Abstract
φSa3ms, a lysogenic bacteriophage encoding the staphylococcal enterotoxins SEA, SEG, and SEK and the fibrinolytic enzyme staphylokinase (Sak), was identified in the unannotated genome sequence of the hypervirulent community-acquired Staphylococcus aureus strain 476. We found that mitomycin C induction of φSa3ms led to increased transcription of all four virulence factors. The increase in sea and sak transcription was a result of read-through transcription from upstream latent phage promoters and an increase in phage copy number. The majority of the seg2 and sek2 transcripts were shown to initiate from the upstream phage cI promoter and hence were regulated by factors influencing cI transcription. The lysogeny module of φSa3ms was shown to have some λ-like features with divergent cI and cro genes. Band shift assays were used to identify binding sites for both CI and Cro within the region between these genes, suggesting a mechanism of control for the φSa3ms lytic-lysogenic switch. Our findings suggest that the production of phage-encoded virulence factors in S. aureus may be regulated by processes that govern lysogeny.
Staphylococcus aureus is a leading cause of nosocomial and community-acquired infections worldwide (42). S. aureus causes a wide range of diseases that vary in severity from mild skin infections, such as boils and furuncles, to life-threatening diseases, like toxic shock syndrome and endocarditis. The ability of S. aureus to cause such a wide variety of diseases is thought to be due in part to its elaboration of a large number of secreted and cell-surface associated virulence factors (1, 5, 21, 36, 48).
Much of the variation between S. aureus strains appears to be attributable to mobile genetic elements, such as plasmids, bacteriophages, pathogenicity islands, transposons, insertion sequences, and the staphylococcal chromosomal cassette (2, 27). The bacteriophages and pathogenicity islands of S. aureus encode many virulence factors (35). The known phage-encoded virulence factors include Panton-Valentine leukocidins (24, 33), exfoliative toxin type A (47), and staphylococcal enterotoxins (SEs) (3). The SEs constitute a family of related proteins whose activities are associated with staphylococcal food poisoning, toxic shock syndrome, and possibly several autoimmune disorders (17, 41). SEs are powerful superantigens that activate subsets of T lymphocytes to liberate various cytokines, including gamma interferon and tumor necrosis factor (41). In addition to these characterized phage-encoded virulence factors, a number of putative S. aureus virulence factors, such as staphylokinase (Sak), are also encoded in phage genomes (25). Staphylokinase, a potent plasminogen activator, has been hypothesized to aid in the dissemination of S. aureus from fibrinous clots and abscesses (1).
The genome sequences of two S. aureus isolates derived from life-threatening community-acquired infections have been determined (2; unpublished data). The methicillin-resistant strain MW2 (sequenced at Juntendo University, Tokyo, Japan) was isolated in 1998 from North Dakota after it caused septic arthritis and fatal septicemia in a 16-month-old girl (2). Methicillin-sensitive strain MSSA476 (sequenced at the Sanger Institute, Cambridge, United Kingdom) was isolated in 1998 from Oxford, United Kingdom after it caused osteomyelitis and bacteremia in a 9-year-old boy. By mining the unannotated MSSA476 genome for phage-like sequences, we identified a prophage that encodes the previously characterized enterotoxins SEA, SEG, and SEK and the fibrinolytic enzyme Sak. Here we describe and characterize this phage, designated φSa3ms, from MSSA476.
Although the physical linkage of S. aureus virulence factors with phage genomes has been recognized for almost two decades (3), it is not known whether the life cycles of the phages influence the expression of the virulence genes and thus S. aureus pathogenicity. Here we show that upon φSa3ms prophage induction, transcription of sea, seg2, sek2, and sak are greatly increased. Phage-encoded factors were found to be required for the increases in sea and sak transcription. Interestingly, the majority of seg2 and sek2 transcripts initiated from the upstream φSa3ms repressor (cI) promoter, indicating that factors that influence cI transcription also influence virulence gene expression. Finally, the location of the putative CI and Cro binding sites suggests a potential mechanism for regulation of φSa3ms lysogeny and hence virulence factor expression.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A list of the bacterial strains and plasmids used in this study is shown in Table 1. Escherichia coli strains were grown in Luria-Bertani broth with ampicillin (100 μg/ml). S. aureus strains were grown in Todd-Hewitt broth (Difco) containing 0.2% (wt/vol) yeast extract (Difco) (THY) or in trypic soy broth (TSB) (Becton Dickinson) as indicated below; media were supplemented with 10 μg of chloramphenicol per ml when appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or Reference |
|---|---|---|
| S. aureus strains | ||
| MSSA476 | Wild type | Sanger Centre |
| RN4220 | Proficient in DNA uptake via transformation | 26 |
| MS79 | MSSA476 (φSa3ms ror::pPS79) | This study |
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| DH5α | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 ΔlacU169 (φ80 lacIqZΔM15) | |
| Plasmids | ||
| pBT9 | E. coli-S. aureus shuttle vector (Ts in S. aureus), Cmr in S. aureus, Ampr in E. coli | 12 |
| pPS79 | pBT9 containing internal fragment of φSa3ms ror | This study |
| pQE-60 | Six-His E. coli expression vector, Ampr | Qiagen |
| pCROX | pQE-60 containing φSa3ms cro | This study |
| pCIX | pQE-60 containing φSa3ms cI | This study |
Bioinformatic analyses.
The currently unannotated MSSA476 genome sequence (produced by the S. aureus Sequencing Group at the Sanger Institute and available at www.sanger.ac.uk/Projects/S_aureus/) was mined for phage-like sequences by performing BLAST searches (available at www.ncbi.nlm.nih.gov/BLAST/) by using known phage-associated sequences, including sequences encoding SEs and integrases. Overlapping regions of interest were obtained from the FTP site, and putative open reading frames (ORFs) were identified by using Frameplot 3.0beta (available at www.nih.go.jp/∼jun/cgi-bin/frameplot-3.0b.pl). BLAST searches were then performed with these putative ORFs to identify orthologues. The method of Dodd and Egan (18) was used to identify putative helix-turn-helix motifs, and possible N-terminal signal sequences were identified by using SignalP V2.0.b2 (available at www.cbs.dtu.dk/services/SignalP-2.0/). Protein sequences were scanned for sites or signatures against the PROSITE database by using PROSCAN (available at http://npsa-pbil.ibcp.fr). Conserved domains were identified using the National Center for Biotechnology Information conserved domain search program (available at www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml).
Construction of a ror null mutant.
PCR primers were created to amplify a central region of ror (5′-GCGGATCCTGGCTATTTAAAGAAAAGAG-3′ and 5′-GCCTGCAGACATGAGGTTTGTATGTTTG-3′), which was cloned directly into pBT9 after restriction digestion of the PCR products at the sites underlined in the primers, creating plasmid pPS79. pBT9 is an E. coli-S. aureus shuttle vector that is temperature sensitive for replication in S. aureus (12). pPS79 was introduced into the restriction-deficient S. aureus strain RN4220 by transformation as previously described (28). pPS79 was then transduced from RN4220 into MSSA476 by using phage 80α as previously described (34). Single crossovers were selected by inoculating 2 ml of TSB containing chloramphenicol with a single colony and incubating the culture for 8 h at 42°C before streaking it onto TSA plates containing chloramphenicol. The plates were then incubated at 42°C overnight. The presence of ror::pPS79 was confirmed by Southern blotting (data not shown). One caveat of this approach is that MS79 (ror::pPS79) must be grown at 42°C to maintain the integrated form of the plasmid. Therefore, in all experiments in which MS79 was utilized, both this strain and wild-type strain MSSA476 were streaked and grown overnight at 42°C. However, after appropriate dilution of overnight cultures, additional incubation was carried out at 37°C. In each experiment an MSSA476 control which had been grown exclusively at 37°C was also included. In all cases there was no difference between the results for MSSA476 grown at 37°C and the results for MSSA476 grown at 42°C.
