Abstract
The high-resolution two-dimensional (2D) protein gel electrophoresis technique combined with matrix-assisted laser desorption ionization-time of flight mass spectrometry was used for identification of proteins whose levels were changed by a mutation in hemB. Cytoplasmic protein extracts obtained from the mutant and the wild type (strain COL) at different stages of growth in tryptone soya broth (exponential, transitional, and stationary growth phases) were separated on 2D protein gels. Comparison of the 2D patterns of the protein extracts of the two strains revealed major differences. Because the electron transport chain of the mutant is interrupted due to the deficiency of heme, this organism should be unable to use oxygen or nitrate as a terminal electron acceptor. Consistent with this hypothesis, proteins involved in the glycolytic pathway and related pathways (glyceraldehyde-3-phosphate dehydrogenase, enolase, and phosphoglycerate kinase) and in fermentation pathways (lactate dehydrogenase, alcohol dehydrogenase, and pyruvate formate lyase) were induced in exponentially growing cells of the mutant. These results strongly indicate that the hemB mutant generates ATP from glucose or fructose only by substrate phosphorylation. Analyses of the fermentation reactions showed that the main product was lactate. Although pyruvate formate lyase (Pfl) and pyruvate dehydrogenase were present, neither ethanol nor acetate was detected in significant amounts. Presumably, Pfl was not activated in the presence of oxygen, and pyruvate dehydrogenase might have very low activity. Transcriptional analysis of citB, encoding the aconitase, revealed that the activity of the citrate cycle enzymes was down-regulated in the hemB mutant. The arginine deiminase pathway was also induced, and it could provide ATP as well. Furthermore, the amounts of most of the extracellular virulence factors were significantly reduced by a mutation in hemB, which is consistent with previous reports.
In the last decade, interest in infections due to Staphylococcus aureus small-colony variants (SCVs) has emerged because an association between the recovery of S. aureus SCVs and persistent and relapsing infections has been established (12, 22, 23, 25, 33, 34). S. aureus SCVs are a slowly growing subpopulation of the species and have characteristics that can be tied together by a common thread (i.e., alterations in electron transport). Specifically, the following features are very likely linked to interruption of electron transport: (i) slow growth, producing pinpoint colonies; (ii) decreased pigment formation because carotenoid biosynthesis requires electron transport; (iii) resistance to gentamicin and other aminoglycosides because uptake of these compounds requires a large membrane potential generated by electron transport; and (iv) mannitol fermentation negative because utilization of a sugar alcohol, such as mannitol, is decreased when electron transport is not present (12, 14, 16, 22, 23, 25, 33, 34).
The morphotype of S. aureus SCVs is often due to auxotrophy for hemin, which is required for the biosynthesis of cytochromes. Supplementation of media with hemin often allows clinical SCVs to grow as rapidly as the parent strain and to exhibit a normal phenotype with regard to antibiotic susceptibility, membrane potential, and exoprotein and pigment production (2, 12, 14, 16, 23, 32, 35). To characterize the phenotype of a genetically defined SCV of S. aureus, a stable mutant with a mutation in in electron transport was generated by interrupting one of the heme biosynthetic genes, hemB, which is a member of the family of genes encoding enzymes of the porphyrin biosynthetic pathway (9).
The site-directed S. aureus hemB mutant has all of the features described for SCVs recovered from clinical specimens (12, 23, 25, 33-35). In addition, this mutant provided the opportunity to investigate a stable SCV phenotype, in contrast to naturally occurring clinical SCV isolates, which are genetically undefined and exhibit a high rate of reversion to the large-colony form. Previous studies showed that the hemB mutant was phagocytosed by cultured endothelial cells but did not lyse these cells. It was assumed that this observation was due to the fact that this mutant, as determined on the protein and transcription levels, produced very little alpha-toxin; consequently, it was concluded that an intracellular location may shield SCVs from host defenses and antimicrobial agents, thus providing one explanation for the difficulty of clearing S. aureus SCVs from host tissues (35).
In this study, the two-dimensional (2D) gel electrophoresis technique was used to characterize the hemB mutant of S. aureus in more detail. During this study we found that proteins involved in fermentation and glycolysis were induced in the mutant. In contrast, the synthesis of several virulence factors was repressed. These results should contribute to an increasingly complete picture of the cell physiology of S. aureus SCVs, as well as a comprehensive understanding of the pathogenicity of these organisms.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. aureus strains used in this study were wild-type S. aureus strain COL (26) and its isogenic hemB mutant IA48 (32). The hemB mutant was constructed by using the plasmids previously described in detail (35). Successful transfer of the ermB cassette into the S. aureus chromosome was indicated by the appearance of colonies having the phenotype of SCVs that are chloramphenicol sensitive and erythromycin resistant. The successful recombination was verified by PCR; the PCR product of the mutant was 1.4 kb larger (the size of the inserted erythromycin cassette) than the product of the wild-type S. aureus strain (35). In addition, S. aureus RN6390 and an isogenic agr mutant of this strain were used (21).
The strains were cultivated with vigorous agitation at 37°C in tryptone soya broth (TSB) (Oxoid). For anaerobic cultures, screw-cap bottles with rubber seals were filled to the top with a cell suspension in the same medium and incubated with shaking.
Preparative 2D gel electrophoresis.
For preparation of cell extracts, bacteria were grown in TSB. At different optical densities, cells from 50 ml of culture were separated from the supernatant by centrifugation (7,000 × g) for 10 min at 4°C, washed twice with Tris-EDTA buffer, and resuspended in Tris-EDTA buffer containing 2 mM phenylmethylsulfonyl fluoride. After incubation for 10 min on ice with lysostaphin (50 μg/ml), the cells were disrupted with a French press. The lysate was centrifuged for 10 min at 9,000 × g (in order to remove cell debris) and then for 30 min at 21,000 × g at 4°C (in order to remove insoluble and aggregated proteins that disturbed the isoelectric focusing of the proteins). The supernatant was stored frozen (8).
