Abstract
Certain oral treponemes express a highly proteolytic phenotype and have been associated with periodontal diseases. The periodontal pathogen Treponema denticola produces dentilisin, a serine protease of the subtilisin family. The two-gene operon prcA-prtP is required for expression of active dentilisin (PrtP), a putative lipoprotein attached to the treponeme's outer membrane or sheath. The purpose of this study was to examine the diversity and structure of treponemal subtilisin-like proteases in order to better understand their distribution and function. The complete sequences of five prcA-prtP operons were determined for Treponema lecithinolyticum, “Treponema vincentii,” and two canine species. Partial operon sequences were obtained for T. socranskii subsp. 04 as well as 450- to 1,000-base fragments of prtP genes from four additional treponeme strains. Phylogenetic analysis demonstrated that the sequences fall into two paralogous families. The first family includes the sequence from T. denticola. Treponemes possessing this operon family express chymotrypsin-like protease activity and can cleave the substrate N-succinyl-alanyl-alanyl-prolyl-phenylalanine-p-nitroanilide (SAAPFNA). Treponemes possessing the second paralog family do not possess chymotrypsin-like activity or cleave SAAPFNA. Despite examination of a range of protein and peptide substrates, the specificity of the second protease family remains unknown. Each of the fully sequenced prcA and prtP genes contains a 5′ hydrophobic leader sequence with a treponeme lipobox. The two paralogous families of treponeme subtilisins represent a new subgroup within the subtilisin family of proteases and are the only subtilisin lipoprotein family. The present study demonstrated that the subtilisin paralogs comprising a two-gene operon are widely distributed among treponemes.
Spirochetes include a large number of human- and animal-associated species. Well-known pathogenic representatives of different spirochetal genera and their associated diseases include Treponema pallidum and syphilis (27), Borrelia burgdorferi and Lyme disease (3), and Leptospira spp. and leptospirosis (19). Spirochetes have long been recognized as members of the subgingival biofilm adjacent to teeth (20, 21). The indigenous spirochetes localize at the interface of subgingival plaque and the gingival tissues and have been shown to have the ability to invade periodontal tissues (2, 20, 26, 30). The cultivable oral spirochetes Treponema denticola, Treponema lecithinolyticum, and Treponema socranskii subsp. socranski have been strongly associated with human periodontal diseases (7, 24, 25, 33, 39). Recent studies have identified over 50 treponeme species-level phylotypes based on sequence analysis of cloned 16S ribosomal DNA amplicons from DNA extracted from subgingival dental plaque (5, 28). Three-fourths of these oral treponemes are currently uncultivated. Relatively little is known about the physiology and virulence of oral treponemes, even for those that can be cultivated.
The readily cultivated T. denticola has served as a convenient model for understanding the putative virulence determinants of oral treponemes (7). Periodontal pockets provide protein-rich, anaerobic environments that select for T. denticola and cohabiting species that survive by peptide metabolism. Treponema denticola has been reported to degrade native human protein substrates, including fibrinogen, fibronectin, keratin, collagen IV, transferrin, and bioactive peptides (22, 31). It expresses a prolyl-phenylalanine-specific chymotrypsin-like protease (CTLP) that has been implicated in the ability to colonize and survive in periodontal pockets and to infect gingival tissues (36). CTLP can directly disrupt junctional complexes between epithelial cells (17), and it can also activate host neutrophil procollagenase MMP-8 (35), promoting the bacterium's penetration of epithelial barriers and facilitating the bacterium's migration through the extracellular matrix (11). This protease is also responsible for the organism's gelatinase activity (13).
The native form of CTLP has a molecular mass of approximately 95 kDa and was found to consist of three polypeptides, the largest being 72 kDa, while the sizes of the two smaller peptides have variously been reported as 27 and 23 kDa (26), 39 and 32 kDa (22), or 43 and 38 kDa (15). N-terminal amino acid sequences from the 43- and 72-kDa proteins enabled the identification of their corresponding genes, which are positioned adjacent to one another in the T. denticola genome (15). The gene encoding the 72-kDa protein is designated prtP, and the gene product, dentilisin, has amino acid similarity to subtilisin-type serine proteases. Mutant strains deficient in dentilisin expression produce no CTLP and aberrantly express a second outer sheath protein complex called the major outer sheath protein (Msp) (9, 16). The PrtP-deficient mutant strain K1 was less able than its parent strain to disrupt epithelial monolayers in vitro (6) and was less virulent in a murine subcutaneous abscess model (16). The K1 strain also demonstrated marked pleiotropic effects as a result of the CTLP knockout: it grew more slowly, in tightly coiled aggregates; it had a disrupted outer sheath that failed to contain the periplasmic flagella; it was less hydrophobic; and it exhibited enhanced coaggregation compared to the wild-type strain with a target strain of Fusobacterium nucleatum (16).
The functions of the 38- and 43-kDa proteins of the CTLP complex are unknown. Initially, the open reading frame (ORF) upstream of prtP was reported to encode only the 43-kDa protein. However, recent experiments demonstrate that both proteins are processed from a common polypeptide, which is encoded by the upstream ORF (18). This upstream gene has been named prcA for protease complex-associated protein. Genetic analysis, employing prtP null mutants, indicates that PrcA is processed only when wild-type PrtP protein is present, strongly suggesting that PrcA is a substrate for the chymotrypsin-like serine protease activity of dentilisin. The exact site of PrcA cleavage has not been determined.
Genes encoding subtilisin-like proteases are widely disseminated among bacteria. It is known that several oral treponemes have dentilisin-like activity (13). The purpose of this study was to examine the diversity and structure of subtilisin-like proteases among oral treponemes in order to better understand these putative virulence factors. Subtilisin genes were examined across a phylogenetically diverse set of treponemes to identify conserved elements, distribution, and mode of transmission.
MATERIALS AND METHODS
Bacterial strains.
Table 1 lists the strains examined in this study and their sources. Strains are from the culture collections of the Institut für Orale Mikrobiologie und Immunologie, University of Zürich, and of the Forsyth Institute. Strains were usually grown in an anaerobic chamber (Coy Laboratory Products Inc., Grass Lake, Mich.) containing 80% N2, 10% H2, and 10% CO2 in medium OMIZ-P4; this is a modification of medium OMIZ-Pat+Hus (38), and its complete listing is available as ATCC medium 2131 at the American Type Culture Collection (ATCC) website. OMIZ-P4 medium is also available from Anaerobe Systems (Morgan Hill, Calif.). For the survey of a wider range of enzyme substrates, T. denticola 35405T was grown for 3 to 4 days (ca. mid-log phase) in modified NOS medium as described previously (4) and “T. vincentii” OMZ 861 was grown for 4 to 7 days in modified NOS medium supplemented with selected vitamins, minerals, fatty acids, and other growth factors of medium OMIZ-P4 (except human serum) (37) that were not already in modified NOS, plus yeast extract (Gibco-BRL) and Bacto-neopeptone solution (Difco 0119); the final concentration of each was 0.5% (wt/vol).
