Abstract
Bordetella pertussis, the causative agent of whooping cough, produces a wide array of factors that are associated with its ability to cause disease. The expression and regulation of these virulence factors are dependent upon the bvg locus, which encodes three proteins: BvgA, a 23-kDa cytoplasmic protein; BvgS, a 135-kDa transmembrane protein; and BvgR, a 32-kDa protein. It is hypothesized that BvgS responds to environmental signals and interacts with BvgA, a transcriptional regulator, which upon modification by BvgS binds to specific promoters and activates transcription. An additional class of genes is repressed by the products of the bvg locus. The repression of these genes is dependent upon the third gene, bvgR. Expression of bvgR is dependent upon the function of BvgA and BvgS. This led to the hypothesis that the binding of phosphorylated BvgA to the bvgR promoter activates the expression of bvgR. We undertook an analysis of the transcriptional activation of bvgR expression. We identified the bvgR transcript by Northern blot analysis and identified the start site of transcription by primer extension. We determined that transcriptional activation of the bvgR promoter in an in vitro transcription system requires the addition of phosphorylated BvgA. Additionally, we have identified cis-acting regions that are required for BvgA activation of the bvgR promoter by in vitro footprinting and in vivo deletion and linker scanning analyses. A model of BvgA binding to the bvgR promoter is presented.
Whooping cough is an acute respiratory disease caused by the small, gram-negative bacterium Bordetella pertussis. B. pertussis expresses several factors that contribute to its ability to cause disease (12-15, 23, 30-34, 46). Several of these factors, including filamentous hemagglutinin (FHA), pertactin, fimbriae, and tracheal colonization factor, contribute to the interaction between the bacterium and host cells. Other virulence factors are exotoxins that impair the function of immune cells or are capable of causing damage to host tissues. These factors include pertussis toxin, adenylate cyclase toxin, dermonecrotic toxin, and tracheal cytotoxin. The expression of these virulence factors, with the exception of tracheal cytotoxin, is activated at the level of transcription by a single locus, referred to as the bvg locus (3, 35, 40, 41, 43).
The bvg locus encodes a two-component regulatory system consisting of a sensor protein, BvgS, and a transcriptional activator, BvgA. Although the relevant signals for regulation of the bvg locus in vivo are unknown, the activity of the bvg locus is repressed when cells are grown in the presence of MgSO4 or nicotinic acid or when the cells are grown at reduced temperature in vitro (22). This bvg-mediated change in the patterns of transcription in response to environmental signals is referred to as phenotypic modulation. Under nonmodulating conditions, autophosphorylation of BvgS on a conserved histidine residue is followed by two intramolecular phosphotransfer reactions and the transfer of the phosphate moiety to a conserved aspartate residue on BvgA (42, 44, 45). Upon phosphorylation by BvgS, BvgA binds to cis-acting sequences in the promoter regions of the bvg-activated genes and activates transcription of those genes (1, 6-8, 10, 18, 21, 35, 47).
Several of the bvg-activated virulence operons are regulated in a differential manner. For example, after a shift in temperature from 25°C to 37°C, the fha operon is expressed within 10 min, while the ptx and cya operons are not expressed for several hours (36). Similarly, under steady-state conditions, the expression of the ptx locus is shut off at lower concentrations of MgSO4 than is the expression of the fha locus (39). The fha promoter contains a high-affinity BvgA binding site consisting of nearly perfect inverted heptanucleotide repeats that are centered at position −88.5 relative to the start of transcription (35). Binding of a BvgA dimer to this site, followed by cooperative binding of two additional BvgA dimers 3′ to the high-affinity site, leads to the activation of fha transcription (4, 5, 7-9).
The ptx promoter also contains a site of initial BvgA binding which extends from approximately positions −167 to −123 relative to the start of transcription. Binding of two BvgA dimers to this site, followed by cooperative binding of multiple BvgA dimers downstream, leads to the activation of ptx transcription (8, 25). These observations are consistent with the hypothesis that differential regulation of the fha and ptx promoters is the result of differences in the sensitivity of each promoter to intracellular concentrations of phosphorylated BvgA (36). The requirement for a higher concentration of phosphorylated BvgA at the ptx promoter can be attributed to the lower affinity of the primary binding site of BvgA and the requirement for the cooperative binding of a larger number of BvgA dimers (8, 25). According to this model, when the temperature is shifted to induce bvgAS expression, the concentration of phosphorylated BvgA increases with time and, under steady-state conditions, the concentration of phosphorylated BvgA is inversely related to the MgSO4 concentration (22, 36, 39).
The function(s) of the bvg-repressed genes is unknown. The fact that these genes are repressed by the locus responsible for the regulation of known virulence genes suggests that they may play a role in the pathogenesis of the bacterium, perhaps by contributing to the late stages in the infectious cycle. Alternatively, inappropriate expression of the bvg-repressed genes may interfere with the bacterium's ability to cause disease (28). The regulation of the bvg-repressed genes has been shown to be dependent upon bvgR (27). The bvgR locus lies immediately downstream of bvgAS and is convergently transcribed relative to bvgAS (26). Expression of bvgR was determined to be dependent upon an intact bvgAS locus and shown to be responsive to modulators of BvgA activity (26). Taken together, these results suggest that bvgR expression is activated by the binding of BvgA to the bvgR promoter. In this study we examined the BvgA-regulated expression of bvgR and BvgA binding to the bvgR promoter.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are presented in Table 1. Escherichia coli strains were grown on L-agar or in L-broth supplemented with antibiotics when appropriate (29). B. pertussis strains were grown on Bordet-Gengou (BG) agar (Difco, Detroit, Mich.) containing 1% proteose peptone (Difco) and 15% defibrinated sheep blood. Unless otherwise noted, the concentrations of antibiotics used were: gentamicin sulfate, 10 μg/ml; kanamycin sulfate, 10 μg/ml; and streptomycin sulfate, 100 μg/ml. E. coli strain DH5α was obtained from Bethesda Research Laboratories (Bethesda, Md.).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant features | Source or reference |
|---|---|---|
| E. coli K-12 | ||
| DH5α | High-efficiency transformation | Invitrogen |
| S17 | Tra functions of IncP plasmids integrated into chromosome | 37 |
| B. pertussis | ||
| Tohama I | Patient isolate | 20 |
| BP338 | Tohama I, Nalr Strr | 46 |
| TM1316 | TohamaI, bvgR-phoA Nalr Strr | 26 |
| TM1316::ΔXhoI | TohamaI, bvgR-phoA, 4-kb deletion of bvgR upstream region, Nalr Strr | This study |
| Plasmids | ||
| pSS2809 | Apr GmroriT, B. pertussis chromosomal DNA, E. coli lac operon | 9 |
| pTM025 | pSS2000, bvg sequences 4711-5991 (bvgR) | 27 |
| pTM093 | bvgR-phoA | 27 |
| pTM193 | pBBRKAN with multiple cloning site inserted | 27 |
| pTE-BVGR | pTE103, PbvgR | This study |
Construction of promoter deletion fusions.