For preparation of extracellular protein extracts, bacteria were grown in the medium described above. At different optical densities cells from 250 ml of culture were separated from the supernatant by centrifugation for 20 min at 4°C and 7,000 × g. The extracellular proteins in the supernatant were precipitated with 10% (wt/vol) trichloroacetic acid at 4°C overnight. The precipitate was harvested by centrifugation (4°C, 7,000 × g, 1.5 h), washed with 96% (vol/vol) ethanol several times, and dried. The protein extracts were resolved in an appropriate volume of a solution containing 8 M urea and 2 M thiourea (38).
Preparative 2D gel electrophoresis was performed by using the immobilized pH gradient technique described by Bernhardt et al. (3). The protein samples were separated by using immobilized pH gradient strips (Amersham Pharmacia Biotech, Piscataway, N.J.) in the pH range from 4 to 7 or from 3 to 10. For identification of proteins by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), protein samples (500 μg) were separated on preparative 2D gels, and the proteins were stained with Coomassie blue R-250. The resulting peptide mass fingerprints were analyzed by using the MS-Fit software (http://prospector.ucsf.edu), GPMAW 4.10 (Lighthouse data), and genome sequences of S. aureus N315, Mu50, and COL.
Database searches were done by using the BLAST program (1).
Northern blot analyses.
Total RNA of S. aureus strains was isolated by using the modified acid-phenol method described by Gertz et al. (8). Northern blot analyses were performed by using the protocol described by Wetzstein et al. (36). Chemiluminescent signals were detected with a Lumi-Imager and were analyzed by using the LumiAnalyst program (Boehringer Mannheim). The sizes of the transcripts were calculated with the help of digoxigenin-labeled RNA Molecular Weight Marker I from Roche.
The specific digoxigenin-labeled RNA probes were prepared by in vitro transcription with T7 RNA polymerase by using the Dig-RNA labeling mixture (Roche) and the appropriate PCR fragments as templates. The PCR fragments were generated by using chromosomal DNA of S. aureus COL isolated with a chromosomal DNA isolation kit according to the protocol of the manufacturer (Promega) and the appropriate oligonucleotides listed in Table 1. The PCRs were carried out by using the following standard protocol: annealing at 50°C for 1 min, extension at 72°C for 1.25 min, and denaturation at 95°C for 1 min. Oligonucleotides complementary to the C-terminal regions of the genes contained the T7 recognition sequence at the 5′ end (8, 11).
TABLE 1.
Oligonucleotides used in this study
| Gene (locus in S. aureus COL) | Primer sequence (5′→3′)a |
|---|---|
| adh (SA0660) | CAGTTGTAACGAAAGATCAC |
| CTAATACGACTCACTATAGGGAGACATCAAGCACTAATCTTGGG | |
| arcB (SA2656) | CCGTATGATTTAAAAGGCAG |
| CTAATACGACTCACTATAGGGAGATTTGTATCATGGAATGCTGG | |
| citB (SA1385) | GGCTGCAAATTTTAAAGAGC |
| CTAATACGACTCACTATAGGGAGAGCTAGTGGTAAATGTTGTAC | |
| fba (SA2117) | GGAAAAGGCTTTATTGCCGC |
| CTAATACGACTCACTATAGGGAGAATGCGTCGTAGATTGATTC | |
| gap (SA0838) | GTTTCACAGGTGAAGTAGAGG |
| CTAATACGACTCACTATAGGGAGAACACGAGTTTGTGTAGCGTC | |
| ldh (SA0222) | CATGCCACACCATATTCTCC |
| CTAATACGACTCACTATAGGGAGATCTGCTTCAGCCATAATATC | |
| pflB (SA0204) | CATGCAACAGCTTGGCAAGG |
| CTAATACGACTCACTATAGGGAGACCTGCTTTAAGGTCACGTTT | |
| RNAIII | AGGAAGGAGTGATTTCAATG |
| CTAATACGACTCACTATAGGGAGAACTCATCCCTTCTTCATTAC |
The underlined sequences are the recognition sequence for T7 RNA polymerase.
Detection of glucose, lactate, and acetate.
The concentrations of glucose, lactate, and acetate were determined by using the test combinations d-glucose, dl-lactate, and acetate (Boehringer Mannheim) according to the manufacturer's instructions. The determinations were based on enzymatic reactions resulting in the formation of NADH (for lactate and acetate) or NADPH (for glucose). The increases in the amounts of NADH and NADPH were determined by measuring the absorbance at 340 nm. The amounts of NADPH and NADH formed in these reactions were stoichiometric with respect to the amounts of glucose and lactate, respectively. For acetate, the amount of NADH formed was not directly proportional to the acetate concentration because of the equilibrium of a preceding indicator reaction. The calculation was done as recommended by the manufacturer.
Assay of intracellular ATP concentration.
A 100-μl portion of a cell suspension was added to 120 μl of lysis solution (50 mM trichloroacetic acid, 15 mM EDTA; pH 7.5), incubated on ice for 15 min, and stored at −80°C. The growth of the culture was monitored simultaneously by measuring the absorption at 540 nm. The ATP concentration was determined by a luciferase assay by using a CLSII ATP bioluminescence kit (Roche) according to the manufacturer's instructions. Dilutions of the extracts were incubated with the luciferase assay reagent. Bioluminescent signals were detected with the Lumi-Imager and were analyzed by using the program LumiAnalyst. Samples of the wild-type culture at an optical density at 540 nm of 0.5 were defined 100% in each experiment.
RESULTS
Growth and pH.
S. aureus COL and the isogenic hemB mutant S. aureus IA48 were grown in TSB with vigorous agitation. Compared to the wild type, the mutant had an impaired growth rate and reached conspicuously lower cell densities (Fig. 1). At the exponential growth phase the levels of acidification of the medium were comparable for the two strains. In contrast, at the transitional phase, the wild-type culture became more alkaline; the supernatant of the hemB mutant, however, remained acidic, indicating that there were changes in the metabolism due to deletion of hemB (Fig. 1).
