Abstract
Hypertrophic Cardiomyopathy (HCM) is a common primary cardiac disorder defined by a hypertrophied left ventricle, is one of the main causes of sudden death in young athletes and has been associated with mutations in most sarcomeric proteins (tropomyosin, Troponin T and I, and actin, etc.). Many of these mutations appear to affect the functional properties of cardiac troponin C (cTnC), i.e., by increasing the Ca2+-sensitivity of contraction, a hallmark of HCM, and surprisingly, prior to this report, cTnC had not been classified as a HCM susceptibility gene. In this study, we show that mutations occurring in the human cTnC (HcTnC) gene (TNNC1) have the same prevalence (~0.4%) as well established HCM-susceptibility genes that encode other sarcomeric proteins. Comprehensive open reading frame/splice site mutation analysis of TNNC1 performed on 1025 unrelated HCM patients over the last 10 years revealed novel missense mutations in TNNC1: A8V, C84Y, E134D, and D145E. Functional studies with these recombinant HcTnC HCM mutations showed increased Ca2+ sensitivity of force development (A8V, C84Y and D145E) and force recovery (A8V and D145E). These results are consistent with the HCM functional phenotypes seen with other sarcomeric HCM mutations (E134D showed no changes in these parameters). This is the largest cohort analysis of TNNC1 in HCM that details the discovery of at least three novel HCM-associated mutations and more strongly links TNNC1 to HCM along with functional evidence that supports a central role for its involvement in the disease. These types of studies may help to further define TNNC1 as an HCM-susceptibility gene that has already been established for the other members of the Troponin complex.
Keywords: troponin C, TnC, hypertrophic cardiomyopathy, HCM, mutation, calcium, genetics
Introduction
The leading cause of sudden cardiac death in the young is hypertrophic cardiomyopathy (HCM) which affects approximately 1 in 500 individuals and is defined clinically as thickening of the left ventricle and septum in the absence of any identifiable cause [1–3]. Through initial linkage studies and subsequent hypotheses that HCM was a disease of the sarcomere, investigations over the past two decades have led to the identification of hundreds of HCM-associated mutations scattered among the various sarcomeric genes [4–10]. This is reflected in the commercially-available clinical genetic tests for HCM which scan for mutations in the genes encoding β-myosin heavy chain, myosin binding protein C, cardiac troponin I, cardiac troponin T, α-tropomyosin, cardiac actin, regulatory myosin light chain, and ventricular myosin light chain. Despite these tremendous advances, approximately 20% of patients with reverse curve-HCM and nearly 90% of the patients with sigmoidal-HCM are genotype negative with respect to the genetic test panel for sarcomeric/myofilament HCM [11–13]. Notably absent from this list is the TNNC1-encoded human cardiac troponin C (HcTnC) which has yet to be firmly associated with HCM [14, 15]. To date, only one mutation in TnC has been linked to a 60 year old HCM patient [16]. In a small cohort based study, the authors did not find the L29Q TnC mutation in any other patient (the number of HCM patients screened was unreported) nor in 96 healthy volunteers and they concluded that additional studies would be necessary to elucidate whether TnC should be considered in fact a disease gene for HCM [16].
Cardiac troponin is a heterotrimeric complex comprised of a Ca2+-binding subunit TnC, an inhibitory subunit troponin I (TnI) encoded by TNNI3, and an elongated troponin T (TnT) encoded by TNNT2. TnC acts as a cytosolic Ca2+ sensor which, when bound to the divalent cation at the single Ca2+-specific binding site, strengthens its interaction with TnI and transversely weakens the inhibitory function of TnI causing its release from actin. The troponin-tropomyosin complex then shifts deeper into the actin groove thereby exposing the myosin binding sites on actin making them available for contraction (for review see [17]). cTnC belongs to the EF-hand superfamily of Ca2+ binding proteins and consists of N and C terminal globular domains that are connected through a flexible linker. Each globular domain has a pair of EF-hand helix-loop helix Ca2+ binding motifs [18, 19]. The C-terminus (also called as structural domain) contains two high affinity Ca2+ binding sites III and IV (~107 M−1) that also binds to Mg2+ competitively with low affinity (~103 M−1) and the N-terminus contains only one functional low affinity “Ca2+-specific” regulatory Ca2+ binding site II (~105 M−1) [20–22]. The N-terminus is considered the regulatory domain since Ca2+ binding initiates muscle contraction [23, 24]. In this manner, TnC represents a critical molecular switch through which defects in the primary sequence of the protein may disrupt the TnC-Ca2+ regulation process.
At least 90% of HCM Tn mutations (TnT and TnI), that have been investigated in situ cause an increase in the Ca2+ sensitivity of force development that would result in increased force at sub-maximal Ca2+ concentrations [25–27]. The same functional phenotype has also been observed in transgenic mice containing Tn mutations related to HCM when compared to the WT. Also there seems to be a correlation between the change in Ca2+ sensitivity of force development and time of onset of disease and prognosis [25–27]. Only one HCM-associated TnC mutation (L29Q) has been functionally studied by two different groups. Schmidtmann et al, showed, using a reconstituted fast skeletal system containing cardiac troponin complex, a decrease in the Ca2+ sensitivity measured by ATPase activity and in vitro motility assays [28]. However, Liang et al have shown an increase in the Ca2+ sensitivity of force development measured in TnC depleted mouse skinned cardiac myocytes reconstituted with recombinant mouse cardiac TnC [29].
Since a seven year span has elapsed since the first report of an HCM-associated mutation in cTnC and no subsequent reports have shown any linkage between HCM and cTnC, we sought to determine whether genetic perturbations in TNNC1 may play a role in the pathogenesis of HCM in a large cohort-based study. We report four novel missense mutations in HcTnC – A8V, C84Y, E134D, and D145E – which at least three alter the Ca2+ sensitivity of contraction when reconstituted into TnC-depleted porcine cardiac muscle fibers. Furthermore, these results suggest that a role for calcium mishandling in the pathogenesis of sarcomeric-HCM and warrants further scrutiny.
