Abstract
Bam35, a 15-kbp double-stranded DNA phage, infects Bacillus thuringiensis. Recently, sequencing of the related Bacillus cereus revealed a 15.1-kbp linear plasmid, pBClin15. We show that pBClin15 closely resembles Bam35 and demonstrate conversion of Bam35 to a prophage. This state is common, as several B. thuringiensis strains release Bam35-related viruses.
Phage Bam35 was originally isolated from Bacillus thuringiensis var. alesti strain 35 (1). As Bam35 could also be isolated from Bam35-resistant colonies of Bacillus megaterium and from a sensitive strain of B. thuringiensis var. entomocidus, a carrier state has been proposed (1). Recently, a clear-plaque mutant (Bam35c) was sequenced and further characterized (25). This work showed that Bam35 is related to PRD1, a linear double-stranded DNA virus with 5′-terminal proteins and an internal lipid membrane (4). PRD1 infects gram-negative bacteria harboring an IncP, -W, or -N conjugative plasmid. Structural analyses of PRD1 have, surprisingly, indicated that this member of the Tectiviridae is similar to the Adenoviridae, Phycodnaviridae, and Iridoviridae, which all infect eukaryotic hosts (9, 24, 33). This has led to the hypothesis that all these viruses belong to the same lineage, with a common ancestor existing before the separation of the three domains of life (5, 6).
Phage Bam35 is closely related to Bacillus cereus plasmid pBClin15.
Bacilli harbor a large variety of plasmids, which include a linear species of ≈15 kb (2, 13, 15, 31, 34). Recently, the genome sequencing of Bacillus cereus ATCC 14579 revealed the sequence of a linear plasmid (pBClin15) of 15,100 bp (19). While studying Bam35, we matched putative genes of the pBClin15 plasmid in independent database searches for the Bam35c coat protein and ATPase. After submission of this note, the nucleotide sequence of a linear B. thuringiensis plasmid, pGIL01, which differs from the Bam35c sequence by only approximately 10 nucleotides was published (32).
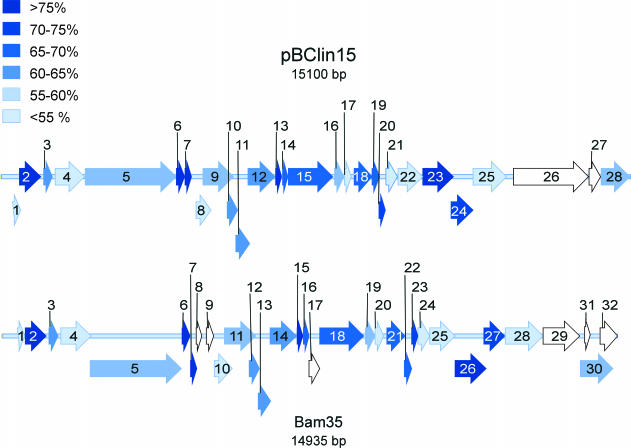
To investigate the relationship of pBClin15 to Bam35c (14,935 bp), their genomes were compared. The organizations of the open reading frames (ORFs) of pBClin15 and those of Bam35c are highly similar (Fig. 1). Moreover, their sequences agree with 45 to 81% identity. The corresponding amino acids have 18 to 88% identity (Table 1). The gene identification of Bam35c (25) was based on the reasonably detailed understanding of the corresponding PRD1 genes (7, 8, 16). A similar annotation shows that pBClin15 is related to Bam35 and PRD1 (Table 1). Among the most conserved genes in pBClin15 are those assigned to viral capsid components. These include the major coat protein 15 (corresponding to PRD1 protein P3) and the unique vertex packaging proteins 12, 14, and 16 (proteins P9, P20, and P22, respectively, in PRD1). Interestingly, the most conserved protein corresponds to a LexA-type transcription regulator (17, 21) homologue found in Bam35 but absent in PRD1.
FIG. 1.
A comparison of the pBClin15 and Bam35 genomes. Open reading frames are depicted by block arrows, shaded to show the level of DNA sequence identity with the corresponding gene in the other phage. Open reading frames with no counterpart are shown in white. The three levels of arrows reflect the different reading frames.
TABLE 1.