RNA extraction and Northern blotting.
Total RNA was isolated from S. aureus cultures by using Qiagen RNeasy mini kits (Qiagen, Valencia, Calif.) as recommended by the manufacturer, except that lysostaphin was substituted for lysozyme. Overnight TSB cultures were diluted 1:200 (see Fig. 2) or 1:400 (see Fig. 4) into fresh TSB and grown with shaking at 37°C to an optical density at 600 nm of 0.1 (see Fig. 2) or 0.14 (see Fig. 4) before extraction of RNA from aliquots (time zero). Each culture was then split into two parts, and mitomycin C was added to one of the parts at a final concentration of 0.3 μg/ml. The cultures were then incubated at 37°C, and RNA was extracted from aliquots of both cultures at several subsequent times. Total RNA was quantified by spectrophotometric analysis (optical density at 260 nm), and equal amounts of RNA were used for Northern blotting performed by using the Ambion NorthernMax-Gly protocol (Ambion, Austin, Tex.). Probes for Northern blotting were created by using an Ambion Strip-EZ kit.
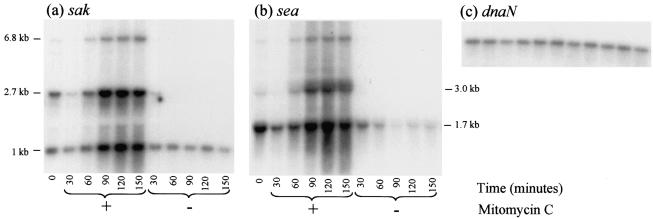
FIG. 2.
Treatment with the prophage-inducing agent mitomycin C increases sak and sea transcript levels. Mitomycin C (300 ng/ml) was added to one of two identical MSSA476 cultures, and RNA was extracted from both cultures every 30 min for 2.5 h. Six micrograms of total RNA was loaded in each lane and subsequently probed with sak (a), sea (b), or dnaN (c). The dnaN blot acted as a control for RNA loading.
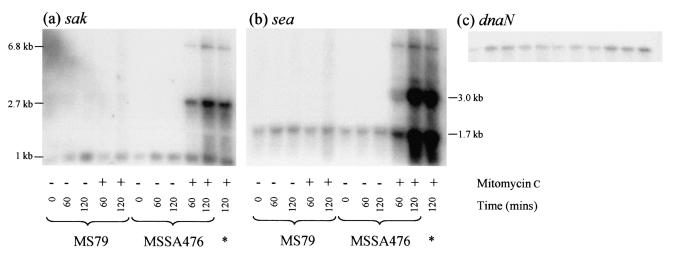
FIG. 4.
Mitomycin C does not induce sak and sea transcription in MS79. Northern blots containing 5 μg of total RNA per lane were probed with sea (a), sak (b), or dnaN (c). The dnaN blot acted as a control for RNA loading. The lane indicated by an asterisk contained RNA from MSSA476 that was grown exclusively at 37°C to show that initial growth of this strain at 42°C had no effect on the subsequent transcription profiles of sea and sak.
Genomic DNA extraction and Southern blotting.
Genomic DNA was isolated as previously described (34). Southern blotting was performed by using standard techniques, and the blots were probed with horseradish peroxidase-labeled probes generated by using an Amersham ECL direct labeling kit (Amersham, Little Chalfont, Buckinghamshire, England).
Phage DNA isolation.
Phage DNA was isolated from supernatants of MSSA476 cultures as previously described (23). Isolated phage DNA was subjected to restriction analysis and separated on standard agarose gels stained with ethidium bromide.
Single-plaque purification of phage and plaque lifts.
Single plaques were isolated by seeding 3 ml of soft THY (supplemented with 5 mM CaCl2) with 10 μl of an overnight RN4220 culture and overlaying the resulting culture onto a plate containing THY (supplemented with 5 mM CaCl2) spread with dilutions of MSSA476 culture supernatants. Plaque lifting was performed by using standard procedures after incubation of the plates for 18 h at 37°C.
Identification of transcriptional start sites by 5′-RACE.
The Invitrogen 5′ rapid amplification of cDNA ends (RACE) system (Invitrogen, Carlsbad, Calif.) was used to identify putative transcriptional start sites for cI and cro according to the manufacturer's instructions. The following primers were used for these assays: CRORA (5′-TTGTTCCATTGTGTCCCTCC-3′), CROXR (5′-CGGGATCCTATAACATGAACTTTTTC-3′), MSREP2 (5′-GCGAATTCTCACTGTCTTTCCAACCAAC-3′), CISF (5′-CCACATGCAATATACGATAC-3′), and SEQR (5′-GAGTAGAAACTTTGTCTGTAG-3′). Briefly, primer SEQR or CRORA was added to total RNA isolated from MSSA476 and used to prime first-strand cDNA synthesis with reverse transcriptase. A poly(C) 3′ tail was added to the cDNA by using terminal transferase, and each of the tailed cDNAs was used as template in a PCR performed with a downstream primer (relative to the directions of the SEQR and CRORA primers) and a primer that ended with a poly(G) sequence (primer AAP; Invitrogen). Control PCRs in which nontailed cDNAs were used as templates were used as controls for AAP primer specificity. Products were visualized on standard agarose gels stained with ethidium bromide. Bands were excised from the gels by using a Qiagen gel extraction kit and were sequenced at the Tufts University School of Medicine Core Sequencing Facility.
Cloning and overexpression of CI and Cro genes.
cI and cro were amplified by PCR by using primers CIXF (CGCCATGGGCCGCGAAAAAGTTTCAAATCGCCTTAAACAC) and CIXR (GCGGATCCCAATACAACTTTGCCCATTAC) for cI and CROXF (CGCCATGGGCTGTTACGACTACTCACGTTTG) and CROXR (CGGGATCCTATAACATGAACTTTTTC) for cro. The PCR products were cloned into the E. coli expression vector pQE-60 (Qiagen), creating pCIX and pCROX, respectively. To enable cloning into pQE-60, the PCR primers for amplifying both cI and cro contained an NcoI site overlapping the start codon and a BamHI site at the 3′ end of the gene. Ligation of the NcoI-BamHI-digested PCR products into pQE-60 generated a six-His tag at the C terminus of each expressed protein. Proteins were expressed in E. coli strain XL1-Blue following standard isopropyl-β-d-thiogalactopyranoside (IPTG) induction. Proteins were purified on Ni-agarose columns (Qiagen) by following the manufacturer's instructions.
Band shift assays.
Three probes were generated by PCR by using 5′-radiolabeled primers. Probe 1 was a 147-bp region of φSa3ms from the region between ORFs 49 and 50 and contained a putative CI binding half-site (primers 5′-CATCTACAACTTTATCTTGC-3′ and 5′-AGTTGAACGTGTTGATGATG-3′); probe 2 was a 99-bp region overlapping the 3′ end of cro and contained a complete CI binding site (primers 5′-TTGTTCCATTGTGTCCCTCC-3′ and 5′-ATACAACTACTAGATATACC-3′); and probe 3 was a 209-bp region overlapping the region between cI and cro and contained two complete putative CI binding sites and a single putative Cro binding site (primers 5′-TTTCTACTATTTTCCCGCTC-3′ and 5′-CTCTCATTTAGTGCACCTCC-3′). After the primers were treated with kinase, unincorporated nucleotides were removed by using a MicroSpin G-25 column (Amersham). Unincorporated primers were removed after PCR by using a Qiagen PCR purification kit. Purified protein, either CI or Cro, was diluted in 1× binding buffer (20 mM Tris-HCl [pH 7.5], 1 mM MgCl2, 5% glycerol, 50 μg of bovine serum albumin per ml); 50 ng of sonicated salmon sperm DNA per μl and 4 mM dithiothreitol were included in the binding reaction mixtures, which were incubated at room temperature for 10 min. Samples were loaded and run on Invitrogen 6% polyacrylamide retardation gels. The gels were dried and then exposed to BioMax MS film (Eastman Kodak Company, Rochester, N.Y.).