Methods
Study Population
Between April 1997 and April 2007, 1025 unrelated patients, evaluated in the Hypertrophic Cardiomyopathy Clinic at Mayo Clinic, Rochester, Minnesota, consented to genetic testing. Following receipt of written consent for this Mayo Foundation Institutional Review Board-approved protocol, DNA was extracted from peripheral blood lymphocytes using the Purgene DNA extraction kit (Gentra, Inc, Minneapolis, MN). Clinical data was collected on all patients including physical examination, pertinent personal and family history, 12-lead electrocardiogram (ECG) analysis, and echocardiographic testing to determine maximum left ventricular wall thickness (MLVWT) and maximum left ventricular outflow tract gradient (MLVOT). Each of the subjects met the clinical diagnostic criteria for HCM of a MLVWT greater than 13 mm in the absence of other confounding diagnoses.
Troponin C Mutational Analysis
All six TNNC1 exons, with flanking intronic regions and splice junction, were amplified by PCR using oligonucleotide primers. Each amplicon was evaluated for mutations using denaturing high performance liquid chromatography (DHPLC, Transgenomic, Omaha, Nebraska), and samples with an abnormal elution profile were directly sequenced (ABI Prism 377, Applied Biosystem, Foster City, CA) to characterize the difference between the wild type and variant alleles. Primer sequences, PCR, and DHPLC conditions are available upon request. Using previously published conditions, TNNC1-positive subjects were analyzed for mutations in 15 established HCM-susceptibility genes including the eight genes that comprise the commercially available genetic test for sarcomeric-HCM.
Site-Directed Mutagenesis, Expression, and Protein Purification of Human Cardiac Troponin C
The cDNA for human cardiac TnC (HcTnC) was cloned previously in our laboratory by RT-PCR using human heart total RNA (Stratagene), sequence specific primers and a Omniscript RT Kit (Qiagen) [30]. The HcTnC-cDNA was used as a template for PCR using primers designed to produce the specific mutants: A8V, C84Y, E134D and D145E. All subcloned DNA sequences were inserted into the pET3d expression plasmid and sequenced to verify that sequences were correct prior to expression and purification. Escherichia coli BL21 (DE3) Codon plus bacterial cells were transformed with pET-3d constructs containing HcTnC. After sonication (Heat Systems XL 2020), the cell lysate containing the HcTnC was centrifuged at 39,000 g for 45 m at 4°C and first applied onto a pre-equilibrated fast flow Q-Sepharose column and eluted in buffer 50 mM Tris-HCl pH 7.8, 6 M Urea, 1 mM EDTA and 1 mM DTT using a 0 – 0.6 M KCl gradient. The cleanest fractions were then dialyzed against 50 mM Tris-HCl pH 7.5, 1 mM CaCl2, 1 mM MgCl2, 50 mM NaCl and 1 mM DTT. After HcTnC was extensively dialyzed, ammonium sulfate was added to a final concentration of 0.5M and the protein was loaded onto a pre-equilibrated Phenyl Sepharose column. Pure HcTnC was directly eluted using a buffer containing 50 mM Tris-HCl, 1 mM EDTA, 1 mM DTT, pH 7.5. Fractions of > 98% purity as determined by SDS-PAGE were pooled, dialyzed extensively against 5 mM ammonium bicarbonate and then lyophilized.
Fiber Preparation and Determination of the Ca2+ Dependence of Force Development
Cardiac tissue from newly slaughtered pigs was obtained from a nearby slaughterhouse. Strips of muscle, 3–5 mm in diameter and ~5 mm in length were dissected from the papillary muscle of the left ventricle and skinned overnight in a 50% glycerol relaxing solution containing low Ca2+ concentration (10−8 M [Ca2+]free, 1 mM [Mg2+]free, 7 mM EGTA, 2.5 mM MgATP2−, 20 mM MOPS (pH 7.0), 20 mM creatine phosphate, and 15 units/ml creatine phosphokinase, I = 150 mM) and 1% Triton X-100 at − 20°C. Fibers were then transferred to a similar solution without Triton X-100 and kept at − 20°C. A small skinned fiber bundle with the diameter of ~ 75 – 100 µm was mounted using stainless steel clips to a force transducer and immersed in a pCa 8.0 relaxation solution (described above). The contraction solution (pCa 4.0) had the same composition as the pCa 8.0 solution except that the Ca2+ concentration was 10−4m and was used to measure the initial force. To analyze the Ca2+ dependence of force development, the skinned fiber tension was tested in intermediate Ca2+ solutions ranging from pCa 8.0 to 4.0 that were calculated using the pCa calculator program developed in our laboratory [31]. Data was analyzed using the following equation: % Change in Force = 100 X [Ca2+]n / ([Ca2+]n + [Ca2+ 50]n) where “[Ca2+50]” is the free [Ca2+] that produces 50% force and “nHill” is the Hill coefficient. All fiber experiments were carried out at room temperature (22°C).
The native cardiac TnC was depleted by incubating the fiber in a CDTA extracting solution (5 mM CDTA and 25 mM Tris-HCl, pH 8.4) for ~ 1.5h. The efficiency of TnC extraction the residual tension was assessed by activating the fiber in pCa 4.0 solution. Residual tensions of 15% or below from the initial maximal force were considered satisfactory for experimentation. After this, fibers were successively incubated for 20 min with 28 µM of mutant or WT HcTnC diluted in pCa 8.0 solution. To ensure exogenous cTnC reconstitution the force recovery was verified using pCa 4.0 solution. We only considered the fiber fully reconstituted when there was no additional increase in the force recovery values obtained using pCa 4.0 after successive TnC incubations. The average time for full recovery of force for all the proteins was ~ 1 – 1.3h (incubation of three to four times 20 min with recombinant protein).