Comparison of Bam35c genes with pBClin15 ORFs at the protein and DNA levelsa
| Bam35c Protein (no. of residues) | pBClin15 protein (no. of residues) | Location on pBClin15 (nt) | Identity (%) | Identity at DNA level (%) | Protein functionb | PRD1 protein (no. of residues) |
|---|---|---|---|---|---|---|
| 1 (58) | 1 (63) | 260-451 | 17.5 | 45.9 | ||
| 2 (167) | 2 (167) | 432-935 | 84.5 | 81.3 | ||
| 3 (74) | 3 (71) | 1010-1225 | 43.2 | 62.2 | ||
| 4 (245) | 4 (233) | 1290-1991 | 31.0 | 50.0 | ||
| 5 (735) | 5 (729) | 2004-4193 | 46.9 | 58.5 | DNA polymerase | P1 (553) |
| 6 (66) | 6 (66) | 4196-4396 | 87.9 | 79.1 | Lex A-type repressor | |
| 7 (50) | 7 (49) | 4412-4561 | 70.0 | 75.8 | ||
| 8 (46) | ||||||
| 9 (57) | ||||||
| 10 (145) | 8 (118) | 4671-5027 | 36.3 | 52.8 | Unique vertex | P6 (166) |
| 11 (252) | 9 (243) | 4838-5569 | 36.0 | 56.5 | Assembly | P10 (203) |
| 12 (80) | 10 (81) | 5415-5660 | 44.4 | 60.2 | ||
| 13 (102) | 11 (106) | 5629-5949 | 59.4 | 64.8 | ||
| 14 (212) | 12 (216) | 5921-6571 | 62.5 | 63.6 | Packaging ATPase | P9 (227) |
| 15 (46) | 13 (46) | 6584-6724 | 78.3 | 78.0 | ||
| 16 (46) | 14 (45) | 6739-6876 | 65.2 | 69.5 | Unique vertex | P20 (42) |
| 17 (84) | ||||||
| 18 (356) | 15 (355) | 6880-7947 | 65.2 | 66.4 | Capsid protein | P3 (395) |
| 19 (76) | 16 (76) | 7988-8218 | 42.9 | 58.7 | Unique vertex | P22 (47) |
| 20 (68) | 17 (57) | 8221-8394 | 32.4 | 52.7 | ||
| 21 (143) | 18 (140) | 8468-8890 | 61.9 | 65.2 | ||
| 22 (58) | 19 (58) | 8891-9067 | 67.2 | 69.5 | ||
| 23 (48) | 20 (48) | 9064-9210 | 72.9 | 71.4 | ||
| 24 (91) | 21 (92) | 9222-9500 | 35.9 | 53.0 | ||
| 25 (207) | 22 (197) | 9513-10106 | 42.4 | 54.3 | Infectivity | P11 (207) |
| 26 (250) | 23 (243) | 10110-10841 | 76.8 | 75.0 | Transglycosylase | P7 (265) |
| 27 (170) | 24 (173) | 10789-11310 | 79.9 | 74.9 | ||
| 28 (304) | 25 (264) | 11323-12117 | 20.2 | 48.3 | ||
| 26 (597) | 12290-14083 | |||||
| 29 (293) | ||||||
| 27 (95) | 14096-14383 | |||||
| 30 (265) | 28 (227) | 14396-15079 | 57.2 | 59.5 | Endolysin | |
| 31 (40) | ||||||
| 32 (141) |
Each ORF was compared with the other whole genome (Align X/ Vector NTI 7.0). The ORFs of pBClin15 and the corresponding genes of Bam35c and their proteins were then compared individually. The known biochemical and structural properties of related PRD1 proteins and proposed functions of Bam35 proteins are listed in the last two columns. The accession number for Bam35c is AY257527. The accession number for pBClin15 is NC_004721. The genome has been reannotated, and the ORF numbering does not correspond to that used in GenBank. Stop codons are included in the given nucleotide coordinates. Location numbering for pBClin15 refers to NC_004721.
From reference 25.
