Abstract
Pseudomonas sp. strain MT1 is capable of degrading 4- and 5-chlorosalicylates via 4-chlorocatechol, 3-chloromuconate, and maleylacetate by a novel pathway. 3-Chloromuconate is transformed by muconate cycloisomerase of MT1 into protoanemonin, a dominant reaction product, as previously shown for other muconate cycloisomerases. However, kinetic data indicate that the muconate cycloisomerase of MT1 is specialized for 3-chloromuconate conversion and is not able to form cis-dienelactone. Protoanemonin is obviously a dead-end product of the pathway. A trans-dienelactone hydrolase (trans-DLH) was induced during growth on chlorosalicylates. Even though the purified enzyme did not act on either 3-chloromuconate or protoanemonin, the presence of muconate cylcoisomerase and trans-DLH together resulted in considerably lower protoanemonin concentrations but larger amounts of maleylacetate formed from 3-chloromuconate than the presence of muconate cycloisomerase alone resulted in. As trans-DLH also acts on 4-fluoromuconolactone, forming maleylacetate, we suggest that this enzyme acts on 4-chloromuconolactone as an intermediate in the muconate cycloisomerase-catalyzed transformation of 3-chloromuconate, thus preventing protoanemonin formation and favoring maleylacetate formation. The maleylacetate formed in this way is reduced by maleylacetate reductase. Chlorosalicylate degradation in MT1 thus occurs by a new pathway consisting of a patchwork of reactions catalyzed by enzymes from the 3-oxoadipate pathway (catechol 1,2-dioxygenase, muconate cycloisomerase) and the chlorocatechol pathway (maleylacetate reductase) and a trans-DLH.
Large amounts of chloroaromatic compounds produced as biocides or formed during incineration of chloride-containing waste have been released into the environment. Although industrial pollution has been considerably reduced over the last few years, new sites contaminated with these xenobiotic compounds are still discovered every year. Chloroaromatic compounds are generally toxic and very stable. However, a broad set of microorganisms capable of mineralizing chloroaromatic compounds have been described in recent decades.
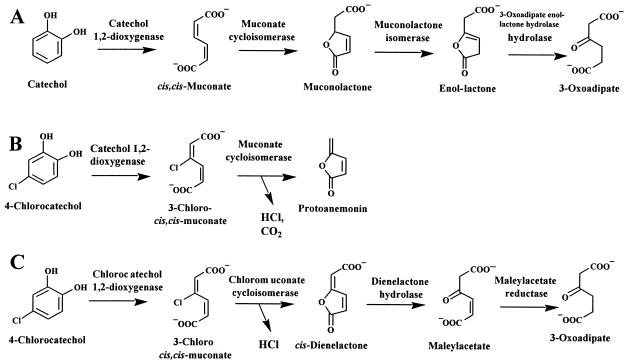
Natural nonhalogenated aromatic compounds, like lignin and its decomposition products, represent a major portion of the earth's carbon cycle. It has been shown that aerobic degradation of aromatic compounds proceeds via a small number of central intermediates, such as gentisate, protocatechuate, and catechol (51). Catechol is degraded either via ortho cleavage and the widespread 3-oxoadipate pathway (14) (Fig. 1A) or via meta cleavage pathways (2).
FIG. 1.
Degradation of catechol by enzymes of the 3-oxoadipate pathway (A) and degradation of 4-chlorocatechol by enzymes of the 3-oxoadipate pathway (B) or the chlorocatechol pathway (C).
Similarly, chloroaromatic compounds are usually mineralized by nonspecific enzymes that activate the aromatic ring via chlorocatechols as central intermediates (13, 39). In most of the cases reported thus far, chlorocatechols are in turn further degraded by enzymes of the chlorocatechol pathway (38, 39), which involves ortho cleavage by a chlorocatechol 1,2-dioxygenase with a high level of activity against chlorocatechols (11), a chloromuconate cycloisomerase with a high level of activity against chloromuconates (47), a dienelactone hydrolase (DLH), which is active against both cis- and trans-dienelactones (4-carboxymethylenebut-2-en-4-olide) (47), and a maleylacetate reductase (MAR) (21) (Fig. 1C). However, the majority of microorganisms that are capable of transforming haloaromatic compounds by nonspecific reactions do not simultaneously harbor enzymes of the chlorocatechol pathway. Therefore, it is not surprising that chlorocatechols often accumulate (13, 39) or are transformed by enzymes of the widespread 3-oxoadipate pathway (5, 50) (Fig. 1B).
Whereas chlorocatechol 1,2-dioxygenases and catechol 1,2-dioxygenases (5, 11) differ only in substrate specificity, chloromuconate cycloisomerases and muconate cycloisomerases (MCIs) also differ in the reactions catalyzed (5, 54, 55). 2-Chloromuconate formed by proteobacterial chloromuconate cycloisomerases is subject to cycloisomerization and dehalogenation, which result in trans-dienelactone (47, 54), whereas proteobacterial MCIs catalyze only cycloisomerization and 2- and 5-chloromuconolactones are the products (55). Also, 3-chloromuconate is subject to cycloisomerization and dehalogenation by chloromuconate cycloisomerase, and cis-dienelactone is the product (47). However, MCIs form protoanemonin (5) as a toxic dead-end product. Hence, transformation of 4-chlorocatechol by enzymes of the 3-oxoadipate pathway constitutes a problematic catabolic misrouting in bacterial communities (4). Protoanemonin is a poor substrate for the DLH of the chlorocatechol pathway (8), and no effective activity against protoanemonin has been characterized thus far.
However, a degradative pathway with protoanemonin as an intermediate has been proposed recently for chlorosalicylate degradation by Pseudomonas sp. strain RW10 (57). In addition to a salicylate 1-hydroxylase, this organism was shown to have a 3-oxoadipate pathway but no chlorocatechol pathway and thus forms protoanemonin from 3-chloromuconate. Wittich et al. (57) assumed that protoanemonin is transformed by a trans-DLH into cis-acetylacrylate, even though previous studies had shown that this enzyme exhibits negligible activity with this substrate (8).
Pseudomonas sp. strain MT1 is the most abundant organism in a four-member community that was isolated by continuous culture enrichment based on the ability to grow on 4-chlorosalicylate as a sole carbon source (32). Like strain RW10, MT1 can grow in monocultures on salicylate or 4- or 5-chlorosalicylate as the sole source of carbon and energy. During growth on chlorosalicylates neither strain expresses enzymes of the chlorocatechol pathway, but both strains contain a high level of trans-DLH. Analogous to RW10, MT1 was proposed to harbor a new 4-chlorocatechol-degradative pathway, which was thought to involve protoanemonin as an intermediate (32). In the present study we obtained evidence that protoanemonin is only a side product of chlorosalicylate degradation in MT1. Furthermore, we found that MT1 indeed uses a previously undescribed metabolic route for chlorosalicylate degradation, which comprises a patchwork of enzymes known to be involved in the 3-oxoadipate pathway and in the chlorocatechol pathway, as well as a trans-DLH.
MATERIALS AND METHODS
Bacterial strains.
The 5- and 4-chlorosalicylate-degrading organism Pseudomonas sp. strain MT1 was isolated by continuous culture enrichment from sediment of the Elbe River in Germany (32). Strain MT1 is the most abundant strain in a stable four-member community.
Culture conditions and preparation of cell extracts.
Liquid cultures were grown in mineral salts medium (10) by using 50 mM phosphate buffer (pH 7.5). The medium was supplemented with different carbon sources, usually at a concentration of 2.5 mM for chlorinated carbon sources and at a concentration of 5 mM for unchlorinated carbon sources. Cells were grown in fluted Erlenmeyer flasks that were incubated at 30°C on a rotary shaker at 150 rpm. Growth was monitored spectrophotometrically at 600 nm. Harvested cells were resuspended in 50 mM Tris-HCl buffer (pH 7.5) supplemented with 2 mM MnCl2 and, after addition of a trace amount of DNase I, were disrupted with a French press (Aminco, Silver Spring, Md.). Cell debris was removed by 30 min of ultracentrifugation at 100,000 × g and 4°C.
Enzyme assays.
Gentisate 1,2-dioxygenase (EC 1.13.11.4), salicylate 1-hydroxylase (EC 1.14.13.1), catechol 2,3-dioxygenase (EC 1.13.11.2), catechol 1,2-dioxygenase (EC 1.13.11.1), chlorocatechol 1,2-dioxygenase, and MCI (EC 5.5.1.1) activities were measured spectrophotometrically as previously described (6, 11, 23, 46, 47, 56). trans-DLH activity was determined by measuring depletion of trans-dienelactone (50 μM; ɛ280 = 15,625 M−1 cm−1 [47] in 50 mM Tris-HCl-2 mM MnCl2 [pH 7.5]). MAR (EC 1.3.1.32) activity was measured by the method of Seibert et al. (49) by using 0.05 mM maleylacetate freshly prepared by alkaline hydrolysis of cis-dienelactone. The activities of trans-DLH with 3-chloromuconate (160 μM) and protoanemonin (100 μM) were determined by high-performance liquid chromatography (HPLC) by using up to 80 U of purified trans-DLH per ml (measured with trans-dienelactone).
