Abstract
The diphtheria toxin repressor, DtxR, is a global iron-dependent regulatory protein in Corynebacterium diphtheriae that controls gene expression by binding to 19-bp operator sequences. To further define the DtxR regulon in C. diphtheriae, a DtxR repressor titration assay (DRTA) was developed and used to identify 10 previously unknown DtxR binding sites. Open reading frames downstream from seven of the newly identified DtxR binding sites are predicted to encode proteins associated with iron or heme transport. Electrophoretic mobility shift assays indicated that DtxR was able to bind to DNA fragments carrying the 19-bp operator regions, and transcriptional analysis of putative promoter elements adjacent to the binding site sequences revealed that most of these regions displayed iron- and DtxR-regulated activity. A putative siderophore biosynthesis and transport operon located downstream from one of the DtxR binding sites, designated sid, is similar to the yersiniabactin synthesis and uptake genes encoded on the Yersinia pestis high pathogenicity island. The siderophore biosynthetic genes in the sid operon contained a large deletion in the C. diphtheriae C7 strain, but the sid genes were unaffected in four clinical isolates that are representative of the dominant strains from the recent diphtheria epidemic in the former Soviet Union. Mutations in the siderophore biosynthetic genes in a clinical strain had no effect on siderophore synthesis or growth in low-iron conditions; however, a mutation in one of the putative transport proteins, cdtP, resulted in reduced growth in iron-depleted media, which suggests that this system may have a role in iron uptake. The findings from this study indicate that C. diphtheriae contains at least 18 DtxR binding sites and that DtxR may affect the expression of as many as 40 genes.
Corynebacterium diphtheriae is a gram-positive bacterium and the causative agent of diphtheria, a disease associated with severe infections of the upper respiratory tract and with skin ulcerations (19). The primary virulence determinant of this organism is diphtheria toxin, a potent exotoxin secreted by virulent toxigenic strains of C. diphtheriae (11). In high-iron conditions, toxin production is repressed due to the activity of the diphtheria toxin repressor protein DtxR, a global iron-dependent regulator that controls the iron regulon in C. diphtheriae (9, 15, 45). While DtxR is functionally similar to the ferric uptake repressor Fur, a global iron-dependent regulator in numerous bacterial species (18), the two proteins have relatively little primary sequence identity. However, the recently determined crystal structure of Fur suggests that the two repressors may have structural and mechanistic similarities (33). Homologs of DtxR are present in other bacterial species, including IdeR in Mycobacterium tuberculosis, which controls the iron regulon (37), and MntR in Bacillus subtilis, which regulates genes involved in Mn homeostasis (36).
Eight DtxR binding sites have been identified on the C. diphtheriae chromosome (22, 35, 42, 48, 49, 54). Promoters regulated by DtxR include the promoters upstream of the tox gene and hmuO, which encodes a heme oxygenase involved in acquisition of iron from heme (41, 42), and six promoters that have been designated IRP1, IRP2, IRP3, IRP4, IRP5, and IRP6. Promoters IRP1 to IRP5 and their associated DtxR binding sites were identified by screening promoter fusion libraries for DNA fragments that contained iron- and DtxR-regulated promoter activity (22, 48), while the IRP6 promoter was obtained by a cycle selection method (35). Open reading frames downstream from IRP1 and IRP6 encode homologs of ABC iron transport systems, and it was recently reported that genes downstream from the IRP6 promoter are involved in the uptake of the C. diphtheriae C7 siderophore corynebactin (35). Downstream from IRP3 is a gene that is predicted to encode an AraC-like regulator, while open reading frames linked to the IRP2, IRP4, and IRP5 binding sites have no significant matches to genes of known function.
Because of the iron-depleted environment of mammalian hosts, the acquisition of iron by invading bacterial pathogens is thought to be important for virulence (58). Numerous bacteria, including C. diphtheriae, have evolved mechanisms for acquiring iron from host compounds such as heme and transferrin (21, 29). Many of these iron utilization systems in bacteria are regulated by iron and include cell surface binding proteins and their associated transport factors and high-affinity siderophore uptake systems. The C. diphtheriae siderophore, corynebactin, is involved in the acquisition of iron from transferrin (41), and the synthesis of corynebactin is regulated by DtxR and iron (45, 53). The structure of the siderophore has not been elucidated, and it tests negative in assays used to detect phenolate or hydroxamate type siderophores (38, 53). However, the C. diphtheriae siderophore is routinely detected with the chrome Azurol S assay (35, 45, 53). The genes encoding the enzymes involved in the biosynthesis of the siderophore have not been identified.
The ferric uptake repressor titration assay (FURTA) was initially developed to identify iron-regulated genes in Escherichia coli through the cloning of DNA fragments that carried Fur binding sites (51). The FURTA system was later used in other bacterial species to identify genes that are part of the fur regulon (5, 55). Recent genomic studies, which identified iron-regulated genes in Bacillus subtilis (2), Pseudomonas aeruginosa (28), Neisseria gonorrhoeae (50), Pasteurella multocida (32), and M. tuberculosis (37), revealed that many of these organisms may possess 20 or more Fur binding sites (or IdeR sites for M. tuberculosis) and that the expression of more than 100 genes in some of these species may be affected by iron levels.
In this study, we developed a DtxR repressor titration assay (DRTA) and identified 10 additional DtxR binding sites on the chromosome of C. diphtheriae. Seven of the operons located downstream of DtxR binding sites are predicted to encode factors involved in either iron or heme transport. One of these operons, designated sid, encodes a putative siderophore biosynthetic and transport system that shows similarities to the Yersinia pestis high pathogenicity island (HPI) and was found in only a specific group of Russian clinical isolates and in the PW8 strain. This observation suggests a possible association between the sid operon and strains that are representative of the dominant clonal group identified in the recent diphtheria epidemic in the former Soviet Union.
MATERIALS AND METHODS
Bacterial strains and media.
The E. coli and C. diphtheriae strains used in this study are listed in Table 1. Luria-Bertani (LB) medium was used for culturing of E. coli, and heart infusion broth (Difco, Detroit, Mich.) containing 0.2% Tween 80 (HIBTW) was used for routine growth of C. diphtheriae strains. Bacterial stocks were maintained in 20% glycerol at −70°C. Antibiotics were added to LB medium at 34 μg/ml for chloramphenicol and at 100 μg/ml for ampicillin and to HIBTW for C. diphtheriae cultures at 2 μg/ml for chloramphenicol and 50 μg/ml for kanamycin. HIBTW was made low in iron by the addition of ethylenediamine di(o-hydroxyphenylacetic acid) (EDDA) at the indicated concentrations.
TABLE 1.
Strains used in this study
| Strain | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | Routine cloning and lacZ expression | Invitrogen |
| TOP10 | Cloning PCR products | Invitrogen |
| C. diphtheriae | ||
| C7(−) | Wild type, Mitis biotype, ET?, tox | 3 |
| C7(β)hm723 | dtxR mutant, tox+ | 20 |
| PW8 | Biotype?, ET?, tox+, siderophore mutant | 30 |
| NCTC13129 | Strain used for genome sequence (used only for sequence analysis), Gravis biotype, ET8 complex, tox+ | Sanger Institute |
| Russian isolates | ||
| 1716 | Gravis biotype, ET8 complex, tox+ | 34 |
| 1718 | Gravis biotype, ET8 complex, tox+ | 34 |
| 1737 | Gravis biotype, ET8 complex, tox+ | 34 |
| 1897 | Gravis biotype, ET8 complex, tox+ | 34 |
| 1751 | Mitis biotype, ET175, tox+ | 34 |
| G4193 | Gravis biotype, ET16, tox | 34 |
ET, electrophoretic type; ET is unknown for C7 and PW8, and a biotype for PW8 has not been described. ET8 complex, dominant clonal group associated with the Russian epidemic.
Several of the Russian clinical strains did not grow in mPGT (53), a medium that is used to measure siderophore production in C. diphtheriae. Alterations to mPGT were done that allowed growth of the clinical strains and measurement of the siderophore. This new growth medium, PGTH, contains 80% mPGT medium (without added iron) and 20% HIBTW medium that had been treated with Chelex-100 (5 g of Chelex-100/100 ml for 4 h at room temperature). After Chelex-100 treatment of the HIBTW medium, the following metals were added: MgSO4 and CaCl2 at 1 mM, and MnCl2, CuSO4, and ZnSO4 at 0.1 μM. The presence of Chelex-100-treated HIBTW in the PGTH medium did not interfere with the assay used for the measurement of siderophore production.
Diphtheria toxin repressor titration assay.