Statistical analysis and three-dimensional modeling
For the clinical studies ANOVA analysis was performed to establish differences between experimental groups. HcTnC mutations were modeled in each functional domain of PDB (1AJ4) using PyMol software. Student’s t test was used to determine the significance of skinned fiber Ca2+ sensitivity and force recovery. P-values less than 0.05 were considered statistically significant.
Results
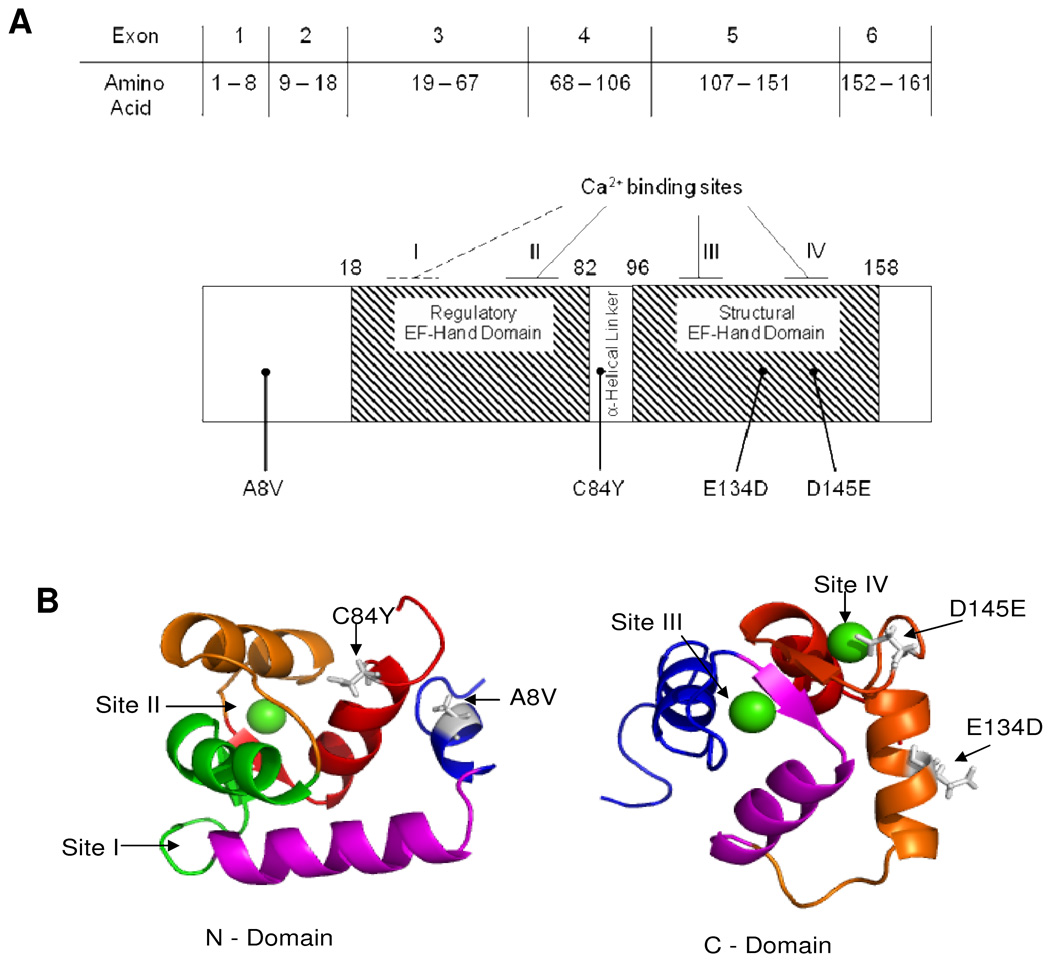
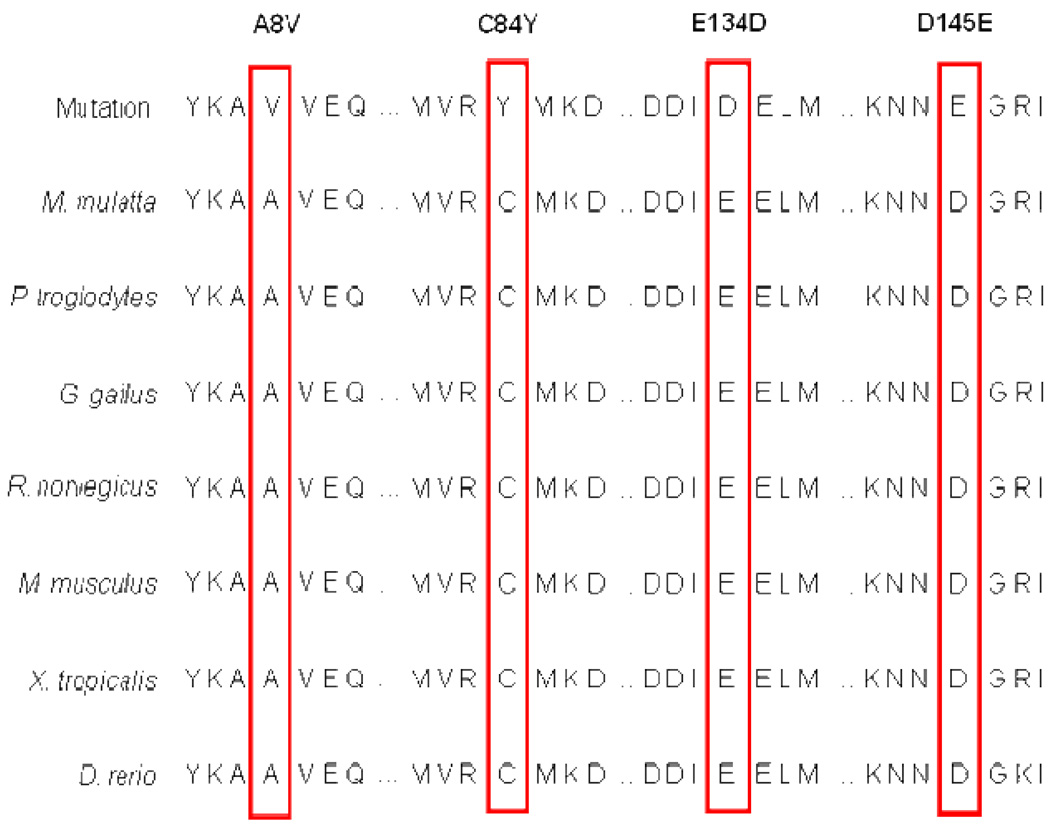
The demographics for one of the largest ever assembled cohorts of unrelated patients with HCM (N=1025) are shown in the first column of Table 1. The mean age at diagnosis was 49.2 ± 18 years with a maximal left ventricle wall thickness (MLVWT) of 22.3 ± 7 mm. Four novel missense mutations in TNNC1 were discovered in four unrelated, Caucasian patients within this cohort: Ala8Val (nucleotide 23, C>T), Cys84Tyr (nucleotide 251, G>A), Glu134Asp (nucleotide 402, G>T), and Asp145Glu (nucleotide 435, C>A). The topographical location for each mutation is shown in Figure 1A as well as the three dimensional position of each residue on the amino- and carboxy-termini of HcTnC Figure 1B. The missense mutations involved residues that were completely conserved across all species queried (Figure 2). No other mutations were detected in these four patients following comprehensive open reading frame/splice site mutational analysis of known HCM-susceptibility genes including the eight genes comprising the commercially available genetic test for sarcomeric HCM. As a control, a panel of 1000 reference alleles derived from 100 African American and 200 Caucasian Coriell Repository (Camden, NJ) DNA samples and 200 Caucasian subjects with normal screening electro- and echo-cardiograms were comprehensively genotyped for TNNC1. Absence of these variants in 1000 reference alleles demonstrates with 95% confidence that the true allelic frequency of these variants is less than 0.003 – statistically excluding the possibility that the mutations are genetic polymorphisms. Furthermore, with the exception of a single synonymous variant (G70G) found in a single reference allele, no sequence variants of any type were observed throughout the coding region and splice sites of all control alleles (data not shown).
Table 1.
Clinical Characteristics of HCM Cohort
| Clinical Characteristic | HCM Cohort | TNNC1 Positive |
|---|---|---|
| No. of Individuals | 1025 | 4 |
| Male/Female | 621/404 | 3/1 |
| Age at Diagnosis (years) | 49.2 ± 18 | 29.4 ± 10.6 |
| Cardiac Symptoms | 55% | 75% |
| Max LVWT (mm) | 22.3 ± 7 | 24.3 ± 2.7 |
| Resting LVOTO (mmHg) | 43.9 ± 44 | 77.3 ± 19.2 |
| Pos. FH for HCM | 277 (27%) | 1 (25%) |
