Abstract
Twenty-six Y-chromosomal short tandem repeat (STR) loci were amplified in a sample of 769 unrelated males from Nepal, using two multiplex polymerase chain reaction (PCR) assays. The 26 loci gave a discriminating power of 0.997, with 59% unique haplotypes, and the highest frequency haplotype occurring 12 times. We identified novel alleles at four loci, microvariants at a further two, and nine examples of amelogenin-Y deletions (1.2%). Comparison with a similarly sized Bhutanese sample typed with the same markers suggested histories of isolation and drift, with drift having a greater effect in Bhutan. Extended (11-locus) haplotypes for the Nepalese samples have been submitted to the Y-STR Haplotype Reference Database (YHRD).
Keywords: Y chromosome, STRs, Microsatellites, Haplotype, Nepal, Bhutan, Himalayas
1. Introduction
The analysis of multiple Y-chromosomal short tandem repeats (STRs) provides informative male-specific DNA profiles in forensic analysis. As well as possessing high discriminating power in distinguishing individuals, haplotypes defined by STRs can provide information about likely geographical origin, since they are often concentrated in particular populations or regions.
Population databases of Y haplotypes [1] are increasing in size and coverage, greatly contributing to the utility of Y-chromosomal analysis in forensic casework. In this study we describe alleles at 26 Y-STRs, and properties of the haplotypes they define, in a large sample of a previously unrepresented population, that of Nepal in the Himalayas. Eleven-locus haplotypes have been submitted to the Y-STR Haplotype Reference Database (YHRD), and full data are available from the authors on request. Our report follows guidelines for the publication of population data [2].
Sampling and Y-chromosomal analysis of 769 Nepalese males was undertaken as part of a larger collaborative project [3] investigating genetic diversity in Himalayan populations within the framework of their cultural and linguistic diversity [4]. Here we describe our initial findings with Y-STRs, treating the Nepalese sample as a single population; future publications will explore genetic relationships between subpopulations of the Himalayas. The sample represents 15 distinct ethnolinguistic groups widely distributed throughout Nepal, with ∼75% of sampled individuals speaking languages belonging to the Tibeto-Burman family, and the remainder speaking Indo-European languages.
In this study, we employ the same set of Y-STRs as that used recently to analyse 856 Bhutanese males [5]. This allows us to carry out a preliminary comparison of diversity and haplotype sharing between these two Himalayan samples.
2. Materials and methods
2.1. DNA samples
Seven hundred and sixty-nine Bhutanese males provided blood samples with informed consent, and DNA was extracted as described [3]. DNA samples from collections of the authors, including Y Chromosome Consortium (YCC) cell lines [6], were used as haplotype reference materials.
2.2. Y-STR multiplexes
Two PCR multiplexes (a 20plex [7] and a partially overlapping 14plex [5]) were used to type 26 Y-STRs, as follows: DYS19, DYS385a/b, DYS388, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS425, DYS426, DYS434, DYS435, DYS436, DYS437, DYS438, DYS439, DYS447, DYS448, DYS460, DYS461, DYS462, YCAIIa/b, and Y-GATA-H4.1. The eleven Y-STR markers in the European ‘extended haplotype’ (http://www.yhrd.org/) are indicated in bold. The 14plex includes the amelogenin sex test. Full details of the protocol are given by Parkin et al. [5].
2.3. Y-STR nomenclature
Allele nomenclature (explained fully in Parkin et al. [5]) was according to Butler et al. [7] and Bosch et al. [8], with the exception of DYS439, DYS448 and Y-GATA-H4.1, where nomenclature was changed for compatibility with ISFG recommendations [9]. Compared to Butler et al. [7], seven repeats were subtracted from DYS439, three subtracted from DYS448, and eight added to Y-GATA-H4.1.
2.4. Calculations
Gene diversity and haplotype diversity were calculated using Arlequin [10]. A median-joining network was constructed using Network 4.0 ([11] http://www.fluxus-engineering.com/sharenet.htm), and the weighting scheme described by Qamar et al. [12].