Fusions of a nested set of bvgR promoter deletions to the phoA gene on a low-copy-number plasmid were constructed as follows. Oligonucleotide B1 (Table 2) was used in combination with oligonucleotides F1, F2, F3, F4, and F5 in a PCR with plasmid pTM093 as a template. Plasmid pTM093 (27) contains a transcriptional fusion of the gene encoding alkaline phosphatase (phoA) to the bvgR promoter. The products of these PCRs were double-stranded DNA fragments containing phoA fused to truncated bvgR promoters with terminal BglII sites. After digestion with BglII, these fragments were inserted into plasmid pTM193 (26) which had been digested with BglII.
TABLE 2.
Sequences of oligonucleotides used to generate promoter deletion and linker substitution mutants
| Primer | Sequence |
|---|---|
| F1 | 5′-AGATCTCCTGCCGGTCCGCCACATCGAGCAGGGCTGCAAC-3′ |
| F2 | 5′-AGATCTCTGGTGGTTTCGATGCGGCCGCTGAAGGCCGCGG-3′ |
| F3 | 5′-AGATCTGCGACCCCGCATTGATCGGTATCGCCGACCTCGG-3′ |
| F4 | 5′-AGATCTCCGGCCACATGCTGGTCACCGATCTGCTCAACAG-3′ |
| F5 | 5′-AGATCTGTTCATTTCGCCAACGTCAGTACTGCTCCATGAT-3′ |
| F6 | 5′-AGATCTACAGCCGGCTGGCCGCCTTCTAAGAGATGGGCGA-3′ |
| F7 | 5′-AGATCTCGCCTTCTAAGAGATGGGCGAACGCGCCACGCAG-3′ |
| F8 | 5′-AGATCTGAGATGGGCGAACGCGCCACGCAGTACGATTGGG-3′ |
| F9 | 5′-AGATCTCGCAGTACGATTGGGAAAAGTACGGCGGACATGT-3′ |
| F10 | 5′-AGATCTCGGCGGACATGTGAGTTGTATCCTATGGCTTGGAT-3′ |
| F11 | 5′-AGATCTAGTTGTATCCTATGGCTTGGATGGCATCGGTCCA-3′ |
| F12 | 5′-AGATCTGGATGGCATCGGTCCATTTCAATATGACGTTGTT-3′ |
| B1 | 5′-AGATCTGGAATGAATTCATCGCGCTTGAGCGCGGCGCGCA-3′ |
| Fs1 | 5′-GGCCGGCCTGGCTTGGATGGCATCGGTCCATTTCAATATG-3′ |
| Fs2 | 5′-GGCCGGCCAACTCACATGTCCGCCGTACTTTTCCCAATCG-3′ |
| Fs3 | 5′-GGCCGGCCAAAGTACGGCGGACATGTGAGTTGTATCCTAT-3′ |
| Fs4 | 5′-GGCCGGCCGTACTGCGTGGCGCGTTCGCCCATCTCTTAGA-3′ |
| Fs5 | 5′-GGCCGGCCTGGGCGAACGCGCCACGCAGTACGATTGGGAA-3′ |
| Fs6 | 5′-GGCCGGCCAAGGCGGCCAGCCGGCTGTTGAGCAGATCGGT-3′ |
Although the entire sequences of the PCR products were not determined, at least two independent PCRs were performed, and the products from each reaction gave the same result, indicating that differences in activity between the truncated DNA fragments generated by PCR and the full-length wild-type fragment are a result of the deletion of required sequences at the termini rather than results of misincorporation of nucleotides during the extension reactions. The resultant plasmids were transformed into E. coli strain S17 and subsequently transferred into B. pertussis strain BP338 by conjugation.
Fusions of a nested set of bvgR promoter deletions to the lacZ gene on the B. pertussis chromosome were constructed as follows. Oligonucleotide B1 (Table 2) was used in combination with oligonucleotides F1 through F12 in PCRs with plasmid pTM025 (27) as the template. Plasmid pTM025 contains the sequences encoding bvgR and the sequence extending 476 bp upstream of the bvgR open reading frame. The products of these PCRs were double-stranded DNA fragments containing truncated bvgR promoters with terminal BglII sites. After digestion with BglII, these fragments were inserted into plasmid pSS2809 (9) which had been linearized by digestion with BglII. The sequence of each PCR product was confirmed by sequence analysis. The resultant plasmids were transformed into E. coli strain S17 and subsequently transferred into B. pertussis strain BP338 by conjugation.
Use of an algorithm to identify BvgA binding sites.
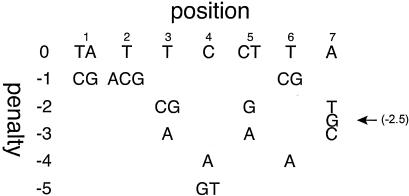
The DNA sequence of the bvgR promoter region was examined for the presence of putative BvgA binding sites through the use of an algorithm derived from the results of a mutational analysis of the high-affinity BvgA primary binding site in the fha promoter. As shown in Fig. 1, for each position in the 7-bp half-site, substitutions were assigned a penalty value depending on their effect on in vitro BvgA binding and/or on fha transcriptional activation in vivo (4). All heptad sequences on both strands of the bvgR promoter region were analyzed and assigned a value. For reference, both heptad sequences in the fha promoter primary binding site have a score of 0, while the most likely primary binding half-sites in the ptx promoter have scores of −4 or −5.
FIG. 1.
Algorithm for the identification of BvgA binding sites. For each position in the consensus half-site, substitutions are assigned a penalty value depending on their effect on in vitro binding and/or in vivo effect on fha transcriptional activation (4).
Construction of linker-substituted promoter fusions.
The linker substitution mutations in the bvgR promoter were generated by a modification of the method of Ho et al. (17). Oligonucleotide F1 was used in combination with oligonucleotides Fs1, Fs3, and Fs5 in PCRs with plasmid pTM025 as the template. The products of these PCRs were double-stranded DNA fragments containing bvgR promoter fragments with a BglII site at the upstream end and an FseI site at the downstream end. These fragments were designated BgFs1, BgFs3, and BgFs5, respectively.
Oligonucleotide B1 was used in combination with oligonucleotides Fs2, Fs4, and Fs6 in PCRs with plasmid pTM025 as the template. The products of these PCRs were double-stranded DNA fragments containing bvgR promoter fragments with a BglII site at the downstream end and an FseI site at the upstream end. These fragments were designated BgFs2, BgFs4, and BgFs6, respectively. After digestion with BglII and FseI, fragments BgFs1 and BgFs2, BgFs3 and BgFs4, and BgFs5 and BgFs6 were combined and inserted into plasmid pSS2809 which had been linearized by digestion with BglII. Each of these three-part ligations resulted in a full-length bvgR promoter fragment containing a precise replacement of 8 bp within the promoter with the 8 bp FseI recognition sequence. The sequence of each linker-substituted promoter was confirmed by sequence analysis. The resultant plasmids were transformed into E. coli strain S17 and subsequently transferred into B. pertussis strain BP338 by conjugation.