TABLE 1.
Sources of strains examined
| Bacterial strain | Sourcea (animal, site) | Location (country) |
|---|---|---|
| “Treponema vincentii” OMZ 858 | Human, ANUG lesion | People's Republic of China |
| “Treponema vincentii” OMZ 860 | Human, ANUG lesion | People's Republic of China |
| “Treponema vincentii” OMZ 861 | Human, ANUG lesion | People's Republic of China |
| “Treponema vincentii” OMZ 862 | Human, ANUG lesion | People's Republic of China |
| Treponema denticola ATCC 35405T | Human, periodontal lesion | Canada |
| “Treponema putidum” OMZ 844 | Human, periodontal lesion | Switzerland |
| Treponema lecithinolyticum OMZ 684T | Human, periodontal lesion | Switzerland |
| Treponema sp. group IV OMZ 839 | Canine, dental plaque | Switzerland |
| Treponema sp. group IV OMZ 840 | Canine, dental plaque | Switzerland |
| Treponema maltophilum OMZ 679T | Human, periodontal lesion | Switzerland |
| Treponema socranskii subsp. socranskii ATCC 35536T | Human, subgingival plaque | United States |
| Treponema socranskii subsp. buccale OMZ 884 | Human, ANUG lesion | People's Republic of China |
| Treponema socranskii subsp. paredis OMZ 882 | Human, ANUG lesion | People's Republic of China |
| Treponema socranskii subsp. 04 VPI D40DR-2 | Human, subgingival plaque | United States |
| Treponema parvum VPI D96NR3 | Human, subgingival plaque | United States |
ANUG, acute necrotizing ulcerative gingivitis.
DNA isolation.
For fresh cultures, DNA was isolated following the MasterPure DNA purification kit (Epicentre Technologies, Madison, Wis.) protocol for bacterial cells. For frozen cell pellets, the following DNA isolation method was used. About 10 μl of cells was removed from the pellet with a loop or spatula and suspended directly in 50 μl of a mixture of 50 mM Tris buffer, pH 7.6, 1 mM EDTA, pH 8.0, 0.5% Tween 20, and 200 μg of proteinase K per ml. The suspension was incubated at 55°C for 2 h, followed by heating at 95°C for 5 min to inactivate the proteinase K.
PCR amplification.
The 16S ribosomal DNA operons were amplified with universal primers F15 and F16 (Table 2). Amplification of the prtP genes was initially performed with degenerate primers K15 and K16. These primers were designed by aligning the sequence of T. denticola dentilisin (GenBank D83264) with subtilisins from other bacterial species in the regions of the conserved histidine and serine catalytic amino acids. PCRs were carried out with an Applied Biosystems GeneAmp PCR System 9700; 50-μl PCR mixtures contained 1 μl of template DNA, 20 pmol of each primer, 40 nmol of deoxynucleoside triphosphates, and 1 U of Taq 2000 polymerase (Stratagene, La Jolla, Calif.) in buffer including Taqstart antibody (Sigma Chemical Co.). The PCRs were performed as follows: initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 45 s, annealing at 50 to 60°C (see Table 2 for primer annealing temperatures) for 45 s, and extension at 72°C for 90 s, with 1 s added to the extension at each cycle, followed by a final extension step at 72°C for 15 min. The PCR products were examined by electrophoresis in a 1% agarose gel. As sequence information accumulated on prtP and prcA genes, additional primers, such as M97 and O72, were designed for PCR amplification of genes in additional organisms from the 3′ end of prcA to K16, encompassing the 5′ two-thirds of prtP. These primers are listed in Table 2.
TABLE 2.
PCR and sequencing primers
| Designation | Target | Positionsa | Temp (°C) | Sequenceb (5′-3′) |
|---|---|---|---|---|
| F15 | 16S rRNA-F | 8-27 | 60 | CCATTGTATGACGTGTG |
| F16 | 16S rRNA-R | 1525-1541 | 60 | AAGGAGGTGWTCCARCC |
| K15 | prtP-F | 772-791 | 50 | CAYGGIACICAYTGYTCIGG |
| K16 | prtP-R | 1334-1352 | 50 | GGIACITCIATGGCIACICC |
| L36 | prtP(II)-F | 865-883 | 50 | ATTTGCGGCGTTTCYCAYG |
| L37 | prtP(II)-R | 1308-1326 | 50 | GCTGWAAATRCTTTCWCCG |
| L76 | prtP(I)-F | 829-844 | 50 | GCMGGCGTYGCRTGGA |
| L77 | prtP(I)-R | 1147-1165 | 50 | CYGTRTARCGSCCYTCGTT |
| M97 | prcA(I)-F | 1759-1777 | 55 | TGGGATATYCARGAYTGGG |
| O72 | prcA(II)-F | 1675-1693 | 55 | TGGGAYATYTTYGAYTGGG |
| R87 | 840prtP1-R | 1552-1574 | 55 | AGCGGCTCGGTAACCTGAACAAG |
| R86 | 840prtP1-F | 903-924 | 55 | TGCAAGAACGCCGGCTGATAAG |
| R88 | 840prcA1-F | 886-902 | 55 | GGCGGCCGGCGTTCTTC |
| R83 | 840prcA1-R | 1634-1657 | 55 | AGGCGACACCCGTTTCATAGTTCA |
| R73 | 839prtP2-R | 1464-1487 | 60 | TCTATTTTATCGGCGGTATGTTCC |
| R76 | 839prtP2-F | 1113-1131 | 60 | GCTTGCGGCGGGTGTTTTA |
| R74 | 839prcA2-F | 733-756 | 60 | GCCGGAAACAGGGTAGAGCGTCAT |
| R75 | 839prcA2-R | 1213-1230 | 60 | GCGGGCGGTTGCGGTAGG |
| R85 | 684prtP1-R | 1088-1110 | 50 | ATAGCGGCCCTCGTTTCCCATAG |
| R84 | 684prtP1-F | 714-735 | 50 | CGACGGGCATGGAACTCACTGT |
| R77 | 684prcA1-F | 1216-1236 | 50 | GCGAATGCGGCAAATCTTACG |
| R78 | 684prcA1-R | 1507-1529 | 50 | ACGCCGGTTGCTTTATCCACTTT |
| R80 | 684prtP2-R | 1353-1372 | 50 | ATTCGGGAGCGGCGGTATTG |
| R79 | 684prtP2-F | 925-947 | 50 | GGCGGCTCTTCGTATTCGGTGTA |
| R81 | 684prcA2-F | 1082-1105 | 50 | ACAATCCGAACTGGCCGTCATACA |
| R82 | 684prcA2-R | 1442-1464 | 50 | GAGTTTTTCGCCGCCGTTCTTTT |
Numbering was based on Escherichia coli for 16S rRNA, T. denticola ATCC 35405T for prtP-I and prcA-I, and T. lecithinolyticum OMZ 684T for prtP-II and prcA-II.