Specific activities were expressed in micromoles of substrate converted or product formed per minute per gram of protein at 25°C. Protein concentrations were determined by the Bradford procedure by using the Bio-Rad Protein assay with bovine serum albumin as the protein standard (7). The protein content of whole cells was estimated as previously described (34).
Resting cell assays.
Cells pregrown on 5-chlorosalicylate were washed twice in 50 mM phosphate buffer (pH 7.4) and resuspended in 10 ml of the same buffer at an optical density at 600 nm of 1. After addition of substrate (1 mM), the cells were incubated at 30°C and 120 rpm for up to 5 h. Cell-free supernatants were analyzed by HPLC.
Transformation by cell extracts.
5-Chlorosalicylate, 4-chlorosalicylate, 4-chlorocatechol, or 3-chloromuconate (200 μM) was incubated in 50 mM Tris-HCl-2 mM MnCl2 (pH 7.5) with extracts of 5-chlorosalicylate-grown cells having final protein contents of 60 to 360 μg of protein/ml. Transformation of 4- or 5-chlorosalicylate was monitored in the presence of NADH (400 μM). Samples were analyzed directly by HPLC analysis.
Analysis of kinetic data.
The Km and kcat of MCI for muconate were determined spectrophotometrically at substrate concentrations up to 1.3 mM. Muconate depletion was monitored at a wavelength of 260 nm (ɛ = 16,800 M−1 cm−1 [11]; substrate concentrations, 50 to 130 μM), 285 nm (ɛ = 3,800 M−1 cm−1; substrate concentrations, 130 to 350 μM), or 295 nm (ɛ = 1,100 M−1 cm−1; substrate concentrations, 350 to 1,300 μM).
Transformation of 3-chloromuconate was monitored by HPLC analysis at substrate concentrations of 75 to 1,500 μM by using 5 mU of purified MCI per ml (measured with cis,cis-muconate). Samples were taken during the reaction, and protoanemonin formation was analyzed directly by HPLC analysis.
The turnover of 4-fluoromuconolactone was analyzed spectrophotometrically at a wavelength of 240 nm by using a reaction coefficient of 4,200 M−1 cm−1 (ɛmaleylacetate = 4,700 M−1 cm−1; ɛ4-fluoromuconolactone = 500 M−1 cm−1 [40]).
Kinetic data for trans-DLH with trans-dienelactone as the substrate were determined spectrophotometrically at substrate concentrations up to 0.5 mM at a wavelength of 280 nm (substrate concentrations, up to 130 μM) or 305 nm (ɛ = 4380 M−1 cm−1; substrate concentrations, 130 to 520 μM).
At least two independent experiments were performed for each value. Km and Vmax were calculated by nonlinear regression to the Michaelis-Menten equation by using the Curve Fit feature of Sigma-Plot 2000. Turnover numbers (kcat values) were calculated by assuming that the subunit molecular mass was 39 ± 4 kDa (trans-DLH) or 40 ± 2 kDa (MCI).
Enzyme purification.
MCI and trans-DLH were purified by using a fast protein liquid chromatography system (Amersham Biosciences, Uppsala, Sweden). Cells were harvested during late exponential growth with 5-chlorosalicylate. Cell disruption and all protein elution steps were performed with 50 mM Tris-HCl-2 mM MnCl2 (pH 7.5).
For simple separation, a cell extract (usually containing between 8 and 20 mg of protein per ml) was applied directly to a MonoQ HR5/5 (anion-exchange) column (Amersham Pharmacia Biotech) or was mixed with 2 M (NH4)2SO4 to obtain a final (NH4)2SO4 concentration of 0.8 M, and after centrifugation, it was applied to a SOURCE 15PHE PE 4.6/100 (hydrophobic interaction) column (Amersham Pharmacia Biotech). At least three independent experiments were performed. Proteins were eluted from the MonoQ HR5/5 column by using a linear gradient of 0 to 0.4 M NaCl over 25 ml with a flow rate of 0.2 ml/min. Proteins were eluted from the SOURCE column by using a linear gradient of (NH4)2SO4 (0.8 to 0 M) over 25 ml with a flow rate of 0.25 ml/min. The fraction volume was 0.5 ml. Hydrophobic interaction chromatography separated MCI [0.06 ± 0.04 M (NH4)2SO4], a novel type of cycloisomerase [0.25 ± 0.04 M (NH4)2SO4], MAR [0 mM (NH4)2SO4], and trans-DLH [0.8 M (NH4)2SO4], which eliminated the possibility that the activities interfered with each other.
For purification of MCI and trans-DLH, up to 400 mg of protein was applied to a MonoQ HR10/10 column (Amersham Pharmacia Biotech). A stepwise gradient of 0 to 60 mM NaCl over 40 ml, 60 to 380 mM NaCl over 120 ml, and 380 to 2,000 mM NaCl over 40 ml was applied. The flow rate was 0.3 ml/min. The eluate was collected in 5-ml fractions. All fractions that eluted at NaCl concentrations of 90 to 330 mM (a total of 90 ml) containing both MCI and trans-DLH were pooled and concentrated to a final volume of 4.25 ml by ultrafiltration by using a Centriprep YM-50 (Millipore, Billerica, Mass.) according to the protocol recommended by the manufacturer. The protein solution was supplemented with a 4 M (NH4)2SO4 solution so that the final (NH4)2SO4 concentration was 0.8 M, and the solution was centrifuged immediately before application of the soluble proteins to the SOURCE column. Aliquots containing 40 mg of protein were separated as described above. Fractions containing MCI or trans-DLH were combined separately and concentrated by using a Centricon YM-50 (Millipore). Further purification was achieved by gel filtration with a Superose 12 HR10/10 column (Amersham Pharmacia Biotech). The proteins were eluted with 15 ml of 50 mM Tris-HCl-2 mM MnCl2 (pH 7.5) (flow rate, 0.2 ml/min; fraction volume, 0.5 ml). The fractions containing high levels of MCI activity (eluting at 11 to 12 ml) or trans-DLH activity (eluting at 13 to 14 ml) were applied to a MonoQ HR5/5 column, which was electrophoresed as described above. While MCI was homogeneous, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the trans-DLH-containing fractions still contained significant amounts of contaminating proteins. Thus, these fractions were desalted by gel filtration, supplemented with 4 M (NH4)2SO4 so that the final (NH4)2SO4 concentration was 1.85 M, and applied to a SOURCE 15PHE PE 4.6/100 column. The proteins were eluted with a stepwise gradient of 1.85 to 0.93 M over 5 ml and 0.93 to 0 M (NH4)2SO4 over 20 ml in 50 mM Tris-HCl-2 mM MnCl2 (pH 7.5). trans-DLH eluted at ammonium sulfate concentrations of 0.42 to 0.47 M. Homogeneity was verified by SDS-PAGE.
Determination of molecular masses.
The molecular masses of MCI and trans-DLH were determined by gel filtration by using a Superose 12 column as described above. The column was calibrated for molecular mass determinations by using ovalbumin (43 kDa), aldolase (158 kDa), catalase (232 kDa), and ferritin (440 kDa) obtained from Bio-Rad.
Electrophoretic methods.
SDS-PAGE was performed with a Bio-Rad Miniprotein II apparatus essentially as previously described (24). The acrylamide (Rotiphorese Gel 30; ROTH) concentration was 10% (wt/vol). The proteins were stained with Coomassie brilliant blue (Serva). A prestained low-range protein marker from Bio-Rad and the peqGOLD protein molecular weight marker from Peqlab were included.
Amino acid sequencing.
N-terminal amino acid and inner protein sequences were determined as described by Heim et al. (15). Sequences were compared with GenBank and SwissProt sequences by using tblastn (BLASTP 2.25 [16 November 2002]).
Transformation of 3-chloromuconate by enzyme mixtures.