Quantitative alkaline phosphatase and β-galactosidase assays.
Bacteria to be assayed were recovered by sterile swab into 3.5 ml of 1 M Tris-HCl, pH 8.0, and the absorbance at 600 nm was measured. For measurement of β-galactosidase activity, 50 μl of cell suspension was added to 1 ml of Z-buffer. Cells were permeabilized by the addition of 30 μl of 0.1% sodium dodecyl sulfate and 30 μl of chloroform, followed by vortexing. The remainder of the assay was performed as described by Miller (29). For measurement of alkaline phosphatase activity, 1 ml of cell suspension was added to 0.5 ml of 1 M Tris-HCl, pH 8.0, the cells were permeabilized as above, and the assay was completed as described by Brickman and Beckwith (16). Units in both cases were calculate by the equation 1,000 × [(A420 − (1.75 × A550)/(T × V × A600)], where T is the incubation time in minutes and V is the volume of permeabilized cells added to the assay in milliliters.
Bacterial conjugations.
Prior to mating, B. pertussis strains were grown for 3 days and E. coli strains were grown overnight at 37°C. Matings between E. coli and B. pertussis strains were performed by swabbing bacteria from fresh plate cultures of each strain onto a BG agar plate supplemented with 10 mM MgCl2. After 3 h of incubation at 37°C, bacteria were swabbed onto BG agar containing the appropriate antibiotics for the selection of exconjugants and incubated at 37°C in screw-top jars.
Northern blot analysis.
Whole-cell RNA was prepared from B. pertussis strain BP338 grown in 1 liter of Stainer-Scholte medium (38) in the absence or presence of 50 mM MgSO4 by the method described by Aiba et al. (2). Total cellular RNA (10 μg per lane) was electrophoresed on a 0.8% agarose gel containing 1.2% formaldehyde as described by Maniatis et al. (24) and transferred to a Nytran membrane (Schleicher and Schuell, Keene, N.H.) as described by the manufacturer. The 1,285-bp XhoI-SalI restriction fragment from bvgR and a 1,158-bp PCR product corresponding to the ompP open reading frame were extracted from agarose gels and labeled with [32P]ATP by nick translation (Lofstrand Laboratories, Gaithersburg, Md.). Labeled DNA fragments were used to probe the RNA on the Nytran membranes with QuikHyb hybridization solution (Stragene, La Jolla, Calif.), in accordance with the manufacturer's directions.
Primer extension analysis.
Whole-cell RNA was prepared from B. pertussis strain BP338 grown in 1 liter of Stainer-Scholte medium (38) in the absence or presence of 50 mM MgSO4 by the method described by Aiba et al. (2). Primer extension analysis was performed with a synthetic oligonucleotide that annealed to the bvgR transcript (5′-GTGCGCCGGGACGGACTGAG-3′). Total RNA (100 μg) and approximately 5 ng of the oligonucleotide were incubated together at 65°C for 20 min, followed by incubation at room temperature for 2 h. PowerScript reverse transcriptase (BD Biosciences, Palo Alto, Calif.) was added, and the mixture was incubated at 42°C for 90 min. The samples were extracted with phenol-chloroform, precipitated with ethanol, resuspended in water, and resolved by electrophoresis on a 6% polyacrylamaide-7 M urea gel. A standard sequencing reaction was performed with the same oligonucleotide primer and pTM025 as the template. The resulting sequence ladder was electrophoresed next to the primer extension products in order to facilitate mapping of the transcription start site.
Footprinting and in vitro transcription.
DNase I footprinting was conducted as described previously (7). DNA probes were obtained as BamHI-SacI fragments from the pBVGR plasmid. This vector was obtained by cloning an EcoRI-PstI fragment containing the putative bvgR promoter (from −186 to + 57) into the pBluescript KS vector digested with EcoRI and PstI. Promoter DNA fragments in which the nontemplate strand was radiolabeled were incubated with purified BvgA in the presence or absence of 20 mM acetylphosphate. In vitro transcription was conducted as described elsewhere (7). The transcription template pTE-BVGR was created by subcloning a BamHI-HindIII fragment into the pTE-103 vector. σ70-saturated E. coli RNA polymerase, when used, was added at 120 nM, while polymerase holoenzyme purified from Bordetella bronchiseptica (7), when used, was added at 700 nM.
RESULTS
Northern blot analysis of bvgR expression.
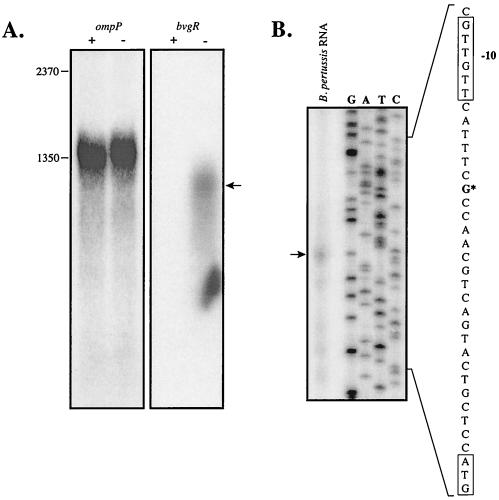
Northern blot analysis (Fig. 2A) demonstrated that an 1,150-base band, corresponding to the bvgR transcript, was present in cells that were grown in the absence of MgSO4. This transcript was undetectable in cells grown in the presence of 50 mM MgSO4. Comparison of the intensities of the bvgR and ompP transcripts indicates that the bvgR transcript is either very poorly expressed or highly unstable. The oligonucleotide probes for bvgR and ompP were labeled with approximately the same specific activity, and the blot probed for bvgR transcript was exposed to film for a time period approximately 10 times longer than that of the blot probed for the ompP transcript. Nevertheless, the bvgR transcript band is much fainter than the porin transcript band, and the bvgR transcript shows clear signs of degradation, as evidenced by the presence of a smear of lower-molecular-weight cross-hybridizing RNAs.
FIG. 2.
Northern blot and primer extension analysis. (A) Total cellular RNA (10 μg per lane), prepared from B. pertussis strain BP338 grown in the absence (−) or presence (+) of 50 mM MgSO4, was electrophoresed on an agarose-formaldehyde gel and transferred to a Nytran membrane. The blots were probed with [32P]ATP-labeled probes specific for the ompP and bvgR transcripts. (B) A 32P-labeled oligonucleotide primer, complementary to the bvgR coding strand, was annealed to 100 μg of total RNA from B. pertussis cells grown on BG agar. The primer was extended with reverse transcriptase, and the products were electrophoresed on a polyacrylamide-urea gel. A DNA sequencing ladder was prepared with the same primer on a recombinant pCR2.1-Topo:bvgR DNA template. The start site of bvgR transcription is indicated with an asterisk.
Identification of the start site of bvgR transcription.