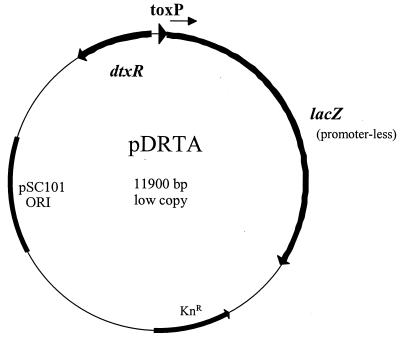
The DRTA system was used to identify DtxR binding sites on cloned DNA fragments, and it is based on the FURTA method that was originally developed to identify Fur binding sites in E. coli (51). The basic concept of the DRTA system is that a DtxR-repressed promoter which is present at low copy number will become activated (or derepressed) in the presence of multiple copies of a second DtxR binding site. The DRTA system was developed in E. coli DH5α carrying plasmids pDRTA and either pKSX65 or pCM2.6 (45). Plasmid pDRTA (Fig. 1), which is derived from the low-copy-number plasmid pWKS130 (Kmr) (57), contains a tox promoter-lacZ fusion, in which the lacZ gene is under transcriptional control of the DtxR-regulated tox promoter. The pDRTA plasmid also encodes the dtxR gene, which is expressed constitutively from its native promoter and is oriented in the opposite direction from that of lacZ (Fig. 1). The pDRTA plasmid was constructed in two steps as follows. A 4.2-kb BglII-ScaI fragment carrying the tox-lacZ fusion region, obtained from plasmid pCMtox (42), was ligated into pWKS130 at the BamHI and EcoRV sites. In the final step, a 1-kb BamHI-BglII fragment carrying the dtxR gene, derived from plasmid pMS297 (45), was ligated into the BamHI site to create pDRTA.
FIG. 1.
Schematic representation of pDRTA. Plasmid pDRTA was used for the DtxR repressor titration assay and is a low-copy-number plasmid derived from pWKS130 (57). pDRTA carries the pSC101 origin of replication, a kanamycin resistance gene (KnR), a tox promoter-lacZ fusion gene cassette, and the dtxR gene.
The second plasmid used for the DRTA system, designated pKSX65, was used for the construction of plasmid libraries and in control studies for the DRTA system. Plasmid pKSX65, a derivative of the high-copy-number vector pBluescript KS (Stratagene, La Jolla, Calif.), was constructed by first excising the 477-bp PvuII fragment from pBluescript KS. A 57-bp XhoI-SpeI fragment, obtained from the pBluescript KS polylinker region, was then ligated into the PvuII site (the 57-bp fragment was made blunt by Klenow prior to ligation). This modification was done to remove the α-complementing lacZ region, so that DH5α harboring pKSX65 would produce white colonies on LB medium with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal).
To demonstrate the feasibility of the DRTA system, DNA fragments encoding several previously identified DtxR binding sites were cloned onto pKSX65 and introduced into DH5α/pDRTA. The sites tested included the DtxR binding sites that overlapped the promoters for IRP1, IRP2, IRP6, tox, and hmuO. DH5α/pDRTA carrying the various binding sites on pKSX65 produced positive results (blue colonies) when plated on LB-X-gal medium. Expression of lacZ (blue colony phenotype) is observed under these conditions, since it is assumed that most of the DtxR protein is sequestered at the newly introduced binding site present on pKSX65 (high copy) and that only minimal amounts of DtxR would be available to repress expression at the tox promoter binding site on plasmid pDRTA (low copy). Positive results in the DRTA were also observed when these DtxR binding sites were tested with plasmid pCM2.6, which replicates at high copy number in E. coli (45). As expected, the vectors alone (pKSX65 and pCM2.6) gave white colonies in DH5α/pDRTA.
The specific procedures for preparing C. diphtheriae chromosomal DNA and the construction of plasmid libraries used for DRTA analysis have been described previously (41). Plasmid libraries containing chromosomal DNA from either C7(−) or from the Russian clinical isolate, 1716, were constructed in pKSX65. One additional plasmid library, which used chromosomal DNA from C7(−), was constructed with the C. diphtheriae shuttle vector pCM2.6. For the construction of libraries in plasmid pKSX65, chromosomal DNA derived from strain C7(−) or 1716 was partially digested with Sau3AI, and DNA fragments in the size range of 0.5 to 3 kb were used for library construction. For libraries constructed in pCM2.6, C7(−) chromosomal DNA was partially digested with MspI, and DNA fragments in the size range of 3 to 7 kb were used for ligation. Plasmid libraries were transformed into DH5α/pDRTA, and transformants were plated onto LB-X-Gal medium and incubated at 37°C for 24 to 48 h.
Plasmid construction and DNA manipulation.
All PCR-generated DNA fragments used in this study were initially cloned into the pCR-Blunt II-Topo vector (Invitrogen), and later excised and moved into the appropriate plasmids. PCR-generated DNA fragments were also used directly for electrophoretic mobility shift assays (EMSAs) or for probes in DNA hybridization studies. The promoter probe vector pCM502 (Cmr) (42), a low-copy-number plasmid that contains a promoterless lacZ gene, was used for the construction and analysis of the lacZ promoter fusions. The PCR primers used to generate the DNA fragments for the promoter fusions are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Clone | Gene designationa | Fragment sizeb (bp) | Sequencec |
|---|---|---|---|---|
| Promoter fusions and EMSA | ||||
| CD6.1-A1 | CD-1 | Zn-dependent alcohol dehydrogenase | 244* | 5′-GGCGTCGACGTTGGATCACTACCAGC-3′ |
| CD6.1-A2 | 5′-GGCGGATCCGGGGAATTCAGTAGCAG-3′ | |||
| CD6.2-A2 | CD-2 | secY | 330* | 5′-GGCGGATCCGCCATGAGTGGCAACTCC-3′ |
| CD6.2-B1 | 5′-GGCGTCGACGCTAAAACCGTGGCTGG-3′ | |||
| CD6.2-C1 | CD-2 | secY | 526 | 5′-GGCGTCGACGCCATGAGTGGCAACTCC-3′ |
| CD6.2-C2 | 5′-GGCGGATCCCCGCATCTCGGAATGCC-3′ | |||
| CD6.3-A1 | CD-3 | Transcriptional regulator of ABC iron transport system | 495 | 5′-GGCGTCGACTTCCTAGAAATAGCAGG-3′ |
| CD6.3-A2 | 5′-GGCGGATCCCCTTCTTAGCGGGAGCC-3′ | |||
| CD6.3-B1 | 243* | 5′-GGCGTCGACCGCTATTGAAGAACTGTG-3′ | ||
| CD6.4-A1 | CD-4 | recA | 569 | 5′-GGCGTCGACGCAGATACTGTACTGCG-3′ |
| CD6.4-A2 | 5′-GGCGGATCCGTTGGTTGCTGCGTCTTAC-3′ | |||
| CD6.4-B1 | 342* | 5′-GGCGTCGACCTGGTTTGATTTGTGCG-3′ | ||
| CD7.4-A1 | CD-7-4 | ywjA | 380* | 5′-GGCGTCGACCGAACATATATGACCGACC-3′ |
| CD7.4-A2 | 5′-GGCGGATCCGATCGCACAGTTGAGTCC-3′ | |||
| CD7.20-A1 | CD-20 | piuB | 150* | 5′-GGCGTCGACCATAACGTTGAAATATAGAC-3′ |
| CD7.20-A2 | 5′-GGCGGATCCCTGCTTCTCGGCGGACG-3′ | |||
| CD7.40-A1 | CD-40 | chtA | 211* | 5′-GGCGTGGACGAGTTGTGGTCTAGGTGG-3′ |
| CD7.40-A2 | 5′-GGCGGATCCCAAAGACGTAAGTGTTGC-3′ | |||
| HtaA-1 | CD-50 | htaA | 319* | 5′-GGCGGATCCGCAGCAGCCATTAGTCC-3′ |
| HtaA-2 | 5′-GGCGTCGACGGGGAGGCTGTGGGCAGG-3′ | |||
| HtaC-1 | CD-50 | htaC | 319 | 5′-GGCGGATCCGGGGAGGCTGTGGGCAGG-3′ |
| HtaC-2 | 5′-GGCGTCGACGCAGCAGCCATTAGTCC-3′ | |||
| FrgA-1 | CD-frg | frgA | 236* | 5′-GGCGGATCCATGATTACAAGGAAAGTG-3′ |
| FrgA-2 | 5′-GGCGTCGACCTTGAGCGTTAGTCGAGG-3′ | |||
| FrgD-1 | CD-frg | frgD | 236 | 5′-GGCGTCGACATGATTACAAGGAAAGTG-3′ |
| FrgD-2 | 5′-GGCGGATCCCTTGAGCGTTAGTCGAGG-3′ | |||
| SidA-1 | CD-sid | sidA | 499* | 5′-GGCGTCGACCGGGGCCGGTGGCGGCG-3′ |
| SidA-2 | 5′-GGCGGATCCGGCGGTGAGAATTTCGG-3′ | |||
| TprT-1 | CD-sid | Transcriptional regulator | 499 | 5′-GGCGGATCCCGGGGCCGGTGGCGGCG-3′ |
| TprT-2 | 5′-GGCGTCGACGGCGGTGAGAATTTCGG-3′ | |||
| Vector integration mutants | ||||
| SidA-C1 | CD-sid | C-terminal sidA mutant | 711 | 5′-GGCGGATCCCGTCCACtACTGAGCAACGAT-3′ |
| SidA-C2 | 5′-GGCGTCGACGGCGAATTCGCATaGTTCCAATTGCGTGCA-3′ | |||
| SidA-2 | CD-sid | N-terminal sidA mutant | 819 | 5′-GGCGTCGACGGCGAATTCGCCATATAgGGCCCCGTCGACCC-3′ |
| SidA-3 | 5′-GGCGGATCCGTCcTAAGTCATCTCGGCTAG-3′ | |||
| SidB-1 | CD-sid | sidB mutant | 810 | 5′-GGCGTCGACGGCGAATTCGAATGCtGATTCCCAGGCATCC-3′ |
| SidB-2 | 5′-GGCGGATCCCACCTaAGCATGTGCGTGCATCGA-3′ | |||
| CdtP-2 | CD-sid | cdtP mutant | 630 | 5′-GGCGTCGACGGCGAATTCCGCTAgATCCACGGACGCTGGCCG-3′ |
| CdtP-3 | 5′-GGCGGATCCGTCCTaACCCACGCGGTTGAACAC-3′ |
The first gene in the operon is specified.