RESULTS
Identification and analysis of phage genomes within S. aureus MSSA476.
Conserved phage genes (e.g., int) and known phage-encoded virulence genes (e.g., sak) were used to search the unfinished MSSA476 database (Sanger Institute) for phage-like sequences. Two putative phage genomes, designated φSa3ms and φSa4ms, were identified by this method. The designations chosen were based on the classification proposed by Baba et al. (2). φSa4ms most closely resembles φ12 from the prototypical S. aureus strain NCTC 8325 (22). However, unlike integration of φ12 in NCTC 8325, φSa4ms is integrated into the MSSA476 genome between the gene encoding a toxic anion resistance protein orthologue and an orthologue of htrA, which encodes a stress response serine protease implicated in the virulence of a number of pathogenic bacteria (15, 37). The attB site is located only 30 bases upstream of the htrA start codon, raising the possibility that φSa4ms integration and excision affect the transcription of htrA. Analysis of each individual ORF within φSa4ms failed to identify any known or putative virulence factors, similar to what has been reported for φ12 (22).
The φSa3ms genome is shown in Fig. 1. φSa3ms encodes the SEs SEA, SEG, and SEK, along with the putative virulence factors SAK and a protein (Orf57) thought to be involved in expression of a fibrinogen binding protein (4, 22). Remarkably, φSa3mw, a phage recently identified in the community-acquired methicillin-resistant S. aureus strain MW2 (2), differs from φSa3ms at only 14 bp over the entire 42,612-bp genome. The 14 bp of sequence divergence results in silent mutations at four sites, single amino acid changes in the Sak, amidase, Orf49, tail, portal, and integrase proteins, and a 4-bp deletion within a putative antirepressor gene resulting in a truncated product in φSa3ms compared to the φSa3mw product. The 4-bp deletion in φSa3ms was shown to be authentic and not a result of a sequencing error by resequencing this region of φSa3ms (data not shown).
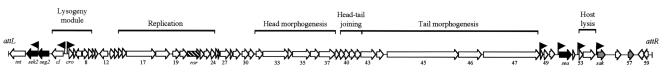
FIG. 1.
Genetic organization and ORF map of the 42,612-bp φSa3ms genome. Putative ORFs are indicated by arrows that show the directions of transcription. The raised arrows indicate the approximate locations of the promoters mapped in this study. Enterotoxin genes are indicated by solid arrows, while other putative virulence factors are indicated by gray arrows. orf21, designated ror, which was inactivated in this study, is indicated by a cross-hatched arrow. Groups of genes whose protein products are functionally interrelated are indicated above the ORF map.
Table 2 shows the closest homologue of each φSa3ms ORF identified by BLASTP searches of the GenBank database (to be more informative, φSa3mw hits were omitted). Remarkably, the sequence encoded by every φSa3ms ORF exhibited at least 90% amino acid identity to the sequences encoded by ORFs in other double-stranded DNA tailed S. aureus phages. The extensive similarity of φSa3ms sequences to the sequences found in other phages presumably reflects recombination among several other phages that occurred during the evolutionary history of φSa3ms (20). As is typical for phage genomes, φSa3ms genes with similar functions are clustered together (Fig. 1). The putative φSa3ms lysogeny module consists of divergent cI and cro homologues, suggesting that there is a mechanism for control of the lysogenic-lytic decision similar to the mechanism in λ. The putative replication module includes homologues of genes encoding the recombination protein RecT, the Holliday junction resolvase Rus, a single-stranded binding protein, a DNA ligase (ORF 22), and the putative replisome organizer Ror (Table 2). ORF 21 was designated ror after BLASTP analysis identified the origin-binding Ror protein from Bacillus subtilis phage SPP1 as the highest-scoring homologue with a defined function (32, 38). Further evidence that Ror has a role in replication was obtained from a National Center for Biotechnology Information conserved domain search analysis that revealed that Ror is similar to the PriA primosome component DnaD (31). The head morphogenesis region contains two terminase subunits, a characteristic of phages that package their genomes via a head-full mechanism (pac-type phage) (29). ORF 31 encodes a putative homing nuclease based on the similarity of the protein to a number of endonucleases belonging to the HNH family (16). The putative head-tail joining, tail morphogenesis, and host lysis regions are similar to those found in previously described S. aureus phages (22).
TABLE 2.
Putative ORFs in φSa3ms
| ORF (gene) | Coding regiona | Probable function or motifsb | % Identity (no. of amino acids)c | Homologyd
|
|
|---|---|---|---|---|---|
| Gene | Source | ||||
| 1 (int) | 139-1176e | Integrase | 99 (345) | int | φ42 |
| 2 (sek2) | 1260-1988e | Enterotoxin | 97 (241) | sek | SaP13 |
| 3 (seg2) | 2012-2740e | Enterotoxin | 98 (242) | entQ | SaP11 |
| 4 (cI) | 2936-3706e | Repressor (cl-like) | 100 (256) | orf31 | φPVL |
| 5 (cro) | 3865-4089 | Repressor (cro-like) | 100 (74) | orf4 | φ13 |
| 6 | 4086-4367 | Regulatory | 98 (93) | orf5 | φ13 |
| 7 | 4391-4930e | Unknown | 100 (179) | suv1995 | φN31 |
| 8 | 4987-5379 | Anti-repressor (C terminus truncated) | 99 (130) | orf7 | φ13 |
| 9 | 5381-5732 | Anti-repressor (N terminus truncated) | 100 (117) | orf7 | φ13 |
| 10 | 5748-5945 | Unknown | 100 (65) | sav1993 | φN315 |
| 11 | 5976-6116 | Unknown | 95 (46) | orf8 | φPV83 |
| 12 | 6131-6763e | Unknown | 100 (211) | sav1992 | φN315 |
| 13 | 6822-7142 | Conserved, unknown | 100 (106) | sav1991 | φN315 |
| 14 | 7139-7300 | Signal sequence | 100 (53) | orf9 | φ13 |
| 15 | 7393-7653 | Replication (unknown) | 100 (86) | orf10 | φ13 |
| 16 | 7662-7925 | Unknown | 100 (87) | sav1987 | φN315 |
| 17 | 7928-9877 | ATP/GTP binding (replication) | 98 (647) | sav1986 | φN315 |
| 18 (rec7) | 9879-10799 | Recombination | 100 (306) | sav1985 | φN315 |
| 19 | 11012-11497 | Metal-dependent hydrolase (β-lactamase superfamily 1) | 97 (161) | sav1984 | φN315 |
| 20 (ssb) | 11498-11968 | Single-strand binding protein | 96 (156) | sal792 | φN315 |
| 21 (ror) | 11998-12891 | Replisome organizer | 97 (297) | saI791 | φN315 |
| 22 | 12898-13116 | DNA ligase | 98 (72) | saI790 | φN315 |
| 23 (rus) | 13125-13529 | Holliday junction resolvase | 91 (134) | sav1980 | φN315 |
| 24 | 13542-13856 | Unknown | 90 (95) | saI788 | φN315 |
| 25 (rinB) | 13870-14019 | int expression | 100 (49) | sas062 | φN315 |
| 26 | 14019-14171 | Serine protease | 100 (50) | orf58 | φPVL |
| 27 | 14178-14828 | Unknown | 100 (216) | orf59 | φPVL |
| 28 | 14828-15028 | Signal sequence | 98 (66) | orf33 | φPV83 |
| 29 | 15051-15521 | Unknown | 99 (133) | orf61 | φPVL |