FIG. 1.
Growth (lines) and pH value (bars) of S. aureus COL (▪ and solid bars) and its isogenic hemB mutant IA48 (□ and open bars) in TSB. The times of sampling for transcriptional analysis are indicated by arrowheads on the growth curves. OD 540, optical density at 540 nm.
Differences in the cytoplasmic protein pattern due to a mutation in hemB.
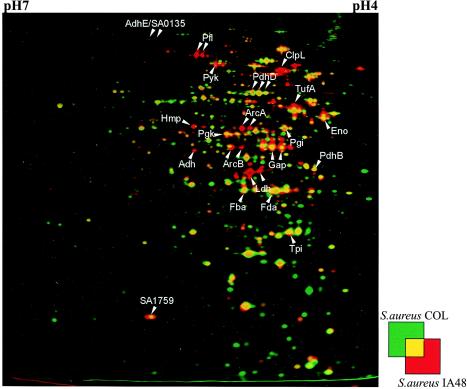
To analyze the influence of the hemB mutation on cellular processes in S. aureus, protein extracts were prepared from cytoplasmic cellular fractions of the mutant and the wild type and compared by 2D gel electrophoresis. For visualization of proteins induced in the hemB mutant we used the dual-channel technique (Fig. 2). The amounts of more than 20 proteins were strikingly different in the hemB mutant (Fig. 2). These proteins were identified by MALDI-TOF MS and included glycolytic enzymes like glyceraldehyde-3-phosphate dehydrogenase (Gap), fructose-1,6-bisphosphate aldolase (Fda), phosphoglycerate kinase (Pgk), and enolase (Eno), indicating that there was higher glycolytic activity in the hemB mutant. Furthermore, proteins involved in fermentation pathways, like lactate dehydrogenase (Ldh), alcohol dehydrogenase (Adh), pyruvate formate lyase (Pfl), arginine deiminase (ArcA), and ornithine carbamylotransferase (ArcB), were found to be exclusively produced in the mutant (Fig. 2 and Table 2).
FIG. 2.
Dual-channel images of 2D gels of cytoplasmic proteins produced with the Delta2D software (Decodon GmbH), showing the differences in the protein patterns of the wild-type S. aureus strain COL (green) and the hemB mutant IA48 (red). The proteins in 100 μg of a crude cell extract from cells grown in TSB at the stationary phase were separated by preparative 2D polyacrylamide gel electrophoresis. The proteins were stained with silver nitrate. The protein spots identified are indicated by arrows. Proteins induced by a mutation in hemB are red.
TABLE 2.
Cytoplasmic proteins identified on 2D gels of S. aureus
| Protein | Function | Locus in S. aureus COL |
|---|---|---|
| Induced by a hemB mutation | ||
| Adh | Energy metabolism: fermentation (alcohol dehydrogenase) | SA0660 |
| ArcA | Energy metabolism: amino acids and amines (arginine deiminase) | SA2657 |
| ArcB | Energy metabolism: amino acids and amines (ornithine carbamoyltransferase) | SA2656 |
| ClpL | ATP-dependent Clp proteinase chain ClpL | SA2563 |
| Eno | Energy metabolism: glycolysis/gluconeogenesis (enolase) | SA0842 |
| Fba | Energy metabolism: glycolysis/gluconeogenesis (fructose bisphosphate aldolase, class II) | SA2117 |
| Fda | Energy metabolism: glycolysis/gluconeogenesis (fructose bisphosphate aldolase, class I) | SA2622 |
| Gap | Energy metabolism: glycolysis (glyceraldehyde-3-phosphate dehydrogenase) | SA0838 |
| Hmp | Unknown function: general (flavohemoprotein, putative) | SA0220 |
| l-Ldh | Energy metabolism: fermentation (l-lactate dehydrogenase) | SA0222 |
| Pgi | Energy metabolism: glycolysis (phosphoglucomutase) | SA0966 |
| Pgk | Energy metabolism: glycolysis (phosphoglycerate kinase) | SA0839 |
| Pfl | Energy metabolism: fermentation (pyruvate formate lyase) | SA0205 |
| Pyk | Energy metabolism: glycolysis (pyruvate kinase) | SA1745 |
| Tpi | Energy metabolism: glycolysis (triose phosphate isomerase) | SA0840 |
| Not influenced by a hemB mutation | ||
| PdhB | Energy metabolism: pyruvate dehydrogenase (pyruvate dehydrogenase complex, E1 component; pyruvate dehydrogenase, beta subunit) | SA1103 |
| PdhD | Energy metabolism: pyruvate dehydrogenase (pyruvate dehydrogenase complex, E3 component; lipoamide dehydrogenase) | SA1105 |
Additionally, the levels of flavohemoglobin Hmp, TufA, ClpL, and SA1759 were also strongly increased by the mutation in hemB (Fig. 2 and Table 2).
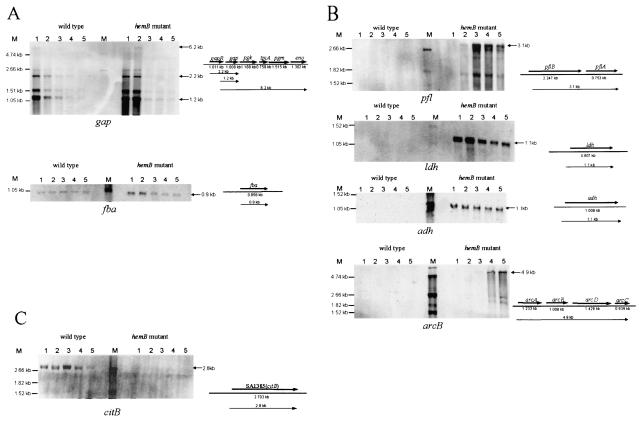
Changes in the amounts of enzymes involved in glycolysis, fermentation, and the tricarbonic acid (TCA) cycle occur at the transcriptional level.