Asterisks indicate PCR product was used in EMSA.
Bold letters identify restriction sites and lowercase letters identify base changes to insert in-frame stop codons.
Mutagenesis techniques.
The procedure for generating site-directed mutations in the C. diphtheriae chromosome by plasmid integration has been described (44). The plasmids used for disruption of the sidA, sidB, and cdtP genes were generated by ligating PCR-derived DNA fragments into the C. diphtheriae shuttle vector pKN2.6 (45). The PCR-derived DNA fragments contain internal portions of the coding region of the targeted gene, and the primers used for PCR had 5′ tail regions that contained relevant restriction sites and also in-frame stop codons to inhibit the formation of fusion proteins that may result after recombination into the chromosome. The primers used for PCR and the sizes of the DNA products that were generated are listed in Table 2. Strains carrying the disrupted gene were characterized by PCR and/or Southern blotting to confirm that the plasmids had integrated into the chromosome at the proper location (not shown).
Electrophoretic mobility shift assays.
The EMSAs were done as described previously (49). PCR-derived DNA fragments containing portions of the various intergenic regions, which included the DtxR binding site, were used for EMSA. The primers used for PCR and the DNA fragment sizes used for EMSA are indicated in Table 2. An approximately 200-bp DNA fragment containing the tox promoter region was derived from plasmid pPOB (49), and the DNA fragment used as a negative control for the DNA binding studies was described previously (43). DNA fragments at 0.5 nM were end labeled at their 3′ termini with [α-32P]dCTP with the Klenow fragment (40). Purified DtxR was prepared as described (47) and used at the indicated concentrations, and CoSO4 was included at 100 μM in all reactions to activate DtxR binding.
LacZ assays.
We inoculated 18-h cultures of C. diphtheriae strains 1:100 into fresh HIBTW medium and grew them for 16 to 18 h at 37°C with shaking. HIBTW medium was made low iron by the addition of 70 μM EDDA. LacZ activity was determined for C. diphtheriae as previously described (46).
Siderophore assays.
Overnight cultures of C. diphtheriae strains grown in HIBTW medium were passaged 1:100 into PGTH medium that contained various supplements as indicated. Cultures were grown for 18 to 20 h, and siderophore measurements were done on 0.5 ml or appropriate dilutions of the culture supernatants with the chrome Azurol S (CAS) assay as described previously (45). Reactions were allowed to develop for 2 h at room temperature, and absorbance at 630 nm was measured. Appropriate dilutions of the culture supernatants were made so that absorbance measurements at 630 nm were between 0.2 and 0.7. Siderophore units were determined as described previously (45) and were normalized for variations in growth. The presence of 1 μM EDDA, which was added to certain cultures, had no effect on the absorbance measurements at 630 nm.
Growth of C. diphtheriae strains in low-iron medium.
Overnight cultures of C. diphtheriae grown in HIBTW were inoculated 1:100 into fresh HIBTW medium containing various concentrations of EDDA as indicated. Cultures were grown for 18 to 20 h with shaking at 37°C, and then optical density measurements at 600 nm were performed.
Computer analysis.
Nucleic acid and amino acid sequences were analyzed with DNA analysis software provided by the Genetics Computer Group Wisconsin package, version 10.3. Amino acid sequence similarity searches were done with the Blast program (1) at the National Center for Biotechnology Information and also with the Blast server provided at the online site for the Sanger Institute (www.sanger.ac.uk/). The unpublished genomic DNA sequences for C. diphtheriae were produced by the C. diphtheriae Sequencing Group at the Sanger Institute and can be obtained from http://www.sanger.ac.uk/Projects/C_diphtheriae.
RESULTS
Diphtheria toxin repressor titration assay.
The DRTA system was used to identify DtxR binding sites on the chromosome of C. diphtheriae (see Materials and Methods). Plasmid libraries used for the DRTA system contained chromosomal DNA obtained from either C7(−) or the clinical strain 1716 (Table 1). The plasmid libraries were transformed into DH5α/pDRTA, and transformants that formed blue colonies on LB medium containing X-Gal were streak purified and subjected to further analysis. The nucleotide sequence was determined for a portion of the cloned insert DNA, and this sequence was used in a Blast search of the C. diphtheriae genome (2.488 Mb), which is available at the Sanger Institute web site (www.sanger.ac.uk/) (the genome sequence has not yet been annotated). Areas of sequence identity were located (designated tagged regions), and the predicted amino acid sequence of open reading frames (ORFs) that flanked the tagged region were used to Blast search the NCBI database to identify homologous genes. Additionally, intergenic sequences located within 7 kb of the tagged regions were analyzed for sequences that were similar to the consensus DtxR binding site.
This analysis identified 10 DRTA clones that contained putative DtxR binding sites that had not been previously reported, and the genetic maps in Fig. 2A and B show the locations of the putative DtxR binding sites relative to nearby ORFs (genes that showed the highest homology in Blast searches to a specific ORF are indicated). All 10 of the newly identified DtxR binding sites described in this study were present in both the C7 strain and 1716. Six of the eight previously known DtxR binding sites, including the sites at tox, hmuO, IRP1, IRP2, IRP3, and IRP4, were identified in this search. Only the binding sites at the IRP5 and IRP6 promoters were not found. Several clones carried duplicates of the 10 newly identified binding sites or of the previously known sites. A detailed summary of the construction of the DRTA libraries as well as the results of the DTRA analysis is presented in Table 3.
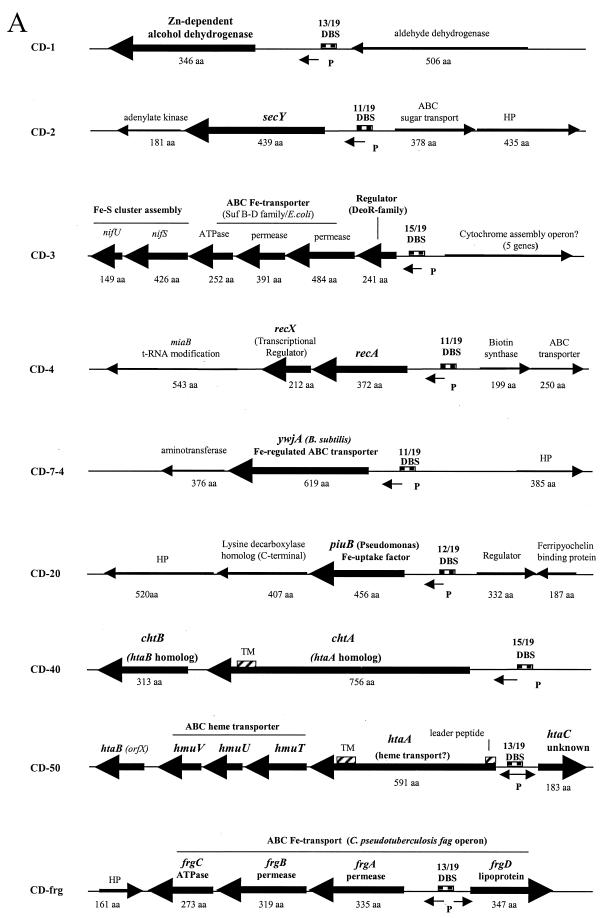
FIG. 2.