3. Results and discussion
3.1. Diversity of alleles
Tables 1 and 2 show the allele frequency distributions for all the Y-STRs studied. Diversities of individual STRs are comparable with those observed in a recently studied Bhutanese sample: DYS385 (when considered as a genotype, Table 2) is the most diverse marker within the Y-STR set, with a gene diversity (h) of 0.915, and the most polymorphic single-locus marker is DYS439 (h = 0.726).
Table 1.
Frequencies of alleles at 22 of the 26 Y-STRs
| Allele | 19 | 388 | 390 | 391 | 392 | 393 | 425 | 426 | 434 | 435 | 436 | 437 | 438 | 439 | 447 | 448 | 460 | 461 | 462 | 389I | 389II-I | H4.1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 0.022 | 0.001 | 0.004 | |||||||||||||||||||
| 8 | 0.003 | 0.001 | ||||||||||||||||||||
| 9 | 0.053 | 0.085 | 0.511 | 0.018 | ||||||||||||||||||
| 10 | 0.563 | 0.776 | 0.072 | 0.003 | 0.087 | 0.001 | 0.168 | 0.151 | 0.270 | 0.008 | 0.001 | 0.003 | ||||||||||
| 11 | 0.004 | 0.164 | 0.261 | 0.009 | 0.001 | 0.826 | 0.870 | 0.982 | 0.001 | 0.003 | 0.719 | 0.248 | 0.176 | 0.140 | 0.217 | 0.014 | ||||||
| 12 | 0.001 | 0.289 | 0.005 | 0.059 | 0.675 | 0.948 | 0.170 | 0.016 | 0.017 | 0.978 | 0.025 | 0.390 | 0.038 | 0.645 | 0.684 | 0.524 | ||||||
| 13 | 0.044 | 0.068 | 0.001 | 0.023 | 0.211 | 0.018 | 0.001 | 0.027 | 0.021 | 0.001 | 0.192 | 0.160 | 0.096 | 0.290 | 0.001 | |||||||
| 14 | 0.644 | 0.020 | 0.497 | 0.100 | 0.386 | 0.016 | 0.027 | 0.001 | 0.165 | |||||||||||||
| 15 | 0.241 | 0.046 | 0.049 | 0.005 | 0.544 | 0.003 | 0.004 | 0.168 | ||||||||||||||
| 16 | 0.066 | 0.008 | 0.016 | 0.065 | 0.003 | 0.550 | ||||||||||||||||
| 17 | 0.004 | 0.001 | 0.001 | 0.001 | 0.003 | 0.046 | 0.189 | |||||||||||||||
| 18 | 0.003 | 0.003 | 0.025 | 0.087 | 0.022 | |||||||||||||||||
| 19 | 0.005 | 0.242 | 0.005 | 0.280 | ||||||||||||||||||
| 20 | 0.614 | 0.586 | ||||||||||||||||||||
| 21 | 0.005 | 0.064 | 0.104 | |||||||||||||||||||
| 22 | 0.051 | 0.057 | 0.003 | 0.008 | ||||||||||||||||||
| 23 | 0.490 | 0.501 | ||||||||||||||||||||
| 24 | 0.321 | 0.144 | ||||||||||||||||||||
| 25 | 0.124 | 0.086 | ||||||||||||||||||||
| 26 | 0.008 | 0.100 | ||||||||||||||||||||
| 27 | 0.001 | 0.072 | ||||||||||||||||||||
| 28 | 0.010 | |||||||||||||||||||||
| 29 | 0.004 | |||||||||||||||||||||
| 21.4 | 0.001 | |||||||||||||||||||||
| 22.4 | 0.013 | |||||||||||||||||||||
| 23.4 | 0.001 | |||||||||||||||||||||
| 9–12 | 0.001 | |||||||||||||||||||||
| 10–11 | 0.004 | |||||||||||||||||||||
| 11–12 | 0.005 | |||||||||||||||||||||
| 12–13 | 0.005 | |||||||||||||||||||||
| 13–14 | 0.001 | |||||||||||||||||||||
| 20–21 | 0.001 | |||||||||||||||||||||
| Null | 0.017 | 0.004 | ||||||||||||||||||||
| ha | 0.521 | 0.593 | 0.639 | 0.368 | 0.673 | 0.490 | 0.040 | 0.289 | 0.235 | 0.036 | 0.043 | 0.551 | 0.447 | 0.726 | 0.702 | 0.553 | 0.633 | 0.536 | 0.476 | 0.614 | 0.626 | 0.567 |
| h(Bh)b | 0.604 | 0.518 | 0.569 | 0.421 | 0.546 | 0.442 | 0.187 | 0.244 | 0.244 | 0.046 | 0.092 | 0.553 | 0.452 | 0.713 | 0.663 | 0.590 | 0.679 | 0.434 | 0.363 | 0.592 | 0.504 | 0.598 |
Calculation of gene diversity, h, excludes null alleles and duplications.