Primer extension analysis of the bvgR transcript was undertaken in order to determine the start site of bvgR transcription (Fig. 2B). RNA was prepared from B. pertussis strain BP338 after growth on BG plates in the presence and absence of MgSO4 and primer extension reactions were performed as described in Materials and Methods. The start site of the bvgR transcript is indicated in Fig. 2B. Two different primer extension experiments, performed on different days and with different RNA preparations, yielded the same start site. It should be noted that detection of the bvgR transcript by this method required exposures for up to 2 weeks.
Determination of the upstream boundary of the bvgR promoter.
Strain TM1316 (26) bears a transcriptional fusion of phoA to bvgR on the chromosome. A chromosomal deletion of the 4-kb XhoI fragment located 476 bp upstream of the start of bvgR transcription was crossed onto the chromosome of strain TM1316 to generate strain TM1316::ΔXhoI. The replacement of the wild-type sequence with the 4-kb deletion of the XhoI fragment was confirmed by Southern blot analysis (data not shown). The expression and regulation of the bvgR-phoA transcriptional fusion were the same in strains TM1316 and TM1316::ΔXhoI, indicating that sequences upstream of the XhoI site at position −476 are not involved in the regulation of bvgR expression (data not shown).
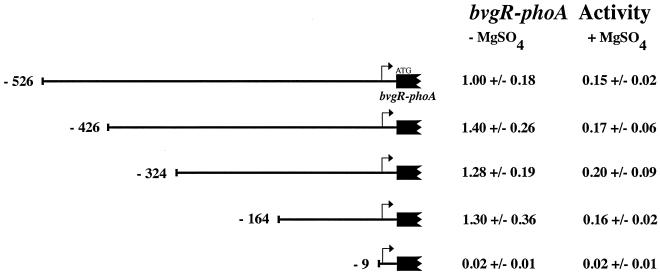
Having determined that the sequences required for the regulated expression of bvgR lie downstream of position −476, we proceeded to generate a nested set of large 5′ deletions of the bvgR promoter fused to phoA. The fusions were transferred into the low-copy-number plasmid pTM193, and the resulting plasmids were transferred into B. pertussis strain BP338 by conjugation. The level of alkaline phosphatase was determined for each plasmid-bearing strain after growth in the absence and presence of 50 mM MgSO4 (Fig. 3). The results demonstrated that the bvgR promoter deleted at the 5′ end as far as position −164 still retained wild-type levels of activity and normal induction. As expected, a deletion that removed all the 5′ sequences up to 9 bp upstream of the start of transcription eliminated expression of bvgR.
FIG. 3.
PbvgR-phoA reporter fusions. The alkaline phosphatase activity of strain BP338 carrying derivatives of plasmid pTM193 with increasing lengths of the bvgR upstream sequence fused to the E. coli phoA gene was determined. The upstream boundary of each promoter deletion, relative to the start of transcription, is indicated. Alkaline phosphatase activities are reported relative to strain BP338, bearing the plasmid with the bvgR sequence extending upstream to the XhoI site (−526) grown in the presence of 50 mM MgSO4 (183 units). All values reported are the averages of at least four independent assays ± the standard deviation.
In vitro transcription of bvgR.
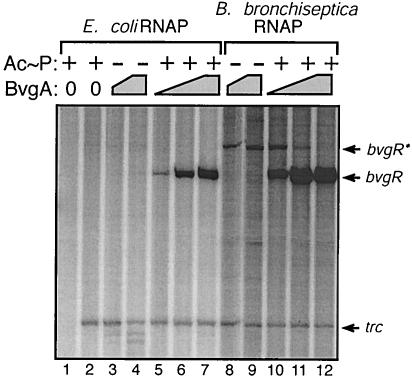
To test the hypothesis that BvgA is directly involved as the transcriptional activator of bvgR, we cloned a 243-bp DNA fragment, containing bvgR-specific sequence from positions −186 to + 57, into plasmid pTE103 (11). The resulting clone, pTE-BVGR, was used for in vitro transcription assays. The presence of phosphorylated BvgA in the transcription reaction mix resulted in transcription from the bvgR promoter, as demonstrated by the presence of bvgR transcript (Fig. 4, lanes 5 to 7 and 10 to 12). No detectable transcript was present with either vector alone (Fig. 4, lane 2) or vector and unphosphorylated BvgA (Fig. 4, lanes 3 to 4 and 8 to 9). These results demonstrate that transcription from the bvgR promoter is dependent upon the presence of phosphorylated BvgA. The addition of increasing levels of phosphorylated BvgA was associated with increasing levels of bvgR transcription when either E. coli RNA polymerase (Fig. 4, lanes 5 to 7) or B. bronchiseptica RNA polymerase (Fig. 4, lanes 10 to 12) was used. Based on the migration of the bvgR transcript relative to an RNA ladder, the location of the transcription start site was determined to be at position −4 relative to the start site determined by primer extension. The error for this analysis is predicted to be ±3 bp
FIG. 4.
In vitro transcription of the bvgR promoter. A single round of transcription was initiated from 20 nM template DNA in reactions containing purified BvgA (lanes 3 to 12) and RNA polymerase (RNAP) holoenzyme purified from either E. coli (lanes 1 to 7) or B. bronchiseptica (lanes 8 to 12). BvgA concentrations were 98 nM (lanes 5 and 10), 195 nM (lanes 3, 6, 8, and 11), and 390 nM (lanes 4, 7, 9, and 12). Additionally, BvgA was incubated either in presence of acetylphosphate (Ac≈P, lanes 1, 2, 5 to 7, and 10 to 12) or in its absence (lanes 3 to 4 and 8 to 9). The expected bvgR transcript, a larger bvgR transcript (bvgR*), and the control trc transcripts are denoted by arrows.
DNase I protection analyses of BvgA binding to the bvgR promoter.
