical isolates obtained from a hospital in the United States (39).
In this report, we show that the VanA resistance encoded on a Tn1546-like transposon was mediated by a pMG1-like plasmid and that this vancomycin resistance pMG1-like plasmid was capable of highly efficient transfer among the enterococci.
Drug resistance of VRE isolates and isolation of vancomycin resistance conjugative plasmids.
The laboratory strains and plasmids used in this study are listed in Table 1. A total of 12 isolates of vancomycin-resistant enterococci (VRE) were used in this study (Table 2). The vancomycin resistance of each strain transferred to E. faecium BM4105RF at a frequency of about 10−5 per donor cell by mating in broth for 4 h at 37°C. The transconjugants of each strain acquired only vancomycin and teicoplanin resistance, indicating that the glycopeptide resistance was transferred during broth mating.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or phenotype | Description; source or reference |
|---|---|---|
| Strains | ||
| E. faecium BM4147 (pIP816 Vanr) | van | 25 |
| E. faecalis FA2-2 | rif fus | Derivative of JH2; 7 |
| E. faecalis JH2SS | str spc | Derivative of JH2; 37 |
| E. faecium BM4105RF | rif fus | Derivative of plasmid-free E. faecium BM4105; 3 |
| E. faecium BM4105SS | str spc | Derivative of plasmid-free E. faecium BM4105; 3 |
| Plasmids | ||
| pMG1 | Gmr | 65.1-kb conjugative plasmid from E. faecium strain; 20 |
| pG200 | Gmr | pMG1-like conjugative plasmid from VRE; 39 |
| pG445 | Gmr | pMG1-like conjugative plasmid from VRE; 39 |
| pG566 | Gmr | pMG1-like conjugative plasmid from VRE; 39 |
| pG700 | Gmr | pMG1-like conjugative plasmid from VRE; 39 |
| pG120 | Gmr | pMG1-like conjugative plasmid from VRE; 39 |
| pAD1 | hly/bac uvr | 59.6-kb pheromone-responsive conjugative plasmid from DS16; 7, 19, 37 |
| pPD1 | bac | 59-kb pheromone-responsive conjugative plasmid from E. faecalis 39-5; 15, 38, 41 |
| pAM373 | tet | 36-kb pheromone-responsive conjugative plasmid; 6 |
| pAMβ1 | erm | 26.5-kb broad-host-range conjugative plasmid from DS5; 8, 24 |
| pIP501 | erm cat | 39.2-kb broad-host-range conjugative plasmid; 2, 13, 18 |
TABLE 2.
Characterizations of vancomycin-resistant enterococcia
| Species | Strain no. | Patientb | Antimicrobial drug resistance level (MIC, μg/ml)c
|
Drug resistance pattern | Vanr plasmid type harbored in strain | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Apc | Erm | Gen | Kan | Str | Tet | Tei | Van | |||||
| E. faecium | FH1 | A | 256 | >128 | 2 | >1,024 | 1,024 | >256 | 32 | 256 | Apc Erm Kan Str Tet Tei Van | α |
| E. faecium | FH2 | B | 256 | >128 | 2 | >1,024 | 512 | >256 | 64 | 256 | Apc Erm Kan Str Tet Tei Van | β |
| E. faecium | FH3 | C | 2 | 4 | 2 | 64 | 64 | 1 | 64 | 512 | Tei Van | β |
| E. faecium | FH4 | D | 128 | >128 | 2 | >1,024 | 256 | >256 | 64 | 256 | Apc Erm Kan Tet Tei Van | β |
| E. faecium | FH5 | E | 64 | >128 | 1 | >1,024 | 1,024 | >256 | 128 | 512 | Apc Erm Kan Str Tet Tei Van | β |
| E. faecium | FH6 | F | 64 | >128 | 2 | >1,024 | 1,024 | >256 | 64 | 256 | Apc Erm Kan Str Tet Tei Van | β |
| E. faecium | FH7 | G | 128 | >128 | >256 | >1,024 | 1,024 | 32 | 128 | 512 | Apc Erm Gen Kan Str Tet Tei Van | γ |
| E. faecium | FH8 | H | 128 | >128 | >256 | >1,024 | >1,024 | 2 | 128 | 512 | Apc Erm Gen Kan Str Tei Van | γ |