In order to verify the proteomic data, Northern blot analyses were performed with RNA harvested from cells at different stages of growth. As expected, during exponential growth the mRNA level of the glycolytic genes was higher in the mutant than in the wild type (Fig. 3A). In the stationary phase, just when the glucose was probably depleted, expression of glycolytic genes, like gap, eno, and fba, was decreased in the wild type and in the mutant strain (Fig. 3A). Transcription of genes coding for enzymes involved in fermentative metabolism (pfl, ldh, and adh) was not found in the wild type but was very strongly increased in the mutant (Fig. 3B). Similar results were obtained for transcription of the arc operon containing four genes probably involved in ATP production from arginine catabolism under anaerobic conditions (15, 31). However, in contrast to ldh and adh, transcription of the arc operon and transcription of the pfl operon encoding pyruvate formate lyase and pyruvate formate lyase-activating enzyme occurred in the hemB mutant only during the stationary phase (Fig. 3B).
FIG. 3.
Northern blot analyses of genes whose transcription was influenced by a mutation in hemB. RNA was isolated from S. aureus COL (wild type) and its isogenic hemB mutant IA48 grown in TSB at 37°C during growth (the sampling times are indicated by arrowheads in the growth curves in Fig. 1). The membranes were hybridized with digoxigenin-labeled RNA probes for genes involved in glycolysis (A), fermentation (B), and the TCA cycle (C).
The levels of the TCA cycle enzymes should have been repressed in the hemB mutant. Analysis of the expression of the aconitase hydratase gene (citB) of S. aureus confirmed this hypothesis. Whereas almost no citB transcript was detectable in the mutant, a strong increase in the amount of the transcript in cells entering the stationary phase of growth was found in the wild type (Fig. 3C).
Analyses of metabolites and ATP in the wild type and the hemB mutant grown in TSB.
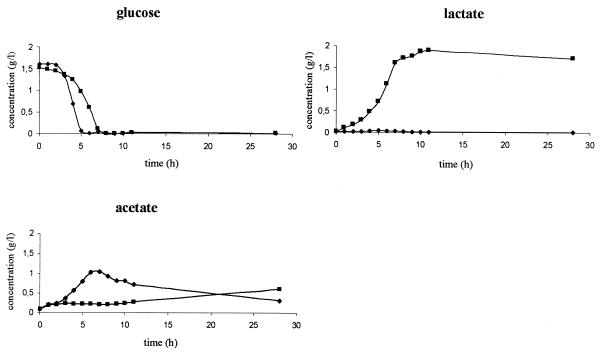
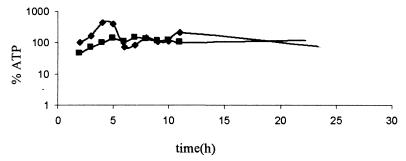
Apparently, most of the proteins present at higher levels in the hemB mutant are involved in glycolysis and anaerobic fermentation product formation. For this reason, the amounts of glucose, lactate, acetate, and ethanol in supernatants of the mutant and the wild type were analyzed (Fig. 4). Simultaneous with consumption of glucose, the wild type (at zero time to 5 h) produced acetate, and the hemB mutant (at zero time to 7 h) produced lactate, which explained the acidification of the growth media. The ATP levels increased in both cases; however, the absolute amount of ATP was significantly higher in wild-type cells (Fig. 5). After the glucose was exhausted (at 5 to 6 h), the ATP level in the wild type dropped dramatically and reached nearly the same low level as the level in mutant cells. After 7 h, the levels of acetate dropped in the medium of the wild-type strain, and this was accompanied by a realkalization of the medium and a slight increase in the amount of ATP. These results indicate that acetate was used as a secondary carbon source after depletion of glucose in the wild-type strain. The hemB mutant was not able to use lactate as an energy source (Fig. 4).
FIG. 4.
Glucose, lactate, and acetate concentrations in the supernatants of a wild-type culture (S. aureus COL) (♦) and a hemB mutant IA48 culture (▪) during growth in TSB.
FIG. 5.
Relative ATP levels in cells of S. aureus COL (♦) and its isogenic hemB mutant (▪) grown under standard conditions in TSB. The ATP concentration in the cells was determined with a Lyciferase assay kit (Roche). The light signals were detected with a Lumi-Imager and were quantified with the LumiAnalyst program from Boehringer Mannheim. The ATP level in the wild type at an optical density of 0.5 was defined as 100%.
Ethanol could not be detected in the growth medium of the wild type under any of the conditions employed; however, very small amounts of ethanol accumulated in the medium of the mutant (data not shown).
MS analysis of pyruvate formate lyase.