| Pos. FH for SCD | 185 (18%) | 0 (0%) |
| Surgical Myectomy | 376 (37%) | 3 (75%) |
| Pacemaker | 176 (17%) | 0 (0%) |
| ICD | 167 (16%) | 1 (25%) |
Values are mean ± SD or n (%). FH, family history; HCM, hypertrophic cardiomyopathy; ICD, implantable cardioverter-defibrillator; LVOTO, left ventricular outflow tract obstruction; LVWT, left ventricular wall thickness.
Figure 1. Mapping and modeling of HCM-susceptibility mutations in TNNC1-encoded cardiac troponin C (HcTnC).
A) The gene and protein linear topology of HcTnC, including exon, splice junction, functional domains, and calcium binding site locations. The location of each of the four mutations is noted respectively. B) N-terminus (left) depicting the location of A8V and C84Y mutations in relationship to the Ca2+ binding sites and HcTnC helices. Ca2+ binding site I is defunct and Ca2+ binding site II shows Ca2+ bound (green sphere); N-helix (blue), A-helix (pink), B-helix (green), C-helix (orange), and D-helix (red). The Ala8 is located in the first helix of TnC in the beginning of the flexible linker connecting the two domains. C-terminus (right) depicting the location of E134D and D145E mutations in relationship to Ca2+ binding sites and helices. Ca2+ binding site III and IV pictured with Ca2+ bound (green spheres); E-helix (blue), F-helix (pink), G-helix (orange), H-helix (red). Glu134 is located in the G-helix between Ca2+ binding sites III and IV and Asp145 is situated at the Z position of Ca2+ coordinating residues of site IV.
Figure 2. Sequence conservation.
The identified mutations in TNNC1 localize to residues completely conserved across all species queried.
The clinical characteristics of the patients with TNNC1-HCM are summarized in Table 1 and 2. The TNNC1 genotype-positive subjects were diagnosed with HCM at 29.4 ± 10.6 years with a MLVWT of 24.3 ± 2.7 mm. As summarized in Table 2, Cases 1, 2, and 3 had no apparent family history of HCM among either first- or second-degree relatives suggesting the possibility of a sporadic de novo mutation or incomplete penetrance. Case 4 has a positive family history consistent with autosomal dominant, familial HCM involving a brother, a maternal grandmother, two maternal uncles, and two daughters of the noted maternal uncles. Lastly, Case 3, a female diagnosed in childhood with HCM died at 22 years of age of an unspecified cause. She had undergone extended surgical myectomy several years previously for management of significant left ventricular outflow tract obstruction (LVOTO) that was refractory to pharmacotherapy. Unfortunately, relatives of all four families have declined participation precluding a molecular determination of co-segregation or sporadicity.