| E. faecium | FH9 | I | 128 | >128 | >256 | >1,024 | >1,024 | 128 | 128 | 512 | Apc Erm Gen Kan Str Tet Tei Van | γ |
| E. faecium | FH10 | J | 128 | >128 | >256 | >1,024 | >1,024 | 128 | 128 | >1,024 | Apc Erm Gen Kan Str Tet Tei Van | γ |
| E. avium | FH11 | C | 16 | 0.25 | 2 | 128 | 64 | 16 | 16 | 256 | Apc Tet Tei Van | β |
| E. avium | FH12 | E | 16 | 0.25 | 1 | 64 | 64 | 256 | 16 | 512 | Apc Tet Tei Van | β |
The isolation of VanA-type VRE from clinical sources is still rare in Japan (i.e., fewer than 30 cases) (16, 33; N. Fujita, M. Yoshimura, T. Komori, K. Tanimoto, and Y. Ike, Letter, Antimicrob. Agents Chemother. 42 :2150, 1998; Y. Ike, K. Tanimoto, Y. Ozawa, T. Nomura, S. Fujimoto, and H. Tomita, Letter, Lancet 353:1854, 1999). The presence of VanA VRE was examined in the feces of a total of 1,699 inpatients obtained by the microbiology division of the clinical microbiology of the university hospital of Fujita Health University School of Medicine, Aichi, Japan, between 1 August 1999 and 31 March 2001.
All strains were isolated from feces of patients. E. faecium FH3 and E. avium FH11, and E. faecium FH5 and E. avium FH12, were isolated from patient C and patient E, respectively. Each of the other strains was isolated from a different patient.
Abbreviations: Apc, ampicillin resistance; Gen, gentamicin resistance; Kan, kanamycin resistance; Str, streptomycin resistance; Tet, tetracycline resistance; Tei, teicoplanin resistance; Van, vancomycin resistance. The drug resistance levels of ampicillin, gentamicin, kanamycin, streptomycin, tetracycline, teicoplanin, and vancomycin were equal to or greater than 16, 64, 1,024, 512, 8, 16, and 64 μg/ml, respectively. Enterococcus strains were grown in Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) throughout this study. Mueller-Hinton broth and Mueller-Hinton agar for the sensitivity disk agar-N (Nissui, Tokyo, Japan) assay were used to test the MICs of antimicrobials. The MICs of the antimicrobials were determined according to the criteria of the National Committee for Clinical Laboratory Standards using Mueller-Hinton agar (32).
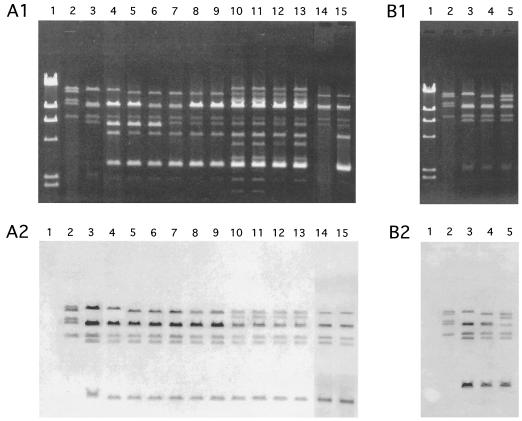
Analysis of agarose gel electrophoresis of restriction fragments of plasmid DNAs of each strain showed many DNA bands, indicating that each of the strains harbored several plasmids (Fig. 1, A1). The conjugative vancomycin resistance plasmid pHTα was identified from the transconjugant of E. faecium FH1 by repeated transfer experiments between E. faecium BM4105 strains. The plasmids isolated from each of the strains were classified into three types, α, β, and γ, with respect to the restriction profiles that hybridized to the type α plasmid pHTα (Fig. 1, A2) (Table 2). The pHTβ and pHTγ plasmids, which were type β and γ plasmids, respectively, were identified from the transconjugants of strains FH4 and FH7, respectively (Fig. 1, B1) (Table 2). Each type of plasmid DNA encoded the VanA gene by PCR analysis with the vanA-specific primer (data not shown) (11, 12, 29). pHTα DNA hybridized to all NdeI and EcoRI fragments of each type of plasmid DNA (Fig. 1, B2). DNA from the conjugative plasmid pMG1 (Gmr; 65.1 kbp) hybridized to specific NdeI or EcoRI fragments (data not shown). Each type of plasmid transferred at a frequency of around 10−3 to 10−5 per donor cell between E. faecium BM4105 or around 10−6 to 10−7 per donor cell between E. faecalis JH2 strains during broth mating.