(A) Genetic maps showing portions of the C. diphtheriae chromosome that flank the various DtxR binding sites identified by DRTA. The names of the various DRTA clones are indicated to the left of each map. Genes that showed the highest homology to C. diphtheriae ORFs are indicated (obtained from Blast searches of the NCBI database). Arrows indicate direction of transcription, and large arrows indicate ORFs that may be regulated by DtxR and iron. Predicted protein sizes for C. diphtheriae genes are indicated in amino acids (aa). Abbreviations: DBS, DtxR binding sites (matches with the 19-bp consensus sequence are shown above each binding site); P, putative promoter elements; TM, putative transmembrane regions in HtaA and ChtA; HP hypothetical protein. (B) Comparison of the sid operon with the Y. pestis HPI. Triangles indicate locations of vector insertion in the various sid operon mutants. Abbreviations: PKS, polyketide synthase; NRPS, nonribosomal peptide synthetase; FBS, Fur binding site. Genetic maps are not to scale.
TABLE 3.
Summary of library construction and DRTA results
| Chromosomal DNA sourcea | Enzymeb | Vectorc | Insert size (kb) | CFU screenedd | DRTA-positive clonese | Clones identifiedf (no.) |
|---|---|---|---|---|---|---|
| C7(−) | Sau3AI | pKSX65 | 0.5-3 | 10,000 | 22 | CD-1 (4), CD-2 (4), CD-3, CD-4, CD-sid (2), IRP3, IRP1 (2), NH (2), NF (5) |
| MspI | pCM2.6 | 3-7 | 3,500 | 13 | CD-7-4 (2), CD-40, CD-frg, IRP1 (4), IRP2 (2), NH (2), NF (1) | |
| 1716 | Sau3AI | pKSX65 | 0.5-3 | 10,000 | 23 | CD-1, CD-3 (3), CD-7-4, CD-20, CD-40 (4), CD-50, CD-sid, IRP4, hmuO, tox (4), NF (5) |
Chromosomal DNA from the indicated C. diphtheriae strain was used for library construction.
Restriction endonuclease used in the construction of the library.
Plasmid vector used for library construction.
Total number of CFU that were screened for the library.
Total number of DRTA-positive clones (blue colonies) that were identified in the library.
Numbers in parentheses indicate the number of additional or duplicate clones that were identified. NH, DNA sequence of the insert had no (or minimal) homology to the genome sequence of C. diphtheriae strain NCTC 13129 or to any sequences in GenBank. NF, a DtxR binding site was not found within 7 kb of the tagged sequence on these DRTA-positive clones.
Electrophoretic mobility shift assays.
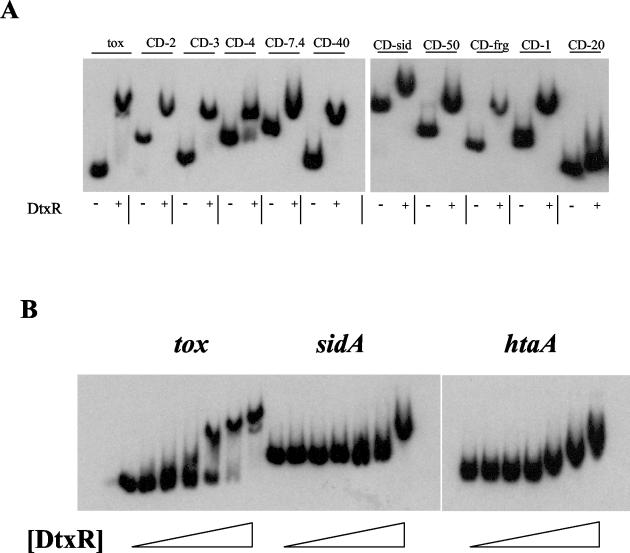
To determine if DtxR was able to bind to the putative binding sites found on the DRTA clones, EMSA studies were done with DNA fragments carrying the 10 putative DtxR binding sites. A DNA fragment carrying the tox promoter was used as a positive control. The electrophoretic mobility of all of the DNA fragments was significantly reduced in the presence of purified DtxR, with the exception of the DNA fragment carried by clone CD-20 (Fig. 3A). This observation suggests that DtxR is able to bind to all of the DtxR binding sites with the possible exception of CD-20, which was only marginally affected by the presence of DtxR. DtxR did not affect the mobility of a DNA fragment used as a negative control (not shown). The DtxR binding sites at the sidA, htaA, and tox promoters were analyzed further in EMSA studies with a range of DtxR concentrations (Fig. 3B). EMSA studies with DNA fragments carrying the sidA and htaA binding sites showed that the htaA binding site was shifted at lower concentrations of DtxR than the sidA binding site, which suggests that the htaA binding site may have a slightly higher affinity for DtxR than the sidA site. The DNA fragment carrying the tox promoter region was shifted at DtxR concentrations below levels required to alter the mobility of either htaA or sidA, which suggests that DtxR binds more strongly to the tox binding site than to the binding sites at sidA and htaA.
FIG. 3.
(A) EMSA analysis with DtxR and DNA fragments carrying DtxR binding sites identified on the 10 DRTA clones and at the tox promoter region. DtxR was present at 2 μM (+) or absent (−). (B) EMSA analysis with a range of DtxR concentrations with the DtxR binding sites sequences at the tox, htaA, and sidA promoter regions. DtxR was present in the reactions (from left to right) at 0, 5 nM, 25 nM, 50 nM, 250 nM, 500 nM, and 2 μM.
An alignment of the 18 known DtxR binding sites, which includes the 10 additional sites identified in this study, is shown in Fig. 4. The sequence of the 19-bp DtxR consensus binding site, which was originally derived from the eight sites that were identified from previous studies (the top eight sites shown in Fig. 4), was not altered by the inclusion of the 10 new sites described in this report (Fig. 4). However, some notable differences were observed: T residues at positions −9 and −8 were present in all 10 of the new sites, whereas T residues at these positions showed some variability in the original eight binding sites: position −8 (5 of 8) and position −9 (6 of 8). Additionally, the consensus C residue at position 0 was found in six out of the eight original binding sites but was present in only two of the 10 new binding sites. Overall, positions that showed the highest sequence variability in the eight original binding sites also showed variability in the 10 new sites, and similarly, the most highly conserved residues in the original eight sites are among the most highly conserved residues in an alignment of all 18 DtxR binding sites (Fig. 4).
FIG. 4.
Alignment of the 18 known DtxR binding sites. The top eight sites were reported previously, and the lower 10 sites were identified in this study. The 19-bp DtxR consensus sequence (35) is indicated below the alignment, and the mostly highly conserved residues are indicated in bold. Matches to the consensus sequence for each position are indicated.
LacZ assays.
Promoter-lacZ fusions were constructed to determine whether the newly identified DtxR binding sites affected promoter activity. DNA fragments carrying the DtxR binding sites and flanking sequences within the various intergenic regions were ligated upstream of the promoterless lacZ gene on pCM502. Any promoter activity in the intergenic regions would drive expression of the lacZ gene on plasmid pCM502. Promoter fusion constructs were placed into C7(−) and into C7βhm723, a dtxR mutant of C7 (20), and β-galactosidase activity was measured after growth in high- and low-iron media (Table 4). Nine of the 12 promoter fusion constructs were shown to have transcriptional activity that was repressed in high-iron medium and regulated by DtxR. Two clones, CD-50 and CD-frg, were shown to have promoter activity that was regulated by iron and DtxR in both orientations, which indicates that a single DtxR binding site is able to control divergently transcribed promoters (Table 4 and Fig. 2A). The nine promoter constructs that were regulated by DtxR and iron showed great variability in the level of expression and in the amount of iron-dependent repression. Promoters located on clones CD-3, CD-4, and CD7-4 were not regulated by DtxR or iron (Table 4), although the DtxR binding site on each of these clones was able to bind DtxR in the EMSA experiments (Fig. 3A). None of the promoter fusions were regulated by iron and DtxR as strongly as the tox promoter (Table 4).
TABLE 4.