I = inosine; R = A or G; S = C or G; W = A or T; and Y = C or T.
DNA sequencing and analysis.
Amplicons were sequenced with an ABI Prism cycle-sequencing kit (BigDye Terminator cycle sequencing kit with AmpliTaq DNA polymerase FS; Perkin-Elmer) on an Applied Biosystems 377 or 3100 DNA sequencer. Half-dye or quarter-dye reactions were used following the manufacturer's instructions. Complete 16S rRNA sequences were determined for each strain with sequencing primers described previously (5). The sequences for the prtP and prcA genes were determined with the primers listed in Table 2 and several custom walking primers that were strain specific (sequences available from corresponding author). Sequences were assembled into contigs with Sequencer (Gene Codes Corp., Ann Arbor, Mich.). Similarity matrices were determined from the aligned sequences, and phylogenetic trees were constructed based on the neighbor-joining method of Saitou and Nei (32).
Lambda libraries.
Genomic DNA was partially digested by Sau3AI, size fractionated (3 to 7 kb) through a 1% agarose gel, and purified with the QIAquick gel extraction kit (Qiagen Inc., Valencia, Calif.); 1 μg of DNA was ligated into the BamHI digested, calf intestine alkaline phosphatase-treated arms of the ZAP Express vector (Stratagene, La Jolla, Calif.) and then packaged into lambda heads with the Gigapack III Gold packaging extract (Stratagene, La Jolla, Calif.). Plaques were formed with the XL1-Blue MRF[prime] strain in NZY top agar (5 g of NaCl, 2 g of MgSO4 · 7H2O, 5 g of yeast extract, 10 g of casein hydrolysate, 0.7% [wt/vol] agarose per liter, pH 7.5) on petri dishes (150 by 15 mm; ≈1,500 plaques/dish). The plaques were transferred and cross-linked to nylon membranes (Roche, Mannheim, Germany).
A prtP probe specific for each strain (see above PCR protocol) was digoxigenin labeled with the digoxigenin High Prime DNA labeling and detection starter kit (Roche, Mannheim, Germany) and hybridized to the DNA from the plaques. Hybridization-positive plaques were detected by enzyme immunoassay with a chemiluminescent substrate specific for alkaline phosphatase with the Roche kit. The area on the plates containing positive hybridizing plaques was eluted into TE buffer, replated, and rehybridized until single positive plaques were obtained. The inserts from these plaques were amplified with a combination of primers specific for prtP and the T3 and T7 promoters, which flanked the BamHI cloning site on the lambda ZAP vector. The PCR amplicons were sequenced with the primers in Table 2 or specifically designed walking primers. Sequences submitted to GenBank were fully sequenced on both strands.
Inverse PCR.
Genomic DNA (1 μg) from individual strains was digested with 6-bp recognition restriction enzymes, e.g., BamHI, PstI, EcoRI, KpnI, PvuI, and SalI, chosen on the basis of lack of recognition sites for that enzyme within a sequenced region. From the digests, 50 ng of DNA was self-ligated and then used in a PCR with primers specifically designed to prime outwards from a region of previously sequenced DNA. High-yield amplicons were then purified with the QIAquick PCR purification kit (Qiagen Inc., Valencia, Calif.) prior to sequencing. Low-yield amplicons were ligated into pCR4-Topo (Invitrogen, Carlsbad, Calif.), transformed into TOP10 cells, and selected on LB plates containing 50 μg of kanamycin per ml. Plasmids containing the correct sized inserts were purified with the QIAprep Spin miniprep kit (Qiagen Inc., Valencia, Calif.) prior to insert sequencing.
Expression analysis.
Expression of prcA and prtP was examined in Treponema sp. group IV OMZ 839, Treponema sp. group IV OMZ 840, and T. lecithinolyticum OMZ 684T. These species contain putative operons of paralogs group I, group II, and both group I and II genes, respectively (see Fig. 2, Fig. 3, and Results and Discussion, below). A 400-μl aliquot of a stationary-phase culture of each treponeme strain was added to 800 μl of RNAprotect bacterial reagent (Qiagen Inc., Valencia, Calif.). After centrifugation, the pellet was resuspended in 100 μl of lysozyme solution (400 μg/ml in 10 mM Tris-HCl pH 8.0; 1 mM EDTA), and RNA was isolated on an RNeasy minicolumn following the protocol detailed in the RNeasy kit instructions. The RNA samples were treated with 4 units of DNase I, amplification grade (Invitrogen, Carlsbad, Calif.), for 15 min at room temperature, followed by 10 min at 65°C to inactivate DNase. cDNA was prepared by reverse transcription with the primers listed in Table 4. Gene specific primers (2 pmol) were incubated for 50 min at 45°C with SuperScript II RNase H-negative reverse transcriptase (200 units). The reverse transcription was terminated by heat inactivation at 70°C for 15 min. To test that a single operon was transcribed, PCR amplification of the prcA and prtP genes was attempted from the cDNA. The PCR primers shown in Table 4 were used with SureStart Taq DNA polymerase (Stratagene, La Jolla, Calif.). The annealing temperatures for PCR were optimized for the gene-specific primer pairs used, as listed in Table 2.
FIG. 2.
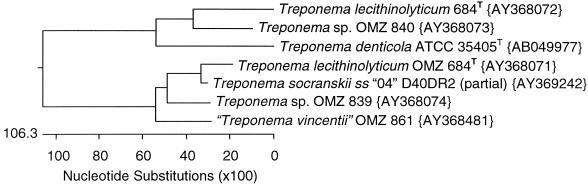
Phylogenetic tree of treponeme prtP genes. Tree based on amino acid comparisons with PAM250. GenBank accession numbers are given in braces.
FIG. 3.
Phylogenetic tree of treponeme prcA genes. Tree based on amino acid comparisons with PAM250. GenBank accession numbers are given in braces.
TABLE 4.
Expression analysis for prcA and prtP operonsa
| Organism | cDNA (primer) | prcA (I)a (primers) | prtP (I)a (primers) | prcA (II)a (primers) | prtP (II)a (primers) |
|---|---|---|---|---|---|
| Treponema sp. strain OMZ 840 | R87 | +, R88/R83 | +, R86/R87 | ||
| Treponema sp. strain OMZ 839 | R73 | +, R74/R75 | +, R76/R73 | ||
| T. lecithinolyticum OMZ 864T (I) | R85 | +, R77/R78 | +, R84/R85 | ||
| T. lecithinolyticum OMZ 864T (II) | R80 | +, R81/R82 | +, R79/R80 |
+ and − are used to indicate that a PCR product was obtained or not with the indicated primers.