FIG. 1.
Agarose gel electrophoresis of restriction endonuclease-digested plasmid DNAs and hybridization with the pHTα probe. Southern hybridization was performed with the digoxigenin-based nonradioisotope system of Boehringer GmbH (Mannheim, Germany), and all procedures were based on the manufacturer's manual and standard protocols (34). (A1) Agarose gel electrophoresis of NdeI-digested plasmid DNAs isolated from vancomycin-resistant E. faecium or E. avium (VRE) isolates. (A2) The gel was Southern blotted and hybridized to pHTα. Lanes of panels A1 and A2: 1, HindIII-digested lambda DNA; 2, NdeI-digested pMG1; 3, NdeI-digested pHTα; 4 to 15, NdeI-digested plasmid DNAs from the strains FH1, FH2, FH3, FH4, FH5, FH6, FH7, FH8, FH9, FH10, FH11 and FH12, respectively. (B1) Agarose gel electrophoresis of NdeI-digested pHTα, pHTβ, and pHTγ plasmid DNA isolated from each transconjugant of FH1, FH4, and FH7, respectively. (B2) The gel was Southern blotted and hybridized to the pHTα probe. Lanes of panels B1 and B2: 1, HindIII-digested lambda DNA; 2, pMG1; 3, pHTα; 4, pHTβ; 5, pHTγ.
The restriction maps of the vancomycin resistance plasmids.
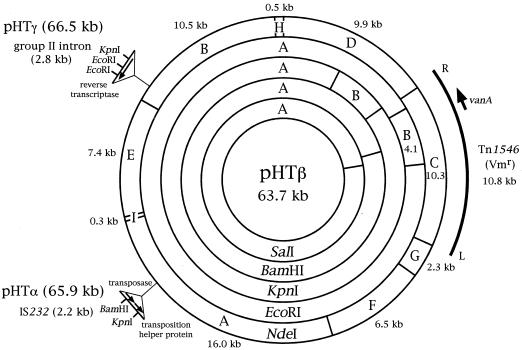
The restriction maps of pHTα (65.9 kbp), pHTβ (63.7 kbp), and pHTγ (66.5 kbp) were constructed (Fig. 2). The molecular sizes of the NdeI A fragment of pHTα and the NdeI B fragment of pHTγ were 18.2 and 13.3 kbp, respectively, which were 2.2 and 2.8 kbp larger than the NdeI A fragments (16 kbp) and NdeI B fragments (10.5 kbp) of pHTβ, respectively.
FIG. 2.
Physical map of the vancomycin resistance conjugative plasmid pHTβ (63.7 kb) and its relation to pHTα (65.9 kb) or pHTγ (66.5 kb). To determine the DNA sequence of the 2.2-kbp fragment of pHTα and the 2.8-kbp fragment of pHTγ and to confirm that these fragments had inserted into the NdeI A and NdeI B fragments of pHTβ, respectively, random fragments of the region of the 2.2-kb fragment or of the 2.8-kb fragment were cloned and sequenced as previously described (38). pHTα resulted from the insertion of the 2.2-kb fragment of IS232 into the region of NdeI fragment A of pHTβ. pHTγ resulted from the insertion of the 2.8-kb fragment of the group II intron into the region of NdeI fragment B of pHTβ. DNA sequence and PCR analysis were carried out to analyze the VanA determinant as described previously (1, 11, 16). The VanA-type determinant of pHTβ was encoded on the transposon Tn1546 or a closely related transposon. The location of the VanA determinant of each plasmid was determined by Southern analysis, PCR, and comparison of the restriction map covering the region of the VanA determinant with that of Tn1546.
The nucleotide sequences showed that the 2.2-kbp (2,156-bp) fragment of pHTα contained two open reading frames of 1,236 bp (412 amino acids) and 759 bp (253 amino acids), which were homologous with the IS232-mediating transposase and the transposition helper protein, respectively (28). The nucleotide sequence of the 2.8-kb (2,748-bp) fragment of the pHTγ plasmid was homologous with that of the group II intron that encodes a reverse transcriptase consisting of 638 amino acids (22, 23, 30, 31). The nucleotide sequences around the 2.2-kbp fragment of the NdeI A fragment of pHTα were completely identical to the nucleotide sequence of the NdeI A fragment of the pHTβ plasmid. Likewise, the nucleotide sequences around the 2.8-kbp fragment of NdeI B framgent of pHTγ were completely identical to that of the NdeI B fragment of the pHTβ plasmid. These results indicated that pHTβ might be the original or wild-type plasmid, and the 2.2-kbp fragment and the 2.8-kbp fragment were inserted into the NdeI A and NdeI B fragments of the pHTβ plasmid, respectively.