Transcriptional analysis of promoter-lacZ fusions in C. diphtheriae C7(−) and C7(β)hm723 (dtxR) in high- and low-iron conditions
| Clone/genea | β-Galactosidase activity, unitsb (SD)
|
|||||
|---|---|---|---|---|---|---|
| C7(−) (dtxR+)
|
C7(β)hm723 (dtxR)
|
|||||
| +Fe | −Fe | Ratio, −Fe/+Fe | +Fe | −Fe | Ratio, −Fe/+Fe | |
| CD-1/Zn-alcohol dehydrogenase | 16.1 (2.6) | 34.8 (3.6) | 2.2 | 63.1 (7.8) | 69.5 (2.5) | 1.1 |
| CD-2/secY | 44.8 (4.6) | 61.1 (3.1) | 1.4 | 58.5 (2.0) | 54.6 (0.5) | 1.1 |
| CD-3/deoR | 12.2 (1.4) | 12.5 (1.3) | 1.0 | 7.6 (1.4) | 6.3 (1.7) | 1.0 |
| CD-4/recA | 23.7 (0.7) | 22.3 (0.9) | 1.0 | 32.5 (1.3) | 34.5 (4.8) | 1.0 |
| CD-7-4/ywjA | 1.7 (0.2) | 1.6 (0.3) | 1.0 | 1.7 (0.1) | 1.7 (0.1) | 1.0 |
| CD-20/piuB | 2.5 (0.4) | 10.5 (0.8) | 4.2 | 6.6 (1.8) | 7.3 (2.2) | 1.1 |
| CD-40/chtA | 8.1 (2.2) | 268.4 (4.2) | 33.5 | 572.4 (8.1) | 582.0 (25.4) | 1.0 |
| CD-50/htaA | 4.6 (0.5) | 26.2 (2.2) | 5.7 | 31.1 (1.7) | 33.2 (2.3) | 1.0 |
| CD-50/htaC | 25.4 (1.3) | 71.6 (4.5) | 2.8 | 103.5 (5.7) | 100.4 (4.0) | 1.0 |
| CD-frg/frgA | 44.2 (4.0) | 76.7 (1.6) | 1.7 | 95.9 (3.2) | 63.8 (10.8) | 0.7 |
| CD-frg/frgD | 89.8 (4.9) | 299.0 (3.8) | 3.3 | 302.1 (23.1) | 329.6 (18.2) | 1.1 |
| CD-sid/sidA | 1.7 (0.4) | 26.1 (1.1) | 15.4 | 8.8 (1.8) | 21.5 (4.1) | 2.4 |
| tox | 0.4 (0.1) | 98.7 (7.6) | 247.0 | 109.4 (10.2) | 114.8 (4.5) | 1.0 |
The first gene in the operon is indicated.
β-Galactosidase units were determined as described previously (46). Values are the mean of three experiments ± standard deviation. +Fe is HIBTW medium, and −Fe is HIBTW with 70 μM EDDA.
Analysis of the C. diphtheriae sid genes.
Downstream from the novel DtxR binding site on clone CD-sid was an operon, which we have designated sid, that shows homology to genes involved in siderophore biosynthesis and transport (Fig. 2B). The sid operon has some similarities to the Yersinia HPI, a mobile genetic element required for virulence that encodes the yersiniabactin high-affinity siderophore transport system (6, 10, 12, 16). The sid operon in C. diphtheriae contains two genes associated with siderophore biosynthesis, sidA and sidB, which are homologous to the Yersinia pestis irp2 and irp1 genes, respectively. Also contained in the sid operon are two genes predicted to encode factors involved in siderophore transport, cdtP and cdtQ, the deduced products of which have homology to the yersiniabactin transport proteins YbtP (33% identity) and YbtQ (30% identity), respectively. The predicted products of the cdtP and cdtQ genes both contain a C-terminal region that has homology to ATPases found in ABC-type metal transporters. The N-terminal regions of these proteins contain putative transmembrane regions characteristic of membrane permeases. Similar structural characteristics are also observed in the Y. pestis YbtP and YbtQ proteins (16).
A comparison of the C. diphtheriae sid operon to the Y. pestis HPI also reveals numerous differences in the genetic organization and in the number of genes between the two systems (Fig. 2B). The Yersinia HPI contains genes for an outer membrane receptor, psn, transcriptional regulator, ybtA, and five additional genes, not present in the sid operon, that are either associated with siderophore biosynthesis or have no known function (10). Both genetic systems are adjacent to phage genes and an insertion sequence element; however the HPI system is also flanked by a phage integrase gene (int) and a tRNA gene, elements that are thought to have a role in the mobility of this region in Yersinia species (10).
DNA hybridization studies of the sid operon.
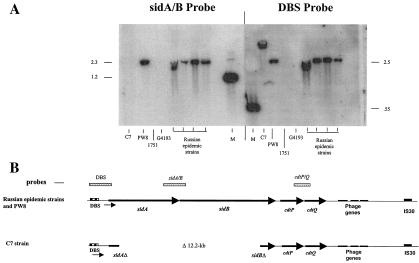
DNA probes specific to various regions of the sid operon were used to hybridize with chromosomal DNA derived from several different C. diphtheriae strains. The results from the hybridization studies revealed that a probe specific for the sidA and sidB genes (sidA/B probe) failed to hybridize with chromosomal DNA from the C7 strain, but the sidA/B probe was able to hybridize with chromosomal DNA from four closely related clinical isolates obtained from the recent Russian diphtheria epidemic (Russian epidemic strains 1716, 1718, 1737, and 1897) and with DNA from the PW8 strain (Fig. 5A). The hybridization results indicate that the C7 strain contained a deletion within the sidA and sidB genes, and this finding was confirmed by PCR analysis of the sid operon (data not shown). Additionally, sequence analysis of clone CD-sid, which is derived from a C7 chromosomal library, determined that the deletion in C7 is approximately 12.2 kb and begins 235 bp from the 5′ end of sidA and extends to 635 bp from the 3′ end of sidB (Fig. 5B). Chromosomal DNA from C7, PW8, and the Russian epidemic strains hybridized with probes carrying the sidA DtxR binding site (DBS probe) (Fig. 5A). The Russian clinical strains G4193 and 1751, which are not members of the dominant clonal group associated with the diphtheria epidemic (see Table 1), contained deletions of sidA, sidB, and the sidA DtxR binding site (Fig. 5A). Chromosomal DNA from all of the strains hybridized with a probe that carried the cdtP and cdtQ region (data not shown).
FIG. 5.
(A) Chromosomal DNA from C. diphtheriae strains was digested with BamHI (sidA/B probe) or HindIII (DBS probe) and hybridized with 32P-labeled DNA fragments carrying portions of the sid operon. Hybridization experiments were done as described previously (40). Size markers are indicated in kilobases. Lane M, size markers. (B) Comparison of the sid operon between the Russian epidemic strains (and PW8) and the C7 strain. Probes used in the hybridization experiments are indicated.
Mutational analysis of the sid operon.
Site-specific mutations were constructed in the chromosomal sidA and sidB genes in the Russian clinical strain 1737 and also in the cdtP gene in both strains C7(−) and 1737. A vector integration method that has been reported previously (44) was used to construct the mutations in the C. diphtheriae strains (see Materials and Methods and Fig. 2B). Two separate mutations were constructed in the 1737 sidA gene; one mutation was placed into the N-terminal region (45 bp from the 5′ end of sidA, 1737/sidAN), and a second mutation was constructed in the C-terminal region (576 bp from the 3′ end of sidA, 1737/sidAC). No difference in siderophore production or regulation was observed between the 1737 sidAN mutant and the wild-type 1737 strain (Table 5).
TABLE 5.
Siderophore production by C. diphtheriae strains
| Supplement to PGTH medium | Siderophore unitsa (SD)
|
||
|---|---|---|---|
| C7 | 1737 | 1737/sidAN | |
| Fe (10 μM) | 6.6 (2.2) | 1.4 (0.5) | 1.9 (1.2) |
| None | 65.5 (3.7) | 5.5 (1.7) | 4.6 (2.3) |
| EDDA (1 μM) | 58.7 (8.7) | 33.0 (2.6) | 29.7 (5.7) |
Siderophore units were determined as described previously (45). Values are the mean of three experiments ± standard deviation.
Additionally, wild-type levels of siderophore were produced in the 1737 sidB mutant and in 1737 sidAC (data not shown). The four Russian epidemic strains, which include 1737, all produced equivalent levels of siderophore and showed similar iron-dependent regulation of siderophore production (not shown). Mutations in the 1737 sidA and sidB genes did not affect the growth of these mutants in low-iron medium relative to the growth of the wild-type 1737 strain (data not shown). As shown in Table 5, the C7 strain produced higher levels of siderophore than the 1737 strain under all conditions examined. PGTH medium is a low-iron medium that was developed to measure siderophore production in the Russian clinical strains (see Materials and Methods). The C7 strain produced maximal levels of siderophore in the absence of added EDDA, whereas maximum siderophore production in 1737 (and also in the three other epidemic strains) was detected only after the addition of 1 μM EDDA, an iron chelator, to the PGTH medium (Table 5 and not shown). This finding suggests that the clinical strains require lower-iron conditions than C7 to produce optimal levels of siderophore. Higher levels of EDDA in the PGTH medium, which would result in lower-iron conditions, did not cause any further increase in siderophore production in any C. diphtheriae strain (data not shown). The C7 strain and all of the Russian epidemic strains gave negative results when tested for the production of phenolate or hydroxamate siderophores (data not shown).