Comparative Bhutanese values from [5].
Table 2.
Frequencies of genotypes at DYS385 and YCAII
| Genotype | DYS385 | YCAII |
|---|---|---|
| 10–14 | 0.005 | |
| 11–11 | 0.017 | |
| 11–12 | 0.007 | |
| 11–13 | 0.004 | |
| 11–14 | 0.070 | |
| 11–15 | 0.001 | |
| 11–16 | 0.001 | |
| 11–18 | 0.003 | |
| 11–19 | 0.001 | |
| 11–20 | 0.001 | |
| 12–12 | 0.004 | |
| 12–13 | 0.003 | |
| 12–14 | 0.009 | |
| 12–15 | 0.001 | |
| 12–16 | 0.027 | |
| 12–17 | 0.029 | |
| 12–18 | 0.014 | |
| 12–19 | 0.007 | |
| 12–20 | 0.012 | |
| 13–13 | 0.022 | |
| 13–14 | 0.022 | |
| 13–15 | 0.003 | |
| 13–16 | 0.027 | |
| 13–17 | 0.056 | |
| 13–18 | 0.182 | |
| 13–19 | 0.177 | |
| 13–20 | 0.060 | |
| 13–21 | 0.014 | |
| 13–22 | 0.001 | |
| 13–23 | 0.003 | |
| 14–14 | 0.004 | |
| 14–15 | 0.007 | |
| 14–16 | 0.010 | |
| 14–17 | 0.013 | |
| 14–18 | 0.044 | |
| 14–19 | 0.031 | |
| 14–20 | 0.027 | |
| 14–22 | 0.001 | |
| 15–15 | 0.004 | |
| 15–16 | 0.009 | 0.009 |
| 15–17 | 0.007 | |
| 15–18 | 0.013 | |
| 15–19 | 0.003 | 0.014 |
| 15–20 | 0.007 | |
| 15–21 | 0.001 | |
| 16–16 | 0.001 | 0.008 |
| 16–17 | 0.004 | 0.003 |
| 16–18 | 0.004 | |
| 16–19 | 0.003 | 0.010 |
| 16–20 | 0.003 | |
| 16–22 | 0.001 | |
| 17–17 | 0.003 | 0.036 |
| 17–18 | 0.029 | |
| 17–19 | 0.606 | |
| 17–20 | 0.113 | |
| 17–21 | 0.091 | |
| 17–22 | 0.001 | |
| 17–23 | 0.001 | |
| 18–18 | 0.012 | |
| 18–19 | 0.001 | 0.025 |
| 18–20 | 0.001 | |
| 19–19 | 0.001 | 0.036 |
| 20–20 | 0.001 | |
| 20–21 | 0.001 | |
| 13–17.2 | 0.001 | |
| 13–18.2 | 0.016 | |
| h | 0.915 | 0.607 |
| h(Bh)a | 0.921 | 0.524 |
Comparative Bhutanese values from [5].
Previously unreported alleles (defined with reference to Butler [13], Parkin et al. [5] and STRBase, http://www.cstl.nist.gov/biotech/strbase/index.htm) were found at four loci, as follows: DYS426 (allele 13), DYS437 (allele 11), DYS439 (allele 15), DYS447 (alleles 17, 18 and 19).
‘Null’ alleles or multiple peaks were reproducibly obtained at a number of loci. For DYS448, three individuals carried null alleles, while one carried both alleles 20 and 21. For DYS461, one individual carried both alleles 13 and 14. As observed previously [5], DYS425 exhibits a relatively high frequency of various nulls and duplications.