Peptidase activity.
All OMZ and ATCC strains in Table 1 were tested for prolyl-phenylalanine-specific peptidase activity by a colorimetric method with the substrate N-succinyl-alanyl-alanyl-prolyl-phenylalanine-p-nitroanilide (SAAPFNA; Bachem). The assay mixture consisted of 100 μl of washed cells (optical density at 550 nm, 0.1) and 100 μl of substrate solution (1 mM), each in buffer A (100 mM NaCl, 50 mM Tris, 1 mM CaCl; pH 8.0) and was incubated aerobically (6 h at 37°C). Enzymatic activities were determined before PCR results were available.
Survey of protein and peptide digestion.
In an attempt to determine a proteolytic phenotype for the group II PrtP family and to identify substrate specificities that may differentiate the two paralogous PrtP families, the proteolytic and peptidolytic activities of T. denticola type strain ATCC 35405T and “T. vincentii” OMZ 861 were compared. Cell-associated enzyme activity was studied under a wide range of conditions, with both washed and unwashed cells suspended to an optical density at 550 nm of either 0.2 or 1.0 (Ultraspec III, Amersham Pharmacia). Cells were washed twice in fresh growth medium, centrifuged at 12,000 × g for 10 min, and resuspended in either phosphate-buffered saline (0.1 M, pH 7.2) or Tris-buffered saline (20 mM, pH 8.0), washed, and resuspended in the same buffer.
Digestion of fibronectin, fibrinogen, acid-soluble type VI collagen, laminin (200 to 400 μg/ml stocks; all from Sigma) and vitrogen (Cohesion Technologies, Palo Alto, Calif.) was determined by examination of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels for intact substrate and its degradation products. Briefly, 20 μl of substrate solution was incubated with 80 μl of either bacterial cell suspension or bacteria-free growth medium or buffer control, at either ambient temperature or 37°C, for either 3 h or overnight. After incubation, the cells were pelleted by centrifugation at 12,000 × g for 10 min, and 50 μl of supernatant was mixed with 50 μl of 2× sample buffer and boiled for 10 min. The samples were run on 10% Tris-glycine Ready Gels (Bio-Rad) at 200 V and stained with 0.25% Coomassie blue in 40% methanol and 10% acetic acid.
In addition to SAAPFNA, T. denticola ATCC 35405T and “T. vincentii” OMZ 861 were compared for their peptidolytic activity against a range of peptide para-nitroanilides (p-NA; Sigma): benzoyl-Arg p-NA, glutaryl-Phe-p-NA, succinyl-Ala-Ala-Pro-Leu p-NA, Ala-Ala-Phe p-NA, benzoyl-Tyr p-NA, succinyl-Ala-Ala-Ala p-NA, Gly-Phe p-NA, and Lys p-NA. Bacterial suspensions were mixed with a substrate solution and Tris-buffered saline 1:1:3, incubated overnight at 37°C, and centrifuged at 12,000 × g for 10 min. The absorbance of the supernatants was read at 405 nm (Bio-Rad model 3550 microplate reader).
Digestion of casein and gelatin was analyzed by zymography with 12% and 10%Ready Gel Zymogram gels (Bio-Rad), respectively. Sonic extracts of the bacterial cells in 20 mM Tris-HCl (pH 8.0) containing 1% CHAPS were prepared as described by Ishihara and coworkers (15) and stored refrigerated overnight. The extracts were mixed 1:1 with 2× β-mercaptoethanol-free sample buffer and loaded in the zymography gels without boiling. Chymotrypsin (Sigma, C4629), 200 μg/ml in 20 mM Tris-HCl, was used as a positive control. After electrophoresis, the gels were washed in 2% Triton X-100 (Sigma) in 20 mM Tris-HCl for 1 h, followed by two quick washes and a 30-min wash in the same buffer. The gels were incubated in Tris-HCl for 3 h at ambient temperature and kept overnight at 4°C. The gels were stained with Coomassie blue as above.
Nucleotide sequence accession numbers.
The GenBank accession numbers for all genes described or discussed in this manuscript are given in the phylogenetic trees in Fig. 1 to 3 and in the gene map in Fig. 4.
FIG. 1.
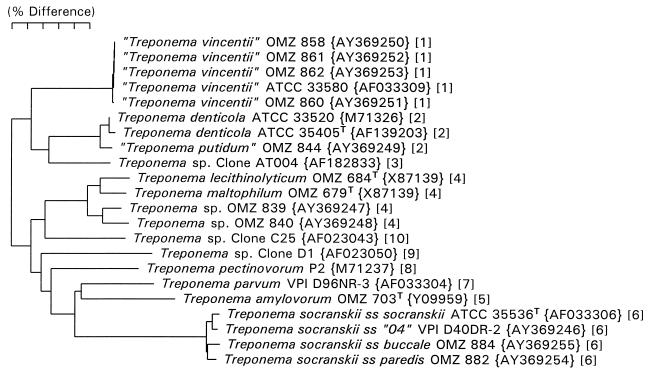
Neighbor-joining phylogenetic tree for treponeme strains examined in this study and named human oral species as determined from 16S rRNA sequence comparisons. GenBank accession numbers are given in braces. Numbers in brackets refer to treponeme phylogenetic groups defined previously (5).
FIG. 4.
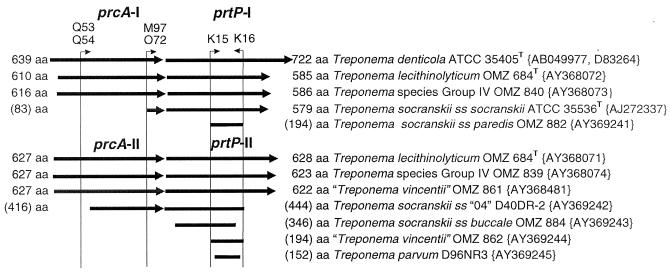
Operon map of sequenced genes. Solid lines with arrowheads indicate sequenced portions of each ORF, drawn to scale. The amino acid length of each ORF is indicated, without parentheses for completed sequences and with parentheses for incomplete sequences. The positions of key degenerate primers used to amplify the ORFs are indicated by vertical lines below small, labeled arrowheads. Paralog names and group numbers are indicated above the appropriate ORFs. Genus, species, and strain designations for each ORF are indicated at the right. GenBank accession numbers are given in braces.
RESULTS
16S rRNA sequencing.