Table 2.
Clinical Phenotype of TNNC1-Positive Patients with HCM
| Case | Mutation | Age (y)/ Sex | Age at Dx (y) | Race | Symptoms at Presentation | Subsequent symptoms | AF | Max LVWT (mm) | Resting LVOTO (mmHg) | FH of HCM | FH of SCD | Treatment |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A8V | 37/M | 33.9 | C | Dyspnea | Dyspnea, chest pain | No | 18 | 117 | No | No | β-blockade, myectomy |
| 2 | C84Y | 17/M | 8.4 | C | Syncope on exertion | Pre-syncope | No | 19 | 32 | No | No | β-blockade |
| 3 | E134D | 22a/F | 16 | C | Chest pain | Dyspnea, chest pain | No | 26 | 60b | No | No | β-blockade, Ca2+-channel blockade, extended myectomy |
| 4 | D145E | 58/M | 57.3 | C | Chest pain | Dyspnea, chest pain | No | 22 | 100 | Yes | No | β-blockade, myectomy |
HCM, hypertrophic cardiomyopathy; M, male; F, female; Dx, diagnosis; C, Caucasian; SBE, subacute bacterial endocarditis; AF, atrial fibrillation; LVWT, left ventricular wall thickness; LVOTO, left ventricular outflow tract obstruction; FH, family history; SCD, sudden cardiac death; ICD, implantable cardioverter-defibrillator; NA; not available
deceased
distal left ventricular gradient with a 15 mmHg gradient at rest and provocation across outflow tract.
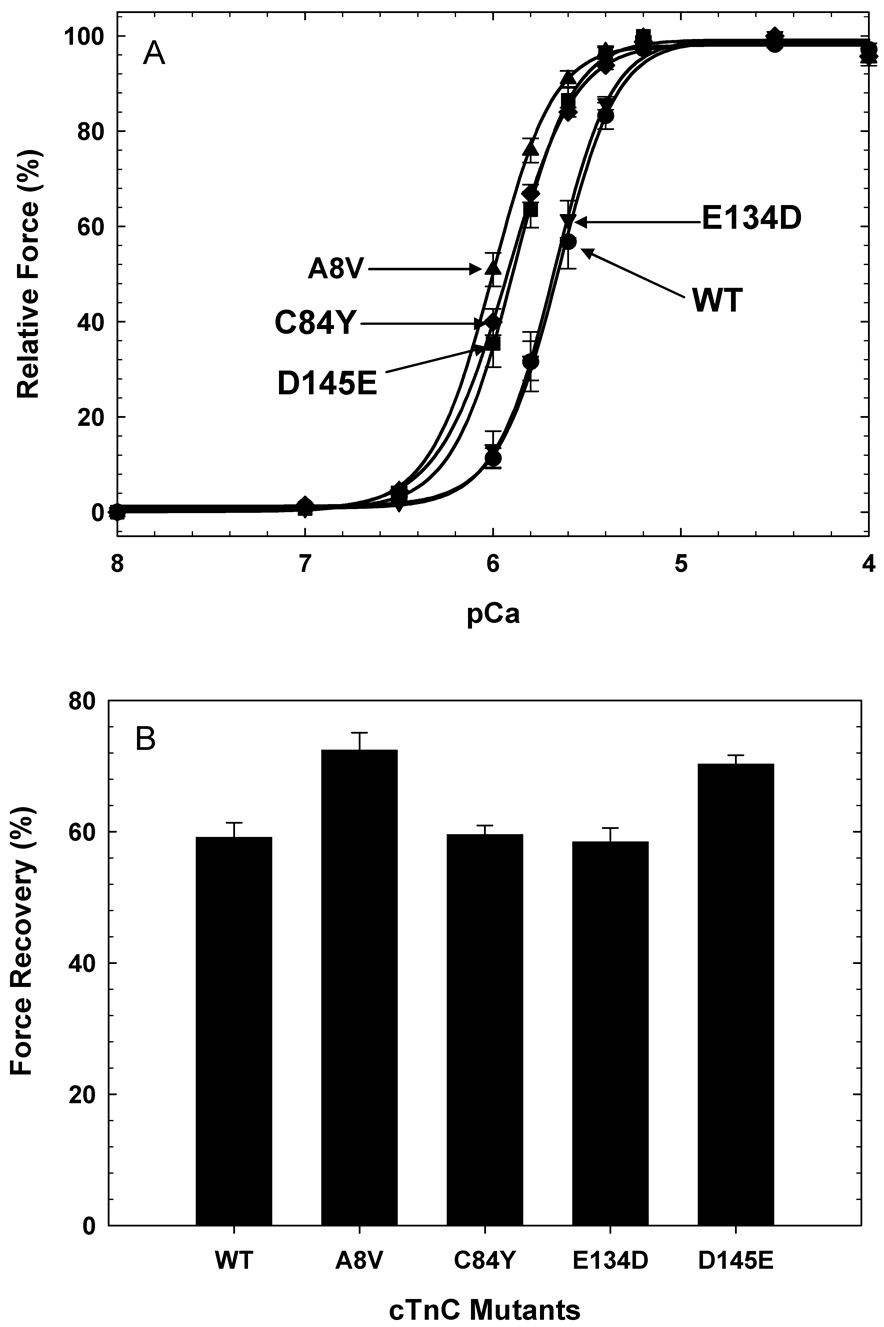
To determine whether these TNNC1 mutations functionally perturb myofilament Ca2+ regulation, the Ca2+ sensitivity of force development and force recovery were evaluated using cTnC-depleted, porcine cardiac skinned fibers reconstituted with each HcTnC mutant. A8V produced the largest leftward shift (~0.4 log units) from wild type, followed by D145E and C84Y (~0.3 log units), while E134D showed no significant difference compared to the wild type (Figure 3A). Furthermore, the force recovery under the same conditions of fiber incubation, i.e., same HcTnC concentration and dissolving buffer (see methods for additional details) was shown to be significantly increased in fibers reconstituted with A8V and D145E (Figure 3B). The pCa50 (defined as the [Ca2+] to reach 50% of the maximal tension), nHill, and the relative force recovery values are summarized in Table 3.