Transformation of 3-chloromuconate by partially purified enzymes was performed in 50 mM Tris-HCl-2 mM MnCl2 (pH 7.5). The reaction mixtures contained 100 μM 3-chloromuconate and 1 to 5 μl of enzyme fraction in a total volume of 0.1 ml. Product formation from 3-chloromuconate catalyzed by purified MCI in the presence of purified trans-DLH was analyzed by HPLC in assays performed at room temperature in 150 μl of 50 mM Tris-HCl-2 mM MnCl2 (pH 7.5) with 100 to 150 μM 3-chloromuconate as the substrate. MCI was always added to give an activity of 0.9 mU/ml (as determined by transformation of 100 μM cis,cis-muconate), corresponding to 2.7 nM MCI, whereas the amounts of trans-DLH added ranged from 0.9 to 1,750 mU/ml (as determined by transformation of 50 μM trans-dienelactone), corresponding to 0.06 to 117 nM trans-DLH.
Influence of enzyme inhibitors on trans-DLH activity.
Five milliunits of trans-DLH per milliliter was incubated in the presence of EDTA (1 or 10 mM), o-phenanthroline (1 mM), HgCl2 (1 mM), ZnCl2 (1 mM), CuCl2 (1 mM), or 4-chloromercurybenzoate (0.02 or 2 mM) in 50 mM Tris-HCl (pH 7.5). Samples of the incubation mixtures were tested for enzyme activity at various times up to 24 h. Specific inactivation was assessed by comparing the activity remaining to the activity of a control incubated without inhibitor.
Analytical methods.
HPLC was performed with a Lichrospher SC 100 RP8 reversed-phase column (125 by 4.6 mm; Bischoff, Leonberg, Germany). Methanol-H2O containing 0.1% (vol/vol) H3PO4 was used as the eluent at a flow rate of 1 ml/min. The column effluent was monitored simultaneously at 210, 260, and 280 nm with a diode array detector (Shimadzu, Kyoto, Japan). The typical retention volumes with methanol-H2O (18:82) were as follows: 3-chloro-cis,cis-muconate, 9.6 ml; cis-dienelactone, 5.2 ml; trans-dienelactone, 1.9 ml; protoanemonin, 4.9 ml; maleylacetate, 0.7 ml; cis-acetylacrylate, 2.3 ml; and trans-acetylacrylate, 2.9 ml. The typical retention volumes with methanol-H2O (30:70) were as follows: 3-fluoro-cis,cis-muconate, 1.8 ml; 4-fluorocatechol, 4.4 ml; 4-fluoromuconolactone, 1.6 ml; and protoanemonin, 2.3 ml. And the typical retention volumes with methanol-H2O (66:34) were as follows: 5-chlorosalicylate, 3.2 ml; 4-chlorosalicylate, 3.6 ml; salicylate, 1.6 ml; and 4-chlorocatechol, 1.0 ml.
Chemicals.
Chemicals were purchased from Riedel de Haen (Seeke, Germany), Baker (Philipsburg, N.Y.), Aldrich Chemie (Munich, Germany), Sigma (St. Louis, Mo.), Fluka AG (Buchs, Switzerland), and Merck AG (Darmstadt, Germany). 4-Chlorocatechol was obtained from Helix Biotech (Dallas, Tex.). 3-Chloro-cis,cis-muconate and 3-fluoro-cis,cis-muconate were freshly prepared from 4-chlorocatechol and 4-fluorocatechol in 50 mM Tris-HCl-2 mM MnCl2 (pH 7.5) by using chlorocatechol 1,2-dioxygenase TetC of Pseudomonas chlororaphis RW71 (35). 4-Fluoromuconolactone was formed by addition of MCI to the 3-fluoro-cis,cis-muconate preparation. Enzymes were removed from the preparation by filtration with a Centricon YM-10 (Millipore). Complete substrate conversion was verified by HPLC. Maleylacetate was freshly prepared by alkaline hydrolysis of cis-dienelactone (49). cis-Dienelactone and 4-fluorocatechol were kindly provided by Walter Reineke (Bergische Universität-Gesamthochschule Wuppertal, Wuppertal, Germany) and Stefan Kaschabeck (Technical University Bergakademie Freiberg, Freiberg, Germany). trans-Dienelactone and protoanemonin were prepared as described previously (5, 40). cis-Acetylacrylate was obtained by isomerization of 5 mM trans-acetylacrylate (Lancaster Synthesis, Morecambe, United Kingdom) overnight under UV irradiation (wavelength, 254 nm) (44). The efficiency of isomerization was checked by HPLC analysis.
RESULTS
Growth of Pseudomonas sp. strain MT1 on chlorosalicylates and enzyme activities in cell extracts.
Pseudomonas sp. strain MT1 grew on 4- and 5-chlorosalicylates and on salicylate as sole sources of energy and carbon with growth rates of 0.05 h−1 (with 2.5 mM 4-chlorosalicylate as the carbon source), 0.16 h−1 (with 2.5 mM 5-chlorosalicylate as the carbon source), and 0.38 h−1 (with 5 mM salicylate as the carbon source). Small amounts of protoanemonin (7% ± 5%) and cis-dienelactone (12% ± 5%) accumulated during growth on 4- and 5-chlorosalicylates. MT1 was not able to use 3-chlorosalicylate as a sole source of carbon and energy.
Cell extracts of MT1 grown on 5-chlorosalicylate (Table 1) exhibited significant catechol 1,2-dioxygenase activity but neither catechol 2,3-dioxygenase nor gentisate dioxygenase activity, indicating that there was metabolism via salicylate 1-hydroxylase and chlorocatechol ortho cleavage. This finding was supported by the presence of an NADH-dependent salicylate- and chlorosalicylate-transforming activity in cell extracts. All monochlorinated salicylates were transformed at rates that were 25 to 50% of the rates of salicylate transformation, and thus the substrate specificity resembled the substrate specificity of salicylate 1-hydroxylase encoded by the nahG gene (25). The rates of transformation of 4-chlorocatechol by both salicylate- and 5-chlorosalicylate-grown cells were 13 to 21% of the rates of transformation of catechol, whereas the activities with 3-chlorocatechol were negligible (<2% of the activities with catechol). This substrate specificity indicated that there was induction of a catechol 1,2-dioxygenase rather than a chlorocatechol 1,2-dioxygenase, an enzyme which usually is highly active against 3-chlorocatechol (11, 38). Consistent with induction of enzymes of the 3-oxoadipate pathway was the observation that there was a muconate-transforming activity, while activity against 2-chloromuconate was absent (47). Cell extracts of both salicylate- and 5-chlorosalicylate-grown cells exhibited high levels of activity against trans-dienelactone but not against cis-dienelactone. As previously described, DLHs involved in the chlorocatechol pathway are active against both the cis and trans isomers (43, 46, 47) or only the cis isomer (26). However, enzymes that transform only the trans isomer (43, 45) have been described previously, and a trans-DLH with an unknown function has been observed in Pseudomonas sp. strain RW10 growing on chlorosalicylate (57). Like the level of activity in Pseudomonas sp. strain RW10, the level of trans-DLH activity in MT1 was higher in 5-chlorosalicylate-grown cells than in salicylate-grown cells (Table 1). The level of activity against maleylacetate was remarkably high and unexpected, because MAR is usually expressed together with other enzymes of the chlorocatechol pathway and Pelz et al. reported that there was almost no activity in strain MT1 (32).
TABLE 1.
Enzyme activities in cell extracts of Pseudomonas sp. strain MT1
| Enzyme | Substrate | Sp act (U/g of protein) after growth with:
|
||
|---|---|---|---|---|
| 5-Chlorosalicylate | Salicylate | Succinate | ||
| Salicylate 1-hydroxylase | Salicylate | 460 | 120 | <5 |
| 3-Chlorosalicylate | 120 | NDa | ND | |
| 4-Chlorosalicylate | 240 | ND | ND | |
| 5-Chlorosalicylate | 190 | ND | ND | |
| Catechol 2,3-dioxygenase | Catechol | <1 | <1 | ND |
| Catechol 1,2-dioxygenase | Catechol | 740 | 390 | <10 |
| 3-Chlorocatechol | 14 | <1 | ND | |
| 4-Chlorocatechol | 160 | 50 | ND | |
| MCI | Muconate | 35 | 370 | <5 |
| DLH | cis-Dienelactone | <5 | <5 | ND |
| trans-Dienelactone | 600 | 100 | <5 | |
| Protoanemonin | <5 | <5 | ND | |
| MAR | Maleylacetate | 1,120 | 510 | ND |
| cis-Acetylacrylate | 60 | 20 | ND | |
| trans-Acetylacrylate | <5 | <5 | <5 | |
ND, not determined.