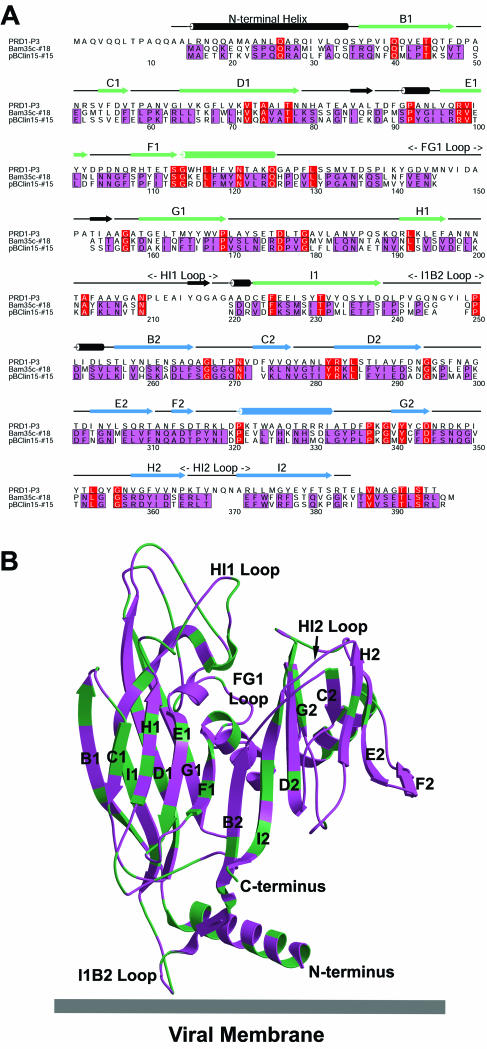
We then explored the relationship between pBClin15, Bam35c, and PRD1 by comparing their major coat proteins. First, the three sequences were aligned (Fig. 2A ). A model was then made of the pBClin15 protein, based on an earlier threading of the corresponding Bam35c sequence onto the high-resolution structure of the PRD1 coat protein, P3 (9, 10; unpublished results) (Fig. 2B). The three proteins clearly have the same fold, although the sequence similarity between Bam35c and PRD1 is very low (12% identity). In contrast, the Bam35c and pBClin15 proteins are very similar (65% identity), with their differences scattered throughout the molecule (Fig. 2B). Of note, two regions in PRD1 P3 (the N terminus and I1B2 loop) that interact with the internal membrane (28, 29) are shorter in Bam35c and pBClin15. The N-terminal helix shows some conservation in key residues but lacks the flexible tip of PRD1 P3 (Fig. 2B).
FIG. 2.
Major capsid proteins. (A) A sequence alignment of the major coat protein, P3, of phage PRD1 with those predicted for Bam35c (protein 18) and pBClin15 (protein 15). The Bacillus phages and prophages show high identity (65%; purple), whereas PRD1 P3 is more distantly related (12%, red). The secondary structural elements for PRD1 P3, determined by X-ray crystallography, are shown above the alignment with α-helices as rods and β-strands as arrows. The two eight-stranded viral jelly rolls that define the structure are shown in green and blue (strands marked B1-I1 and B2-I2 [9]). Note that the deletions in the Bacillus phage/prophage relative to PRD1 occur in the loops connecting the strands of the jelly rolls. These affect the loops at the top of the molecule forming the viral surface (FG1, HI1, and HI2) and the I1B2 loop at the base (see below). (B) A model of the pBClin15 coat protein based on a threading of the corresponding Bam35c sequence onto the PRD1 P3 crystal structure with the alignment as a guide. The residues that are identical in the Bam35c andpBClin15 proteins are shown in purple, and the ones that differ are in green. Interactions with the membrane occur through residues in the N-terminal helix and I1B2 loop at the base of the molecule. Both features are shorter in Bam35c and pBClin15 than in PRD1.
Plasmid pBClin15 lacks inverted terminal repeats.
Microbial extrachromosomal linear elements so far characterized are divided into two groups: those carrying covalently closed ends (hairpins) and those with covalently attached 5′-terminal proteins, similar to those in viral genomes (22, 26). Bacteriophage PRD1 contains inverted terminal repeats, has covalently linked proteins at its 5′ ends, and replicates using a protein-primed mechanism (3, 7, 11, 30). Bam35c also has inverted terminal repeats and may have terminal proteins, as suggested by the finding that the migration of Bam35c DNA in agarose gels is dependent on protease treatment (25). The 5′ ends of the nearly identical pGIL01 DNA are also protected by terminal proteins: in addition to its similar protease-dependent gel migration, pGIL01 is only degraded by exonuclease III (a 3′-nuclease) but not by λ nuclease (a 5′-nuclease) (32). Comparison of Bam35c and pBClin15 genome termini revealed similar 5′ noncoding regions (over 70% identity at DNA level) of about equal length, but pBClin15 lacks the inverted terminal repeats.