Full 16S rRNA sequences were determined for each strain in order to definitively identify them. A phylogenetic tree, based on the 16S rRNA sequence analysis, is shown in Fig. 1. The GenBank accession number for each strain is included in the figure. Treponema strains OMZ 839 and OMZ 840 both fall in treponeme group IV as defined previously (5). This tree provides a phylogenetic context for comparison with the prcA and prtP gene trees described below.
Distribution of subtilisin homologs in oral treponemes.
The conserved regions around the protease active-site residues histidine and serine were targeted for amplification with degenerate PCR primers K15 and K16, respectively. Amplicons were obtained for 14 of 15 strains examined, as shown in Table 3. The amplicons were sequenced and aligned based on translated amino acid sequences with Clustal V. Phylogenetic analysis demonstrated that the sequences clustered into two distinct groups (A phylogenetic tree based on partial sequences is not shown, as a tree based on complete sequences is presented below,) Group I contained T. denticola, Treponema sp. OMZ 840, T. lecithinolyticum, T. socranskii subsp. socranskii, and T. socranskii subsp. paredis. A second group comprised “T. vincentii” strains, T. lecithinolyticum, T. socranskii subsp. buccale, T. socranskii subsp. 04, Treponema sp. OMZ 839, and Treponema parvum VPI D96NR3. The three strains of “T. vincentii” formed a tight cluster (98 to 100% similarity of translated amino acid sequence) but were only 40 to 45% similar to that of T. denticola PrtP. Strains did not necessarily cluster strictly according to species, as indicated above for the T. socranskii subspecies. Also, the phylogenetically closely related group IV species T. maltophilum and T. lecithinolyticum showed zero and two prtP genes, respectively.
TABLE 3.
Subtilisin gene homologs in oral treponemes
| Bacterial strain | Subtilisina | Group Ib | Group IIc | SAAPFNA activity |
|---|---|---|---|---|
| “Treponema vincentii” OMZ 858 | + | − | + | − |
| “Treponema vincentii” OMZ 861 | + | − | + | − |
| “Treponema vincentii” OMZ 862 | + | − | + | − |
| “Treponema vincentii” OMZ 860 | + | − | + | − |
| Treponema denticola ATCC 35405T | + | + | − | + |
| “Treponema putidum” OMZ 844 | + | − | + | − |
| Treponema lecithinolyticum OMZ 684T | + | + | + | + |
| Treponema sp. group IV OMZ 839 | + | − | + | − |
| Treponema sp. group IV OMZ 840 | + | + | − | + |
| Treponema maltophilum OMZ 679T | − | − | − | − |
| Treponema socranskii subsp. socranskii ATCC 35536T | +d | +d | − | +d |
| Treponema socranskii subsp. buccale OMZ 884 | + | − | + | − |
| Treponema socranskii subsp. paredis OMZ 882 | + | + | − | + |
| Treponema socranskii subsp. 4 VPI D40DR-2 | + | − | + | NDe |
| Treponema parvum VPI D96NR3 | − | − | + | NDf |
PCR amplicon with subtilisin active primers K15 and K16.
PCR amplicon with group I paralog primers L76 and L77.
PCR amplicon with group II paralog primers L36 and L37.
Results from Heuner et al. (13).
ND, not determined.
Activity is negative for T. parvum strain OMZ 684.
Based on aligned sequences, two sets of primers were designed to specifically amplify either prtP-I (L36/L37) or prtP-II (L76/L77) and used to survey the oral treponemes. One, the other, or both sets of primers produced amplicons for all strains that had previously produced an amplicon with the K15/K16 subtilisin primers (Table 3). In the case of T. parvum, a group II amplicon was obtained even though the initial K15/K16 amplification had been negative. The K15 and K16 primers were based on alignment of phylogenetically diverse subtilisin sequences with the dentilisin of T. denticola. Based on sequence information obtained in the course of this study, we realized that these primers did not contain sufficient degeneracy to optimally amplify all known treponeme prtP genes.
Complete sequences for prtP gene paralogs.
Efforts were made to obtain full sequences from representatives of both paralogous gene groups. The complete prtP(I) and prtP(II) genes from T. lecithinolyticum OMZ 684T, “T. vincentii” OMZ 861, and Treponema sp. group IV strains OMZ 839 and OMZ 840 were obtained through a combination of techniques. Lambda libraries of each species were constructed that contained partial Sau3AI genomic inserts. The libraries were screened with probes amplified from the paralog discriminating primer sets L36/L37 and L76/L77. A complete prtP gene could not be obtained from any of the three libraries, but from the sequence information obtained, primers were designed for inverse PCR-based gene walking. Complete sequences were obtained and aligned based on translated amino acid sequences with Clustal V. A phylogenetic tree based on the aligned sequences is shown in Fig. 2. The GenBank accession number for each sequence is given in the tree.
Substrate specificity of PrtP paralogs.
The Treponema denticola protease dentilisin was identified by its ability to cleave the para-nitroanilide-conjugated peptide SAAPFNA (36). Dentilisin belongs to paralog group I. All members of paralog group I examined in this study were able to cleave SAAPFNA (Table 3). In contrast, Treponema species encoding paralog group II, including all “T. vincentii” strains, T. socranskii OMZ 882, and group IV OMZ strain 839, lacked a chymotrypsin-like activity, as indicated by the inability of whole-cell suspensions to cleave the peptide substrate SAAPFNA. T. denticola ATCC 35405T expressed a wide range of proteolytic and peptidolytic activities. When the digestion of matrix proteins was analyzed by SDS-PAGE, the activity of T. denticola cells yielded greatly diminished native protein band intensity and evidence for lower mass cleavage products for fibronectin, fibrinogen, type VI collagen, and laminin; T. denticola tested negative for bovine serum albumin and vitrogen.
By zymography, T. denticola showed clear digestion of both casein and gelatin. Among the peptide-nitroanilide substrates, T. denticola yielded strong positive peptidase activity against SAAPFNA, Pro-Leu p-NA, and Ala-Ala-Phe p-NA, for which the chymotrypsin solution control was also positive, and against benzoyl-Tyr p-NA, Lys p-NA, and benzoyl-Arg-Pro p-NA, for which the chymotrypsin control was negative. The latter peptidase activities are known to be due to T. denticola enzymes other than PrtP (8). In contrast to T. denticola, “T. vincentii” OMZ 861 failed to digest any of the protein substrates, nor did it cleave any of the peptides with the exception of Lys p-NA for which there was a strong positive reaction. “Treponema vincentii” even cleaved Lys p-NA in the presence of the serine protease inhibitor phenylmethylsulfonyl fluoride, which is known to inhibit the proteolytic and SAAPFNA-degrading activity of T. denticola. Therefore, the biological activity and potential substrates of the prcA-prtP gene product(s) of paralog group II remain unidentified.
Full sequences for prcA gene paralogs.