It has been proposed previously that MT1, like RW10, has a new metabolic route for chlorosalicylate degradation in which protoanemonin is a central intermediate (32, 57) and is formed from 4-chlorocatechol by the combined activities of catechol 1,2-dioxygenase and MCI (5). However, significant activities against protoanemonin have been observed in neither of these organisms. Similarly, we did not observe any protoanemonin-transforming activity in cell extracts of MT1, raising doubts about the hypothesis that protoanemonin is an intermediate. Wittich et al. postulated that protoanemonin could be hydrolyzed to give cis-acetylacrylate, possibly by the action of trans-DLH (57). However, as shown for the trans-DLH of RW10 (8), the activity of purified trans-DLH of MT1 with protoanemonin was <1% of the activity with trans-dienelactone. Despite the absence of a level of protoanemonin-hydrolyzing activity in cell extracts high enough to explain the observed growth rates, there was a significant NADH-dependent cis-acetylacrylate-transforming activity in MT1. Partial protein purification indicated that this activity was a side activity of MAR (data not shown). Cell extracts of 5-chlorosalicylate-grown cells exhibited significantly higher specific activities of most key enzymes analyzed than cell extracts of salicylate-grown cells exhibited. An exception to this was the muconate-cycloisomerizing activity, which was 10-fold higher in salicylate-grown cells than in 5-chlorosalicylate-grown cells (Table 1). Cell extracts of succinate-grown MT1 did not contain a significant level of any of the activities tested. Therefore, all of these activities were inducible.
Transformation of chlorosalicylates by resting cells.
Incubation of 5-chlorosalicylate-grown cells with 5-chlorosalicylate, 4-chlorosalicylate, or 4-chlorocatechol revealed that in all cases approximately 7% ± 5% of the substrate applied was excreted as protoanemonin and approximately 10% ± 5% of the substrate applied was excreted as cis-dienelactone. No significant further metabolism of these compounds was observed. Small amounts (<10% of the applied substrate) of both 4-chlorocatechol and 3-chloromuconate were excreted transiently during transformation of 4- and 5-chlorosalicylates, indicating that these compounds are intermediates of the metabolic pathway.
Metabolites formed by cell extracts incubated with 5-chlorosalicylate, 4-chlorosalicylate, 4-chlorocatechol, or 3-chloromuconate.
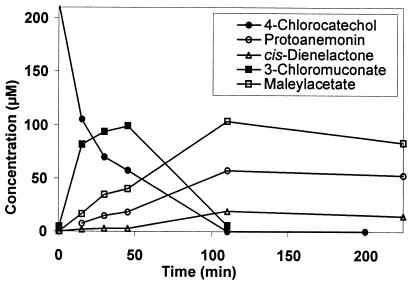
When 4-chlorocatechol was transformed by cell extracts of 5-chlorosalicylate-grown cells (corresponding to 60 μg of protein per ml), there was fast depletion along with concomitant formation of 3-chloromuconate. 3-Chloromuconate was only a transient intermediate, and both cis-dienelactone and protoanemonin were found to be transformation products. Even though 40% ± 10% of the substrate applied accumulated as protoanemonin (30% ± 5%) and cis-dienelactone (10% ± 5%), the transformation was not stoichiometric. In fact, formation of a third product was observed; as determined by HPLC analysis, this product coeluted with authentic maleylacetate and exhibited an absorption spectrum identical to that of maleylacetate. Assuming that this product was maleylacetate, it accounted for 55% ± 10% of the of the initial substrate concentration (Fig. 2). This compound was further transformed when NADH was added to the reaction mixture.
FIG. 2.
Transformation of 4-chlorocatechol by cell extract of 5-chlorosalicylate-grown Pseudomonas sp. strain MT1. The experiment was performed in 50 mM Tris-HCl-2 mM MnCl2 (pH 7.5) with 360 μg of cell extract per ml. The substrate and product concentrations were analyzed by HPLC.
Similarly, 40% ± 20% of the substrate applied accumulated as cis-dienelactone (20% ± 10%) and protoanemonin (25% ± 10%) when 5- or 4-chlorosalicylate was incubated with cell extracts (60 to 360 μg of protein/ml). As salicylate hydroxylase activity requires the presence of NADH, significant amounts of maleylacetate did not accumulate during transformation, but this compound was probably further transformed into 3-oxoadipate, which could not be quantified by HPLC by using UV detection. Thus, it can be assumed that in addition to protoanemonin and cis-dienelactone, maleylacetate is a major product of chlorosalicylate degradation in MT1. All three products were produced (protoanemonin, 35% ± 20%; maleylacetate, 45% ± 20%; cis-dienelactone, 20% ± 5%) when 3-chloromuconate was transformed by cell extracts (60 to 360 μg of protein/ml).
Transformation of 3-chloromuconate by partially purified enzymes.
In order to characterize enzymes involved in the formation of maleylacetate from 3-chloromuconate, extracts of 5-chlorosalicylate-grown cells were fractioned by fast protein liquid chromatography, and fractions were tested for 3-chloromuconate-transforming activity by HPLC analysis. Astonishingly, two distinct 3-chloromuconate-transforming activities were observed after hydrophobic chromatography. Fractions eluting at 0.05 ± 0.04 M (NH4)2SO4 converted 3-chloromuconate predominantly into protoanemonin (80% ± 5%). The corresponding enzyme activity thus resembled an MCI and was later used for detailed analyses (referred to as MCI below). Fractions that eluted at 0.25 ± 0.04 M (NH4)2SO4 formed approximately equal amounts of protoanemonin (50% ± 3%) and cis-dienelactone (47% ± 5%) and thus had a product spectrum intermediate between the product spectra of MCI and chloromuconate cycloisomerase (the enzyme activity referred to below as MCIB). However, in contrast to transformation by cell extracts, only negligible amounts of maleylacetate were observed, and the reaction products detected under these conditions were dead-end metabolites for MT1. Because both 3-chloromuconate-converting activities formed considerable amounts of protoanemonin and hardly any maleylacetate, we considered the possibility that another enzyme in the cell extract might convert an intermediate product resulting from 3-chloromuconate conversion into maleylacetate.
To evaluate this possibility, cell extracts were fractioned by a different method, anion-exchange chromatography. Again two distinct activities against 3-chloromuconate were observed. Enzymes that eluted at 0.4 ± 0.02 M NaCl converted 3-chloromuconate to a mixture containing approximately equal amounts of protoanemonin and cis-dienelactone, as described above. However, 3-chloromuconate was transformed into maleylacetate and protoanemonin by fractions that eluted at 0.26 ± 0.04 M NaCl, demonstrating that these fractions contained all of the proteins necessary for maleylacetate formation.
It has been assumed previously that conversion of 3-chloromuconate to protoanemonin should proceed via 4-chloromuconolactone as a reaction intermediate (5), and it can be postulated that hydrolysis of 4-chloromuconolactone leads to maleylacetate. 4-Fluoromuconolactone has previously been described as a rather stable product of 3-fluoromuconate cycloisomerization by MCI (44). Furthermore, Schlömann et al. suggested that 4-fluoromuconolactone might be converted by DLH to maleylacetate (44). Thus, it could be speculated that trans-DLH acts on 4-chloromuconolactone, formed transiently during cycloisomerization of 3-chloromuconate, and is therefore responsible for the formation of maleylacetate. In fact, trans-DLH of MT1 coeluted with MCI during anion-exchange chromatography.
Characterization of homogeneous MCI.
To prove the hypothesis that maleylacetate was formed from 3-chloromuconate by the combined action of MCI and trans-DLH, both proteins were purified to homogeneity. The molecular mass of the native MCI was estimated by gel filtration to be 255 ± 20 kDa, and a single band at 40 ± 2 kDa was observed by SDS-PAGE. Thus, it can be assumed that this enzyme, like MCI of Pseudomonas putida PRS2000 (16) or chloromuconate cycloisomerase of Ralstonia eutropha JMP 134 (18), is a homo-octamer.
The N-terminal sequence of the MCI from MT1 (TQAKIESIETILVDLPTVRPHKLAM) exhibited significant similarity to the CatB protein sequence of Acinetobacter calcoaceticus (accession number Q43931; 14 identical amino acids) and to the CatB sequence of P. putida PRS2015 (accession number P08310; 10 identical amino acids), which also exhibits significant similarity (amino acids 213 to 231) to an internal sequence of the strain MT1 MCI (ACKVLGDNGLDLLEKPLSR) (14 identical amino acids).
The substrate spectrum and kinetic parameters of MT1 MCI were only partially similar to those of previously described proteobacterial MCIs. Both 2-chloromuconate and 2,4-dichloromuconate were poor substrates for the enzyme (Table 2), whereas muconate and 3-chloromuconate were transformed at relatively high rates. The Km value for muconate (1.73 ± 0.35 mM) was significantly higher than the Km values previously reported for MCIs of P. putida PRS2000 (0.04 mM) (53), A. calcoaceticus (0.13 mM) (53), and Pseudomonas sp. strain B13 (0.056 mM) (47); however, the maximal turnover numbers were the same order of magnitude (53). Surprisingly, a comparison of kcat/Km values indicated that 3-chloromuconate is preferred over muconate as a substrate by strain MT1 MCI, and activities approaching those reported here have been obtained only for two site-directed mutants of P. putida MCI by Vollmer et al. (53).