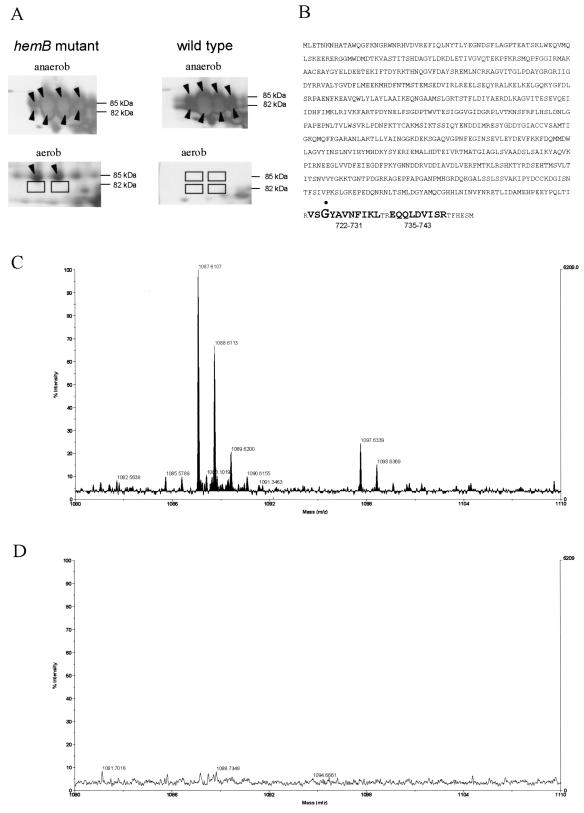
While the pyruvate formate lyase spot was one of the most abundant protein spots (Fig. 2 and 6A), no formate was produced during aerobic growth of the hemB mutant (data not shown). In Escherichia coli, the pyruvate formate lyase acts as a homodimer and is activated by introduction of a free radical to a glycine residue in one subunit of the enzyme. In the presence of oxygen (for instance, during aerobic protein extraction), the radicalization causes peptide bond cleavage near the C terminus of the modified monomer (30). Thus, activated Pfl in E. coli appears in two bands on one-dimensional protein gels; these bands represent the mature form and the cleaved form. Both in wild-type cells of S. aureus COL and in the hemB mutant grown under anaerobic conditions, Pfl spots at two different molecular masses were identified (85 and 82 kDa) (Fig. 6A). MALDI-TOF analyses of the trypsin-digested protein spots indicated that C-terminal peptides 735-743 and 722-731 (Fig. 6B) were missing in the 82-kDa protein (Fig. 6D), while both of these peptides were detected in the 85-kDa protein spot (Fig. 6C). These results may indicate that under anaerobic conditions the C terminus of one subunit of Pfl in the S. aureus wild type and the hemB mutant was cleaved in the presence of oxygen (during protein preparation). Therefore, we presume that in S. aureus grown under anaerobic conditions the protein was activated by a mechanism very similar to the mechanism in E. coli. In the hemB mutant grown under aerobic conditions only the 85-kDa protein spot representing Pfl was observed on the gel (Fig. 6A), demonstrating that the protein was synthesized in the mutant under aerobic conditions; however, it was not activated.
FIG. 6.
MALDI-TOF MS analyses of Pfl. (A) Sectors of 2D gels containing the region where Pfl is located. Protein extracts of the S. aureus hemB mutant and the wild type were isolated under anaerobic (anaerob) and aerobic (aerob) conditions. The proteins were stained with silver nitrate. (B) Amino acid sequence of the pyruvate formate lyase of S. aureus. The Gly residue at position 724 that is probably radicalized is indicated by a dot. C-terminal peptide fragments 735-743 and 722-731 obtained after trypsin digestion of Pfl are indicated by boldface type. The theoretical molecular weights (monoisotopic, [MH+]) for peptides 735-743 and 722-731 are 1,086.566 and 1,096.591, respectively. (C) Sector of the MALDI-TOF MS spectrum of the 85-kDa Pfl spot covering the region for peptide 735-743 and peptide 722-731.(D) Sector of the MALDI-TOF MS spectrum of the 82-kDa Pfl spot covering the region for peptide 735-743 and peptide 722-731.
Synthesis of extracellular proteins and regulation of RNAIII in the mutant strain.
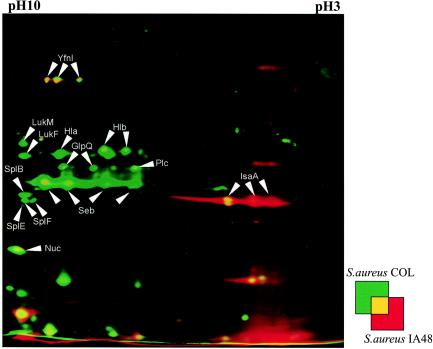
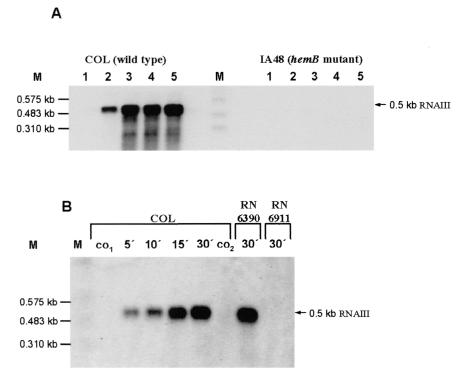
A comparative proteomic analysis of the wild type and the mutant strain also revealed very strong differences in the extracellular protein patterns of these strains in the stationary growth phase. Most of the known virulence factors expressed during the late exponential phase were not found in the mutant or were present at low levels (Fig. 7). These factors include α-hemolysin (Hla), β-hemolysin (Hlb), enterotoxin B (Seb), glycerophosphoryl diester phosphodiesterase (GlpQ), 1-phosphatidylinositol phosphodiesterase (Plc), thermonuclease (Nuc), leukocidins F (LukF) and M (LukM), and the serine proteases SplB, SplE, and SplF. Immunodominant antigen A (IsaA), an extracellular protein synthesized at low cell densities, was present only in the mutant. Simultaneously, RNAIII, the regulator of late virulence factor production, could not be detected in the mutant strain (Fig. 8A). When we compared the optical densities of the two strains during the stationary phase, the cell density of the mutant culture was significantly lower (Fig. 1). Supplementation of the mutant culture with the supernatant of wild-type cells at an optical density of 6.5 allowed transcription of RNAIII in the mutant. At the same time, supernatant of an agr mutant (S. aureus RN6911) lacking the quorum-sensing signal peptide did not affect the RNAIII transcription.
FIG. 7.
Dual-channel images of 2D gels of extracellular proteins produced with the Delta2D software (Decodon GmbH), showing the differences in the protein patterns of the wild-type S. aureus strain COL (green) and the hemB mutant IA48 (red). The proteins in 100 μg of the supernatant from cells in the stationary growth phase were separated by preparative 2D polyacrylamide gel electrophoresis. The proteins were stained with silver nitrate. Proteins induced by a mutation in hemB are red, and proteins present only in the wild-type strain are green. The following proteins that were identified are indicated by arrows: glycerophosphoryl diester phosphodiesterase (GlpQ), α-hemolysin (Hla), β-hemolysin (Hlb), immunodominant antigen A (IsaA), leucocidine F (LukF), leucocidine M (LukM), thermonuclease (Nuc), 1-phosphatidylinositol phosphodiesterase (Plc), enterotoxin B (Seb), serinproteases (SplB and SplF), and a hypothetical protein (YfnI) (38).