The prcA gene upstream of prtP was shown by Lee et al. (18) to encode the 38- and 43-kDa proteins of the CTLP complex in T. denticola. Gene walking upstream of prtP revealed homologues of the T. denticola prcA gene in each case. In order to better understand the structure and function of prcA genes, the full sequences were determined for five species by inverse PCR gene walking. The five complete prcA sequences were aligned with the T. denticola prcA sequence (AY069957) based on their translated amino acid sequences with Clustal V. A phylogenetic tree for the aligned sequences is shown in Fig. 3. As seen with the prtP sequences, the prcA sequences fell into two paralogous clusters.
prcA-prtP operon.
A map of all the sequenced prcA and prtP genes is shown in Fig. 4. This map gives the deduced number of amino acids in each full-length protein. To determine if prcA and prtP are transcribed as a single operon in diverse species, mRNA was isolated from cultures of three strains and cDNA was produced by reverse transcription with the prtP 3′ reverse primers listed in Table 4. Treponema sp. OMZ 840 contains a paralogous group I prcA-prtP operon. Treponema sp. OMZ 839 contains a paralogous group II prcA-prtP operon. Treponema lecithinolyticum OMZ 684T contains genes for both types of putative prcA-prtP operons. PCR was then performed to determine if only prtP or both prcA and prtP amplicons could be produced from the appropriate cDNA. In each case, both prcA and prtP amplicons were obtained, indicating that prcA-prtP is transcribed as a single operon. Amplicons were obtained for both paralogous operons in T. lecithinolyticum.
DISCUSSION
Dentilisin is a purported virulence factor of T. denticola. This study was initiated to examine the diversity and structure of dentilisin homologs present in oral treponemes. The conserved amino acid residues at the active site of the subtilisins allowed the amplification of an internal fragment of the prtP genes from the majority of species and strains examined, including “T. vincentii,” T. socranskii, T. lecithinolyticum, and two canine Treponema group IV species. Thus, dentilisin homologs are widespread among oral treponemes.
Complete prtP gene sequencing showed that a homolog of prcA was always found immediately upstream of the prtP gene. The stop codons of the prcA genes were separated by 11 to 24 bases from the methionine start codon of the prtP genes. In four of seven operons for which we sequenced the junction of prcA and prtP, the genes are in different reading frames. Expression analysis indicated that both genes are cotranscribed from a single mRNA. In the case of T. lecithinolyticum, both operons are expressed. Fluorescent in situ hybridization analysis or similar technique will need to be performed to distinguish between all cells simultaneously transcribing both operons and half the cells transcribing one operon and the other half transcribing the second operon. The dendrograms for prcA and prtP (Fig. 2 and 3) branch essentially identically, suggesting that they evolved as an operon and have been exchanged as a unit.
Phylogenetic analysis of either the prcA or prtP gene shows that they fall into two paralogous groups. The sequences of the prcA and prtP genes of paralog group I are similar to those of T. denticola. The prtP gene sequences in paralog group II are distinct from that of T. denticola but more similar to T. denticola prtP than to any other subtilisin gene. While this work was in progress, Heuner et al. (13) reported the existence of two copies of prtP in T. lecithinolyticum. The current work confirms and significantly extends their findings by providing complete sequences of the prtP and prcA genes.
Closely related treponeme species or subspecies do not necessarily express the same paralogous prcA-prtP operon, as demonstrated by comparing T. socranskii subsp. socranskii, T. socranskii subsp. paredis (paralog group I), and T. socranskii subsp. buccale and T. socranskii subsp. 04 (paralog group II, Table 3). The random distribution of either no prcA-prtP operon, paralog group I operon, paralog group II operon, or both operons, compared to the 16S rRNA-based phylogeny of treponemes, suggests that the operons can be lost or obtained by horizontal exchange. However, this suggestion is speculative, as natural genetic exchange in treponemes has not been documented. Treponema denticola strains ATCC 33521 and ST10A are SAAPFNA negative (unpublished observation), suggesting that dentilisin may not be actively expressed in all T. denticola strains. However, these strains have not been examined by PCR and sequencing of prcA-prtP genes to determine if the genes are missing or inactivated. Since dozens of oral treponemal phylotypes usually cocolonize the surface of dense biofilms and translocate in the confined medium between the biofilm and host tissues, the environment would be expected to support frequent contact and possible genetic exchange.
A number of strains representing species in spirochete genera other than Treponema were PCR negative with the K15 and K16 primers (unpublished results). While we know from examination of the complete genomes of T. pallidum, Borrelia burgdorferi, and Leptospira interrogans that these species lack subtilisin homologs, it is conceivable that there were false negatives in our screen and that prcA-prtP homologs are present in species of spirochetes outside the genus Treponema.
The strains which fall into paralog group I all possess chymotrypsin-like protease activity (i.e., in a whole-cell assay they cleave the substrate SAAPFNA, while those strains that fall into paralog group II all lack such activity). Despite examination of numerous substrates, including proteins and small chromogenic peptides, we have been unable to determine the biological activity and specificity of the group II subtilisin paralogs.
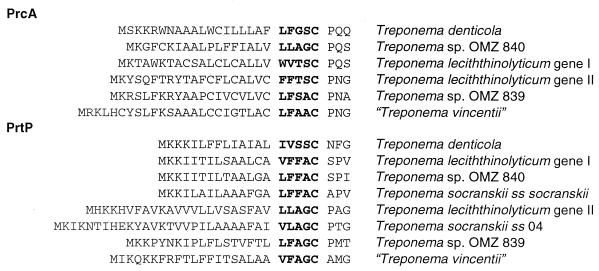
In T. denticola, dentilisin is located in the cell's outer sheath and is a part of a three protein complex. Lee et al. (18) demonstrated that both the 38-kDa protein and the 43-kDa protein are encoded by the prcA gene. They have postulated that PrcA is cleaved by the chymotrypsin-like activity of PrtP into two peptides PrcA1 (38 kDa protein) and PrcA2 (43 kDa protein). The cleavage of PrcA most likely occurs after PrcA and PrtP are both transported to the outer membrane and have associated. Our analysis shows that there is only a single peptide lipobox at the amino terminus of prcA; therefore PrcA2 would not be sorted to the outer sheath if cleaved from PrcA1 cytoplasmically. Lee et al. (18) postulated two potential processing sites in PrcA, as indicated with arrows in Fig. 5. Alignment of PrcA amino acid sequences for T. denticola with T. lecithinolyticum and Treponema sp. strain OMZ 840 indicates that the first site is not conserved; it contains an aspartate (D) in place of a proline (P) at the −2 position. The second site is conserved, containing only a conservative substitution of a tyrosine (Y) for a phenylalanine (F) at the −1 position. Alignment of prcA group II paralogs with group I paralogs revealed a conserved phenylalanine-histidine (PH) at the second potential cleavage site. However, at present it is not known if the group II PrcA proteins are cleaved.