| 30 | 15636-16088 | Unknown | 99 (150) | orf62 | φPVL |
| 31 | 16104-16448 | HNH nuclease | 100 (114) | orf63 | φPVL |
| 32 | 16577-17044 | Terminase small subunit | 100 (155) | orf1 | φPVL |
| 33 | 17047-18741 | Terminase large subunit | 100 (564) | orf38 | φPV83 |
| 34 | 18755-18955 | Signal sequence | 98 (66) | orf39 | φPV83 |
| 35 | 18961-20211 | Portal protein | 99 (416) | orf4 | φPVL |
| 36 | 20204-20788 | Prohead protease | 100 (194) | orf41 | φPV83 |
| 37 | 20876-22123 | Head | 99 (415) | orf42 | φPV83 |
| 38 | 22159-22317 | Unknown | 100 (52) | orf43 | φPV83 |
| 39 | 22326-22658 | DNA packaging | 100 (109) | orf44a | φPV83 |
| 40 | 22645-22980 | Head-tail joining | 100 (110) | orf10 | φPVL |
| 41 | 22980-23357 | Head-tail joining | 100 (125) | orf47 | φPV83 |
| 42 | 23354-23734 | Head-tail joining | 99 (126) | orf38 | φ13 |
| 43 | 23735-24688 | Tail with immunoglobulin-like domain 2 | 100 (317) | orf39 | φ13 |
| 44 | 24752-25199 | Unknown | 100 (148) | orf40 | φ13 |
| 45 | 25437-30086 | Tape measure | 98 (1,550) | orf41 | φ13 |
| 46 | 30086-31576 | Unknown | 96 (496) | orf42 | φ13 |
| 47 | 31592-35374 | Tail | 99 (1,260) | savI953 | φN315 |
| 48 | 35367-35519 | Unknown | 100 (50) | savI952 | φN315 |
| 49 | 35565-35852 | Unknown | 100 (95) | savI951 | φN315 |
| 50 | 35908-36282 | Transmembrane region | 100 (124) | savI950 | φN315 |
| 51 (sea) | 36655-37428 | Enterotoxin | 100 (257) | sep | φN315 |
| 52 | 37579-37713 | Unknown | 100 (44) | savI947 | φN315 |
| 53 | 37925-38179 | Holin | 100 (84) | savI946 | φN315 |
| 54 | 38191-38946 | Amidase | 99 (251) | orf47 | φ13 |
| 55 (sak) | 39137-39628 | Staphylokinase | 100 (163) | sak | φN315 |
| 56 | 40318-40614 | Truncated amidase | 100 (98) | ‘lytA | φN315 |
| 57 | 41126-41476 | fib expression (signal sequence) | 100 (116) | savI942 | φN315 |
| 58 | 41768-42076e | Unknown | 99 (94) | saI753 | φN315 |
| 59 | 42100-42279e | Unknown | 100 (59) | savI940 | φN315 |
Nucleotide positions from the start codon to the stop codon.
Motifs and potential functions were determined by bioinformatic analysis as described in Materials and Methods.
Level of amino acid identity between each ORF and the best hit, excluding φSa3mw ORFs (number of amino acids over which identity exists).
The best hit (excluding φSa3mw ORFs) as determined by BLASTP analysis and its source.
Transcription occurs on the complementary strand (from right to left in Fig. 1).
Integration of φSa3ms disrupted the β-hemolysin gene of MSSA476. Such negative conversion of a potential virulence gene has been observed after integration of a number of S. aureus phages (14). Studies of the regulation of integration and excision of S. aureus phages have identified either xis homologues or an ORF adjacent to the integrase gene termed ORFC, which is thought to function like xis and facilitate phage excision (13). However, no such gene is present upstream of the putative φSa3ms int gene, raising the question of how φSa3ms integration and excision are controlled. φSa3ms, like all previously described S. aureus phages, contains a conserved hairpin loop upstream of int (13). A number of S. aureus phages also contain a second conserved hairpin sequence upstream of int (22). These putative hairpin sequences have been hypothesized to be important in int gene regulation (13, 22). The putative hairpin sequence identified by Carroll and colleagues (13) even occurs upstream of both the staphylococcal pathogenicity island 1 (SaPI1) and SaPI3 int genes.
Both φSa3ms and φSa4ms form phage particles.
We initially examined whether φSa3ms and φSa4ms were defective prophages by assaying supernatants of mitomycin C-treated MSSA476 cultures for the presence of phage DNA. After the isolated phage DNA was subjected to restriction analysis, bands of the expected sizes for φSa3ms and φSa4ms DNA were detected at nearly equal intensities (data not shown). Southern blot analysis of isolated phage DNA performed with a φSa3ms-specific sea probe provided further evidence that φSa3ms was capable of producing phage particles (data not shown). No hybridizing signal was observed when a chromosomally encoded gene was used as a probe (data not shown). Single-plaque purification of phage derived from mitomycin C-treated MSSA476, followed by DNA isolation and restriction analysis, revealed that while φSa4ms was capable of producing infectious particles (plaques), no φSa3ms plaques were observed (data not shown). Plaque lift assays with the sea probe were also negative, suggesting either that φSa3ms produces phage particles that are noninfectious, that the indicator strain RN4220 lacks the necessary φSa3ms receptor, or that the conditions used were not conducive for φSa3ms infection.
Prophage induction greatly increases transcription of sea and sak from their native promoters and from upstream latent phage promoters.
Northern blot analyses of RNA isolated from MSSA476 cultures at various times after mitomycin C treatment were carried out to investigate the effects of prophage induction on sea and sak transcript levels. In the absence of mitomycin C, a single approximately 1.0-kb sak transcript was present at relatively constant levels during the course of the experiment (Fig. 2). The levels of the single ∼1.7-kb sea transcript in uninduced cultures also remained relatively constant during the experiment (Fig. 2). The 1.7-kb transcript likely originated from a previously characterized promoter immediately upstream of sea (8). Ninety minutes after mitomycin C treatment there were marked increases in both the 1.7-kb sea and 1.0-kb sak transcripts (Fig. 2). Mitomycin C treatment also resulted in the production of two higher-molecular-weight mRNAs for both genes (Fig. 2). The fact that the highest-molecular-weight bands for both sea and sak were the same size (approximately 6.8 kb), as well as the relative organization of these two genes in the φSa3ms genome, suggested that the 6.8-kb mRNA encodes both virulence factors.
Reprobing these Northern blots with a series of probes specific for genes 5′ of sea (orf47, orf49, and orf50) established that the 6.8-kb transcript likely initiates from a region between orf47 and orf49 (data not shown). The likely start site of the 3-kb sea transcript also mapped to this region. A probe for the φSa3ms holin gene showed that the 2.7-kb transcript containing sak originates 5′ of the phage holin gene (data not shown). These data confirm that sea and sak are expressed from their own promoters in uninduced cultures. However, following prophage induction, previously repressed phage promoters become active and contribute to increased sea and sak expression. To our knowledge, this is the first demonstration that there is linkage between prophage induction and increased virulence factor transcription in S. aureus.