Phage Bam35c can convert to a prophage.
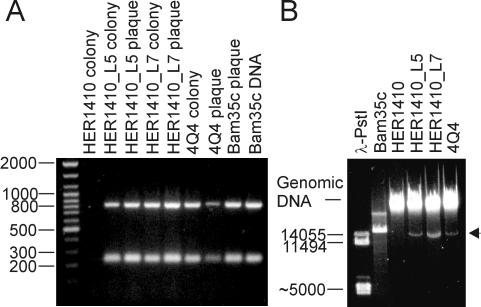
We investigated whether Bam35c can establish a carrier state, as proposed by Ackermann et al. (1). Lysogenic cell lines were obtained by picking microcolonies from the centers of plaques, as well as from confluently lysed plates of B. thuringiensis serovar israelensis HER1410 (obtained from the Félix d'Herelle Reference Center for Bacterial Viruses, Laval University, Quebec, Canada) infected with Bam35c. To eliminate remaining free phage particles, single-colony isolations were performed (a total of eight passages). Two of the cell lines obtained, named HER1410_L5 and HER1410_L7, were shown to contain Bam35c-specific sequences by PCR from single bacterial colonies (for method, see reference 20) with specific primers hybridizing to the ends of Bam35c genes 6 and 14 (GenBank accession no. AY257527) (Fig. 3A).
FIG. 3.
Detection of Bam35-related genomes. (A) PCR amplification of Bam35-specific sequences from lysogenic cell lines and plaques with specific primers against Bam35c ORFs 6 and 14, producing PCR products of 235 bp and 810 bp, respectively. Lane 1 contains Generuler DNA ladder mix (MBI Fermentas). Lanes 2, 3, 5, and 7 contain PCR products obtained by colony PCR of host strain HER1410, lysogenic cell lines HER1410_L5 and HER1410_L7, and strain 4Q4, respectively. Lanes 4, 6, 8, and 9 show PCR products obtained by plaque PCR of cell lines HER1410_L5 and HER1410_L7, strain 4Q4, and a Bam35c plaque, respectively. Lane 10 contains the PCR products obtained from purified Bam35c DNA (control). (B) The lysogenic cell lines HER1410_L5 and HER1410_L7 and strain 4Q4 carry ≈15-kbp DNA molecules that cannot be found in the original Bam35 host strain HER1410. Lanes 1 and 2 contain purified PstI-digested λ DNA and purified Bam35c DNA, respectively. Lanes 3 to 6 contain purified DNA (Wizard Genomic DNA purification kit; Promega) from host strain HER1410, lysogenic cell lines HER1410_L5 and HER1410_L7, and strain 4Q4, respectively. The arrowhead depicts the ≈15-kbp DNA element found in the lysogenic cell lines and strain 4Q4.
The isolated lysogenic cell lines released viruses into culture supernatants (typically 102 to 103 PFU/ml after 8 h of growth of cells in Luria-Bertani (27). The phage were shown to arise from Bam35c by PCR from single plaques (for method, see reference 18) with specific primers as described above (Fig. 3A). The virus-producing cell lines now carried ≈15-kbp DNA elements, the Bam35c prophage, that were not present in the original HER1410 (Fig. 3B).
Bam35-like prophage are common in bacilli.
To check the distribution of similar prophages, we investigated several B. thuringiensis strains from the Bacillus Genetic Stock Center (Ohio State University, Columbus). Of seven strains tested, four released viruses into the culture supernatant that were detectable on the Bam35 host strain HER1410. One of these, B. thuringiensis serovar israelensis 4Q4 (WHO2013-9), was identified as carrying a Bam35c-related prophage by colony and plaque PCR as described above (Fig. 3A). As with the virus-producing cell lines isolated previously, 4Q4 contains a specific ≈15-kbp DNA element not found in HER1410 (Fig. 3B).
Is the Bam35 carrier state maintained by protein-primed replication?