Analysis of the pMG1 traA gene.
The traA gene of pMG1, which encodes a 287-amino-acid protein, is involved in the tra gene system for conjugation and is specific to pMG1 (36). Each plasmid was examined to determine whether traA was conserved in each of these plasmids by sequence analysis of the PCR product for traA.
The nucleotide sequence and the deduced amino acid sequence of the open reading frame in 945-bp PCR products analyzed in pHTα, pHTβ, and pHTγ were completely identical to those of traA of pMG1, with the exception of eight nucleotide substitutions and six amino acid substitutions (i.e., V19F, S23N, R26S, V84M, A102V, and K237E). The nucleotide sequence and the deduced amino acid sequence of the gentamicin resistance pMG1-like plasmids (39) pG200, pG445, pG560, pG700, and pG120 were completely identical to those of pMG1 traA.
Based on the differences observed in the nucleotide sequence of traA, these results indicated that the traA gene of pMG1 was conserved in pMG1-like plasmids and that there was no direct connection between the gentamicin resistance pMG1 plasmid (including pMG1-like plasmids) and the vancomycin resistance pHT plasmids.
Incompatibility of vancomycin resistance plasmids and pMG1 and Southern analysis with other reported plasmids.
The transfer frequency of each of the vancomycin resistance plasmids to the recipient cell carrying pMG1 was lower than that when the recipient was plasmid free (Table 3). All transconjugants were vancomycin resistant (conferred by the incoming plasmid), but they had lost gentamicin resistance (encoded by the resident plasmid). These results indicate that each of the vancomycin resistance plasmids and pMG1 were incompatible. Southern analysis showed that the pHTβ plasmid did not contain any sequence homologous with those of the pheromone-responsive plasmids (Table 1) (4-7, 10, 15, 19, 38, 41) and the broad-host-range plasmids (Table 1) (2, 8, 13, 18) (data not shown).
TABLE 3.
Transfer frequencies of vancomycin resistance plasmids from donor strains to recipients carrying the pMG1 plasmida
| Plasmid from donor cells of E. faecium BM4105SS | Transfer frequency in broth mating with recipient E. faecium BM4105RF carrying:
|
Transfer frequency in filter mating with recipient E. faecium BM4105RF carrying:
|
||
|---|---|---|---|---|
| pMG1 | None | pMG1 | None | |
| pHTα | <1 × 10−7 | 1 × 10−4 | 2 × 10−3 | >1 × 100 |
| pHTβ | <1 × 10−7 | 3 × 10−4 | 5 × 10−3 | >1 × 100 |
| pHTγ | <1 × 10−7 | 2 × 10−4 | 3 × 10−3 | >1 × 100 |
The mating experiments were carried out as previously described (14, 20). The mating times of broth mating and filter mating were 3 and 18 h, respectively. The transconjugants were examined after 48 h of incubation of the selective agar plates at 37°C. Throughout the mating experiments, the antibiotic concentration used for the selection of gentamicin- or vancomycin-resistant transconjugants was 100 or 12.5 μ/ml, respectively. The selection of rifampin- and fuscidic acid-resistant recipient strains was carried out at a concentration of 25 μg/ml each, while selection of streptomycin- and spectinomycin-resistant recipient strains was carried out at concentrations of 500 and 250 μg/ml, respectively.
Gentamicin and kanamycin resistance determinants on pMG1.
pMG1 was examined to determine whether the gentamicin and kanamycin resistance determinants also reside on a transposon. The nucleotide sequence revealed that the EcoRI B fragment of pMG1 encoded a Tn4001-like transposon (4,523 bp) (17, 26). The composite transposon Tn4001 (4,566 bp) carries the gentamicin and kanamycin resistance gene aacA-aphD, which is flanked by two 1,324-bp inverted repeats, IS256L and IS256R (26). The nucleotide sequence of the Tn4001-like transposon was completely identical to that of the original Tn4001 transposon, except that the resistance gene aacA-aphD was flanked by two 1,324-bp (IS256) direct repeats and there was deletion of a 43-bp sequence upstream from the end of IS256R.