φSa3ms replication contributes to mitomycin C induction of sea and sak transcription.
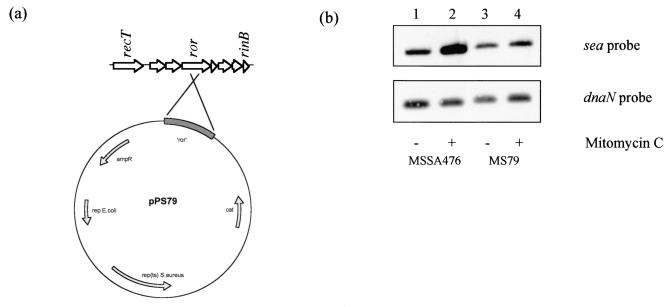
At least two processes may contribute to the large increases in the amounts of the sea and sak transcripts observed after mitomycin C treatment of MSSA476. First, the data presented above suggest that mitomycin C treatment leads to activation of latent prophage promoters that can initiate transcription of sea and sak. Second, mitomycin C treatment likely leads to φSa3ms excision and replication, thereby increasing the amount of phage DNA template available for transcription. A replication-deficient φSa3ms prophage was constructed (Fig. 3) to investigate the contribution of φSa3ms replication to sea and sak transcription. To construct the replication-deficient prophage, we insertionally inactivated the putative replisome organizer (ror) gene through homologous recombination with pPS79 (Fig. 3a), a conditionally replication-defective plasmid containing a 719-bp internal fragment of ror.
FIG. 3.
Creation of replication-deficient φSa3ms. (a) An internal fragment of ror was cloned into the temperature-sensitive shuttle vector pBT9, creating pPS79, and used to insertionally inactivate ror through plasmid integration into the MSSA476 chromosome. (b) Southern blot analysis comparing the amounts of φSa3ms DNA (sea probe) and chromosomal DNA (dnaN probe) in MSSA476 and MS79 (ror::pPS79) in the absence and presence of mitomycin C.
Southern blot analyses revealed that mitomycin C treatment of MSSA476 resulted in a large increase in the φSa3ms copy number, as indicated by the increased intensity of the sea hybridizing band (Fig. 3b, lanes 1 and 2). A probe for a chromosomal gene, dnaN, was used as a loading control for these experiments. Insertional inactivation of ror in MS79 eliminated the increase in φSa3ms replication seen after mitomycin C treatment (Fig. 3b, lanes 3 and 4). These results demonstrate that mitomycin C treatment leads to φSa3ms replication and that pPS79 integration into ror eliminates φSa3ms replication.
In marked contrast to the pronounced increases in sea and sak transcription in MSSA476, there were no detectable increases in sea or sak transcription after mitomycin C treatment of MS79 cultures (Fig. 4), suggesting that φSa3ms replication is critical for the enhanced sea and sak transcription observed following prophage induction. An alternative explanation is that the integration of pPS79 into ror blocked the expression of an activator of sea and sak transcription. The absence of any higher-molecular-weight sea or sak transcripts in mitomycin C-treated MS79 cultures suggests that in addition to the replication deficit in the MS79 φSa3ms mutant, activation of latent phage promoters does not occur in this background. The relative contributions of replication and latent phage promoter activation to the increases in sea and sak transcription that accompany prophage induction cannot be determined from analysis of MS79. Presumably, both processes contribute to the increases in sea and sak transcription that occur upon φSa3ms induction.
φSa3ms induction leads to increases in seg2 and sek2 transcripts.
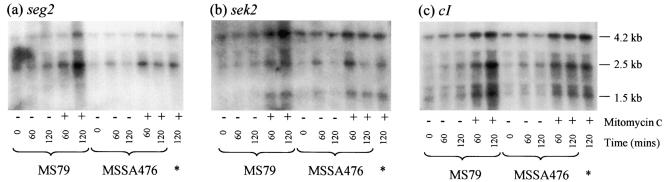
We also investigated whether mitomycin C treatment of MSSA476 affected the levels of seg2 and sek2 transcripts. The direction of transcription of these two virulence factors is opposite the direction of transcription of sea and sak, and seg2 and sek2 are located at the opposite end of the φSa3ms prophage (Fig. 1). Two seg2 hybridizing species and three sek2 hybridizing species were detected in Northern blots of RNA isolated from uninduced MSSA476 cultures (Fig. 5a and b). Addition of mitomycin C to MSSA476 cultures increased the levels of all hybridizing species of both seg2 and sek2 (Fig. 5), although the magnitude of the increase was not as great as the magnitude of the increase observed with sea and sak. Unlike the lack of an effect of mitomycin C on sea and sak transcription in MS79, mitomycin C treatment elevated seg2 and sek2 transcript levels to similar degrees in MS79 and MSSA476 (Fig. 5a and b). These findings suggest that the levels of seg2 and sek2 expression are not influenced by phage replication or latent promoter activation.
FIG. 5.
Enterotoxin genes seg2 and sek2 are cotranscribed with the φSa3ms repressor gene cI. The Northern blot shown in Fig. 4 was stripped and reprobed with either seg2 (a), sek2 (b), or cI (c).
The Northern blots probed with seg2 and sek2 probes both showed that 4.2- and 2.5-kb hybridizing fragments were present (Fig. 5a and b). This observation, along with the fact that seg2 and sek2 are adjacent and are transcribed in the same direction, suggested that these two transcripts contain both enterotoxin genes. Since both genes together span only ∼1.6 kb, these two transcripts are also likely to include either the upstream cI gene or the downstream int gene (or both in the case of the 4.2-kb mRNA). To investigate these possibilities, the Northern blots described above were stripped and reprobed with a cI probe. As shown in Fig. 5c, 2.5- and 4.2-kb cI-encoding mRNAs were detected, in addition to a new 1.5-kb transcript, suggesting that there is transcriptional coupling of cI, seg2, and sek2. To further investigate this, 5′ RACE was used to map the 5′ transcriptional start site of seg2. We found that cDNA synthesized with a primer within seg2 (SEQR [Fig. 6a]) extended beyond the 5′ end of cI (Fig. 6b, lane 3), indicating that at least some of the transcription of seg2 initiates upstream of cI. The three cI transcripts that were different sizes, all of which presumably initiate from the same site, could arise by inefficient termination, antitermination of RNA polymerase, or posttranscriptional RNA processing. The relative locations of the 4.2- and 2.5-kb transcripts are shown in Fig. 6a. The presence of a sek2 transcript that does not contain seg2 (the 1.5-kb transcript [Fig. 5b]) implies that there is a functional promoter upstream of sek2. Since these data show that the enterotoxin genes seg2 and sek2 are mostly transcribed from the upstream cI promoter, it follows that factors effecting transcription of cI also regulate seg2 and sek2 transcription.
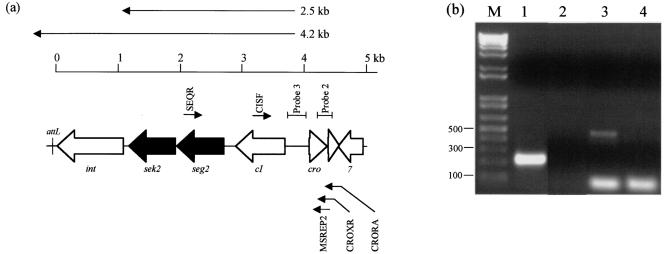
FIG. 6.