The Bacillus anthracis phage AP50 (23) is also related to Bam35 and PRD1 (4). We have shown here that B. thuringiensis strains carry Bam35-like prophages and that B. cereus plasmid pBClin15 is closely related to Bam35. Obviously, these three bacilli carry related phage/prophage systems. The likely mechanism by which the prophage state is maintained is intriguing. The Bam35c genome, analogously to PRD1, may contain terminal proteins (25). These proteins are used as primers for initiating replication, and protein-primed replication mechanisms occur in lytic bacteriophages, such as PRD1 and φ29, and also in adenovirus and linear plasmids (26). The idea that protein-primed replication can also operate in the carrier state is novel, as this mechanism has so far not been reported for prophages (12, 14). As pBClin15 does not contain inverted terminal repeats, it may be a degenerating prophage that cannot give rise to virus particles. These observations open interesting avenues for future research in the Bam35-like virus-plasmid system.
Acknowledgments
Sari Korhonen, Sara Nummi and Anna Rantala are greatly thanked for their skillful and swift technical assistance.
This investigation was supported by research grants 1201964 (J.K.H.B., Academy of Finland), 172621 and 1202108 (D.H.B., Finnish Center of Excellence Program [2002-2005] from the Academy of Finland), AI-17270 and CA-09171 (R.M.B., National Institutes of Health), and RG320/2001 (D.H.B. and R.M.B., Human Frontiers Science Program).
REFERENCES
- 1.Ackermann, H.-W., R. Roy, M. Martin, M. R. V. Murthy, and W. A Smirnoff. 1978. Partial characterization of a cubic Bacillus phage. Can. J. Microbiol. 24:986-993. [DOI] [PubMed] [Google Scholar]
- 2.Andrup, L., G. B. Jensen, A. Wilcks, L. Schmidt, L. Hoflack, and J. Mahillon. 2003. The patchwork nature of rolling-circle plasmids: comparison of six plasmids from two distinct Bacillus thuringiensis serotypes. Plasmid 49:205-232. [DOI] [PubMed] [Google Scholar]
- 3.Bamford, D. H., and L. Mindich. 1984. Characterization of the DNA-protein complex at the termini of the bacteriophage PRD1 genome. J. Virol. 50:309-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bamford, D. H. and H.-W. Ackermann. 2000. Family Tectiviridae, p. 111-116. In M. H. V. van Regenmortel et al. (ed.), Virus taxonomy: classification and nomenclature of viruses. Academic Press, San Diego, Calif.
- 5.Bamford, D. H., R. M. Burnett, and D. I. Stuart. 2002. Evolution of viral structure. Theor. Pop. Biol. 61:461-470. [DOI] [PubMed] [Google Scholar]
- 6.Bamford, D. H. 2003. Do viruses form lineages across different domains of life? Res. Microbiol. 154:231-236. [DOI] [PubMed] [Google Scholar]
- 7.Bamford, J. K. H., A.-L. Hänninen, T. M. Pakula, P. M. Ojala, N. Kalkkinen, M. Frilander, and D. H. Bamford. 1991. Genome organization of PRD1, a membrane-containing bacteriophage infecting E. coli. Virology 183:658-676. [DOI] [PubMed] [Google Scholar]
- 8.Bamford, J. K. H., J. Cockburn, J. Diprose, J. M. Grimes, G. Sutton, D. I. Stuart, and D. H. Bamford. 2002. Diffraction quality crystals of PRD1, a 66 MDa ds DNA virus with an internal membrane. J. Struct. Biol. 139:103-112. [DOI] [PubMed] [Google Scholar]
- 9.Benson, S. D., J. K. H. Bamford, D. H. Bamford, and R. M. Burnett. 1999. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98:825-833. [DOI] [PubMed] [Google Scholar]
- 10.Benson, S. D., J. K. H. Bamford, D. H. Bamford, and R. M. Burnett. 2002. The X-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 Å resolution. Acta Crystallograph. D 58:39-59. [DOI] [PubMed] [Google Scholar]
- 11.Caldentey, J., L. Blanco, D. H. Bamford, and M. Salas. 1993. In vitro replication of bacteriophage PRD1 DNA. Characterisation of the protein-primed initiation site. Nucleic Acids Res. 21:3725-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brüssow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlson, C. R., A. Grønstad, and A.-B. Kolstø. 1992. Physical maps of the genomes of three Bacillus cereus strains. J. Bacteriol. 174:3750-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277-300. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, J. M., Jr., and B. C. Carlton. 1980. Patterns of plasmid DNA in crystalliferous and acrystalliferous strains of Bacillus thuringiensis. Plasmid 3:92-98. [DOI] [PubMed] [Google Scholar]
- 16.Grahn, A. M., S. J. Butcher, J. K. H. Bamford, and D. H. Bamford. 2003. PRD1—dissecting the genome, structure and entry. In R. Calendar (ed.), The bacteriophages. Oxford University Press, Oxford, England, in press.