Conclusions.
The pheromone-independent gentamicin resistance plasmid pMG1 and pMG1-like plasmids are found in E. faecium and are widely disseminated in vancomycin-resistant E. faecium isolates in the United States (39). The data shown in this report suggest that pMG1-like plasmids without any resistance gene or any other selectable determinant must be prevalent in E. faecium, and there is the possibility that a mobile genetic element encoding drug resistance or another determinant might insert onto them. As shown by this study, there is now evidence that in addition to gentamicin and kanamycin resistance transposon Tn4001-like elements, vancomycin resistance transposon Tn1546-like elements and other mobile genetic elements, such as IS232 and the group II intron, are capable of insertion onto pMG1-type plasmids.
Nucleotide sequence accession numbers. The nucleotide sequence data reported here have been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession numbers AB091473, AB105542, and AB105543
Acknowledgments
This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science, and Technology [Tokuteiryoiki, Kiban (B)] and Japanese Ministry of Health, Labor, and Welfare (H15-Shinko-9).
We thank Elizabeth Kamei for helpful advice.
REFERENCES
- 1.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behnke, D., and M. S. Gilmore. 1981. Location of antibiotic resistance determinants, copy control, and replication functions on the double-selective streptococcal cloning vector pGB301. Mol. Gen. Genet. 184:115-120. [DOI] [PubMed] [Google Scholar]
- 3.Carlier, C., and P. Courvalin. 1990. Emergence of 4′, 4′′-aminoglycoside nucleotidyltransferase in Enterococcus. Antimicrob. Agents Chemother. 34:1565-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clewell, D. B. 1993. Bacterial sex pheromone-induced plasmid transfer. Cell 73:9-12. [DOI] [PubMed] [Google Scholar]
- 5.Clewell, D. B. 1993. Sex pheromones and the plasmid-encoded mating response in Enterococcus faecalis, p. 349-367. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Press, New York, N.Y.
- 6.Clewell, D. B., F. Y. An, B. A. White, and C. Gawron-Burke. 1985. Streptococcus faecalis sex pheromone (cAM373) also produced by Staphylococcus aureus and identification of a conjugative transposon (Tn918). J. Bacteriol. 162:1212-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clewell, D. B., P. K. Tomich, M. C. Gawron-Burke, A. E. Franke, Y. Yagi, and F. Y. An. 1982. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clewell, D. B., Y. Yagi, G. M. Dunny, and S. K. Schultz. 1974. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J. Bacteriol. 117:283-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis; evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunny, G. M., B. A. B. Leonard, and P. J. Hedberg. 1995. Pheromone-inducible conjugation in Enterococcus faecalis: interbacterial and host-parasite chemical communication. J. Bacteriol. 177:871-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutka-Malen, S., S. R. Leclercq, V. Coutant, J. Doval, and P. Courvalin. 1990. Phenotypic and genotypic heterogeneity of glycopeptide resistance determinants in gram-positive bacteria. Antimicrob. Agents Chemother. 34:1875-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, R. P., and F. L. Macrina. 1983. Streptococcal R plasmid pIP501: endonuclease site map, resistance determinant location, and construction of novel derivatives. J. Bacteriol. 154:1347-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franke, A., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimoto, S., H. Tomita, E. Wakamatsu, K. Tanimoto, and Y. Ike. 1995. Physical mapping of the conjugative bacteriocin plasmid pPD1 of Enterococcus faecalis and identification of the determinant related to the pheromone response. J. Bacteriol. 177:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto, Y., K. Tanimoto, Y. Ozawa, T. Murata, and Y. Ike. 2000. Amino acid substitutions in the VanS sensor of the VanA-type vancomycin-resistant enterococcus strains result in high-level vancomycin resistance and low-level teicoplanin resistance. FEMS Microbiol. Lett. 185:247-254. [DOI] [PubMed] [Google Scholar]