Figure 3. The Ca2+ dependence of force development and maximal relative force in reconstituted muscle fibers.
A) Ca2+ dependence of force development in (●) WT, (▲) A8V, (♦) C84Y, (▼) E134D, and (■) D145E. B) Relative force recovery measured after HcTnC reconstitution normalized to the initial force. Data in each experiment are the average of 7–9 experiments and are expressed as mean ± S.E. in Table 3.
Table 3.
Summary of Skinned Cardiac Fibers Reconstituted with Mutant Cardiac Troponin C
| HcTnC | pCa50 | ΔpCa50 | nHill | Force Recovery (%) | # of experiments |
|---|---|---|---|---|---|
| WT | 5.657 ± 0.012 | 0 | 2.74 ± 0.19 | 59.1 ± 2.3 | 8 |
| A8V | 6.017 ± 0.011* | + 0.36 | 2.68 ± 0.18 | 72.4 ± 2.7* | 9 |
| C84Y | 5.929 ± 0.011* | + 0.272 | 2.42 ± 0.15* | 59.5 ± 3.7 | 8 |
| E134D | 5.679 ± 0.009 | + 0.022 | 2.82 ± 0.16 | 58.4 ± 2.1 | 7 |
| D145E | 5.898 ± 0.008* | + 0.241 | 2.73 ± 0.17 | 70.3 ± 1.4* | 8 |
a ΔpCa50 = WT pCa50 – HCM TnC pCa50
P < 0.05 comparing HCM TnC with WT TnC values.
Discussion
Since the identification of the first mutation in MYH7, HCM has been viewed conceptually as a disease of the sarcomere. This is reflected in the commercially-available clinical genetic test which is comprised of a Panel A (β-myosin heavy chain, myosin binding protein C, cardiac troponin I, cardiac troponin T, and α-tropomyosin) and a Panel B (cardiac actin, regulatory myosin light chain, ventricular myosin light chain). Absent from this is cardiac troponin C which has yet to be established as a sarcomeric-HCM susceptibility gene in a cohort-based study. To this end, we report the novel discovery of 4 missense mutations, A8V, C84Y, E134D, and D145E, in a cohort of 1025 patients with HCM. In addition, our cohort demonstrates that the prevalence of TNNC1 mutations among unrelated patients with HCM is approximately 0.4% (4/1025) which is comparable in frequency to both α-tropomyosin-HCM (~0.5%) and actin-HCM (~0.3%), previously elucidated in our original Mayo cohort that comprised 388 unrelated patients with HCM [11]. Accordingly, it seems reasonable for TNNC1 to be added to the panel of HCM-susceptibility genes.
In the functional studies, surprisingly, the A8V and D145E mutations, located in two different functional regions of HcTnC, demonstrated a nearly equivalent increase in Ca2+ sensitivity of force development and force recovery. The A8V mutation is located in the N-Helix of the amino-terminal domain of HcTnC, a region known to affect the Ca2+ affinity of the regulatory EF-hand domain. Indeed, deletion of residues 1–14 in skeletal TnC, corresponding to 1–11 in HcTnC, produced alterations in the Ca2+ sensitivity of force development and maximal force [32, 33]. Conversely, the D145E mutation may disrupt or lessen the affinity of Ca2+/Mg2+ site IV for its divalent cation (Figure 1). This possibility is supported by prior work demonstrating that Asp to Ala mutations in HcTnC at the Z-coordinated position of the Ca2+ binding loop disrupt Ca2+ binding to sites III and/or IV resulting in an increase in the Ca2+ sensitivity of force development [34]. Importantly, the effect of this mutation on Ca2+-sensitivity would likely be indirect, as the binding of Ca2+ at site IV plays a structural role in HcTnC. It is possible that the effects seen on force recovery with the A8V and D145E mutations may be due to their intrinsic properties in modulating the ratio between strong and weak crossbridges or due to alterations in their affinity for the thin filament consequently intervening in the protein incorporation. In any event, the altered force recovery seen with these mutants may be an important new phenotype not observed previously.
In case 2 (C84Y), the cysteine 84 is situated at the beginning of the central helix and has been shown to affect Ca2+ regulation and maximal force generation, as this position is involved with changes in the orientation between the central helix and the N-terminal domain [35, 36]. Ca2+ binding to the N-terminal portion of HcTnC induces separation of the C-Helix from the central helix allowing greater exposure of Cys 84 to solvent [37]. The substitution of a bulky tyrosine at this position could modify the angle between these two domains mimicking an intermediate HcTnC Ca2+ open state and consequently cause an increase in the Ca2+ sensitivity of force development. C84Y was the only mutation that showed a slight but significant decrease in the cooperativity of force development. However, it is known that variation in active TnC content or TnC Ca2+ affinity can modify cooperativity [38, 39].
While the E134D HcTnC mutant had no effect on the parameters measured, the mutation may indirectly induce misregulation of another physiological system. For example, the only reported HCM HcTnC mutation to date, L29Q, appears to diminish the effects of PKA phosphorylation of cardiac troponin I [28]. Additionally, despite its rarity and species conservation, it is possible that E134D is not a pathogenic, HCM-causing mutation but simply a functionally/clinically insignificant rare variant.