5′ RACE analysis of cro and seg2 transcription start sites. (a) Physical map showing the organization of the left end of φSa3ms. The relative positions of the primers used in the gel shown in panel b are indicated, as are the positions of the 2.5- and 4.2-kb transcripts (Fig. 5) and probes 2 and 3 (Fig. 8). CRORA and SEQR were used in reverse transcription reactions to create cro and seg2-cI cDNA, respectively. (b) Ethidium bromide-stained gel showing the products of 5′ RACE reactions. Lanes 1 and 2, PCRs performed with primers MSREP2 and AAP with cro cDNA as the template (tailed in lane 1 and not tailed in lane 2); lanes 3 and 4, PCRs performed with primers CISF and AAP with seg2-cI cDNA as the template (tailed in lane 3 and not tailed in lane 4); lane M, DNA size marker.
Mapping the cI and cro transcription initiation sites.
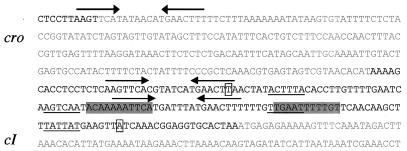
As induction of φSa3ms into the lytic cycle led to significant increases in sea, sak, seg2, and sek2 transcription, we investigated the genetic switch controlling the φSa3ms lytic-lysogenic cycle. We concentrated on the φSa3ms cI and cro homologues, two critical mediators of the λ genetic switch (39). In λ, CI production results in establishment of lysogeny and repression of the majority of the λ genes. Conversely, λ Cro production results in repression of cI expression, derepression of phage-encoded genes, and induction of λ into lytic growth. As a first step toward understanding how the φSa3ms cI and cro homologues are regulated, we identified the putative transcriptional start sites of cI and cro by 5′ RACE. To identify the cI start site, the 450-bp PCR product discussed above (Fig. 6b, lane 3) was sequenced. The cro transcriptional start site was identified by sequencing the 200-bp PCR product from Fig. 6b, lane 1. Figure 7 shows the putative transcriptional start sites for cI and cro and their corresponding −10 and −35 promoter sequences. The −10 and −35 sequences are consistent with typical staphylococcal promoters (34).
FIG. 7.
Locations of putative CI and Cro binding sites with respect to the likely cI and cro transcriptional start sites and promoter regions. Coding sequences are indicated by gray type, and noncoding sequences are indicated by black type. The transcription start sites of the divergently transcribed cI and cro genes are indicated by boxes. Putative −10 and −35 promoter sequences are underlined. The putative Cro and CI inverted repeat binding sites are shaded and are below converging arrows, respectively.
CI and Cro binding sites are present in the region between cI and cro.
Inspection of the sequence of the region between the divergent cI and cro genes resulted in identification of two different inverted repeat sequences that might serve as CI and/or Cro binding sites (Fig. 7). One inverted repeat sequence is present once at the 3′ end of cro and twice in the cI-cro intergenic region; since one of these sequences overlaps the putative cro promoter, it was a good candidate CI operator for repression of cro transcription. The putative Cro binding site occurs once in the φSa3ms genome and overlaps the −35 sequence of the cI promoter, again suggesting a mechanism of action, this time for Cro prevention of cI transcription.
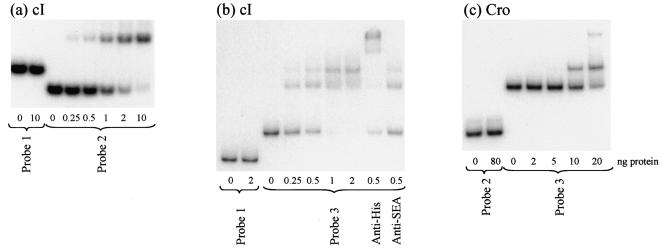
φSa3ms CI and Cro were overexpressed and purified from E. coli as C-terminal His-tagged fusion proteins and tested for the ability to bind to different regions of the φSa3ms genome in band shift assays. CI bound to a region of the φSa3ms genome that contained a single copy of the putative CI binding site (probe 2) but did not bind to a region containing only a half-site from the orf49-orf50 intergenic region (probe 1) (Fig. 8a). Probe 3 consisted of the cI-cro intergenic region and thus contained a single putative Cro binding site and two putative CI binding sites (Fig. 7). Figures 8b and c show that both CI and Cro bound to probe 3 but not to control probes lacking the corresponding inverted repeat sequences. Two different DNA-protein complexes were seen when either CI or Cro was incubated with probe 3. Since probe 3 contained two putative CI binding sites, sequential loading of CI complexes onto the probe could account for the observed pattern. As there was only a single putative Cro inverted repeat binding site in this probe, the presence of two shifted complexes suggests that Cro monomers may be able to bind to individual half-sites; alternatively there may be more than one sequence to which Cro binds in this region. Supershift experiments performed with an anti-His antibody (Fig. 8b and data not shown) confirmed that the shifted DNA protein complexes were due to CI or Cro binding.
FIG. 8.
φSa3ms CI and Cro specifically bind to specific regions of φSa3ms DNA. Bandshift assays were performed to test binding of purified C-terminal His-tagged CI (a and b) and Cro (c) to three φSa3ms-derived probes. Probe 1 was a 147-bp region of φSa3ms from the region between ORFs 49 and 50 that lacked the putative inverted repeat binding sites of CI and Cro. Probe 2 was a 99-bp region of φSa3ms that overlapped the 3′ end of cro and contained a single putative CI binding site (see Fig. 6a). Probe 3 was a 209-bp region of φSa3ms that overlapped the cI-cro intergenic region and contained two putative CI binding sites and one putative Cro binding site (see Fig. 6a). Anti-His antibody supershifted the cI-DNA binding complexes in panel b, whereas a negative control antibody (anti-SEA) did not.
DISCUSSION
Phage-encoded toxins were first recognized more than 50 years ago (19); however, very little is known about the biology and role in pathogenesis of the many phages that encode virulence factors. Here we identified and annotated a prophage genome in the incomplete genome of the methicillin-sensitive S. aureus strain MSSA476, an organism that has caused severe community-acquired S. aureus infections (Nick Day, personal communication). This phage, φSa3ms, contains three enterotoxin genes, sea, seg2, and sek2, along with a gene encoding the fibrinolytic enzyme staphylokinase. The 42,612-bp φSa3ms genome exhibits extensive similarity to the genomes of other S. aureus-derived bacteriophages. φSa3ms was inducible with mitomycin C, and its DNA was detected in supernatants of mitomycin C-treated MSSA476 cultures; however, we did not identify suitable conditions for infecting the nonlysogenic S. aureus strain RN4220.
Mitomycin C treatment of MSSA476 led to increased transcription of four φSa3ms-encoded virulence factors. In the absence of prophage induction, sea and sak were transcribed at low levels. Prophage induction with mitomycin C led to marked increases in the amounts of these transcripts, as well as the appearance of new sea and sak transcripts from activation of latent upstream phage promoters. Putative sites for the two latent phage promoters were determined. One site was upstream of the φSa3ms holin gene; the activation of this promoter led to transcriptional read-through of the downstream sak gene (2.7-kb mRNA in Fig. 2). The second promoter was in a region upstream of orf49 and downstream of orf47, the activation of which led to production of two virulence factor-encoding mRNAs, a 6.8-kb transcript containing both sea and sak and a 3-kb transcript containing only sea (Fig. 2). The mechanisms governing the production of transcripts of different sizes from these latent φSa3ms promoters are unknown. The possible mechanisms include inefficient termination, transcriptional antitermination, and RNA processing. An interesting observation from Fig. 2 is the presence of late gene phage-specific transcripts at time zero. This may be attributable to a stationary-phase-dependent decrease in prophage stability, as previously described for φLC3 in lactococci and P22 in Salmonella enterica serovar Typhimurium (30, 40). Evidence in support of this hypothesis comes from the lack of phage transcripts at time zero in Fig. 4, as the RNA for this analysis was isolated from cultures that had undergone a greater number of doublings.