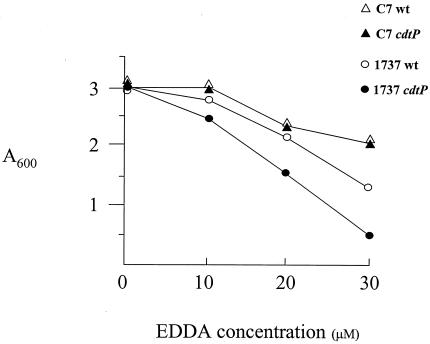
Mutations in the C7 cdtP gene had no effect on growth in low-iron medium relative to the wild-type C7 strain (Fig. 6). However, the identical mutation in the 1737 cdtP gene resulted in reduced growth in low-iron medium: a difference of approximately twofold in cell density was observed between the mutant and wild-type strains at 30 μM EDDA (Fig. 6). This observation suggests that the 1737 cdtP gene may have a role in iron transport; however, the absence of an observable phenotype with mutations in the 1737 sidA and sidB genes suggests that the growth defect in the 1737 cdtP mutant may not be associated with the transport of the putative siderophore encoded in the sid operon. It has not been determined what other iron sources might be utilized by the CdtP permease. The 1737 cdtP mutant and the wild-type 1737 strain produced similar levels of siderophore, as determined by the CAS assay (data not shown).
FIG. 6.
Growth analysis of C. diphtheriae strains in HIBTW medium containing various EDDA concentrations. C. diphtheriae strains C7 and 1737 and their isogenic cdtP mutants were grown for 18 to 20 h in HIBTW medium that contained the iron chelator EDDA at the concentrations indicated. Culture densities were measured at an absorbance of 600 nm. The values shown are the means of three independent experiments, and the results for each data point varied from the mean by less than 10%.
DISCUSSION
In this article, we describe a DtxR repressor titration system, DRTA, which was used to identify 10 previously unknown DtxR binding sites on the chromosome of C. diphtheriae. Although six of the previously known DtxR binding sites were identified in the DRTA system, two previously characterized sites, IRP5 and IRP6, were not found. The inability of the DRTA system to identify all known sites suggests that the chromosomal libraries used in this study were not representative of the complete C. diphtheriae genome and that additional novel binding sites may be present on the C. diphtheriae chromosome. This possibility is supported by the observation that a test clone which carried the IRP6 binding site gave a positive result when examined in the DRTA system (see Materials and Methods). This finding suggests that the failure to detect IRP6 in DRTA was not due to an inability to detect a positive result for clones carrying the IRP6 binding site, but to the absence (or very low level) of clones encoding the IRP6 binding site in these libraries. The use of different restriction enzymes to construct future plasmid libraries may provide more extensive coverage of the C. diphtheriae chromosome and allow the identification of additional DtxR binding sites.
Four of the DRTA-positive clones derived from the C7(−) libraries contained tagged sequences that had no homology to the genome sequence from C. diphtheriae strain NCTC 12139 (Table 3, NH) and only limited homology to sequences in GenBank. No DtxR binding sites were identified on these four clones, and it appears that these sequences are unique to the C7 strain. A previous report had noted that the C7-encoded IRP1 and IRP2 promoters and their associated downstream genes are not present in the genome of strain NCTC 12139 (35), and in this report we show that the sidA and sidB genes are deleted in the C7 strain. These findings suggest that numerous differences may exist in the genome sequences between the C7 strain and the Russian clinical strains.
Eleven additional DRTA-positive clones (designated NF, Table 3) in which no DtxR binding site was found but sequences homologous to the tagged region were present in the genome were identified. Seven of these clones contain genes that are predicted to encode proteins involved in iron binding: ferredoxins (two clones) or cytochromes (five clones). The overproduction of iron-binding proteins was previously reported to provide a positive signal in FURTA (51). Of the four remaining clones, no binding sites were found, nor were any ORFs identified that are predicted to encode products involved in iron binding or iron metabolism. It is possible that these clones as well as the sequences on the four clones described above that are unique to the C7 strain, designated NH, may contain DtxR binding sites that exhibit weak homology to the consensus sequence.
Seven of the 10 DtxR binding sites found in this study were adjacent to ORFs that have homology to genes involved in either heme or iron transport. While the function of most of these genetic systems is not known, one of the unexpected findings from the DRTA analysis was the identification of a DtxR binding site upstream of C. diphtheriae htaA, the promoter-proximal gene in an operon that also contains an ABC heme transport system encoded by the hmuTUV genes (14, 44). Earlier studies that examined gene expression in C. diphtheriae and in Corynebacterium ulcerans showed that the hmuTUV genes were transcribed at low constitutive levels and there was no evidence for iron regulation (44). The function of HtaA in C. diphtheriae is not known; however, an htaA mutant of C. ulcerans exhibits reduced ability to utilize heme as an iron source, suggesting that htaA may have a role in heme transport (M. P. Schmitt, unpublished observation).
While the genetic organization of the heme transport operons of C. diphtheriae and C. ulcerans is similar, a striking difference was observed between the predicted products for the htaA genes in these two closely related species. The HtaA protein from C. diphtheriae contains an additional 181 amino acids at its N terminus, and unlike the C. ulcerans protein, HtaA in C. diphtheriae also harbors a putative leader peptide that may be recognized by a type I signal peptidase. Both HtaA proteins are predominantly hydrophilic and contain a possible transmembrane region in their C terminus, which may function to anchor the proteins to the cell membrane.
The frg genes (CD-frg) have homology to the Corynebacterium pseudotuberculosis fag operon, an ABC metal transporter that is iron regulated and required for virulence (7). Mutations in the C. pseudotuberculosis fag genes had no effect on iron transport, and the function of the frg genes in C. diphtheriae is not known. The operon found downstream of the binding site on clone CD-3 contained several genes, including those with homology to ABC iron transporters and also to genes involved in the assembly of iron-sulfur clusters (Fig. 2A). The association between the ABC-type iron transport system on CD-3 and iron-sulfur cluster assembly is not known; however, a similar Fur-regulated operon exists in E. coli (31) and in the plant pathogen Erwinia chrysanthemi, where it has been proposed that the ABC transporter encoded by sufB, sufC, and sufD is only indirectly involved in iron transport and that its primary function may be in providing energy for the repair of iron-sulfur clusters that are damaged under oxidative stress conditions (25).
A gene downstream from the CD-7-4 binding site showed the highest homology to the iron-regulated ywjA gene from Bacillus subtilis, which is predicted to encode a protein that is similar to ABC transporters (2). Downstream from the CD-20 binding site is an open reading frame that is predicted to encode a protein homologous to an uncharacterized iron uptake factor, PiuB (accession number AF051690), which was identified by a cycle selection method in a search for Fur- and iron-regulated genes in Pseudomonas aeruginosa (27). An open reading frame designated chtA is downstream from the CD-40 DtxR binding site, and this gene is predicted to encode a protein that shows homology to the heme transport-associated protein HtaA (clone CD-50). HtaA and ChtA have homology over an approximately 200-amino-acid region in their N termini. ChtA, like HtaA, is predicted to be predominantly hydrophilic and to contain a transmembrane segment in its C-terminal region. These structural characteristics are shared by the Streptococcus pyogenes Shr (or Shp) protein, which is able to bind heme and various heme proteins (4, 23). The chtA gene was strongly regulated by DtxR and iron and contained the most highly expressed promoter of all those examined in this study (Table 4). Downstream from chtA is a gene designated chtB, which shows homology to the htaB gene (previously orfX), a gene of unknown function that is associated with the hmuTUV heme transport operon (14).
Downstream from the DtxR binding sites on CD-1, CD-2, and CD-4 are open reading frames whose predicted products include a Zn-dependent alcohol dehydrogenase, SecY, and RecA, respectively (Fig. 2A). These factors are not typically associated with iron metabolism and would not necessarily be expected to be iron regulated. However, some recent reports suggest that iron levels may affect the expression of these genes. A Fur binding site was found upstream of the secY gene in Neisseria gonorrhoeae, which suggests that secY may be Fur and iron regulated (50). While expression of recA is not known to be directly affected by iron or Fur, recombination functions are regulated by iron in some bacteria (50). Moreover, recent genomic studies have found that iron and Fur regulate the expression of genes encoding certain dehydrogenase enzymes, although alcohol dehydrogenases have not yet been specifically shown to be iron regulated (28, 32).
Transcriptional analysis of the promoter regions in clones CD-3, CD-4, and CD7-4 indicated that they were not regulated by iron in C. diphtheriae, and it is unclear what role if any the DtxR binding site located in this region has in the regulation of these genes. While the presence of a DtxR binding site upstream of these genes may have occurred by chance, it seems more likely either that these genes are regulated by a mechanism that may be more complex than direct repression by DtxR or that the regulation involves environmental signals other than iron. A previous study, which examined the Fur regulon in B. subtilis, noted that several genes that contained Fur-like binding sites failed to be regulated by Fur (2).
The newly independent states of the former Soviet Union experienced a significant outbreak of diphtheria throughout the 1990s (56). The causes of the epidemic were not clear, although several factors are thought to have contributed to the outbreak, including reduced vaccination rates among children, waning immunity in adults, population movements following the dissolution of the Soviet Union, and the emergence of a closely related group of C. diphtheriae strains. Epidemiological studies of C. diphtheriae strains associated with the outbreak identified a group of closely related isolates that emerged during the early stages of the epidemic (34). This group of strains became the dominant isolates that were identified during the peak of the epidemic and were characterized as belonging to a unique electrophoretic type, known as the ET8 complex, which includes strains of the Gravis biotype and of the G1 or G4 ribotype (34). The reason strains of the ET8 complex were dominant in the epidemic has not been determined; however, it is possible that this group of strains may encode factors that have provided them with an advantage in either colonization or dissemination within the population.