FIG. 8.
Northern blot analyses of RNAIII. (A) RNA was isolated from S. aureus COL (wild type) and from its isogenic hemB mutant IA48 grown in TSB at 37°C during the growth phase (the sampling times are indicated by arrowheads in Fig. 1). (B) At the stationary phase (optical density at 540 nm, 2) 20-ml portions of the hemB mutant culture were supplemented with 20-ml portions of supernatants from wild-type S. aureus strain COL, S. aureus RN6390, and the agr mutant RN6911 (all at an optical density at 540 nm of 6.5). RNA was isolated from the original culture of the mutant at zero time (co1), 30 min later (co2), and at the times indicated after addition of the supernatants to the mutant culture. The membranes were hybridized with a digoxigenin-labeled RNA probe for RNAIII.
DISCUSSION
In bacteria, heme is a prosthetic group of cytochromes, which are essential components of the electron transport chain. Under aerobic conditions, protons are transferred from NADH to the electron transport chain, generating H2O and a membrane potential which is used to synthesize ATP. If oxygen or alternative electron acceptors are absent, NADH must be recycled by fermentation; otherwise, the NAD+ pool is depleted in a very short time. It is reasonable to hypothesize that a block in heme synthesis should impair the ability to use oxygen or nitrate as an electron acceptor and that the only way to oxidize NADH might be reduction of metabolic intermediates by fermentation. Consistent with this view, components of fermentative pathways, like lactate dehydrogenase, alcohol dehydrogenase, and pyruvate formate lyase, were induced in a hemB mutant of S. aureus during exponential growth under aerobic conditions (Fig. 2 and 3 and Table 2). At the same time, significant amounts of lactate were detected in the growth medium, indicating that NADH was mainly oxidized by reducing pyruvate to lactate (Fig. 4). As described previously, lactate could not be used as an alternative carbon source after glucose consumption (2). Surprisingly, in E. coli the situation is quite different. In a hemA mutant, enolase and phosphoglycerate kinase are repressed and simultaneously citrate synthase is induced. The synthesis of fermentative enzymes seems to be limited to anaerobic growth conditions in this organism (24).
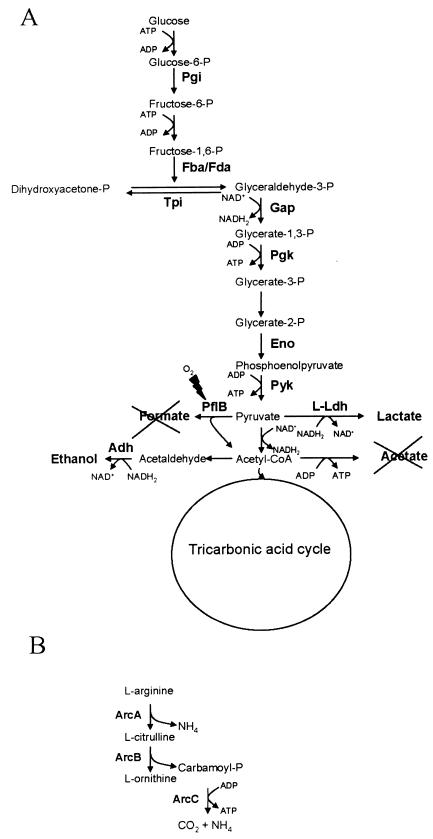
The S. aureus wild type secreted large amounts of acetate during exponential growth in TSB (Fig. 4), indicating that there was overflow metabolism under glucose-excess conditions. Pyruvate dehydrogenase activity is probably responsible for this process. At the same time, the TCA cycle activity was very low and was needed only for anabolism (Fig. 3). When glucose has been consumed, acetate can be used as an alternate carbon source and is oxidized via the TCA cycle. In contrast, the conversion of pyruvate to acetate via acetyl coenzyme A (acetyl-CoA) was found to be very restricted in the hemB mutant, although pyruvate formate lyase (Pfl) and components of the pyruvate dehydrogenase were present (Fig. 2). The pyruvate dehydrogenase activity is probably very low as it is inhibited by the high NADH level. The Pfl converts pyruvate and CoA to formate and acetyl-CoA under anaerobic conditions (Fig. 9A). As in E. coli, the Pfl in S. aureus might form a homodimeric enzyme complex that is synthesized in an inactive form. The enzyme should be activated posttranslationally via an iron-dependent activating enzyme that introduces a glycyl radical to one monomer in the enzyme complex. In the hemB mutant, either the protein was not activated or the organism was able to protect Pfl from oxygenolytic cleavage by reversible removal of the free radical by Pfl deactivase activity, as described previously for E. coli and Streptococcus sanguis (13, 28, 37). The main electron acceptor from NADH for NAD+ recycling in the mutant is pyruvate, which leads to lactate secretion (Fig. 4 and 9A). In order to compensate partially for the loss of most of the energy available from glucose by fermentation, glycolytic activity was up-regulated in the mutant, as indicated by the increased amounts of glyceraldehyd-3-phosphate dehydrogenase, enolase, and 3-phosphate-glycerol kinase (Fig. 2, 3, and 9A and Table 2). Simultaneously, the synthesis of citrate cycle enzymes, like succinate dehydrogenase, fumarate hydratase, and succinyl-CoA synthetase, was down-regulated in the hemB mutant.
FIG. 9.