TABLE 2.
Substrate specificities of chloromuconate cycloisomerase and MCI from different strains and of trans-DLH from Pseudomonas sp. strain MT1
| Strain | Enzyme | Substrate | Activity with 0.1 mM substrate (U/mg)a | Km (μM)b | kcat (min−1)b | kcat/Km (min−1 μM−1)b |
|---|---|---|---|---|---|---|
| Pseudomonas sp. strain MT1 | MCI | Muconate | 9.9 ± 1.2 | 1,730 ± 350 | 7,500 ± 350 | 4.3 |
| 3-Chloromuconate | 35 ± 6 | 410 ± 75 | 7,200 ± 520 | 17.6 | ||
| 2-Chloromuconate | 0.09 ± 0.02 | NDc | ND | ND | ||
| 2,4-Dichloromuconate | <0.001 | ND | ND | ND | ||
| Pseudomonas sp. strain MT1 | trans-DLH | trans-Dienelactone | 710 ± 37 | 480 ± 20 | 159,000 ± 8,500 | 330 |
| 4-Fluoromuconolactone | 190 ± 4 | 1,200 ± 20 | 98,000 ± 2,200 | 82 | ||
| R. eutropha JMP 134 | CMCI (= TfdDI)d | Muconate | 1.6 | 996 ± 35 | 700 ± 10 | 0.71 |
| 3-Chloromuconate | 48 | 98 ± 3 | 3,770 ± 70 | 38 | ||
| 2-Chloromuconate | 3 | 79 ± 3 | 215 ± 5 | 2.7 | ||
| 2,4-Dichloromuconate | 57.5 | 23 ± 1 | 2,810 ± 40 | 120 | ||
| Pseudomonas sp. strain B13 | CMCI | Muconate | 3.3 | 270 ± 55 | 480 ± 70 | 1.8 |
| 3-Chloromuconate | 21.7 | 160 ± 55 | 2,240 ± 480 | 14 | ||
| 2-Chloromuconate | 8.5 | 42 ± 6 | 480 ± 30 | 11 | ||
| 2,4-Dichloromuconate | 7.9 | 26 ± 2 | 390 ± 10 | 15 | ||
| P. putida PRS2000 | MCI | Muconate | 223 | 40 ± 4 | 12,600 ± 400 | 310 |
| 3-Chloromuconate | 0.38 | 320 ± 200 | 64 ± 30 | 0.20 | ||
| 2-Chloromuconate | 0.76 | 59 ± 5 | 49 ± 2 | 0.83 | ||
| 2,4-Dichloromuconate | 0.06 | 26 ± 3 | 3.3 ± 0.1 | 0.13 |
The data for strains JMP 134, B13, and PRS2000 were calculated from data in references 52 and 53.
The data for strains JMP 134, B13, and PRS2000 are from references 52 and 53.
ND, not determined.
CMCI, chloromuconate cycloisomerase.
3-Fluoromuconate was transformed by MCI from strain MT1 into a new product, which, as described for 4-fluoromuconolactone (44), the product of 3-fluoromuconate cycloisomerization by MCI of R. eutropha 335, showed no maximum UV absorption at wavelengths between 200 and 350 nm and which exhibited negligible absorbance (ɛ = <500 M−1 cm−1) at wavelengths of >240 nm.
Characterization of trans-DLH.
The molecular mass of native trans-DLH was determined to be 85 ± 30 kDa by gel filtration. As a single band at 39 ± 4 kDa was visible on SDS-PAGE gels, it can be assumed that trans-DLH of Pseudomonas sp. strain MT1 is a dimeric enzyme. In contrast, the DLHs of Pseudomonas sp. strain B13 (31), P. putida strain 87 (52), and Rhodococcus opacus 1CP (26), as well as cis-DLH of Burkholderia cepacia (45), have been shown to be monomeric proteins. Partial protein sequences (N-terminal sequence TDTSKSLPTYKQLLERKDAPPGSSWGLFGK and inner sequence VSQGVTLEPGDAVLLR) were compared with sequences in the SwissProt and GenBank databases, but no significant similarity was observed. Also, no significant similarity was found when 21 selected DLH protein sequences from the SwissProt database and the unpublished N-terminal sequence of trans-DLH of R. eutropha 335 (17) were compared with the N-terminal sequence of MT1 trans-DLH. The Km for trans-dienelactone (480 μM) (Table 2) was higher than the values reported for DLHs of the chlorocatechol pathway (43, 47) for this substrate. trans-DLH also exhibited significantly higher turnover values than the DLH of Pseudomonas sp. strain B13 (1,800/min) (29) and even a higher turnover value than cis-DLH with cis-dienelactone (46,000/min) (45). No trans-DLH activity was observed with 3-chloromuconate, protoanemonin, or cis-dienelactone (the detection limit was <0.05% of the activity with 0.1 mM trans-dienelactone). Thus, while trans-DLH obviously interferes with the reaction catalyzed by MCI, presumably preventing the formation of protoanemonin and supporting the formation of maleylacetate (see below), the enzyme acts on neither the substrate (3-chloromuconate) nor the product (protoanemonin) of this reaction.
As 4-chloromuconolactone, the postulated intermediate of 3-chloromuconate cycloisomerization (5, 22), was not available, transformation of the rather stable analogue 4-fluoromuconolactone (44) by trans-DLH was tested. HPLC revealed that there was quantitative transformation of 4-fluromuconolactone into a single product with a retention behavior and absorption spectrum identical to those of maleylacetate. Spectrophotometric analysis showed that the product exhibited an absorption maximum under physiological conditions of 243 nm, in accordance with identification of the compound as maleylacetate (33, 49). According to the kcat/Km values, trans-dienelactone was the preferred substrate and was transformed approximately four times more efficiently than 4-fluoromuconolactone (Table 2).
trans-DLH was significantly inhibited by the chelating agents EDTA and o-phenanthroline (Table 3). In contrast to DLHs from Pseudomonas sp. strain B13 (29) and R. opacus 1CP (26) and trans-DLH of R. eutropha 335 (43), which are strongly inhibited by 4-chloromercuribenzoic acid, the trans-DLH activity of MT1 was not influenced by this inhibitor; a similar result was obtained for cis-DLH of B. cepacia (45). However, heavy metals had strong effects on the trans-DLH activity.
TABLE 3.
Effects of inhibitors on the trans-DLH activity of Pseudomonas sp. strain MT1
| Enzyme inhibitor | Concn (mM) | Relative enzyme activity (%) after incubation fora:
|
||
|---|---|---|---|---|
| 5 min | 60 min | 240 min | ||
| EDTA | 1 | 27 ± 3 | 17 ± 3 | NDb |
| 10 | 13 ± 3 | 7 ± 3 | ND | |
| o-Phenanthroline | 1 | 55 ± 4 | 18 ± 3 | ND |
| 4-Chloromercuribenzoate | 0.02 | >90 | >90 | >90 |
| 2 | >90 | >90 | >90 | |
| HgCl2 | 1 | 6 ± 3 | ND | ND |
| ZnCl2 | 1 | <5 | ND | ND |
| CuCl2 | 1 | 34 ± 5 | 26 ± 4 | ND |
Enzyme activity is expressed as a percentage based on a comparison with the activity of a control incubated without inhibitor.
ND, not determined.
In situ transformation of 3-chloromuconate to maleylacetate by combined action of MCI and trans-DLH.
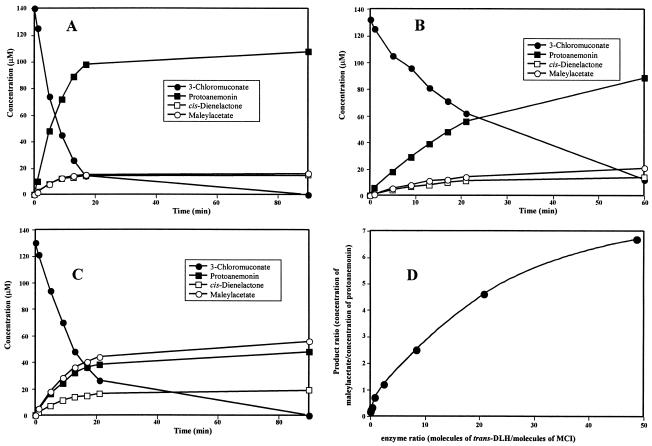
When purified MCI was incubated with 3-chloromuconate, protoanemonin was the major reaction product (>75%), and only minor amounts of cis-dienelactone (8% ± 4%) and maleylacetate (10% ± 5%) were detected as side products (Fig. 3A). However, when trans-DLH was added simultaneously to the reaction mixture, an increase in maleylacetate formation and a decrease in protoanemonin formation were observed (Fig. 3B to D). Monitoring the transformation of 3-chloromuconate over time with different enzyme ratios showed that all the products were continuously formed and that the 3-chloromuconate was transformed quantitatively into protoanemonin, cis-dienelactone, and maleylacetate. No other compounds accumulated during the reaction.