Microvariants (partial alleles) were observed at two loci (Tables 1 and 2) and confirmed in uniplex assays after initial detection in multiplexes. Those at DYS385 were not investigated further, but those at DYS447 were analysed by sequencing, and shown to result from a deletion of 1 bp within the pentanucleotide repeat array [5].
3.2. AMELY deletion chromosomes
Nine chromosomes showed absence of the amelogenin Y (AMELY) peak in electropherograms. Analysis of sequence-tagged sites revealed that these chromosomes carry interstitial deletions of Yp including the AMELY locus (data not shown); none showed null Y-STR alleles, however, which is consistent with the size and location of known AMELY deletions with respect to the position of Y-STR loci [14]. A previous study has found AMELY deletions at a frequency of ∼2% in India [15], so our finding of deletions at 1.2% frequency in Nepal is not unexpected; in contrast, however, none were found in our previous study of Bhutan [5]. These AMELY deletion chromosomes form part of a large set that is currently being characterised, and will be described fully elsewhere.
3.3. Diversity of haplotypes
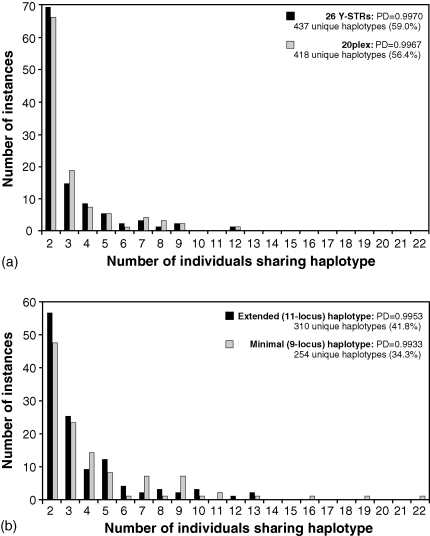
Haplotype diversity (equivalent to power of discrimination, PD) was calculated, omitting chromosomes carrying null alleles and duplications. This provided a sample size of 741. For the full set of 26 Y-STRs, there are 437 unique haplotypes (59.0%), and PD is 0.9970. The corresponding values for the 20plex [7], extended (11-locus) haplotype and minimal (9-locus) haplotype are shown in Fig. 1.
Fig. 1.
Haplotype diversity for (a) all 26 STRs and the 20plex, and (b) the extended and minimal haplotypes. Histograms show the frequency distributions of haplotypes present more than once in the dataset.
Fig. 1 also shows the distribution of haplotypes present more than once in the dataset. Despite the large number of loci used here, in the 741 males one 26-locus haplotype is shared by 12 individuals (Fig. 1a), and a further 13 haplotypes are shared by between 5 and 9 individuals; notably, all these common haplotypes are restricted to particular subpopulations, illustrating the influence of drift. Reduction to 11-locus extended haplotypes allows a global search within the YHRD (release 18): this fails to find matches for three of the six most common Nepalese extended haplotypes (frequency ≥10), consistent with isolation and drift.
3.4. Comparison of Y-STR datasets on Nepal and Bhutan
The availability of large Y-STR haplotype datasets on Nepalese and Bhutanese samples allows us to make comparisons between the frequencies and distributions of alleles and haplotypes in these two Himalayan populations.
Allele distributions at individual loci are similar between the Nepalese and Bhutanese samples, but this gives little information about population relationships. Particular rare and distinctive alleles may carry more information, because they probably reflect identity-by-descent: a good example of this is the sharing of microvariants at DYS447 [5], but apart from this there is little evidence for specific inter-population sharing.
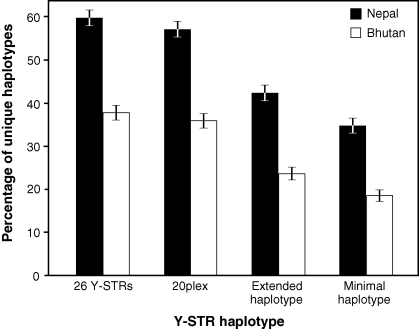
Comparison of haplotype distributions reveals a striking difference between the two populations. The proportion of unique haplotypes in the Nepalese sample is significantly greater than that in the Bhutanese, for all four haplotype resolutions considered (Fig. 2). For example, for the extended haplotype there are 41.8% (±1.8%) unique haplotypes in Nepal, but only 23.3% (±1.5%) in Bhutan. This is explained by the presence of several common haplotypes at high frequency in Bhutan: in the Nepalese dataset, the most common extended haplotypes are each present in 13 individuals, while in the Bhutanese there are haplotypes present in 15, 16, 24 (two instances) and 27 individuals [5].