Due to a lack of studies relating TnC to HCM, not much information was available in the literature about the possible functional consequences of such a mutation. However, two groups have been investigating the possible molecular mechanism of the only mutation described in cTnC that is linked to HCM (L29Q). In contrast to our results, Schmidtmann, et al. using reconstituted fast skeletal muscle myosin, actin and tropomyosin combined with cardiac troponin, reported a decrease in the Ca2+ sensitivity measured by ATPase activity and in vitro motility studies [28]. The contractile machinery has a great deal of cooperativity between proteins and the presence of proteins from different muscle systems may mask effects of the mutations on the cardiac system. The paper from Liang, et al. corresponded with our results and showed a leftward shift in the Ca2+ sensitivity of force development in a more physiological system, i. e. mouse skinned fibers reconstituted with mouse cTnC [29]. The disparate results from the two groups demonstrate that results from functional studies can differ, possibly due to the dissimilar systems in which the experiments were conducted.
The effect of the HcTnC mutations on Ca2+ sensitivity and force recovery may also be caused by alterations in myosin crossbridge interactions with the thin filament. An explanation of this phenomenon is that the ratio between strongly and weakly bound crossbridges affects the Ca2+ sensitivity of force development. In addition, changes in TnC conformation may occur depending on the number and type of attached crossbridges and conversely TnC may control the kinetic parameters of crossbridges in skinned fibers [40, 41]. These reports indicate the existence of a bi-directional mechanism of TnC and crossbridges that could explain some of the results. More structural and functional studies need to be performed to elucidate how these mutations alter thin filament regulation.
Importantly, these observations warrant increased scrutiny of calcium mishandling as a novel pathogenic mechanism of disease in HCM based on the functional results that demonstrate that changes in Ca2+ sensitivity of contraction can also be caused by mutations in cTnC. Changes in the Ca2+ sensitivity of contraction may lead to an altered state of activation/relaxation of muscle contraction which is phenotypically manifested by thickening of the ventricular walls and eventually leads to diastolic dysfunction [42]. Mutations in genes encoding calcium-handling or calcium-sensitive proteins are a newly established pathogenic mechanism for HCM. Indeed, mutations in the promoter and coding regions of phospholamban, a regulator of the sarcoplasmic reticulum calcium ATPase (SERCA2a) and a modulator of calcium flux within the cell, have been identified in HCM and dilated cardiomyopathy (DCM) respectively [43–45]. Recently, mutations in junctophilin-2 (JPH2), a putative structural cardiac protein, confer susceptibility for HCM through disruption of the cardiac dyad and perturbed calcium-induced calcium release of the contracting cardiocyte [46]. Despite all the investigations, it is still poorly understood how these mutations in proteins related to Ca2+ cell homeostasis could lead to drastic cardiac remodeling.
We are reporting for the first time the relationship between newly discovered HcTnC mutations and the substantial effects they have on key functional parameters that may lead to an explanation of the mechanism of disease in these HCM patients. TnC is a major intracellular Ca2+ buffer and the sarcomeric protein responsible for triggering muscle contraction. There is also growing evidence that many of the sarcomeric HCM mutations (tropomyosin, Troponin T and I, and actin, etc.) ultimately work through TnC as these mutations affect the Ca2+ affinity of cTnC, making it a good target for the development of new therapeutics. To this end, we have detailed the discovery of 4 novel missense mutations in TNNC1: A8V, C84Y, E134D, and D145E, in a cohort of 1025 patients with HCM. Derived from the largest assembled cohort of unrelated patients with HCM, we provide molecular and functional evidence suggesting that mutations in TNNC1 may be a novel pathogenic basis for HCM. In conclusion, this report, shows that TNNC1-HCM occurs at a similar frequency to two of the eight genes that currently comprise the commercially available genetic test for sarcomeric-HCM and indicate that the TNNC1 should be routinely included in the genetic tests that screen for HCM mutations.
Acknowledgments
Supported by NIH Grants HL-67415 (JDP) and HL-42325 (JDP) and the Windland Smith Rice Comprehensive Sudden Cardiac Death Program (MJA)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maron BJ, Roberts WC, McAllister HA, Rosing DR, Epstein SE. Sudden death in young athletes. Circulation. 1980;62:218–229. doi: 10.1161/01.cir.62.2.218. [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ, Epstein SE, Roberts WC. Causes of sudden death in competitive athletes. J Am Coll Cardiol. 1986;7:204–214. doi: 10.1016/s0735-1097(86)80283-2. [DOI] [PubMed] [Google Scholar]
- 3.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 4.Thierfelder L, Watkins H, MacRae C, Lamas R, McKenna W, Vosberg HP, et al. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell. 1994;77:701–712. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 5.Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, et al. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 6.Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 7.Kimura A, Harada H, Park JE, Nishi H, Satoh M, Takahashi M, et al. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat Genet. 1997;16:379–382. doi: 10.1038/ng0897-379. [DOI] [PubMed] [Google Scholar]