FIG. 5.
Potential cleavage sites of PrcA. The aligned amino acid sequences for six PrcA genes are shown above. The two potential cleavage sites in PrcA proposed by Lee et al. (18) are indicated with arrows. The positions of the amino acids at the cleavage sites are numbered from the initial methionine with subscripts. The T. socranskii subsp. 04 is a partial sequence, and the numbering of amino acids refers to the start of the partial sequence. The second site is more conserved in the PrcA sequences. While it is not known if PrcA-II is cleaved, there is a conserved PH amino acid sequence at the corresponding position.
While the prcA-prtP operon is necessary for proper processing of Msp, the major outer sheath protein of T. denticola (8), T. maltophilum possesses an Msp homolog (14) but does not possess a prcA-prtP operon. Similarly, T. pallidum appears to express a family of 12 proteins (TprA through TprL) homologous to Msp without possessing a prcA-prtP operon. Treponema lecithinolyticum possesses both paralogous operons. Thus, having zero, one, or two prcA-prtP operons appears to be compatible with survival in the subgingival oral habitat. Since PrtP is an extracellular protease whose apparent function is to provide peptide substrates for transport, perhaps community survival is enhanced by or depends on the dissemination of a prcA-prtP operon among a sufficient number of community members in the biofilm. Because the prcA-prtP operon is necessary for proper expression of the Msp in T. denticola, it will be interesting to determine if group I prcA-prtP operons from other treponemes can rescue prcA-prtP mutants, such as strains K1 and PNE (15, 18).
The prtP gene in T. denticola encodes a protein almost 140 amino acids longer than that of other group I PrtP proteins and the carboxy-terminal extension may be involved in specific protein-protein interactions with Msp. If that is true, then msp, prcA, and prtP may have evolved as a unit and cannot be transferred independently. However, examination of the unfinished genome of T. denticola (www.TIGR.org) shows that the msp and prcA-prtP genes are on different contigs and probably are not located close together on the chromosome.
Each of the prcA and prtP genes for which we have the complete 5′-end sequence possesses a typical treponeme peptide lipobox, as shown in Fig. 6 (12). The difficulty investigators have had in cloning the 5′ end of either prcA or prtP may be due to recognized difficulties of cloning lipoproteins into Escherichia coli (23). Similarly, the finding that the 38-kDa protein was blocked is consistent with lipoprotein modification (15). Examination of the T. denticola sequence with Blast at TIGR indicates that all of the putative lipoprotein secretion and processing genes of T. pallidum and Borrelia burgdorferi (ffh, secA, secD, secE, secF, secY, lgt, lsp, and lnt) are present (12). A number of other T. denticola membrane proteins have typical lipoboxes, such as oppA (AF042861) and outer membrane hemin-binding proteins (AF196837 and AF332358). Analysis of the B. burgdorferi genome (10) indicated that it possesses 105 lipoproteins, substantially more than found in other bacterial genomes. Haake has suggested that a majority of proteins attached to the outer membrane of spirochetes are lipoproteins (12). Among these lipoproteins are many of the virulence factors, antigens, and vaccine targets for spirochete pathogens. Future efforts to understand the role of PrcA in the proper function of PrtP may deepen our understanding of targeting and maturation of treponeme lipoproteins.
FIG. 6.
Putative lipoboxes at the amino termini of PrcA and PrtP. The cysteine in the aligned sequences occurs 19 to 34 amino acids from the initial methionine.
Dentilisin is a member of the subtilisin family of serine proteases; MEROPS, S08.024 (29); and PFAM, PF00082 (1). Six families of subtilisin-like serine proteases have been described by Siezen and Leunissen (34). Based on alignments around the conserved catalytic aspartate, histidine, and serine residues (see Fig. 7) the treponeme proteases appear to differ sufficiently from the known families to represent a novel family most closely related to the subtilisin and thermitase subfamilies (34). As shown in Fig. 7, there is substantial conservation around the catalytic triad for both PrtP-I and PrtP-II. There is nothing in the active-site alignments that would suggest that PrtP-II should lack activity (Fig. 7). Additional alignments and trees of the subtilisin family of proteases can be found at the MEROPS web site (http://merops.sanger.ac.uk). The unique status of the treponeme subtilisins is supported by their being the only known lipoprotein subtilisins.
FIG. 7.
Comparison of PrtP deduced amino acid sequences around the active-site residues. The active-site aspartate (D), histidine (H), and serine (S) are boldfaced and underlined.
The present study demonstrated that there is wide distribution in oral treponemes of two-gene subtilisin operons. There are two paralogous operon families, one with dentilisin or chymotrypsin-like protease activity, and a second with a currently unknown specificity or function. The prcA gene, which previously had no homologs in GenBank, now has six complete sequences. The availability of multiple prcA and prtP gene sequences for comparison should facilitate further studies to determine the function of PrcA and the evolutionary and ecological implications of its dissemination among the many oral treponemes.
Acknowledgments
This work was supported by grant DE-10374 from the National Institute of Dental and Craniofacial Research (F.E.D.), grant MGP-5619, and a group grant from the Canadian Institutes of Health Research (R.P.E.).
We thank V. Zängerle for expert technical assistance and D. A. Grove for the phenotypic characterization of proteolytic and peptidolytic activity.