The four Russian epidemic strains examined in this study (members of the ET8 complex) all contained the complete sid operon, a genetic system that has similarities to the HPI found in pathogenic Yersinia species (10). The Yersinia HPI encodes proteins involved in the synthesis and uptake of the siderophore yersiniabactin and is required for virulence (10). Two Russian clinical strains that were not members of the ET8 complex, 1751 and G4193, and the C7(−) strain contained various deletions within the sid operon. The wild-type C7(β) (tox+) strain and two mutant derivatives, HC1 and HC6 (13, 38), also contained similar deletions within the sid operon (C. A. Kunkle and M. P. Schmitt, unpublished observations). These findings, although preliminary, suggest an association between the Russian epidemic strains that belong to the ET8 complex and the presence of the sid operon. Additional studies with a larger set of C. diphtheriae strains will be needed to determine more precisely the linkage between the presence of the sid operon and strains of the ET8 complex.
Surprisingly, the PW8 strain, an isolate used for vaccine production and an organism that is deficient in siderophore synthesis (39), showed the same hybridization profile with probes of the sid operon as that seen with the Russian epidemic strains. The defect in siderophore production in PW8 is thought to be due to a spontaneous mutation that has not been characterized genetically (39). The biotype, electrotype, and ribotype of the PW8 strain have not been determined, and the relationship between PW8 and strains of the ET8 complex is not well defined (26).
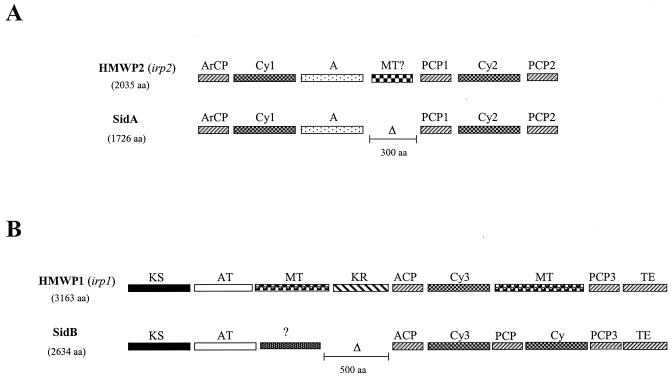
The synthesis of yersiniabactin in pathogenic Yersinia species involves the utilization of a nonribosomal peptide synthetase (NRPS) and a polyketide synthase (PKS). These enzymes together contain a total of 15 distinct functional domains and synthesize the siderophore in an assembly line process, where the precursor siderophore molecule is tethered to these enzymes and is then subsequently modified in an ordered manner by the various catalytic domains contained in the NRPS-PKS modules (12). The Y. pestis irp2 gene encodes HMWP2, an NRPS that contains six functional domains involved in the biosynthesis of yersiniabactin (there is also a seventh region that has sequence similarity to methyltransferases, but a function for this domain in yersiniabactin synthesis has not been shown) (52). SidA has a domain organization similar to that of HMWP2 but lacks the region that shows similarity to the methyltransferase (Fig. 7A). This deletion of the methyltransferase region in SidA appears to account for the 300-amino-acid size difference between these two proteins. The predicted amino acid sequences of SidA and HMWP2 are approximately 29% identical and have 38% similarity.
FIG. 7.
Comparison of the organization of the catalytic domains between SidA and HMWP2 (A) and SidB and HMWP1 (B). Blast alignments were done with the NCBI pairwise Blast alignment tool. Each catalytic domain from HMWP2 and HMWP1 (24) was aligned with the predicted amino sequence from SidA and SidB, respectively. Abbreviations of catalytic domains are as follows and are from Miller and Walsh (24): ArCP, aryl carrier protein; Cy, cyclization/condensation; A, adenylation; MT, methyltransferase; PCP, peptidyl carrier proteins; KS, ketosynthase; AT, acyltransferase; KR, ketoreductase; ACP, acyl carrier protein; TE, thioesterase. Δ, deleted region.
The irp1 gene encodes HMWP1, which contains nine distinct functional domains: the five domains at the N terminus are homologous to PKS modules, while the remaining domains contain an NRPS module (Fig. 7B). While C. diphtheriae SidB shows significant similarity to HMWP1, there are some notable differences in both the PKS module and the NRPS module (Fig. 7B). SidB does not contain either of the methyltransferase domains and is lacking the ketoreductase domain in the PKS module, and this deletion of approximately 500 amino acids appears to account for the size difference between these two proteins. In the SidB protein, the methyltransferase domain in the PKS module has been replaced by a region that has no significant homology to any sequences in GenBank, while in the NRPS module, the methyltransferase domain has been replaced by a peptidyl carrier protein domain and a cyclization/condensation domain (Fig. 7B). Overall, the predicted amino acid sequences of SidB and HMWP1 are 27% identical and 34% similar.
Mutations in the sidA and sidB genes had no effect on siderophore production or growth in low-iron medium in the C. diphtheriae epidemic isolate 1737. C. diphtheriae C7, which contains a partial deletion in the sid operon, produces the siderophore corynebactin (38). These findings clearly indicate that the putative siderophore biosynthetic genes encoded in the sid operon are not involved in the production of corynebactin and presumably are also not required for synthesis of the siderophore detected in the 1737 sid mutants, which is assumed to be corynebactin. The C. diphtheriae sid operon encodes only two genes that are predicted to be involved in siderophore biosynthesis, sidA and sidB, while the Yersinia HPI encodes six genes involved in siderophore production (17), and a seventh gene, ybtD, located outside the HPI, encodes a phosphopantetheinylate transferase that is also required for yersiniabactin synthesis (8). Based on what is known regarding the synthesis of siderophores that utilize NRPS enzymes, it is unlikely that the predicted products encoded by sidA and sidB alone can produce an active siderophore, and it seems almost certain that genes that encode a phosphopantetheinylate transferase (ybtD/entD) and an adenylation activating enzyme, similar to ybtE/entE, would be needed to initiate siderophore biosynthesis (12, 24). Interestingly, a search of the C. diphtheriae genome revealed the presence of a single gene with significant homology to ybtD/entD and three genes that showed similarity to ybtE/entE. Future studies will be needed to determine if any of these genes have a role in siderophore biosynthesis.
Although there is, as yet, no evidence that the genes in the sid operon are involved in the production of a functional siderophore, it is nevertheless possible that the Russian epidemic clones produce two siderophores, one produced by the sid operon and corynebactin. Bacterial pathogens that produce two siderophores include certain virulent E. coli strains that synthesize the siderophore enterobactin, which maintains iron homeostasis in the cell, and aerobactin, which is required for virulence (59). If a similar situation exists in the Russian epidemic strains, then the siderophore produced by the sid operon may be masked by the secretion of corynebactin. This possibility could explain why no change in siderophore secretion is detected in the sid mutants, especially if corynebactin is either more strongly expressed or a more efficient chelator. Another possible explanation for why no alteration in siderophore production was seen in the sid mutants is that mutations in the sidA or sidB gene may result in increased production of corynebactin, which may occur in order to maintain a constant level of siderophore synthesis. It is clear that additional studies will be needed to demonstrate if two siderophores are produced by the Russian epidemic strains and also to determine the association between the sid operon and strains of the ET8 complex.
Acknowledgments
We thank Tanja Popovic for providing the C. diphtheriae clinical strains. We are also grateful to Clare Schmitt, Karen Meysick, and Scott Stibitz for critical reading of the manuscript.