Fig. 2.
Percentage of unique haplotypes in Nepal compared to Bhutan. Haplotypes containing null or duplicated alleles are omitted, giving total sample sizes of 741 and 802, respectively. The error bars represent plus or minus one binomial standard error.
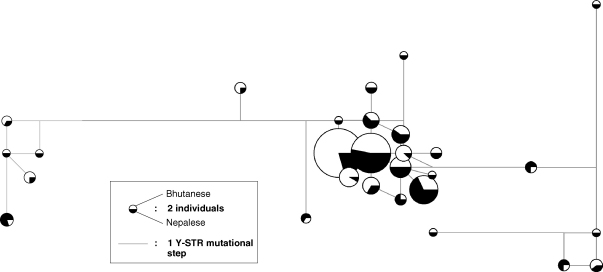
There are no 26-locus haplotypes shared between Nepal and Bhutan, indicating an absence of very recent gene flow. However, forty extended haplotypes are shared between the two samples, and their relationships (omitting the bilocal marker DYS385) are illustrated in a median-joining network in Fig. 3. Most of them fall into one large cluster, with haplotypes linked by single mutational steps, probably representing a common Y-SNP haplogroup. Other shared haplotypes are more widely spread, and may represent several different haplogroups.
Fig. 3.
Median-joining network of haplotypes shared between Nepal and Bhutan. Note that the 40 shared extended haplotypes described in the text are reduced to 28 when the bilocal Y-STR DYS385 is removed for network construction. Circles represent Y-STR haplotypes (based on DYS19, DYS389I, DYS389II-I, DYS390, DYS391, DYS392, DYS393, DYS438, DYS439), with area proportional to number of instances. Lines represent Y-STR mutational steps.
To ask if these shared extended haplotypes are more generally common and widespread, we sought matches for the six most predominant examples (combined frequency >10) within the YHRD. Three of the six haplotypes find a total of six exact matches, all within populations originating from China or the Indian subcontinent. We also find a total of 30 one-step mutational neighbours for five of the six haplotypes, all of Asian origin. One haplotype finds neither exact matches nor one-step neighbours. Thus, the common haplotypes shared between Nepal and Bhutan are Asian-specific, but not generally frequent.
3.5. Concluding remarks
Our study emphasises the discriminating power of high-resolution Y-STR typing, and provides the first substantial dataset on a Nepalese sample. The comparison of Nepalese and Bhutanese datasets reveals an interesting overall picture of isolation and drift within these Himalayan populations, with drift having a greater effect in Bhutan than Nepal. Haplotype sharing provides evidence of some gene flow between Nepal and Bhutan, or possibly of gene flow into both from some other population. Further light will be thrown on these relationships when Y-SNP data become available.
Acknowledgements
We thank all DNA donors, and the many organisations and volunteers of indigenous language communities in Nepal who gave us their assistance. The research was conducted in association with the Centre for Nepal and Asian Studies (CNAS) at Kirtipur under the Bilateral Agreement for Academic Cooperation between Tribhuvan University and Leiden University. As part of the European Science Foundation EUROCORES Programme OMLL, this work was supported by the Arts and Humanities Research Council and the EC Sixth Framework Programme under Contract no. ERAS-CT-2003-980409. T.K., G.L.v.D. and P.dK. were supported by funds from the Netherlands Organisation for Scientific Research (NWO grant number 231-70-001). M.A.J. was supported by a Wellcome Trust Senior Fellowship in Basic Biomedical Science (grant no. 057559).