The role of φSa3ms replication in virulence factor production was investigated by using MS79 (ror::pPS79). ror was chosen for insertional inactivation because the Ror homologue in B. subtilis phage SPP1 is essential for replication (38). In contrast to the transcript levels in MSSA476, the sea and sak transcript levels were not affected by mitomycin C treatment of MS79. While MS79 had a φSa3ms replication-deficient phenotype (Fig. 3b), the lack of φSa3ms lytic cycle transcripts (Fig. 4) suggests that activation of latent lytic cycle promoters does not occur in MS79. We hypothesize that the pPS79 integration in φSa3ms may have had a polar effect on expression of an activator (possibly rinB, which has a role in activation of int transcription in φ11 [49]) required for late gene transcription. As φSa3ms::pPS79 is defective both in replication and in transcriptional activation, the relative contributions of each of these factors to sea and sak transcription cannot be deciphered from analysis of MS79.
While the majority of S. aureus phage-encoded virulence factors are located downstream of phage lysis genes, φSa3ms and φSa3mw are unique due to the positions of virulence factors between the repressor and integrase genes of these phages. The increases in seg2 and sek2 transcription following prophage induction were less pronounced than the increases in sea and sak transcription. The majority of the seg2 and sek2 transcripts were found to initiate from the promoter upstream of the φSa3ms repressor gene cI, showing that cI, seg2, and sek2 are part of an operon. The transcript sizes and the orientation of genes downstream of cI suggest that the 4.2-kb cI transcript also contains the downstream int gene. The identical transcriptional profiles of cI, seg2, and sek2 from MSSA476 and MS79 cultures (Fig. 5) revealed that neither φSa3ms replication nor latent promoter activation plays a role in the transcription of these genes. Furthermore, these data suggest that factors which regulate cI transcription also regulate transcription of seg2, sek2, and int. Read-through transcription of SE genes and int from a repressor gene was suggested by the work of Yarwood and colleagues (48) in which they showed by microarray analysis that int, seq, and sek of SaPI3 have the same transcriptional profiles as the surrounding genes, with the first gene in the operon encoding a CI-like repressor (48). The constant transcription of int in a lysogen has important implications for the regulation of φSa3ms integration and excision.
Since the life cycle of φSa3ms determines transcription of its encoded virulence factors so dramatically, the putative lysogeny module of φSa3ms was analyzed. The φSa3ms CI and Cro proteins bound the region between the divergently transcribed cI and cro genes (Fig. 8), and the locations of the putative CI and Cro binding sites suggest that Cro binding prevents cI transcription and vice versa. This putative transcriptional repression through CI or Cro binding may be similar to that described for phage λ (39). φSa3ms appears to differ from λ, however, by the presence of separate CI and Cro operator sequences and by the increase in φSa3ms cI expression that occurs after mitomycin C treatment. The latter observation is difficult to reconcile with the increases in cro levels that occur with prophage induction (data not shown). Further investigation is required to understand how the φSa3ms lytic switch functions; it appears that the molecular details of this switch differ from those of the λ switch.
The work described here showed that φSa3ms prophage induction leads to transcriptional up-regulation of four φSa3ms-encoded virulence factors. Whether the mitomycin C-stimulated increases in sak, sea, seg2, and sek2 transcripts lead to increases in the levels of the corresponding proteins is currently unknown. However, the results of initial experiments exploring SEA production following mitomycin C treatment did not reveal parallel dramatic increases in SEA levels mirroring the increases observed in sea transcripts after φSa3ms induction (data not shown). Additional evidence in support of the idea that posttranscriptional regulation influences SEA production comes from the work of Borst and Betley (6-8), who suggested that factors in addition to sea mRNA levels affected SEA production.
The list of phage-encoded virulence factors continues to grow (for reviews see references 9 and 45) as the number of sequenced bacterial genomes expands. Lagging behind this expanding genomic information is an understanding of the biology of these prophages. Given the recent discovery that at least in one case phage biology can play an essential role in pathogenicity (45), this gap in our knowledge needs to be addressed. Studies with Shiga toxin-producing E. coli (44) and Streptococcus pyogenes (10) revealed that prophage induction leads to an increase in production of phage-encoded virulence factors. There is accumulating evidence that prophage induction occurs in vivo (11, 43, 44, 46), raising the possibility that host factors that promote prophage induction may play an important role in pathogenesis. The increased transcription of sea and sak upon prophage induction and the location of these genes interspersed with the lysis genes are similar to the increased transcription and location of the stxAB genes in Shiga toxin-producing E. coli. However, seg2 and sek2 differ from stx in both their location within the phage genome and in their level of transcriptional up-regulation upon phage induction. The different effects of φSa3ms induction on transcription of sea and sak and on transcription of seg2 and sek2 may be explained by their relative insertion sites in the φSa3ms genome. Insertion of seg2 and sek2 downstream of the repressor gene guarantees constitutive expression of these genes during lysogeny due to the absence of an efficient terminator downstream of cI, while the clustering of sea and sak with the φSa3ms lysis genes enables a burst of expression upon φSa3ms late gene transcription. It is tempting to speculate that the sites of insertion of virulence genes in phage genomes may be subject to selection that optimizes the phage control of expression of the genes for pathogenicity.
Acknowledgments
We thank Tim Foster, John Iandolo, Jean Lee, and Kimberly Jefferson for strains and plasmids. We also thank the NEMC GRASP Digestive Disease Center for media, Anne Kane for purification of CI and Cro, Nick Day for information about MSSA476, and the Sanger Centre for release of the MSSA476 genome sequence before publication. Finally, we thank Harvey Kimsey and Anne Kane for critical comments on the manuscript.
This work was supported by NIH grant AI42347 and by the Howard Hughes Medical Institute.