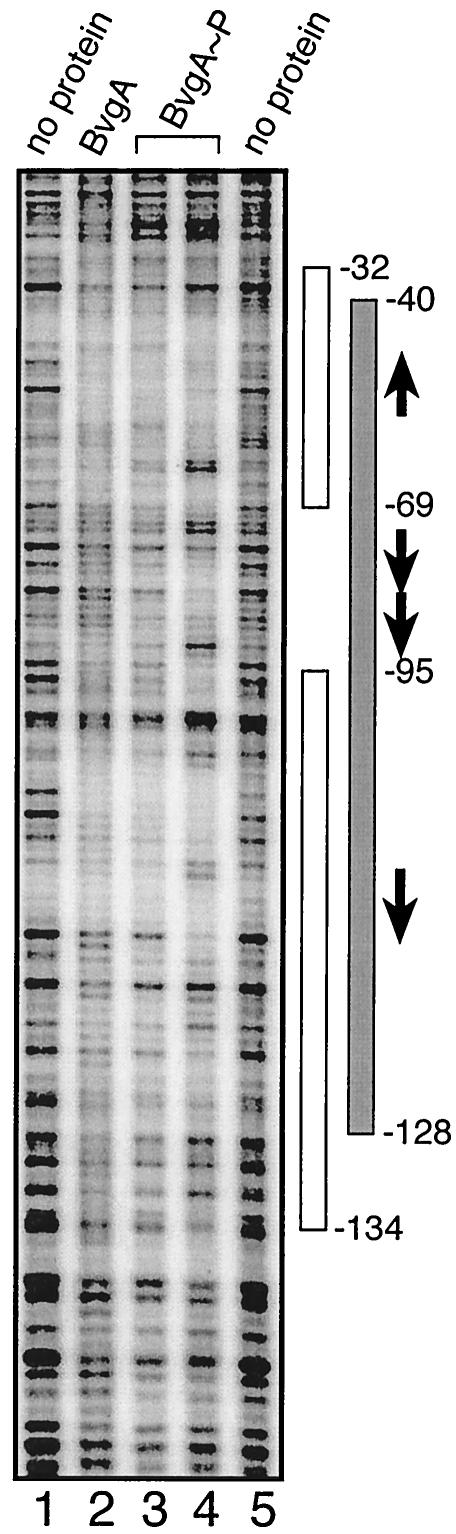
We proceeded to perform DNase I protection analyses in order to determine whether BvgA binds the bvgR promoter, and if so, whether that binding is dependent upon the phosphorylation of BvgA. The DNase I footprinting was performed on the same DNA fragments that were used in the in vitro transcription analysis. Protection of two regions, encompassing nucleotides −32 to −69 and nucleotides −88 to −134, was observed in the presence of unphosphorylated BvgA (Fig. 5, lane 2). In the absence of phosphorylation, BvgA failed to protect the region between these two binding sites. Upon the addition of phosphorylated BvgA, the entire region between positions −40 and −128 was protected, and three prominent hypersensitive sites, at positions −63 and 64, −71 and 72, and −86, and a less prominent hypersensitive site at position −108 were identified (Fig. 5, lanes 3 and 4). Therefore, in the presence of phosphorylated BvgA, the pattern of protection changes to include protection of the region between the two strong BvgA binding sites, and the upstream- and downstreammost boundaries of protection are shifted slightly towards the center of the BvgA binding region.
FIG. 5.
DNase I footprinting of the bvgR promoter. Radiolabeled promoter fragment (0.25 nM) was incubated in the absence of protein (lanes 1 and 5), in the presence of 390 nM unphosphorylated BvgA (lane 2) or 195 nM phosphorylated BvgA (lane 3), and in the presence of 390 nM phosphorylated BvgA (lane 4; BvgA≈P). Regions of protection affected by binding of unphosphorylated BvgA are designated by open rectangles. Protection afforded by BvgA≈P is shown by solid rectangles. Arrows designate putative BvgA binding site heptamers (see text).
Deletion and linker-scanning analysis of the bvgR promoter.
DNase I footprinting of BvgA binding to the bvgR promoter identified three regions of BvgA binding: −32 to −69, −69 to −88, and −88 to −134. Examination of the sequences within these regions, with an algorithm developed by the mutational analysis of the primary BvgA binding site found in the fha promoter, revealed the presence of at least one strong BvgA binding half-site consensus sequence centered within each region (Fig. 1 and 6A). In order to assess the contribution of these BvgA binding sites to the activity of the bvgR promoter, we undertook a deletion and linker-scanning analysis. A nested set of deleted fragments of the bvgR promoter was constructed, and three linker substitutions were independently introduced into the bvgR promoter. In order to determine the activities of these promoter derivatives in vivo, they were inserted into promoter assay vector pSS2809 (9). This vector has a cloning site for promoter fragments upstream of a promoterless lac operon. It also has multiple tandem transcription terminators upstream of the cloning sites to minimize transcriptional readthrough. A 2-kb fragment of B. pertussis genomic DNA enables insertion of the plasmid via homologous recombination into the B. pertussis chromosome at a site distant from the bvg locus.
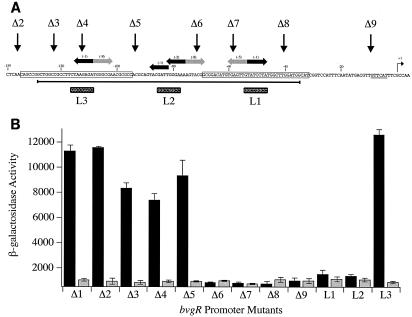
FIG. 6.
Deletion and linker substitution mutations in the bvgR promoter. (A) The nucleotide sequence of the bvgR promoter is shown. The endpoints of each deletion of the bvgR promoter are indicated (Δ2 to Δ9). The endpoint of promoter deletion Δ1 is located at position −526 relative to the start of transcription (not shown in this figure). The sequences replaced with an FseI linker in each of the linker substitution mutations (L1 to L3) are indicated by the black boxes under the sequence. The strong matches to the BvgA binding half-site consensus sequence are indicated by the horizontal black arrows, and the weak matches are indicated by the horizontal gray arrows. The score for each half-site, as determined by the algorithm presented in Fig. 1, is indicated in parentheses above each arrow. The start site of transcription is indicated (+1), and a putative −10 region is underlined. The bases protected by unphosphorylated BvgA are boxed. The dark horizontal line under the sequence identifies the sequences that are protected by phosphorylated BvgA. (B) The β-galactosidase activities (in Miller units) of the bvgR-lacZ fusions, present in single copy on the chromosome, were determined (described in Materials and Methods). The black and gray bars indicate activity in the indicated strains grown in the absence of MgSO4 and in the presence of 50 mM MgSO4, respectively. The values shown are the averages of at least four experiments.
This analysis demonstrated that deletion of all of the sequences upstream of position −135 does not affect the regulated expression from the bvgR promoter (Fig. 6, constructs Δ1 and Δ2). This position corresponds to the upstream boundary of the region protected by unphosphorylated BvgA. Promoter deletions Δ3, Δ4, and Δ5 removed progressively larger fragments of the most promoter-distal BvgA binding site. These truncated promoter constructs had a level of activity that was 65 to 83% of that observed with the full-length promoter fragment (fragment Δ1). Expression from each of these promoter constructs was not observed upon growth in the presence of MgSO4. Deletion of the sequences up to position −71 resulted in a complete loss of bvgR promoter activity, indicating that sequences between −93 and −71 are essential for promoter activity (Fig. 6, constructs Δ6 to Δ9).
Three linker substitutions within the bvgR promoter were constructed. Each substitution replaced eight nucleotides within the identified BvgA binding site consensus sequences with the sequence recognized by the restriction enzyme FseI. The two linker substitutions that affected the BvgA binding sites proximal to the start of transcription eliminated bvgR promoter activity (Fig. 6, constructs L1 and L2). The linker substitution in the predicted BvgA binding site most distal to the start of transcription had no effect on bvgR promoter activity (Fig. 6, construct L3).