FIG. 3.
Transformation of 3-chloromuconate by mixtures of MCI and trans-DLH of Pseudomonas sp. strain MT1. The reaction mixtures contained 50 mM Tris-HCl-2 mM MnCl2 (pH 7.5) and 120 to 140 μM 3-chloromuconate. The substrate and product concentrations were analyzed by HPLC. (A) Transformation catalyzed by 2.1 nM MCI. (B) Transformation catalyzed by a mixture of 0.7 nM MCI and 0.18 nM trans-DLH. (C) Transformation catalyzed by a mixture of 2.1 nM MCI and 5.4 nM trans-DLH. (D) Ratio of maleylacetate to protoanemonin formed from 3-chloromuconate by mixtures of MCI (2.1 nM) and various amounts of trans-DLH (0 to 102 nM).
DISCUSSION
Until recently, aerobic degradation of chlorocatechols was assumed to be due exclusively to enzymes of the chlorocatechol pathway, including chlorocatechol dioxygenase, chloromuconate cycloisomerase, DLH, and MAR. However, recent studies have indicated that bacterial routes for channeling chlorocatechol into central metabolic pathways are more diverse than previously thought. In 1995, Arensdorf and Focht (1) described the degradation of 4-chlorobenzoate via 4-chlorocatechol and a meta cleavage pathway, and similarly, Hollender et al. (19) proposed that degradation of 4-chlorophenol via meta cleavage of 4-chlorocatechol occurs in a Comamonas isolate. P. putida GJ31 grows on chlorobenzene via meta cleavage of 3-chlorocatechol (27). This strain was shown to contain a novel chlorocatechol 2,3-dioxygenase, which transforms 3-chlorocatechol into 2-hydroxymuconate, a central intermediate in classical meta cleavage routes, and thus obviously rapidly hydrolyzes the initially formed acylchloride (20), which otherwise was suggested to be a suicide substrate for catechol 2,3-dioxygenases (3).
Another route of 3-chlorocatechol degradation has recently been reported for R. opacus (28). This organism has an MCI which is highly active with the 3-chlorocatechol ring cleavage product 2-chloromuconate but, like proteobacterial MCIs, is not capable of dehalogenating. In fact, the organism recruits an enzyme whose sequence is similar to the sequences of muconolactone isomerases to dehalogenate the cycloisomerization product 5-chloromuconolactone. The ability to dehalogenate 5-chloromuconolactone has also been reported for muconolactone isomerases of the 3-oxoadipate pathway (37) and seems to be an ability that is shared generally by muconolactone isomerases (36).
Protoanemonin, a highly toxic intermediate, was recently shown to be the dominant product formed from 3-chloromuconate by MCIs (5). Since in Pseudomonas sp. strain MT1, as well as Pseudomonas sp. strain RW10, only enzymes of the 3-oxoadipate pathway have been observed, it has been suggested that these organisms have a new route for 4-chlorocatechol degradation in which protoanemonin is an intermediate (32, 57). However, in these organisms no protoanemonin-hydrolyzing activity formed the postulated pathway intermediate cis-acetylacrylate (9, 57; this study).
In this study we found that the amount of protoanemonin formed from 3-chloromuconate depends on whether single purified cycloisomerases or combinations of enzymes are used for conversion. Thus, 3-chloromuconate conversion by a purified MCI of Pseudomonas sp. strain MT1 resulted in high concentrations of protoanemonin (Fig. 3A), as shown previously for MCIs from Pseudomonas sp. strains B13 and RW10 (5). The simultaneous presence of MCI and trans-DLH of MT1, in contrast, yielded considerably lower protoanemonin concentrations but larger amounts of maleylacetate. Similarly, the presence of trans-DLH during turnover of 3-chloromuconate by MCIB resulted in decreased concentrations of protoanemonin and elevated concentrations of maleylacetate (data not shown). Thus, the possibility that protoanemonin is not an intermediate of the degradative pathway must be considered. Most interestingly, like strain MT1, Pseudomonas sp. strain RW10 does not form quantitative amounts of protoanemonin when a cell extract of chlorosalicylate-grown cells is confronted with 4-chlorocatechol (5). Probably, as in MT1, protoanemonin is not the only product of 4-chlorocatechol and 3-chloromuconate transformation.
MT1 obviously contains two 3-chloromuconate-cycloisomerizing activities. One of these activities resembles proteobacterial MCIs with respect to product formation and therefore was designated an MCI. However, this activity had a higher specificity constant for 3-chloromuconate than for muconate (Table 2). At the same time the N-terminal sequence was found to exhibit only moderate similarity to the MCIs of other pseudomonads (10 to 14 of 25 amino acids). Thus, the MT1 cycloisomerase may be specialized for 3-chloromuconate conversion and may not have the ability to dehalogenate. This clearly sets the MT1 cycloisomerase apart from known proteobacterial chloromuconate cycloisomerases which form cis-dienelactone from 3-chloromuconate and obviously have the ability to eliminate chloride (5, 22, 47).
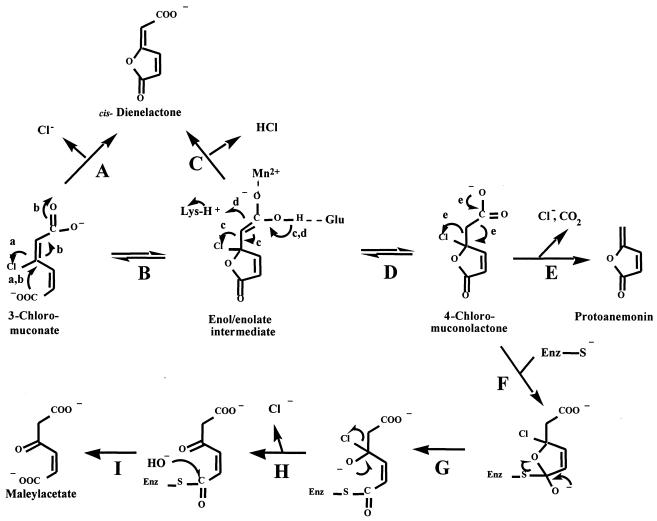
Studies of the reaction mechanisms of MCI and mandelate racemase have suggested that the reaction proceeds via an enol/enolate (Fig. 4, reaction B), to which a proton is added to form muconolactone (Fig. 4, reaction D) (12). Similarly, the formation of protoanemonin from 3-chloro-cis,cis-muconate involves a protonation reaction, as two hydrogen atoms are present on the exocyclic carbon. In contrast, it has been proposed that in the reaction of chloromuconate cycloisomerases with 3-chloromuconate, the corresponding enol/enolate intermediate is not protonated but rather loses the negative charge by chloride abstraction (Fig. 4, reaction C) (22). Replacement of Lys169 of P. putida MCI, which is known to provide the proton for the protonation reaction (12, 41), by alanine resulted in mutants that were not able to form protoanemonin but rather formed cis-dienelactone (22). Thus, a protonation reaction was shown to be necessary for protoanemonin formation but not for cis-dienelactone formation, and, as proposed by Blasco et al. (5), 4-chloromuconolactone is the intermediate of MCI-catalyzed cycloisomerization, from which decarboxylation and chloride elimination to form protoanemonin (Fig. 4, reaction E) occurs. Whether the latter process is spontaneous or enzyme catalyzed has not been directly investigated. If, however, 4-chloromuconolactone is in fact used as a substrate by trans-DLH, then the findings suggest that 4-chloromuconolactone is the final product of the cycloisomerizing enzyme and that protoanemonin formation from this compound may be a spontaneous reaction.
FIG. 4.
Proposed mechanism for the formation of maleylacetate from 3-chloromuconate by the combined action of MCI and trans-DLH. Either 3-chloromuconate is subject to direct dehalogenation (reaction A) or chloride is eliminated from a metal-stabilized enol/enolate intermediate (reactions B and C). Alternatively, a proton, provided by a lysine residue, is added to the enol/enolate to form 4-chloromuconolactone (reaction D). Decarboxylation and chloride elimination from 4-chloromuconolactone give rise to protoanemonin (reaction E). Free 4-chloromuconolactone can be subject to nucleophilic attack by trans-DLH (reaction F), giving rise to a halohydrin (reaction G), which spontaneously eliminates the halogenide ion (reaction H). Hydrolysis (reaction I) then yields maleylacetate.