Carbohydrate (A) and arginine (B) metabolic pathways in the hemB mutant under aerobic conditions. The following enzymes were found at higher levels in the hemB mutant: phosphoglucomutase (Pgi), fructose bisphosphate aldolase (Fba/Fda), glyceraldehyde dehydrogenase (Gap), phosphoglycerate kinase (Pgk), enolase (Eno), pyruvate kinase (Pyk), l-lactate dehydrogenase (L-Ldh), pyruvate formate lyase (PflB), alcohol dehydrogenase (Adh), arginine deiminase (ArcA), ornithine carbamoyltransferase (ArcB), and carbamate kinase (ArcC).
Since it was not able to use the citric acid cycle and electron transport, the hemB mutant had decreased levels of ATP (Fig. 5). To compensate for the loss, the arginine deiminase pathway, with ArcA and ArcB (Fig. 2 and 3 and Table 2), was stimulated in the hemB mutant, which produced ATP (Fig. 9B). This system occurs in a variety of bacteria (5) and appears to be induced by arginine under anaerobic conditions and to be regulated by catabolite repression (6), which might explain our observation that the system was not expressed until glucose was depleted (Fig. 3B). Activation of this pathway allows bacteria to grow anaerobically in the presence of arginine (10, 15, 31). ATP is synthesized in the reaction catalyzed by carbamate kinase (Fig. 9B). As in oral streptococci, this system can protect bacterial cells against the damaging effects of acid environments by releasing ammonia during arginine metabolism (Fig. 9B) (4). Both the synthesis of ATP under anaerobic conditions and protection against acid damage would be beneficial to S. aureus SCVs. Very recently, it was shown that transcription of the arc operon was stimulated in an agr- and cell density-dependent manner (7).
In the related gram-positive bacterium Bacillus subtilis, both aerobic respiration and anaerobic respiration are regulated by the two-component ResDE system (17, 18, 20, 27). This system is activated by an incompletely characterized redox-sensing mechanism (27). Activated ResE increases the transcription of nasDE, the flavohemoglobin hmp, and the redox regulatory gene fnr (19, 20, 27). In S. aureus there is a system that is very similar to resDE in B. subtilis, and a mutation in srhSR led to changes in cell morphology, the expression of proteins involved in energy metabolism, and other metabolic processes, including arginine catabolism (29). Under anaerobic conditions, ResE activates the synthesis of fermentative enzymes, like Adh and Ldh, and at the same time it represses the synthesis of the TCA cycle enzymes aconitase, succinyl-CoA synthetase, and fumarase (29). Interestingly, synthesis of some of these proteins was also impaired in the hemB mutant (Fig. 2 and 3), indicating that the possible ResDE homologue in S. aureus, SrhSR, might be active in the mutant under aerobic conditions. However, no data are available concerning direct SrhSR regulation of the transcription of these genes or whether there is an additional regulator whose synthesis is influenced by the two-component system. An Fnr-like protein like that described in other bacteria is not present in S. aureus (D. Bates, P. McNamara, and R. A. Proctor, unpublished data).
In accordance with previously published results (32, 35), the synthesis of some extracellular proteins, like α-hemolysin, was strongly repressed in the hemB mutant (Fig. 7). Moreover, the serine proteases SplB, SplE, and SplF were not found in the mutant (Fig. 7). At the same time RNAIII, the major regulator of these proteins, was not detected either in exponentially growing cells or during the stationary phase (Fig. 8). Our results strongly indicate that the concentration of the quorum-sensing peptide synthesized by the hemB mutant itself was too low for activation of the agr system. This might have been due either to the lower cell densities reached in the mutant culture and/or to a block in synthesis of the peptide. The loss of hemolysins and leucocidines might be crucial for the ability of the hemB mutant to avoid lysis and to survive inside eukaryotic cells.
Acknowledgments
C. Kohler and C. von Eiff contributed equally to this work.
We thank R. Gloger for excellent technical assistance.
This work was supported by grants from BMBF (031U107A and Pathogenomic Network) and the Fonds der chemischen Industrie to M.H., by grants from BMBF (Pathogenomic Network and EI119912) to C.V.E., and by grant AI42072 from the National Institutes of Health to R.A.P.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Baumert, N., C. von Eiff, F. Schaaff, G. Peters, R. A. Proctor, and H. G. Sahl. 2002. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 8:253-260. [DOI] [PubMed] [Google Scholar]
- 3.Bernhardt, J., K. Büttner, C. Scharf, and M. Hecker. 1999. Dual channel imaging of two-dimensional electropherograms in Bacillus subtilis. Electrophoresis 20:2225-2240. [DOI] [PubMed] [Google Scholar]
- 4.Casiano-Colon, A., and R. E. Marquis. 1988. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl. Environ. Microbiol. 54:1318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunin, R., N. Glansdorff, A. Pierard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curran, T. M., Y. Ma, G. C. Rutherford, and R. E. Marquis. 1998. Turning on and turning off the arginine deiminase system in oral streptococci. Can. J. Microbiol. 44:1078-1085. [DOI] [PubMed] [Google Scholar]
- 7.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of sigmaB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558-566. [DOI] [PubMed] [Google Scholar]
- 9.Granick, S., and S. I. Beale. 1978. Hemes, chlorophylls, and related compounds: biosynthesis and metabolic regulation. Adv. Enzymol. Relat. Areas Mol. Biol. 46:33-203. [DOI] [PubMed] [Google Scholar]
- 10.Hartmann, R., H. D. Sickinger, and D. Oesterhelt. 1980. Anaerobic growth of halobacteria. Proc. Natl. Acad. Sci. USA 77:3821-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jorgensen, E. D., R. K. Durbin, S. S. Risman, and W. T. McAllister. 1991. Specific contacts between the bacteriophage T3, T7, and SP6 RNA polymerases and their promoters. J. Biol. Chem. 266:645-651. [PubMed] [Google Scholar]