REFERENCES
- 1.Arvidson, S. 2000. Extracellular enzymes, p. 379-385. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Betley, M. J., and J. J. Mekalanos. 1985. Staphylococcal enterotoxin A is encoded by phage. Science 229:185-187. [DOI] [PubMed] [Google Scholar]
- 4.Boden, M. K., and J. I. Flock. 1994. Cloning and characterization of a gene for a 19 kDa fibrinogen-binding protein from Staphylococcus aureus. Mol. Microbiol. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 5.Bohach, G. A., and T. J. Foster. 2000. Staphylococcus aureus exotoxins, p. 367-378. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 6.Borst, D. W., and M. J. Betley. 1993. Mutations in the promoter spacer region and early transcribed region increase expression of staphylococcal enterotoxin A. Infect. Immun. 61:5421-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borst, D. W., and M. J. Betley. 1994. Phage-associated differences in staphylococcal enterotoxin A gene (sea) expression correlate with sea allele class. Infect. Immun. 62:113-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borst, D. W., and M. J. Betley. 1994. Promoter analysis of the staphylococcal enterotoxin A gene. J. Biol. Chem. 269:1883-1888. [PubMed] [Google Scholar]
- 9.Boyd, E. F., and H. Brussow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. [DOI] [PubMed] [Google Scholar]
- 10.Broudy, T. B., V. Pancholi, and V. A. Fischetti. 2002. The in vitro interaction of Streptococcus pyogenes with human pharyngeal cells induces a phage-encoded extracellular DNase. Infect. Immun. 70:2805-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broudy, T. B., V. Pancholi, and V. A. Fischetti. 2001. Induction of lysogenic bacteriophage and phage-associated toxin from group A streptococci during coculture with human pharyngeal cells. Infect. Immun. 69:1440-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruckner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 13.Carroll, D., M. A. Kehoe, D. Cavanagh, and D. C. Coleman. 1995. Novel organization of the site-specific integration and excision recombination functions of the Staphylococcus aureus serotype F virulence-converting phages φ13 and φ42. Mol. Microbiol. 16:877-893. [DOI] [PubMed] [Google Scholar]
- 14.Coleman, D. C., D. J. Sullivan, R. J. Russell, J. P. Arbuthnott, B. F. Carey, and H. M. Pomeroy. 1989. Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of beta-lysin, staphylokinase and enterotoxin A: molecular mechanism of triple conversion. J. Gen. Microbiol. 135:1679-1697. [DOI] [PubMed] [Google Scholar]
- 15.Cortes, G., B. de Astorza, V. J. Benedi, and S. Alberti. 2002. Role of the htrA gene in Klebsiella pneumoniae virulence. Infect. Immun. 70:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crutz-Le Coq, A. M., B. Cesselin, J. Commissaire, and J. Anba. 2002. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148:985-1001. [DOI] [PubMed] [Google Scholar]
- 17.Dinges, M. M., P. M. Orwin, and P. M. Schlievert. 2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13:16-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freeman, V. 1951. Studies on the virulence of bacteriophage-infected strains of Corynebacterium diphtheriae. J. Bacteriol. 61:675-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hook, M. F., and T. J. Foster. 2000. Staphylococcal surface proteins, p. 386-391. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 22.Iandolo, J. J., V. Worrell, K. H. Groicher, Y. Qian, R. Tian, S. Kenton, A. Dorman, H. Ji, S. Lin, P. Loh, S. Qi, H. Zhu, and B. A. Roe. 2002. Comparative analysis of the genomes of the temperate bacteriophages φ11, φ12 and φ13 of Staphylococcus aureus 8325. Gene 289:109-118. [DOI] [PubMed] [Google Scholar]
- 23.Ikebe, T., A. Wada, Y. Inagaki, K. Sugama, R. Suzuki, D. Tanaka, A. Tamaru, Y. Fujinaga, Y. Abe, Y. Shimizu, H. Watanabe, and the Working Group for Group A Streptococci in Japan. 2002. Dissemination of the phage-associated novel superantigen gene speL in recent invasive and noninvasive Streptococcus pyogenes M3/T3 isolates in Japan. Infect. Immun. 70:3227-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaneko, J., T. Kimura, S. Narita, T. Tomita, and Y. Kamio. 1998. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage φPVL carrying Panton-Valentine leukocidin genes. Gene 215:57-67. [DOI] [PubMed] [Google Scholar]
- 25.Kondo, I., and K. Fujise. 1977. Serotype B staphylococcal bacteriophage singly converting staphylokinase. Infect. Immun. 18:266-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 27.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 28.Lee, J. C. 1995. Electrotransformation of staphylococci. Methods Mol. Biol. 47:209-216. [DOI] [PubMed] [Google Scholar]
- 29.Le Marrec, C., D. van Sinderen, L. Walsh, E. Stanley, E. Vlegels, S. Moineau, P. Heinze, G. Fitzgerald, and B. Fayard. 1997. Two groups of bacteriophages infecting Streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl. Environ. Microbiol. 63:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lunde, M., J. M. Blatny, D. Lillehaug, A. H. Aastveit, and I. F. Nes. 2003. Use of real-time quantitative PCR for the analysis of φLC3 prophage stability in lactococci. Appl. Environ. Microbiol. 69:41-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsin, S., S. McGovern, S. D. Ehrlich, C. Bruand, and P. Polard. 2001. Early steps of Bacillus subtilis primosome assembly. J. Biol. Chem. 276:45818-45825. [DOI] [PubMed] [Google Scholar]
- 32.Missich, R., F. Weise, S. Chai, R. Lurz, X. Pedre, and J. C. Alonso. 1997. The replisome organizer (G38P) of Bacillus subtilis bacteriophage SPP1 forms specialized nucleoprotein complexes with two discrete distant regions of the SPP1 genome. J. Mol. Biol. 270:50-64. [DOI] [PubMed] [Google Scholar]
- 33.Narita, S., J. Kaneko, J. Chiba, Y. Piemont, S. Jarraud, J. Etienne, and Y. Kamio. 2001. Phage conversion of Panton-Valentine leukocidin in Staphylococcus aureus: molecular analysis of a PVL-converting phage, φSLT. Gene 268:195-206. [DOI] [PubMed] [Google Scholar]
- 34.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 35.Novick, R. P. 2003. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid 49:93-105. [DOI] [PubMed] [Google Scholar]
- 36.Novick, R. P. 2000. Pathogenicity factors and their regulation, p. 392-407. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 37.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26:209-221. [DOI] [PubMed] [Google Scholar]
- 38.Pedre, X., F. Weise, S. Chai, G. Luder, and J. C. Alonso. 1994. Analysis of cis and trans acting elements required for the initiation of DNA replication in the Bacillus subtilis bacteriophage SPP1. J. Mol. Biol. 236:1324-1340. [DOI] [PubMed] [Google Scholar]
- 39.Ptashne, M. 1992. A genetic switch. Cell Press, Cambridge, Mass.
- 40.Ramirez, E., M. Schmidt, U. Rinas, and A. Villaverde. 1999. RecA-dependent viral burst in bacterial colonies during the entry into stationary phase. FEMS Microbiol. Lett. 170:313-317. [DOI] [PubMed] [Google Scholar]
- 41.Schlievert, P. M. 1993. Role of superantigens in human disease. J. Infect. Dis. 167:997-1002. [DOI] [PubMed] [Google Scholar]
- 42.Steinberg, J. P., C. C. Clark, and B. O. Hackman. 1996. Nosocomial and community-acquired Staphylococcus aureus bacteremias from 1980 to 1993: impact of intravascular devices and methicillin resistance. Clin. Infect. Dis. 23:255-259. [DOI] [PubMed] [Google Scholar]
- 43.Voyich, J. M., D. E. Sturdevant, K. R. Braughton, S. D. Kobayashi, B. Lei, K. Virtaneva, D. W. Dorward, J. M. Musser, and F. R. DeLeo. 2003. Genome-wide protective response used by group A streptococcus to evade destruction by human polymorphonuclear leukocytes. Proc. Natl. Acad. Sci. USA 100:1996-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner, P. L., M. N. Neely, X. Zhang, D. W. Acheson, M. K. Waldor, and D. I. Friedman. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 183:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner, P. L., and M. K. Waldor. 2002. Bacteriophage control of bacterial virulence. Infect. Immun. 70:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi, T., T. Hayashi, H. Takami, K. Nakasone, M. Ohnishi, K. Nakayama, S. Yamada, H. Komatsuzawa, and M. Sugai. 2000. Phage conversion of exfoliative toxin A production in Staphylococcus aureus. Mol. Microbiol. 38:694-705. [DOI] [PubMed] [Google Scholar]
- 48.Yarwood, J. M., J. K. McCormick, M. L. Paustian, P. M. Orwin, V. Kapur, and P. M. Schlievert. 2002. Characterization and expression analysis of Staphylococcus aureus pathogenicity island 3. Implications for the evolution of staphylococcal pathogenicity islands. J. Biol. Chem. 277:13138-13147. [DOI] [PubMed] [Google Scholar]
- 49.Ye, Z. H., and C. Y. Lee. 1993. Cloning, sequencing, and genetic characterization of regulatory genes, rinA and rinB, required for the activation of staphylococcal phage φ11 int expression. J. Bacteriol. 175:1095-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]