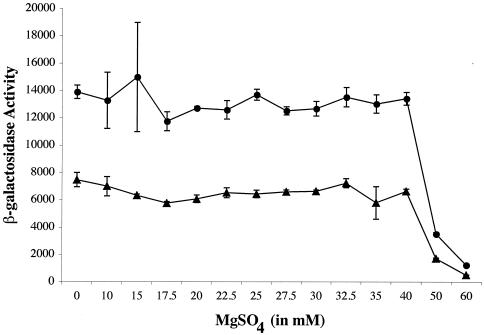
Titration of bvgR promoter activity.
In order to determine the placement of bvgR in the hierarchy of the differentially regulated BvgA-dependent genes, we examined the level of steady-state expression from the bvgR and fha promoters upon growth in the presence of different concentrations of MgSO4 (Fig. 7). The results indicated that the bvgR promoter is induced under the same conditions as the fha promoter. However, the bvgR promoter is significantly stronger than the fha promoter, as demonstrated by the higher levels β-galactosidase activity (Fig. 7). Although both the fha and bvgR promoters were cloned into the same reporter plasmid and mated into the same bacterial strain, the strain bearing the bvgR-lacZ fusion had higher levels of β-galactosidase activity than the strain bearing the fha-lacZ fusion under all of the conditions examined. Both the bvgR-lacZ and fha-lacZ fusion strains were rederived and assayed, and the results were the same for both derivatives.
FIG. 7.
Titration of BvgA activation of the bvgR and fha promoters. Fusions of the bvgR and fha promoters to lacZ on plasmid pSS2809 were crossed onto the chromosome of strain BP338, and the β-galactosidase activity of each strain was determined after growth in the presence of increasing amounts of MgSO4. The absolute activity of the bvgR (circles) and fha (triangles) promoters (in Miller units) observed in cells grown at various concentrations of MgSO4 was determined. Each point represents the average activity measured for three isolates grown at the indicated concentration of MgSO4. The data shown are representative of three independent experiments.
DISCUSSION
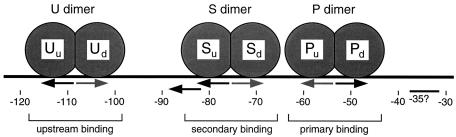
In vitro DNase I protection studies revealed the existence of a region of BvgA binding in the bvgR promoter extending from approximately position −32 to position −134 (Fig. 5 and 6). Significantly, the bases between positions −32 to −69 and between positions −88 to −134 were protected upon the addition of unphosphorylated BvgA. The entire region between positions −35 and −128 was protected when phosphorylated BvgR was added. These results indicate the presence of three BvgA binding sites in the bvgR promoter, located at positions −32 to −69, at positions −69 to −88, and at positions −88 to −134. We have designated these putative BvgA binding sites the P (primary), S (secondary), and U (upstream) BvgA binding sites, respectively (Fig. 8).
FIG. 8.
Model of BvgA binding at the bvgR promoter. u, upstream monomer; d, downstream monomer.
Examination of a number of BvgA-activated promoters resulted in the identification of a BvgA binding site consensus sequence (4, 18, 19, 25, 35). The consensus binding site consists of an inverted repeat of seven nucleotides with the sequence 5′-T/A-T-T-C-C/T-T-A-3′. An algorithm developed through the mutational analysis of the primary BvgA binding site in the fha promoter facilitates the identification of putative BvgA binding sites. The application of this algorithm to the bvgR promoter sequence revealed the presence of matches to the BvgA binding site consensus in each of the three regions protected by BvgA in the DNase I footprint studies. Interestingly, each of these predicted BvgA binding sites consists of a very high scoring half-site in association with a low-scoring half-site. The observation that sites U and P were bound by unphosphorylated BvgA suggests that these sites bind BvgA with high affinity. Binding of site S occurs only in the presence of high levels of phosphorylated BvgA, suggesting that BvgA binds this site with lower affinity.
Deletion analysis of the bvgR promoter demonstrated that all of the sequences required for regulated expression of bvgR were downstream of position −135 (Fig. 6). The three deletions that removed site U, either in part or in its entirety, reduced bvgR promoter activity only slightly. This result indicates that this binding site is not required for the regulated activity of the bvgR promoter but does contribute to promoter strength. The deletions that extended into or removed site S eliminated promoter activity, indicating either that the central binding site is essential for promoter activity or that a minimum of two BvgA binding sites are required to initiate transcription.
Linker substitutions that individually replaced each of the predicted BvgA binding sites demonstrated that both sites P and S are essential for promoter activity (Fig. 6). Replacement of the strong half-site at either of these binding sites completely eliminated promoter activity. The observation that site U does not contribute significantly to promoter activity may reflect an inability of BvgA bound at this site to engage in productive interactions with BvgA bound at sites S and P.
The examination of BvgA binding to the fha promoter indicated that BvgA binds to promoter DNA as a dimer that occupies 22 bp (34) along the DNA helix. On the bvgR promoter, site P is centered at position −53.5, site S is centered at −75.5, and site U is centered at −108.5. Sites P and S are centered 22 bp apart, which would facilitate contact between the BvgA dimers bound at those two sites. Sites S and U are centered 33 bp apart. A BvgA dimer bound at site U would therefore probably not form productive interactions with the BvgA dimer bound at site S. We conclude that the core bvgR promoter is contained within the sequences downstream of position −93 and consists of two BvgA binding sites (sites P and S) and the RNA polymerase binding site.
Binding of BvgA to the promoter-proximal BvgA binding site (site P) does not appear to be sufficient for the activation of transcription from the bvgR promoter. This conclusion is based on the observation that addition of unphosphorylated BvgA was not sufficient to induce transcription from the bvgR promoter in in vitro transcription assays (Fig. 4) and the observation that either deletion of or linker insertion into site S eliminated promoter activity (Fig. 6). The addition of phosphorylated BvgA resulted in occupation of site S and activation of bvgR transcription in vitro, indicating that formation of an active transcription complex at the bvgR promoter may require occupation of both site P and site S by phosphorylated BvgA.
The presence of multiple BvgA binding sites is a striking characteristic shared by all BvgA-activated promoters, including the bvgR promoter. The nature and organization of those binding sites in the bvgR promoter are, however, significantly different from those seen at previously characterized BvgA-activated promoters. For example, the BvgA-activated promoters at the fha, ptx, prn, cya, and bip loci all contain an upstream primary BvgA binding site and one or more secondary binding sites downstream of the primary site (1, 8, 10, 18, 21, 35, 47). We are defining primary binding as binding that does not require cooperative interactions with other BvgA dimers and secondary binding as binding that occurs only in the presence of BvgA dimers bound to adjacent sites on the DNA helix.