References
- 1.Roewer L., Krawczak M., Willuweit S., Nagy M., Alves C., Amorim A., Anslinger K., Augustin C., Betz A., Bosch E., Caglià A., Carracedo A., Corach D., Dobosz T., Dupuy B.M., Füredi S., Gehrig C., Gusmão L., Henke J., Henke L., Hidding M., Hohoff C., Hoste B., Jobling M.A., Kärgel H.J., de Knijff P., Lessig R., Liebeherr E., Lorente M., Martínez-Jarreta B., Nievas P., Nowak M., Parson W., Pascali V.L., Penacino G., Ploski R., Rolf B., Sala A., Schmidt U., Schmitt C., Schneider P.M., Szibor R., Teifel-Greding J., Kayser M. Online reference database of Y-chromosomal short tandem repeat (STR) haplotypes. Forensic Sci. Int. 2001;118:103–111. doi: 10.1016/s0379-0738(00)00478-3. [DOI] [PubMed] [Google Scholar]
- 2.Lincoln P., Carracedo A. Publication of population data of human polymorphisms. Forensic Sci. Int. 2000;110:3–5. [PubMed] [Google Scholar]
- 3.Kraayenbrink T., de Knijff P., van Driem G.L., Opgenort J.R.M.L., Jobling M.A., Parkin E.J., Tyler-Smith C., Carvalho-Silva D.R., Tshering K., Barbujani G., Dupanloup I., Bertorelle G., Tuladhar N.M. 2006. Language and Genes of the Greater Himalayan Region.http://www.le.ac.uk/genetics/maj4/Himalayan_OMLLreport.pdf OMLL Volume. [Google Scholar]
- 4.van Driem G.L. 2 vols. Brill; Leiden: 2001. (Languages of the Himalayas: An Ethnolinguistic Handbook of the Greater Himalayan Region, Containing an Introduction to the Symbiotic Theory of Language). [Google Scholar]
- 5.Parkin E.J., Kraayenbrink T., van Driem G.L., Tshering K., de Knijff P., Jobling M.A. 26-locus Y-STR typing in a Bhutanese population sample. Forensic Sci. Int. 2005 doi: 10.1016/j.forsciint.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler J.M., Schoske R., Vallone P.M., Kline M.C., Redd A.J., Hammer M.F. A novel multiplex for simultaneous amplification of 20 Y chromosome STR markers. Forensic Sci. Int. 2002;129:10–24. doi: 10.1016/s0379-0738(02)00195-0. [DOI] [PubMed] [Google Scholar]
- 8.Bosch E., Lee A.C., Calafell F., Arroyo E., Henneman P., de Knijff P., Jobling M.A. High resolution Y chromosome typing: 19 STRs amplified in three multiplex reactions. Forensic Sci. Int. 2002;125:42–51. doi: 10.1016/s0379-0738(01)00627-2. [DOI] [PubMed] [Google Scholar]
- 9.Gusmão L., Butler J.M., Carracedo A., Gill P., Kayser M., Mayr W.R., Morling N., Prinz M., Roewer L., Tyler-Smith C., Schneider P.M. DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Forensic Sci. Int. 2006:187–197. doi: 10.1016/j.forsciint.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 10.S. Schneider, D. Roessli, L. Excoffier, Arlequin ver. 2.0: A software for population genetics data analysis, 2.0 ed., Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland, 2000.
- 11.Bandelt H.-J., Forster P., Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 12.Qamar R., Ayub Q., Mohyuddin A., Helgason A., Mazhar K., Mansoor A., Zerjal T., Tyler-Smith C., Mehdi S.Q. Y-chromosomal DNA variation in Pakistan. Am. J. Hum. Genet. 2002;70:1107–1124. doi: 10.1086/339929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler J.M. Recent developments in Y-short tandem repeat and Y-single nucleotide polymorphism analysis. Forensic Sci. Rev. 2003;15:91–111. [PubMed] [Google Scholar]
- 14.Lattanzi W., Di Giacomo M.C., Lenato G.M., Chimienti G., Voglino G., Resta N., Pepe G., Guanti G. A large interstitial deletion encompassing the amelogenin gene on the short arm of the Y chromosome. Hum. Genet. 2005;116:395–401. doi: 10.1007/s00439-004-1238-z. [DOI] [PubMed] [Google Scholar]
- 15.Thangaraj K., Reddy A.G., Singh L. Is the amelogenin gene reliable for gender identification in forensic casework and prenatal diagnosis? Int. J. Legal. Med. 2002;116:121–123. doi: 10.1007/s00414-001-0262-y. [DOI] [PubMed] [Google Scholar]