- 12.Kahl, B., M. Herrmann, A. S. Everding, H. G. Koch, K. Becker, E. Harms, R. A. Proctor, and G. Peters. 1998. Persistent infection with small colony variant strains of Staphylococcus aureus in patients with cystic fibrosis. J. Infect. Dis. 177:1023-1029. [DOI] [PubMed] [Google Scholar]
- 13.Kessler, D., I. Leibrecht, and J. Knappe. 1991. Pyruvate-formate-lyase-deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett. 281:59-63. [DOI] [PubMed] [Google Scholar]
- 14.Looney, W. J. 2000. Small-colony variants of Staphylococcus aureus. Br. J. Biomed. Sci. 57:317-322. [PubMed] [Google Scholar]
- 15.Maghnouj, A., T. F. de Sousa Cabral, V. Stalon, and C. Vander Wauven. 1998. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor, argR. J. Bacteriol. 180:6468-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNamara, P. J., and R. A. Proctor. 2000. Staphylococcus aureus small colony variants, electron transport and persistent infections. Int. J. Antimicrob. Agents 14:117-122. [DOI] [PubMed] [Google Scholar]
- 17.Nakano, M. M., Y. P. Dailly, P. Zuber, and D. P. Clark. 1997. Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth. J. Bacteriol. 179:6749-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano, M. M., and Y. Zhu. 2001. Involvement of ResE phosphatase activity in down-regulation of ResD-controlled genes in Bacillus subtilis during aerobic growth. J. Bacteriol. 183:1938-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano, M. M., Y. Zhu, M. Lacelle, X. Zhang, and F. M. Hulett. 2000. Interaction of ResD with regulatory regions of anaerobically induced genes in Bacillus subtilis. Mol. Microbiol. 37:1198-1207. [DOI] [PubMed] [Google Scholar]
- 20.Nakano, M. M., P. Zuber, P. Glaser, A. Danchin, and F. M. Hulett. 1996. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J. Bacteriol. 178:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng, H. L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proctor, R. A., J. M. Balwit, and O. Vesga. 1994. Variant subpopulations of Staphylococcus aureus as cause of persistent and recurrent infections. Infect. Agents Dis. 3:302-312. [PubMed] [Google Scholar]
- 23.Proctor, R. A., P. van Langevelde, M. Kristjansson, J. N. Maslow, and R. D. Arbeit. 1995. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 20:95-102. [DOI] [PubMed] [Google Scholar]
- 24.Rompf, A., R. Schmid, and D. Jahn. 1998. Changes in protein synthesis as a consequence of heme depletion in Escherichia coli. Curr. Microbiol. 37:226-230. [DOI] [PubMed] [Google Scholar]
- 25.Seifert, H., C. von Eiff, and G. Fatkenheuer. 1999. Fatal case due to methicillin-resistant Staphylococcus aureus small colony variants in an AIDS patient. Emerg. Infect. Dis. 5:450-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shafer, W. M., and J. J. Iandolo. 1979. Genetics of staphylococcal enterotoxin B in methicillin-resistant isolates of Staphylococcus aureus. Infect. Immun. 25:902-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun, G., E. Sharkova, R. Chesnut, S. Birkey, M. F. Duggan, A. Sorokin, P. Pujic, S. D. Ehrlich, and F. M. Hulett. 1996. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J. Bacteriol. 178:1374-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi, N., K. Abbe, S. Takahashi-Abbe, and T. Yamada. 1987. Oxygen sensitivity of sugar metabolism and interconversion of pyruvate formate-lyase in intact cells of Streptococcus mutans and Streptococcus sanguis. Infect. Immun. 55:652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Throup, J. P., F. Zappacosta, R. D. Lunsford, R. S. Annan, S. A. Carr, J. T. Lonsdale, A. P. Bryant, D. McDevitt, M. Rosenberg, and M. K. Burnham. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 40:10392-10401. [DOI] [PubMed] [Google Scholar]
- 30.Unkrig, V., F. A. Neugebauer, and J. Knappe. 1989. The free radical of pyruvate formate-lyase. Characterization by EPR spectroscopy and involvement in catalysis as studied with the substrate-analogue hypophosphite. Eur. J. Biochem. 184:723-728. [DOI] [PubMed] [Google Scholar]
- 31.Vander Wauven, C., A. Pierard, M. Kley-Raymann, and D. Haas. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaudaux, P., P. Francois, C. Bisognano, W. L. Kelley, D. P. Lew, J. Schrenzel, R. A. Proctor, P. J. McNamara, G. Peters, and C. Von Eiff. 2002. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small-colony variant phenotypes. Infect. Immun. 70:5428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Eiff, C., K. Becker, D. Metze, G. Lubritz, J. Hockmann, T. Schwarz, and G. Peters. 2001. Intracellular persistence of Staphylococcus aureus small-colony variants within keratinocytes: a cause for antibiotic treatment failure in a patient with darier's disease. Clin. Infect. Dis. 32:1643-1647. [DOI] [PubMed] [Google Scholar]
- 34.von Eiff, C., D. Bettin, R. A. Proctor, B. Rolauffs, N. Lindner, W. Winkelmann, and G. Peters. 1997. Recovery of small colony variants of Staphylococcus aureus following gentamicin bead placement for osteomyelitis. Clin. Infect. Dis. 25:1250-1251. [DOI] [PubMed] [Google Scholar]
- 35.von Eiff, C., C. Heilmann, R. A. Proctor, C. Woltz, G. Peters, and F. Gotz. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179:4706-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wetzstein, M., U. Völker, J. Dedio, S. Löbau, U. Zuber, M. Schiesswohl, C. Herget, M. Hecker, and W. Schumann. 1992. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J. Bacteriol. 174:3300-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamada, T., S. Takahashi-Abbe, and K. Abbe. 1985. Effects of oxygen on pyruvate formate-lyase in situ and sugar metabolism of Streptococcus mutans and Streptococcus sanguis. Infect. Immun. 47:129-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziebandt, A. K., H. Weber, J. Rudolph, R. Schmid, D. Höper, S. Engelmann, and M. Hecker. 2001. Extracellular proteins of Staphylococcus aureus and the role of SarA and sigma B. Proteomics 1:480-493. [DOI] [PubMed] [Google Scholar]