The model proposed for BvgA-dependent activation of these promoters proposes that BvgA first binds with high affinity to the primary binding site. Cooperative interactions drive the occupation of the secondary sites by BvgA, ultimately resulting in direct interaction between RNA polymerase and BvgA. According to this model, the relative strengths of the BvgA-activated promoters and the kinetics of their induction are determined primarily by a combination of the affinity of BvgA binding to the primary site and the number of secondary binding sites between the primary BvgA and RNA polymerase binding sites. In contrast to the promoters that have been described previously, the bvgR promoter appears to be characterized by the presence of a primary BvgA binding site adjacent to the RNA polymerase binding site (site P) and a secondary binding site upstream of the primary binding site (site S). Activation of transcription from the bvgR promoter requires occupation of both BvgA binding sites. These observations suggest that binding of a single BvgA dimer adjacent to the RNA polymerase binding site is not sufficient for transcriptional activation by BvgA.
We propose that the formation of an active initiation complex at Bvg-activated promoters requires the binding of two BvgA dimers immediately upstream of the RNA polymerase binding site. Recently, Boucher et al. reported that during the BvgA-dependent binding of RNA polymerase to the fha promoter, the carboxyl-terminal domains of the two RNA polymerase alpha subunits bind promoter sequences occupied by BvgA dimers. This simultaneous binding of the same sites on the DNA helix by BvgA and the alpha subunit is achieved through the binding of opposite faces of the DNA helix (5). It seems likely that the requirement for two BvgA dimers immediately upstream of the RNA polymerase binding site reflects the need to form productive interactions with the carboxyl-terminal domains of both alpha subunits of RNA polymerase.
Both the Northern blot and the primer extension analyses indicate that very low levels of bvgR transcript are present in the cell. This suggests that the bvgR transcript either is expressed at low levels or is very unstable. We addressed the question of bvgR promoter strength and the timing of induction of bvgR transcription by comparing the steady-state levels of bvgR and fha promoter activity when cells were grown on medium with various concentrations of MgSO4 (Fig. 7). This analysis indicated that the bvgR promoter is significantly stronger than the fha promoter. Such a high level of expression is unusual for a transcriptional regulator. Taken together with the results of the Northern blot analysis, these results suggest that the bvgR transcript is very unstable and is rapidly degraded in the cell after synthesis.
We propose that the strength of the bvgR promoter reflects a need to maintain sufficient levels of BvgR protein in the cell to establish and sustain adequate repression of the bvg-repressed genes despite the rapid turnover of bvgR transcripts. The high level of bvgR transcription combined with the rapid degradation of the bvgR transcript would appear to be designed to ensure repression of the bvg-repressed genes under repressing conditions while allowing rapid induction of expression of the bvg-repressed genes upon modulation of BvgAS activity. The comparison of bvgR and fha promoter activities also demonstrated that the timing of induction of the bvgR promoter is the same as that of the fha promoter (Fig. 7). It has been suggested that fha expression is induced early upon activation of BvgAS because of the important role of FHA in the initial colonization of the host. Based on this rationale, it could be argued that early expression of BvgR is required in order to repress the expression of a gene product(s) that interferes with colonization.
Our analysis of the activation of the bvgR promoter by BvgA revealed interesting differences between the bvgR promoter and previously described BvgA-activated promoters. First, the BvgA binding sites in the bvgR promoter are not symmetrical. All three BvgA binding sites on the bvgR promoter consist of a good match to only the consensus half-site. Second, the order of loading of BvgA dimers on the bvgR promoter is different from that observed for other BvgA-activated promoters. At previously characterized BvgA-activated promoters, BvgA first binds to the most upstream site and subsequently occupies the more downstream sites which are adjacent to the RNA polymerase. At the bvgR promoter, BvgA appears to occupy the site adjacent to the RNA polymerase binding site first and subsequently occupies the more upstream binding site.
A more detailed examination of the interactions of BvgA and RNA polymerase at the bvgR promoter and at other BvgA-activated promoters should lead to a better understanding of the mechanism of BvgA-mediated transcriptional activation. The analysis presented here has provided important insights into the dynamics of BvgR expression and will be the basis for future studies of the regulation of the bvg-repressed genes by BvgR.
Acknowledgments
We thank Gopa Raychaudhuri, Michael Schmitt, and Karen Meysick for critical reading of the manuscript.
REFERENCES
- 1.Adler, E., I. Barak, and P. Stragier. 2001. Bacillus subtilis locus encoding a killer protein and its antidote. J. Bacteriol. 183:3574-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256:11905-11910. [PubMed] [Google Scholar]
- 3.Arico, B., J. F. Miller, C. Roy, S. Stibitz, D. Monack, S. Falkow, R. Gross, and R. Rappuoli. 1989. Sequences required for expression of Bordetella pertussis virulence factors share homology with prokaryotic signal transduction proteins. Proc. Natl. Acad. Sci. 86:6671-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher, P., M. S. Yang, and S. Stibitz. 2001. Mutational analysis of the high-affinity BvgA binding site in the fha promoter of Bordetella pertussis. Mol. Microbiol.. 40:991-999. [DOI] [PubMed] [Google Scholar]
- 5.Boucher, P. E., A. E. Maris, M. Yang, and S. Stibitz. 2003. The response regulator BvgA and RNA Polymerase α subunit C-terminal domain bind simultaeously to different faces of the same segment of promoter DNA. Mol. Cell 11:163-173. [DOI] [PubMed] [Google Scholar]
- 6.Boucher, P. E., F. D. Menozzi, and C. Locht. 1994. The modular architecture of bacterial response regulators: insights into the activation mechanism of the BvgA transactivator of Bordetella pertussis. J. Mol. Biol. 241:363-377. [DOI] [PubMed] [Google Scholar]
- 7.Boucher, P. E., K. Murakami, A. Ishihama, and S. Stibitz. 1997. Nature of DNA binding and RNA polymerase interaction of the Bordetella pertussis BvgA transcriptional activator at the fha promoter. J. Bacteriol. 179:1755-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boucher, P. E., and S. Stibitz. 1995. Synergistic binding of RNA polymerase and BvgA phosphate to the pertussis toxin promoter of Bordetella pertussis. J. Bacteriol. 177:6486-6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boucher, P. E., M. S. Yang, D. M. Schmidt, and S. Stibitz. 2001. Genetic and biochemical analyses of BvgA interaction with the secondary binding region of the fha promoter of Bordetella pertussis. J. Bacteriol. 183:536-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deora, R., H. J. Bootsma, J. F. Miller, and P. A. Cotter. 2001. Diversity in the Bordetella virulence regulaon: transcriptional control of a Bvg-intermediate phase gene. Mol. Microbiol. 40:669-683. [DOI] [PubMed] [Google Scholar]