- 17.Hodel-Christian, S. L., and B. E. Murray. 1991. Characterization of the gentamicin resistance transposon Tn5281 from Enterococcus faecalis and comparison to staphylococcal transposons Tn4001 and Tn4031. Antimicrob. Agents Chemother. 35:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horodniceanu, T., D. Bouanchaud, G. Bieth, and Y. Chabbert. 1976. R plasmids in Streptococcus agalactiae (group B). Antimicrob. Agents Chemother. 10:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ike, Y., and D. B. Clewell. 1984. Genetic analysis of the pAD1 pheromone response in Streptococcus faecalis, using transposon Tn917 as an insertional mutagen. J. Bacteriol. 158:777-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ike, Y., K. Tanimoto, H. Tomita, K. Takeuchi, and S. Fujimoto. 1998. Efficient transfer of the pheromone-independent Enterococcus faecium plasmid pMG1 (Gmr) (65.1 kilobases) to Enterococcus strains during broth mating. J. Bacteriol. 180:4886-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jett, B. D., M. M. Huyche, and M. S. Gilmore. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambowitz, A. M. 1989. Infectious introns. Cell 56:323-326. [DOI] [PubMed] [Google Scholar]
- 23.Lambowitz, A. M., and M. Belfort. 1993. Introns as mobile genetic elements. Annu. Rev. Biochem. 62:587-622. [DOI] [PubMed] [Google Scholar]
- 24.Le Blanc, D. L., and L. N. Lee. 1984. Physical and genetic analysis of streptococcal plasmid pAMβ1 and cloning of its replication region. J. Bacteriol. 157:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leclercq, R., E. Derlot, J. Duval, and P. Courvalin. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157-161. [DOI] [PubMed] [Google Scholar]
- 26.Lyon, B. R., J. W. May, and R. A. Skurray. 1984. Tn4001: a gentamicin and kanamycin resistance transposon in Staphylococcus aureus. Mol. Gen. Genet. 193:554-556. [DOI] [PubMed] [Google Scholar]
- 27.Macrina, F. L., and G. L. Archer. 1993. Conjugation and broad host range plasmids in streptococci and staphylococci, p. 313-329. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Press, New York, N.Y.
- 28.Menou, G., J. Mahillon, M. M. Lecadet, and D. Lereclus. 1990. Structural and genetic organization of IS232, a new insertion sequence of Bacillus thuringiensis. J. Bacteriol. 172:6689-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miele, A., M. Bandera, and B. P. Goldstein. 1995. Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in VanA isolates. Antimicrob. Agents Chemother. 39:1772-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mills, D. A., D. A. Manias, L. L. McKay, and G. M. Dunny. 1997. Homing of a group II intron from Lactococcus lactis subsp. lactis ML3. J. Bacteriol. 179:6107-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mills, D. A., L. L. McKay, and G. M. Dunny. 1996. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J. Bacteriol. 178:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 33.Ozawa, Y., K. Tanimoto, T. Nomura, M. Yoshinaga, Y. Arakawa, and Y. Ike. 2002. Vancomycin resistant enterococci (VRE) in humans and imported chickens in Japan. Appl. Environ. Microbiol. 68:6457-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Schaberg, D. R., and M. J. Zervos. 1986. Intergenetic and interspecies gene exchange in gram-positive cocci. Antimicrob. Agents Chemother. 30:817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanimoto, K., and Ike, Y. 2002. Analysis of the conjugal transfer system of the pheromone-independent highly transferable enterococcus plasmid pMG1: identification of a tra gene (traA) up-regulated during conjugation. J. Bacteriol. 184:5800-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomich, P. K., F. Y. An, and D. B. Clewell. 1980. Properties of erythromycin-inducible transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 141:1366-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomita, H., S. Fujimoto, K. Tanimoto, and Y. Ike. 1997. Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J. Bacteriol. 179:7843-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomita, H., C. Pierson, S. K. Lim, D. B. Clewell, and Y. Ike. 2002. Possible connection between a widely disseminated conjugative gentamicin resistance (pMG1-like) plasmid and the emergence of vancomycin resistance in Enterococcus faecium. J. Clin. Microbiol. 40:3326-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trieu-Cuot, P., C. Carlier, and P. Courvalin. 1988. Conjugative plasmid transfer from Enterococcus faecalis to Escherichia coli. J. Bacteriol. 170:4388-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yagi, Y., R. Kessler, J. Shaw, D. Lopatin, F. An, and D. B. Clewell. 1983. Plasmid content of Streptococcus faecalis strain 39-5 and identification of a pheromone (cPD1)-induced surface antigen. J. Gen. Microbiol. 129:1207-1215. [DOI] [PubMed] [Google Scholar]