REFERENCES
- 1.Altschul, S. F., G. Warren, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 3.Barksdale, W. L., and A. M. Pappenheimer, Jr. 1954. Phage-host relationships in nontoxigenic and toxigenic diphtheria bacilli. J. Bacteriol. 67:220-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates, C. S., G. E. Montanez, C. R. Woods, R. M. Vincent, and Z. Eichenbaum. 2003. Identification and characterization of a Streptococcus pyogenes operon involved in binding of hemoproteins and acquisition of iron. Infect. Immun. 71:1042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumler, A. J., R. M. Tsolis, A. W. van der Velden, I. Stojiljkovic, S. Anic, and F. Heffron. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183:207-213. [DOI] [PubMed] [Google Scholar]
- 6.Bearden, S. W., J. D. Fetherston, and R. D. Perry. 1997. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 65:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billington, S. J., P. A. Esmay, J. G. Songer, and H. B. Jost. 2002. Identification and role in virulence of putative iron acquisition genes from Corynebacterium pseudotuberculosis. FEMS Microbiol. Lett. 208:41-45. [DOI] [PubMed] [Google Scholar]
- 8.Bobrov, A. G., V. A. Geoffroy, and R. D. Perry. 2002. Yersiniabactin production requires the thioesterase domain of HMWP2 and YbtD, a putative phosphopantetheinylate transferase. Infect. Immun. 70:4204-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd, J. M., O. N. Manish, and J. R. Murphy. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. USA 87:5968-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carniel, E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3:561-569. [DOI] [PubMed] [Google Scholar]
- 11.Collier, R. J. 2001. Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon 39:1793-1803. [DOI] [PubMed] [Google Scholar]
- 12.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cryz, S. J., L. M. Russell, and R. K. Holmes. 1983. Regulation of toxinogenesis in Corynebacterium diphtheriae: mutations in the bacterial genome that alter the effects of iron on toxin production. J. Bacteriol. 154:245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drazek, E. S., C. A. Hammack, and M. P. Schmitt. 2000. Corynebacterium diphtheriae genes required for acquisition of iron from hemin and hemoglobin are homologous to ABC hemin transporters. Mol. Microbiol. 36:68-84. [DOI] [PubMed] [Google Scholar]
- 15.Feese, M. D., E. Pohl, R. K. Holmes, and W. G. J. Hol. 2001. Iron-dependent regulators, p. 85-863. In A. Messerschmidt, R. Huber, T. Poulos, and K. Wieghardt (ed.), Handbook of metalloproteins. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 16.Fetherston, J. D., V. J. Bertolino, and R. D. Perry. 1999. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol. 32:289-299. [DOI] [PubMed] [Google Scholar]
- 17.Geoffroy, V. A., J. D. Fetherstone, and R. D. Perry. 2000. Yersinia pestis YbtU and YbtT are involved in synthesis of the siderophore yersiniabactin but have different effects on regulation. Infect. Immun. 68:4452-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 19.Holmes, R. K. 2000. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis. 181:56-67. [DOI] [PubMed] [Google Scholar]
- 20.Kanei, C., T. Uchida, and M. Yoneda. 1977. Isolation from Corynebacterium diphtheriae C7(β) of bacterial mutants that produce toxin in medium with excess iron. Infect. Immun. 18:203-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, C. B. 1995. Quelling the red menace: hemin capture by bacteria. Mol. Microbiol. 18:383-390. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J. H., T. Wang, K. Ault, J. Liu, M. P. Schmitt, and R. K. Holmes. 1997. Identification and characterization of three new promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. Infect. Immun. 65:4273-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei, B., L. M. Smoot, H. M. Menning, J. M. Voyich, S. V. Kala, F. R. Deleo, S. D. Reid, and J. M. Musser. 2002. Identification and characterization of a novel heme-associated cell surface protein made by Streptococcus pyogenes. Infect. Immun. 70:4494-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, D. A. C. T. Walsh. Yersiniabactin synthetase: probing the recognition of carrier protein domains by the catalytic heterocyclization domains, cy1 and cy2, in the chain-initiating HMWP2 subunit. Biochemistry 40:5313-5321. [DOI] [PubMed]
- 25.Nachin, L., L. Loiseau, D. Expert, and F. Barras. 2003. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 22:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakao, H., I. K. Mazurova, T. Glushkevich, and T. Popovic. 1997. Analysis of heterogeneity of Corynebacterium diphtheriae toxin gene, tox, and its regulatory element, dtxR, by direct sequencing. Res. Microbiol. 148:45-54. [DOI] [PubMed] [Google Scholar]
- 27.Ochsner, U. A., and M. L. Vasil. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosoa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. USA 93:4409-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosoa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 29.Otto, B. R., V.-V. Vught, A. M. Verweij-van Vught, and D. M. MacLaren. 1992. Transferrins and heme compounds as iron sources for pathogenic bacteria. Crit. Rev. Microbiol. 18:217-233. [DOI] [PubMed] [Google Scholar]
- 30.Park, W. H., and A. W. Williams. 1896. The production of diphtheria toxin. J. Exp. Med. 1:164-185. [DOI] [PMC free article] [PubMed]
- 31.Patzer, S. I., and K. Hantke. 1999. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J. Bacteriol. 181:3307-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paustian, M. L., B. J. May, and V. Kapur. 2001. Pasteurella multocida gene expression in response to iron limitation. Infect. Immun. 69:41009-44115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pohl, E., J. C. Haller, A. Mijovilovich, W. Meyer-Klaucke, E. Garman, and M. L. Vasil. 2003. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47:903-915. [DOI] [PubMed] [Google Scholar]
- 34.Popovic, T., S. Y. Kombarova, M. W. Reeves, H. Nakao, I. K. Mazurova, M. Wharton, I. K. Wachsmuth, and J. D. Wenger. 1996. Molecular epidemiology of diphtheria in Russia, 1985-1994. J. Infect. Dis. 174:1064-1072. [DOI] [PubMed] [Google Scholar]
- 35.Qian, Y., J. H. Lee, and R. K. Holmes. 2002. Identification of a DtxR-regulated operon that is essential for siderophore-dependent iron uptake in Corynebacterium diphtheriae. J. Bacteriol. 184:4846-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Que, Q., and J. D. Helmann. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35:1454-1468. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell, L. M., S. J. Cryz, and R. K. Holmes. 1984. Genetic and biochemical evidence for a siderophore-dependent iron transport system in Corynebacterium diphtheriae. Infect. Immun. 45:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell, L. M., and R. K. Holmes. 1985. Highly toxinogenic but avirulent Park-Williams 8 strain of Corynebacterium diphtheriae does not produce siderophore. Infect. Immun. 47:575-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 41.Schmitt, M. P. 1997. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenase and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt, M. P. 1997. Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect. Immun. 65:4634-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitt, M. P. 2002. Analysis of a DtxR-like metalloregulatory protein, MntR, from Corynebacterium diphtheriae that controls expression of an ABC metal transporter by an Mn2+-dependent mechanism. J. Bacteriol. 184:6882-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitt, M. P., and E. S. Drazek. 2001. Construction and consequences of directed mutations affecting the hemin receptor in pathogenic Corynebacterium species. J. Bacteriol. 183:1476-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt, M. P., and R. K. Holmes. 1991. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect. Immun. 59:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitt, M. P., and R. K. Holmes. 1991. Characterization of a defective diphtheria toxin repressor (dtxR) allele and analysis of dtxR transcription in wild-type and mutant strains of Corynebacterium diphtheriae. Infect. Immun. 59:3903-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitt, M. P., and R. K. Holmes. 1993. Analysis of diphtheria toxin repressor-operator interactions and characterization of a mutant repressor with decreased binding activity for divalent metals. Mol. Microbiol. 9:173-181. [DOI] [PubMed] [Google Scholar]
- 48.Schmitt, M. P., and R. K. Holmes. 1994. Cloning, sequence and footprint analysis of two promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor and iron. J. Bacteriol. 176:1141-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitt, M. P., E. M. Twiddy, and R. K. Holmes. 1992. Purification and characterization of the diphtheria toxin repressor. Proc. Natl. Acad. Sci. USA 89:7576-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sebastian, S., S. Agarwal, J. R. Murphy, and C. A. Genco. 2002. The gonococcal Fur regulon: Identification of additional genes involved in major catabolic, recombination, and secretory pathways. J. Bacteriol. 184:3965-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 52.Suo, Z., C. T. Walsh, and D. A. Miller. 1999. Tandem heterocyclization activity of the multidomain 230 kDa HMWP2 subunit of Yersinia pestis Yersiniabactin synthetase: Interaction of the 1-1382 and 1383-2035 fragments. Biochemistry 38:14023-14035. [DOI] [PubMed] [Google Scholar]
- 53.Tai, S.-P. S., A. E. Krafft, P. Nootheti, and R. K. Holmes. 1990. Coordinate regulation of siderophore and diphtheria toxin production by iron in Corynebacterium diphtheriae. Microb. Pathog. 9:267-273. [DOI] [PubMed] [Google Scholar]
- 54.Tao, X., and J. R. Murphy. 1992. Binding of the metalloregulatory protein DtxR to the diphtheria tox operator requires a divalent heavy metal ion and protects the palindromic sequence from DNase I digestion. J. Biol. Chem. 267:21761-21764. [PubMed] [Google Scholar]
- 55.Tsolis, R. M., A. J. Baumler, I. Stojiljkovic, and F. Heffron. 1995. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J. Bacteriol. 177:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vitek, C. R., and M. Wharton. 1998. Diphtheria in the former Soviet Union: reemergence of a pandemic disease. Emerg. Infect. Dis. 4:539-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 58.Weinberg, E. D. 1978. Iron and infection. Microbiol. Rev. 42:45-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams, P. H., and N. H. Carbonetti. 1986. Iron, siderophores, and the pursuit of virulence: independence of the aerobactin and enterochelin iron uptake systems in Escherichia coli. Infect. Immun. 51:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]