The proposal that trans-DLH acts on 4-chloromuconolactone was supported by the ability of the enzyme to act on the substrate analogue 4-fluoromuconolactone, which is known to be a rather stable product (44). Dehalogenation of 4-chloromuconolactone by MT1 trans-DLH may be expected to resemble the 4-fluoromuconolactone conversion observed with 3-oxoadipate enol-lactone hydolases and proteobacterial DLHs (42, 43). Some of these enzymes were found to transform 4-fluoromuconolactone mainly to maleylacetate, and based on the reaction mechanism suggested by Cheah et al. (9) for dienelactone hydrolysis by cis/trans-DLH of Pseudomonas sp. strain B13, one may expect that the enzyme nucleophile attacks the lactone carbonyl group (Fig. 4, reaction F), giving rise to a halohydrin (Fig. 4, reaction G), which should spontaneously eliminate the halogenide ion (Fig. 4, reaction H).
The relatedness of MT1 trans-DLH to other hydrolytic enzymes, like those with an α/β-hydrolase fold (30), including the cis/trans-DLH family usually involved in chloroaromatic pathways, is not known, and consequently the conclusions concerning dehalogenation mechanisms are highly speculative. In fact, the high level of susceptibility of trans-DLH to chelators (Table 3) suggests that this enzyme differs from other lactone hydrolases in its basic biochemical properties.
In R. eutropha 335 a trans-DLH has been reported to be induced during growth of the strain with 4-fluorobenzoate (46). This enzyme may play a dual role in the degradation and be responsible for direct conversion of 4-fluoromuconolactone, as well as conversion of trans-dienelactone formed from 4-fluoromuconolactone as a by-product by 3-oxoadipate enol-lactone hydrolase (42). So far, the natural function of the R. eutropha trans-DLH is unclear. This enzyme is highly unlikely to have been selected especially for 4-fluorobenzoate catabolism and thus seems to have been recruited for this process by accidental induction. Similarly, the MAR of R. eutropha 335 (MacA; accession number AF130250), which also is not part of a specialized gene cluster, is obviously induced and thus recruited for 4-fluorobenzoate degradation (48).
In Pseudomonas sp. strain MT1 trans-DLH may also be recruited by chance induction. However, it may also be coinduced with MCIB, which, like trans-DLH, is induced to a significantly greater extent in 5-chlorosalicylate-grown cells than in salicylate-grown cells (data not shown). If this were true, then these enzymes might constitute core elements of another highly unusual chlorocatechol degradative pathway. Future studies of the genetic background of MT1 cycloisomerase and trans-DLH should help answer this question.
Acknowledgments
This work was supported by DFG-European Graduate College 653.
We thank Rita Gezlaff (GBF) for N-terminal and inner protein amino acid sequencing and Iris Plumeier for help with protein purification.
REFERENCES
- 1.Arensdorf, J. J., and D. D. Focht. 1995. A meta cleavage pathway for 4-chlorobenzoate, an intermediate in the metabolism of 4-chlorobiphenyl by Pseudomonas cepacia P166. Appl. Envion. Microbiol. 61:443-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assinder, S., and P. Williams. 1990. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv. Microb. Physiol. 31:1-69. [DOI] [PubMed] [Google Scholar]
- 3.Bartels, I., H.-J. Knackmuss, and W. Reineke. 1984. Suicide inactivation of catechol 2,3-dioxygenase from Pseudomonas putida mt-2 by 3-halocatechols. Appl. Environ. Microbiol. 47:500-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco, R., M. Mallavarapu, K. N. Timmis, and D. H. Pieper. 1997. Evidence that formation of protoanemonin from metabolites of 4-chlorobiphenyl degradation negatively affects the survival of 4-chlorobiphenyl-cometabolizing microorganisms. Appl. Environ. Microbiol. 63:427-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blasco, R., R. M. Wittich, M. Mallavarapu, K. N. Timmis, and D. H. Pieper. 1995. From xenobiotic to antibiotic. Formation of protoanemonin from 4-chlorocatechol by enzymes of the 3-oxoadipate pathway. J. Biol. Chem. 270:29229-29235. [DOI] [PubMed] [Google Scholar]
- 6.Bosch, R., E. R. B. Moore, E. Garcia Valdes, and D. H. Pieper. 1999. NahW, a novel, inducible salicylate hydroxylase involved in mineralization of naphthalene by Pseudomonas stutzeri AN10. J. Bacteriol. 181:2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Brückmann, M., R. Blasco, K. N. Timmis, and D. H. Pieper. 1998. Detoxification of protoanemonin by dienelactone hydrolase. J. Bacteriol. 180:400-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheah, E., G. W. Ashley, J. Gary, and D. Ollis. 1993. Catalysis by dienelactone hydrolase: a variation on the protease mechanism. Protein Struct. Funct. Genet. 16:64-78. [DOI] [PubMed] [Google Scholar]
- 10.Dorn, E., M. Hellwig, W. Reineke, and H.-J. Knackmuss. 1974. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 99:61-70. [DOI] [PubMed] [Google Scholar]
- 11.Dorn, E., and H.-J. Knackmuss. 1978. Chemical structure and biodegradability of halogenated aromatic compounds. Two catechol 1,2-dioxygenases from a 3-chlorobenzoate-grown pseudomonad. Biochem. J. 174:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerlt, J. A., and P. G. Gassmann. 1992. Understanding enzyme-catalyzed proton abstraction from carbon acids: details of stepwise mechanisms for β-elimination reactions. J. Am. Chem. Soc. 114:5928-5934. [Google Scholar]
- 13.Häggblom, M. M. 1992. Microbial breakdown of halogenated aromatic pesticides and related compounds. FEMS Microbiol. Rev. 103:29-72. [DOI] [PubMed] [Google Scholar]
- 14.Harwood, C. S., and R. E. Parales. 1996. The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 15.Heim, S., M. Del Mar Lleo, B. Bonato, C. A. Guzman, and P. Canepari. 2002. The viable but nonculturable state and starvation are different stress responses of Enterococcus faecalis, as determined by proteome analysis. J. Bacteriol. 184:6739-6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helin, S., P. C. Kahn, B. L. Guha, D. G. Mallows, and A. Goldman. 1995. The refined X-ray structure of muconate lactonizing enzyme from Pseudomonas putida PRS2000 at 1.85 Å resolution. J. Mol. Biol. 254:918-941. [DOI] [PubMed] [Google Scholar]
- 17.Hinner, I. 1998. Biochemische und molekularbiologische Untersuchungen zu Lacton-Hydrolasen des bakteriellen Aromaten-und Halogenaromaten-Abbaus. Ph.D. thesis.Universität Stuttgart, Stuttgart, Germany.