- 17.Horii, T., T. Ogawa, and H. Ogawa. 1981. Nucleotide sequence of the LexA gene of E. coli. Cell 23:689-697. [DOI] [PubMed] [Google Scholar]
- 18.Huiskonen, J. T., L. Laakkonen, M. Toropainen, M. Sarvas, D. H. Bamford, and J. K. H. Bamford. 2003. Probing the ability of the coat and vertex protein of the membrane-containing bacteriophage PRD1 to display a meningococcal epitope. Virology 310:267-279. [DOI] [PubMed] [Google Scholar]
- 19.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 20.Kivelä, H. M., R. H. Männistö, N. Kalkkinen, and D. H. Bamford. 1999. Purification and protein composition of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262:364-374. [DOI] [PubMed] [Google Scholar]
- 21.Little, J. W., and J. E. Harper. 1979. Identification of the lexA gene product of Escherichia coli K-12. Proc. Natl. Acad. Sci. 76:6147-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meinhardt, F., R. Schaffrath, and M. Larsen. 1997. Microbial linear plasmids. Appl. Microbiol. Biotechnol. 47:329-336. [DOI] [PubMed] [Google Scholar]
- 23.Nagy, E. 1974. A highly specific phage attacking Bacillus anthracis strain Sterne. Acta Microbiol. Acad. Sci. Hung. 21:257-263. [PubMed] [Google Scholar]
- 24.Nandhagopal, N., A. A. Simpson, J. R. Gurnon, X. Yan, T. S. Baker, M. V. Graves, J. L. Van Etten, and M. G. Rossmann. 2002. The structure and evolution of the major capsid protein of a large, lipid-containing, DNA-virus. Proc. Natl. Acad. Sci. 99:14758-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravantti, J. J., A. Gaidelyte, D. H. Bamford, and J. K. H. Bamford. 2003. Comparative analysis of bacterial viruses Bam35, infecting a gram-positive host, and PRD1, infecting gram-negative hosts, demonstrates a viral lineage. Virology 313:401-414. [DOI] [PubMed]
- 26.Salas, M. 1991. Protein-priming of DNA replication. Annu. Rev. Biochem. 60:39-71. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.San Martín, C., R. M. Burnett, F. de Haas, R. Heinkel, T. Rutten, S. D. Fuller, S. J. Butcher, and D. H. Bamford. 2001. Combined EM/X-ray imaging yields a quasi-atomic model of the adenovirus-related bacteriophage PRD1, and shows key capsid and membrane interactions. Structure 9:917-930. [DOI] [PubMed] [Google Scholar]
- 29.San Martín, C., J. T. Huiskonen, J. K. H. Bamford, S. J. Butcher, S. D. Fuller, D. H. Bamford, and R. M. Burnett. 2002. Minor proteins, mobile arms, and membrane-capsid interactions in the bacteriophage PRD1 capsid. Nat. Struct. Biol. 9:756-762. [DOI] [PubMed] [Google Scholar]
- 30.Savilahti, H., and D. H. Bamford. 1993. Protein-primed replication: role of inverted terminal repeats in the Escherichia coli bacteriophage PRD1 life cycle. J. Virol. 67:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Titok, M. A., J. Chapuis, Y. V. Selezneva, A. V. Lagoditch, V. A. Prokulevitch, S. D. Ehrlich, and L. Jannière. 2003. Bacillus subtilis soil isolates: plasmid replicon analysis and construction of a new theta-replicating vector. Plasmid 49:53-62. [DOI] [PubMed] [Google Scholar]
- 32.Verheust, C., G. Jensen, and J. Mahillon. 2003. pGIL01, a linear tectiviral plasmid prophage originating from Bacillus thuringiensis serovar israelensis. Microbiology 149:2083-2092. [DOI] [PubMed] [Google Scholar]
- 33.Yan, X., N. H. Olson, J. L. Van Etten, M. Bergoin, M. G. Rossmann, and T. S. Baker. 2000. Structure and assembly of large lipid-containing dsDNA viruses. Nat. Struct. Biol. 7:101-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zawadzki, P., M. A. Riley, and F. M. Cohan. 1996. Homology among nearly all plasmids infecting three Bacillus species. J. Bacteriol. 178:191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]