REFERENCES
- 1.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carranza, F. A. Jr., R. Saglie, M. G. Newman, and P. L. Valentin. 1983. Scanning and transmission electron microscopic study of tissue-invading microorganisms in localized juvenile periodontitis. J. Periodontol. 54:598-617. [DOI] [PubMed] [Google Scholar]
- 3.Coyle, P. K. 2002. Lyme disease. Curr. Neurol. Neurosci. Rep. 2:479-487. [DOI] [PubMed] [Google Scholar]
- 4.Dawson, J. R., and R. P. Ellen. 1990. Tip-oriented binding of Treponema denticola to fibronectin. Infect. Immun. 58:3924-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewhirst, F. E., M. A. Tamer, R. E. Ericson, C. N. Lau, V. A. Levanos, S. K. Boches, J. L. Galvin, and B. J. Paster. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol. Immunol. 15:196-202. [DOI] [PubMed] [Google Scholar]
- 6.Ellen, R. P., K. S.-C. Ko, C.-M. Lo, D. A. Grove, and K. Ishihara. 2000. Insertional inactivation of the prtP gene of Treponema denticola confirms dentilisin's disruption of epithelial junctions. J. Mol. Microbiol. Biotechnol. 2:581-586. [PubMed] [Google Scholar]
- 7.Fenno, J. C., and B. C. McBride. 1998. Virulence factors of oral treponemes. Anaerobe 4:1-7. [DOI] [PubMed] [Google Scholar]
- 8.Fenno, J. C., S. Y. Lee, C. H. Bayer, and Y. Ning. 2001. The opdB locus encodes the trypsin-like peptidase activity of Treponema denticola. Infect. Immun. 69:6193-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenno, J. C., G. W. K. Wong, P. M. Hannam, and B. C. McBride. 1998. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol. Lett. 163:209-215. [DOI] [PubMed] [Google Scholar]
- 10.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 11.Grenier, D., V-J. Uitto, and B. C. McBride. 1990. Cellular location of a Treponema denticola chymotrypsinlike protease and importance of the protease in migration through the basement membrane. Infect. Immun. 58:347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haake, D. A. 2000. Spirochaetal lipoproteins and pathogenesis. Microbiology 146:1491-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heuner, K., I. Bergmann, K. Heckenbach, and U. B. Göbel. 2001a. Proteolytic activity among various oral Treponema species and cloning of a prtP-like gene of Treponema socranskii subsp. socranskii. FEMS Microbiol. Lett. 201:169-176. [DOI] [PubMed] [Google Scholar]
- 14.Heuner, K., U. Meltzer, B. K. Choi, and U. B. Göbel. 2001b. Outer sheath associated proteins of the oral spirochete Treponema maltophilum. FEMS Microbiol. Lett. 197:187-193. [DOI] [PubMed]
- 15.Ishihara, K., T. Miura, H. K. Kuramitsu, and K. Okuda. 1996. Characterization of the Treponema denticola prtP gene encoding a prolyl-phenylalanine-specific protease (dentilisin). Infect. Immun. 64:5178-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishihara, K., T. Miura, H. K. Kuramitsu, and K. Okuda. 1998. Dentilisin activity affects the organization of the outer sheath of Treponema denticola. J. Bacteriol. 180:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko, KS-C., C-M. Lo, J. Ferrier, P. Hannam, M. Tamura, B. C. McBride, and R. P. Ellen. 1998. Cell-substrate impedance analysis of epithelial cell shape and micromotion upon challenge with bacterial proteins which perturb extracellular matrix and cytoskeleton. J. Microbiol. Methods 34:125-132. [Google Scholar]
- 18.Lee, S. Y., X-L. Bian, G. W. K. Wong, P. M. Hannam, B. C. McBride, and J. C. Fenno. 2002. Cleavage of Treponema denticola PrcA polypeptide to yield protease complex-associated proteins PrcA1 and PrcA2 is dependent on PrtP. J. Bacteriol. 184:3864-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levett, P. N. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Listgarten, M. A. 1965. Electron microscopic observation the bacterial flora of acute necrotizing ulcerative gingivitis. J. Periodontol. 36:328-339. [DOI] [PubMed] [Google Scholar]
- 21.Listgarten, M. A. 1976. Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J. Periodontol. 47:1-18. [DOI] [PubMed] [Google Scholar]
- 22.Mäkinen, P. L., K. K. Mäkinen, and S. A. Syed. 1995. Role of the chymotrypsin-like membrane associated protease from Treponema denticola ATCC 35405T in inactivation of bioactive peptides. Infect. Immun. 63:3567-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin, B., G. Alloing, C. Boucraut, and J. P. Claverys. 1989. The difficulty of cloning Streptococcus pneumoniae mal and ami loci in Escherichia coli: toxicity of malX and amiA gene products. Gene 80:227-238. [DOI] [PubMed] [Google Scholar]
- 24.Moore, W. E., L. H. Moore, R. R. Ranney, R. M. Smibert, J. A. Burmeister, and H. A. Schenkein. 1991. The microflora of periodontal sites showing active destructive progression. J. Clin. Periodontol. 18:729-739. [DOI] [PubMed] [Google Scholar]
- 25.Moter, A., C. Hoenig, B. K. Choi, B. Riep, and U. B. Göbel. 2000. Molecular epidemiology of oral treponemes associated with periodontal disease. J. Clin. Microbiol. 36:1399-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noiri, Y., L. Li, and S. Ebisu. 2001. The localization of periodontal-disease-associated bacteria in human periodontal pockets. J. Dent. Res. 80:1930-1934. [DOI] [PubMed] [Google Scholar]
- 27.Norris, S. J., D. L. Cox, and G. M. Weinstock. 2001. Biology of Treponema pallidum: correlation of functional activities with genome sequence data. J. Mol. Microbiol. Biotechnol. 3:37-62. [PubMed] [Google Scholar]
- 28.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawlings, N. D., E. O'Brien, and A. J. Barrett. 2002. MEROPS: the protease database. Nucleic Acids Res. 30:343-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riviere, G. R., K. S. Weisz, D. F. Adams, and D. D. Thomas. 1991. Pathogen-related oral spirochetes from dental plaque are invasive. Infect. Immun. 59:3377-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosen, G., R. Naor, E. Rahamim, R. Yishai, and M. N. Sela. 1995. Proteases of Treponema denticola outer sheath and extracellular vesicles. Infect. Immun. 63:3973-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 33.Sela, M. N. 2001. Role of Treponema denticola in periodontal diseases. Crit. Rev. Oral Biol. Med. 12:399-413. [DOI] [PubMed] [Google Scholar]
- 34.Siezen, R. J., and J. A. Leunissen. 1997. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 6:501-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorsa, T., T. Ingman, K Suomalainen, M. Haapasalo, Y. T. Konttinen, O. Lindy, H. Saari, and V.-J. Uitto. 1992. Identification of proteases from periodonto-pathogenic bacteria as activators of latent human neutrophil and fibroblast-type interstitial collagenases. Infect. Immun. 60:4491-4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uitto V.-J., D. Grenier, E. C. Chan, and B. C. McBride. 1988. Isolation of a chymotrypsinlike enzyme from Treponema denticola. Infect. Immun. 56:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyss, C. 1992. Growth of Porphyromonas gingivalis, Treponemas denticola, T. pectinovorum, T. socranskii, and T. vincentii in a chemically defined medium. J. Clin. Microbiol. 30:2225-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyss, C., B.-K. Choi, P. Schüpabach, B. Guggenheim, and U. B. Göbel. 1997. Treponema amylovorum sp. nov., a saccharolytic spirochete of medium size isolated from an advanced human periodontal lesion. Int. J. Syst. Bacteriol. 47:842-845. [DOI] [PubMed] [Google Scholar]
- 39.Wyss, C., B. K. Choi, P. Schüpbach, A. Moter, B. Guggenheim, and U. B. Göbel. 1999. Treponema lecithinolyticum sp. nov., a small saccharolytic spirochaete with phospholipase A and C activities associated with periodontal diseases. Int. J. Syst. Bacteriol. 49:1329-1339. [DOI] [PubMed] [Google Scholar]