- 11.Elliott, T., and E. P. Geiduscheck. 1984. Defining a bacteriophage T4 late promoter: absence of a “−35” region. Cell. 36:211-219. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez, R. C., and A. A. Weiss. 1994. Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect. Immun. 62:4727-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn, T. M., R. Shahin, and J. J. Mekalanos. 1991. Characterization of vir-activated TnPhoA gene fusions in Bordetella pertussis. Infect. Immun. 59:3273-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn, T. M., and L. A. Stevens. 1995. Tracheal colinization factor: a Bordetella pertussis secreted virulence determinant. Mol. Microbiol. 16:625-634. [DOI] [PubMed] [Google Scholar]
- 15.Hewlett, E. L., V. M. Gordon, J. D. McCaffery, W. M. Sutherland, and M. C. Gray. 1989. Adenylate cyclase toxin from Bordetella pertussis. Identification and purification of the holotoxin molecule. J. Biol. Chem. 264:19379-19384. [PubMed] [Google Scholar]
- 16.Heyduk, T., and J. C. Lee. 1989. Escherichia coli cAMP receptor protein: evidence for three protein conformational states with different promoter binding affinities. Biochemistry 28:6914-6924. [DOI] [PubMed] [Google Scholar]
- 17.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension with the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 18.Karimova, G., J. Bellalou, and A. Ullmann. 1996. Phosphorylation-dependent binding of BvgA to the upstream region of the cyaA gene of Bordetella pertussis. Mol. Microbiol. 20:489-496. [DOI] [PubMed] [Google Scholar]
- 19.Karimova, G., and A. Ullmann. 1997. Characterization of DNA binding sites for the BvgA protein of Bordetella pertussis. J. Bacteriol. 179:3790-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasuga, T., Y. Nakase, K. Ukishima, and K. Takatsu. 1954. Studies on Haemophilus pertussis. V. Relation between the phase of bacilli and the progress of the whooping-cough. Arch. Exp. Med. 27:57-62. [PubMed] [Google Scholar]
- 21.Kinnear, S. M., P. E. Boucher, S. Stibitz, and N. H. Carbonetti. 1999. Analysis of BvgA activation of the pertactin gene promoter in Bordetella pertussis. J. Bacteriol. 181:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacey, B. W. 1960. Antigenic modulation of Bordetella pertussis. J. Hyg. 31:423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livey, I., and A. Wardlow. 1984. Production and properties of Bordetella pertussis heat-labile toxin. J. Med. Microbiol. 17:91-103. [DOI] [PubMed] [Google Scholar]
- 24.Maniatis, T., E. F. Fritsch, and J. E. Sambrook. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Marques, R. R., and N. H. Carbonetti. 1997. Genetic analysis of pertussis toxin promoter activation in Bordetella pertussis. Mol. Microbiol. 24:1215-1224. [DOI] [PubMed] [Google Scholar]
- 26.Merkel, T. J., C. Barros, and S. Stibitz. 1998. Characterization of the bvgR locus of Bordetella pertussis. J. Bacteriol. 180:1682-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merkel, T. J., and S. Stibitz. 1995. Identification of a locus required for the regulation of bvg-repressed genes in Bordetella pertussis. J. Bacteriol. 177:2727-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merkel, T. J., S. Stibitz, J. M. Keith, M. Leef, and R. Shahin. 1998. Contribution of regulation by the bvg locus to respiratory infection of mice by Bordetella pertussis. Infect. Immun. 66:4367-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Mooi, F. R., W. H. Jansen, H. Brunings, H. Gielen, H. G. J. van der Heide, H. C. Walvoort, and P. A. M. Guinee. 1992. Construction and analysis of Bordetella pertussis mutants defective in the production of fimbriae. Microb. Pathag. 12:127-135. [DOI] [PubMed] [Google Scholar]
- 31.Munoz, J. J., H. Arai, R. K. Bergman, and P. L. Sadowski. 1981. Biological activities of crystalline pertussigen from Bordetella pertussis. Infect. Immun. 33:820-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pittman, M. 1979. Pertussis toxin: the cause of the harmful effects and prolonged immunity of whooping cough. A hypothesis. Rev. Infect. Dis. 1:401-412. [DOI] [PubMed] [Google Scholar]
- 33.Relman, D. A., M. Domenighini, E. T. Tuomanen, R. Rappuoli, and S. Falkow. 1989. Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc. Natl. Acad. Sci. 86:2634-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts, M. F. N., N. F. Fairweather, E. Leininger, D. Pickard, Hewlett. E. L., A. Robinson, C. Hayward, G. Dougan, and I. G. Charles. 1991. Construction and characterization of Bordetella pertussis mutants lacking the vir-regulated P. 69 outer membrane protein. Mol. Microbiol. 5:1393-1404. [DOI] [PubMed] [Google Scholar]
- 35.Roy, C. R., and S. Falkow. 1991. Identification of Bordetella pertussis regulatory sequences required for transcriptional activation of the fhaB gene and autoregulation of the bvgAS operon. J. Bacteriol. 173:2385-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scarlato, V., B. Arico, A. Prugnola, and R. Rappuoli. 1991. Sequential activation and environmental regulation of virulence genes in Bordetella pertussis. EMBO J. 10:3971-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/technology 1:784-789. [Google Scholar]
- 38.Stainer, D. W., and M. J. Scholte. 1971. A simple chemically defined medium for the production of phase I Bordetella pertussis. J. gentamicin sulfate. Microbiol. 63:211-220. [DOI] [PubMed] [Google Scholar]
- 39.Stibitz, S. 1998. Mutations affecting the α subunit of Bordetella pertussis RNA polymerase suppress growth inhibition conferred by short C-terminal deletions of the response regulator BvgA. J. Bacteriol. 180:2484-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stibitz, S., W. Aaronson, D. Monack, and S. Falkow. 1989. Phase-variation in Bordetella pertussis by frameshift mutation in a gene for a novel two-component system. Nature 338:226-229. [DOI] [PubMed] [Google Scholar]
- 41.Stibitz, S., and J. F. Miller. 1994. Coordinate Regulation of Virulence in Bordetella pertussis mediated by the vir (bvg) Locus, p. 407-422. In V. L. Miller, J. B. Kaper, D. A. Portney, and R. R. Isberg (ed.), Molecular genetics of bacterial pathogenesis. American Society for Microbiology, Washington, D.C.
- 42.Uhl, M. A., and J. F. Miller. 1994. Autophosphorylation and phosphotransfer in the Bordetella pertussis BvgAS signal transduction cascade. Proc. Natl. Acad. Sci. 91:1163-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uhl, M. A., and J. F. Miller. 1995. Bordetella pertussis BvgAS virulence control system, p. 333-349. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 44.Uhl, M. A., and J. F. Miller. 1996. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component phosphorelay. J. Biol. Chem. 271:176-180. [DOI] [PubMed]
- 45.Uhl, M. A., and J. F. Miller. 1996. Integration of multiple domains in a two-component sensor protein: the Bordetella pertussis BvgAS phosphorelay. EMBO J. 15:1028-1036. [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss, A. A., E. L. Hewlett, A. Meyers, and S. Falkow. 1983. Tn5-induced mutations affecting virulence factors of Bordetella pertussis. Infect. Immun. 42:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zu, T., R. Mannetti, R. Rappouli, and V. Scarlato. 1996. Differential binding of BvgA to two classas of virulence genes of Bordetella pertussis directs promoter sensitivity to RNA polymerase. Mol. Microbiol. 21:557-565. [DOI] [PubMed] [Google Scholar]