- 18.Hoier, H., M. Schlömann, A. Hammer, J. P. Glusker, H. L. Carrell, A. Goldman, J. J. Stezowski, and U. Heinemann. 1994. Crystal structure of cloromuconate cycloisomerase from Alcaligenes eutrophus JMP134 (pJP4) at 3 Å resolution. Acta Crystallogr. 50:75-84. [DOI] [PubMed] [Google Scholar]
- 19.Hollender, J., J. Hopp, and W. Dott. 1997. Degradation of 4-chlorophenol via the meta cleavage pathway by Comamonas testosteroni JH5. Appl. Environ. Microbiol. 63:4567-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaschabek, S. R., T. Kasberg, D. Müller, A. E. Mars, D. B. Janssen, and W. Reineke. 1998. Degradation of chloroaromatics: purification and characterization of a novel type of chlorocatechol 2,3-dioxygenase of Pseudomonas putida GJ31. J. Bacteriol. 180:296-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaschabek, S. R., and W. Reineke. 1992. Maleylacetate reductase of Pseudomonas sp. strain B13: dechlorination of chloromaleylacetates, metabolites in the degradation of chloroaromatic compounds. Arch. Microbiol. 158:412-417. [DOI] [PubMed] [Google Scholar]
- 22.Kaulmann, U., S. R. Kaschabek, and M. Schlömann. 2001. Mechanism of chloride elimination from 3-chloro- and 2,4-dichloro-cis,cis-muconate: new insight obtained from analysis of muconate cycloisomerase variant CatB-K169A. J. Bacteriol. 183:4551-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lack, L. 1959. The enzymatic oxidation of gentisic acid. Biochim. Biophys. Acta 34:117-123. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lehrbach, P. R., J. Zeyer, W. Reineke, H.-J. Knackmuss, and K. N. Timmis. 1984. Enzyme recruitment in vitro: use of cloned genes to extend the range of haloaromatics degraded by Pseudomonas sp. strain B13. J. Bacteriol. 158:1025-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maltseva, O. V., I. P. Solyanikova, L. A. Golovleva, M. Schlömann, and H.-J. Knackmuss. 1994. Dienelactone hydrolase from Rhodococcus erythropolis 1 CP: purification and properties. Arch. Microbiol. 162:368-374. [Google Scholar]
- 27.Mars, A. E., T. Kasberg, S. R. Kaschabek, M. H. van Agteren, D. B. Janssen, and W. Reineke. 1997. Microbial degradation of chloroaromatics: use of the meta-cleavage pathway for mineralization of chlorobenzene. J. Bacteriol. 179:4530-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moiseeva, O. V., I. P. Solyanikova, S. R. Kaschabek, J. Gröning, M. Thiel, L. A. Golovleva, and M. Schlömmann. 2002. A new modified ortho cleavage pathway of 3-chlorocatechol degradation by Rhodococcus opacus 1CP: genetic and biochemical evidence. J. Bacteriol. 184:5282-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngai, K.-L., M. Schlömann, H.-J. Knackmuss, and L. N. Ornston. 1987. Dienelactone hydrolase from Pseudomonas sp. strain B13. J. Bacteriol. 169:699-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ollis, D. L., E. Cheah, M. Cygler, B. Dijkstra, F. Frolow, S. M. Franken, M. Harel, S. J. Remington, I. Silman, J. Schrag, et al. 1992. The alpha/beta hydrolase fold. Protein Eng. 5:197-211. [DOI] [PubMed] [Google Scholar]
- 31.Pathak, D., K.-L. Ngai, and D. Ollis. 1988. X-ray crystallographic structure of dienelactone hydrolase at 2.8 Å. J. Mol. Biol. 204:435-445. [DOI] [PubMed] [Google Scholar]
- 32.Pelz, O., M. Tesar, R. M. Wittich, E. R. B. Moore, K. N. Timmis, and W. R. Abraham. 1999. Towards elucidation of microbial community metabolic pathways: unravelling the network of carbon sharing in a pollutant-degrading bacterial consortium by immunocapture and isotopic ratio mass spectrometry. Environ. Microbiol. 1:167-174. [DOI] [PubMed] [Google Scholar]
- 33.Pieper, D. H., K. Pollmann, P. Nikodem, B. Gonzalez, and V. Wray. 2002. Monitoring key reactions in degradation of chloroaromatics by in situ H-1 nuclear magnetic resonance: solution structures of metabolites formed from cis-dienelactone. J. Bacteriol. 184:1466-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pieper, D. H., W. Reineke, K.-H. Engesser, and H.-J. Knackmuss. 1988. Metabolism of 2,4-dichlorophenoxyacetic acid, 4-chloro-2-methylphenoxyacetic acid and 2-methylphenoxyacetic acid by Alcaligenes eutrophus JMP 134. Arch. Microbiol. 150:95-102. [Google Scholar]
- 35.Potrawfke, T., J. Armengaud, and R. M. Wittich. 2001. Chlorocatechols substituted at positions 4 and 5 are substrates of the broad-spectrum chlorocatechol 1,2-dioxygenase of Pseudomonas chlororaphis RW71. J. Bacteriol. 183:997-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prucha, M., A. Peterseim, and D. H. Pieper. 1997. Evidence for an isomeric muconolactone isomerase involved in the metabolism of 4-methylmuconolactone by Alcaligenes eutrophus JMP134. Arch. Microbiol. 168:33-38. [DOI] [PubMed] [Google Scholar]
- 37.Prucha, M., A. Peterseim, K. N. Timmis, and D. H. Pieper. 1996. Muconolactone isomerase of the 3-oxoadipate pathway catalyzes dechlorination of 5-chloro-substituted muconolactones. Eur. J. Biochem. 237:350-356. [DOI] [PubMed] [Google Scholar]
- 38.Reineke, W. 1998. Development of hybrid strains for the mineralization of chloroaromatics by patchwork assembly. Annu. Rev. Microbiol. 52:287-331. [DOI] [PubMed] [Google Scholar]
- 39.Reineke, W., and H.-J. Knackmuss. 1988. Microbial degradation of haloaromatics. Annu. Rev. Microbiol. 42:263-287. [DOI] [PubMed] [Google Scholar]
- 40.Reineke, W., and H.-J. Knackmuss. 1984. Microbial metabolism of haloaromatics: isolation and properties of a chlorobenzene-degrading bacterium. Appl. Environ. Microbiol. 47:395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schell, U., S. Helin, T. Kajander, M. Schlömann, and A. Goldman. 1999. Structural basis for the activity of two muconate cycloisomerase variants toward substituted muconates. Protein Struct. Funct. Genet. 34:125-136. [PubMed] [Google Scholar]
- 42.Schlömann, M. 1988. Die verschiedenen Typen der Dienlacton-Hydrolase und ihre Rolle beim bakteriellen Abbau von 4-Fluorbenzoat. Ph.D. thesis. Universität Stuttgart, Stuttgart, Germany.
- 43.Schlömann, M. 1994. Evolution of chlorocatechol catabolic pathways. Conclusions to be drawn from comparisons of lactone hydrolases. Biodegradation 5:301-321. [DOI] [PubMed] [Google Scholar]
- 44.Schlömann, M., P. Fischer, E. Schmidt, and H.-J. Knackmuss. 1990. Enzymatic formation, stability, and spontaneous reactions of 4-fluoromuconolactone, a metabolite of the bacterial degradation of 4-fluorobenzoate. J. Bacteriol. 172:5119-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlömann, M., K.-L. Ngai, L. N. Ornston, and H.-J. Knackmuss. 1993. Dienelactone hydrolase from Pseudomonas cepacia. J. Bacteriol. 175:2994-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlömann, M., E. Schmidt, and H.-J. Knackmuss. 1990. Different types of dienelactone hydrolase in 4-fluorobenzoate-utilizing bacteria. J. Bacteriol. 172:5112-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt, E., and H.-J. Knackmuss. 1980. Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem. J. 192:339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seibert, V. 1997. Evolution des bakteriellen Abbaus von chlorierten Aromaten—molekularbiologische Untersuchungen zur Rekrutierung der Maleylacetat-Reduktasen. Ph.D. thesis. Universität Stuttgart, Stuttgart, Germany.
- 49.Seibert, V., K. Stadler-Fritzsche, and M. Schlömann. 1993. Purification and characterization of maleylacetate reductase from Alcaligenes eutrophus JMP134(pJP4). J. Bacteriol. 175:6745-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skiba, A., V. Hecht, and D. H. Pieper. 2002. Formation of protoanemonin from 2-chloro-cis,cis-muconate by the combined action of muconate cycloisomerase and muconolactone isomerase. J. Bacteriol. 184:5402-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, M. R. 1990. The biodegradation of aromatic hydrocarbons by bacteria. Biodegradation 1:191-206. [DOI] [PubMed] [Google Scholar]
- 52.Solyanikova, L. P., O. V. Maltseva, and L. A. Golovleva. 1995. A modified ortho-cleavage pathway in Pseudomonas putida strain 87: purification and properties of dienelactone hydrolase. Biochemistry (English Translation of Biokhimya) 60:945-951. [Google Scholar]
- 53.Vollmer, M. D., H. Hoier, H. J. Hecht, U. Schell, J. Gröning, A. Goldman, and M. Schlömann. 1998. Substrate specificity of and product formation by muconate cycloisomerases: an analysis of wild-type enzymes and engineered variants. Appl. Environ. Microbiol. 64:3290-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vollmer, M. D., and M. Schlömann. 1995. Conversion of 2-chloro-cis,cis-muconate and its metabolites 2-chloro- and 5-chloromuconolactone by chloromuconate cycloisomerase of pJP4 and pAC27. J. Bacteriol. 177:2938-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vollmer, M. K., P. Fischer, H.-J. Knackmuss, and M. Schlömann. 1994. Inability of muconate cycloisomerases to cause dehalogenation during conversion of 2-chloro-cis,cis-muconate. J. Bacteriol. 176:4366-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wallis, M. G., and S. K. Chapman. 1990. Isolation and partial characterization of an extradiol non-haem iron dioxygenase which preferentially cleaves 3-methylcatechol. Biochem. J. 266:605-609. [PMC free article] [PubMed] [Google Scholar]
- 57.Wittich, R. M., C. Strömpl, E. R. B. Moore, R. Blasco, and K. N. Timmis. 1999. Interaction of Sphingomonas and Pseudomonas strains in the degradation of chlorinated dibenzofurans. J. Ind. Microbiol. Biotechnol. 23:353-358. [DOI] [PubMed] [Google Scholar]