- 8.Mogensen J, Klausen IC, Pedersen AK, Egeblad H, Bross P, Kruse TA, et al. Alpha-cardiac actin is a novel disease gene in familial hypertrophic cardiomyopathy. J Clin Invest. 1999;103:R39–R43. doi: 10.1172/JCI6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson TM, Doan TP, Kishimoto NY, Whitby FG, Ackerman MJ, Fananapazir L. Inherited and de novo mutations in the cardiac actin gene cause hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2000;32:1687–1694. doi: 10.1006/jmcc.2000.1204. [DOI] [PubMed] [Google Scholar]
- 10.Poetter K, Jiang H, Hassanzadeh S, Master SR, Chang A, Dalakas MC, et al. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat Genet. 1996;13:63–69. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- 11.Van Driest SL, Ellsworth EG, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Prevalence and spectrum of thin filament mutations in an outpatient referral population with hypertrophic cardiomyopathy. Circulation. 2003;108:445–451. doi: 10.1161/01.CIR.0000080896.52003.DF. [DOI] [PubMed] [Google Scholar]
- 12.Van Driest SL, Vasile VC, Ommen SR, Will ML, Tajik AJ, Gersh BJ, et al. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:1903–1910. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 13.Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clin Proc. 2005;80:463–469. doi: 10.1016/S0025-6196(11)63196-0. [DOI] [PubMed] [Google Scholar]
- 14.Liew CC, Dzau VJ. Molecular genetics and genomics of heart failure. Nat Rev Genet. 2004;5:811–825. doi: 10.1038/nrg1470. [DOI] [PubMed] [Google Scholar]
- 15.Alcalai R, Seidman JG, Seidman CE. Genetic basis of hypertrophic cardiomyopathy: from bench to the clinics. J Cardiovasc Electrophysiol. 2008;19:104–110. doi: 10.1111/j.1540-8167.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann B, Schmidt-Traub H, Perrot A, Osterziel KJ, Gessner R. First mutation in cardiac troponin C, L29Q, in a patient with hypertrophic cardiomyopathy. Hum Mutat. 2001;17:524. doi: 10.1002/humu.1143. [DOI] [PubMed] [Google Scholar]
- 17.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 18.Herzberg O, James MN. Structure of the calcium regulatory muscle protein troponin-C at 2.8 A resolution. Nature. 1985;313:653–659. doi: 10.1038/313653a0. [DOI] [PubMed] [Google Scholar]
- 19.Herzberg O, James MN. Refined crystal structure of troponin C from turkey skeletal muscle at 2.0 A resolution. J Mol Biol. 1988;203:761–779. doi: 10.1016/0022-2836(88)90208-2. [DOI] [PubMed] [Google Scholar]
- 20.Holroyde MJ, Robertson SP, Johnson JD, Solaro RJ, Potter JD. The calcium and magnesium binding sites on cardiac troponin and their role in the regulation of myofibrillar adenosine triphosphatase. J Biol Chem. 1980;255:11688–11693. [PubMed] [Google Scholar]
- 21.Potter JD, Gergely J. The calcium and magnesium binding sites on troponin and their role in the regulation of myofibrillar adenosine triphosphatase. J Biol Chem. 1975;250:4628–4633. [PubMed] [Google Scholar]
- 22.Johnson JD, Collins JH, Robertson SP, Potter JD. A fluorescent probe study of Ca2+ binding to the Ca2+-specific sites of cardiac troponin and troponin C. J Biol Chem. 1980;255:9635–9640. [PubMed] [Google Scholar]
- 23.Zot AS, Potter JD. Structural aspects of troponin-tropomyosin regulation of skeletal muscle contraction. Ann Rev Biophys Biophys Chem. 1987;16:535–559. doi: 10.1146/annurev.bb.16.060187.002535. [DOI] [PubMed] [Google Scholar]
- 24.Zot HG, Potter JD. A structural role for the Ca2+-Mg2+ sites on troponin C in the regulation of muscle contraction. Preparation and properties of troponin C depleted myofibrils. J Biol Chem. 1982;257:7678–7683. [PubMed] [Google Scholar]
- 25.Gomes AV, Potter JD. Cellular and molecular aspects of familial hypertrophic cardiomyopathy caused by mutations in the cardiac troponin I gene. Mol Cell Biochem. 2004;263:99–114. doi: 10.1023/B:MCBI.0000041852.42291.aa. [DOI] [PubMed] [Google Scholar]
- 26.Gomes AV, Potter JD. Molecular and cellular aspects of troponin cardiomyopathies. Ann N Y Acad Sci. 2004;1015:214–224. doi: 10.1196/annals.1302.018. [DOI] [PubMed] [Google Scholar]
- 27.Miller T, Szczesna D, Housmans PR, Zhao J, de Freitas F, Gomes AV, et al. Abnormal contractile function in transgenic mice expressing a familial hypertrophic cardiomyopathy-linked troponin T (I79N) mutation. J Biol Chem. 2001;276:3743–3755. doi: 10.1074/jbc.M006746200. [DOI] [PubMed] [Google Scholar]
- 28.Schmidtmann A, Lindow C, Villard S, Heuser A, Mugge A, Gessner R, et al. Cardiac troponin C-L29Q, related to hypertrophic cardiomyopathy, hinders the transduction of the protein kinase A dependent phosphorylation signal from cardiac troponin I to C. Febs J. 2005;272:6087–6097. doi: 10.1111/j.1742-4658.2005.05001.x. [DOI] [PubMed] [Google Scholar]
- 29.Liang B, Chung F, Qu Y, Pavlov D, Gillis TE, Tikunova SB, et al. The familial hypertrophic cardiomyopathy related cardiac troponin C mutation L29Q affects Ca2+ binding and myofilament contractility. Physiol Genomics. 2008;33:257–266. doi: 10.1152/physiolgenomics.00154.2007. [DOI] [PubMed] [Google Scholar]
- 30.Lang R, Gomes AV, Zhao J, Housmans PR, Miller T, Potter JD. Functional analysis of a troponin I (R145G) mutation associated with familial hypertrophic cardiomyopathy. J Biol Chem. 2002;277:11670–11678. doi: 10.1074/jbc.M108912200. [DOI] [PubMed] [Google Scholar]
- 31.Dweck D, Reyes-Alfonso A, Potter JD. Expanding the range of free calcium regulation in biological solutions. Anal Biochem. 2005;347:303–315. doi: 10.1016/j.ab.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 32.Chandra M, da Silva EF, Sorenson MM, Ferro JA, Pearlstone JR, Nash BE, et al. The effects of N helix deletion and mutant F29W on the Ca2+ binding and functional properties of chicken skeletal muscle troponin. J Biol Chem. 1994;269:14988–14994. [PubMed] [Google Scholar]
- 33.Smith L, Greenfield NJ, Hitchcock-DeGregori SE. The effects of deletion of the amino-terminal helix on troponin C function and stability. J Biol Chem. 1994;269:9857–9863. [PubMed] [Google Scholar]
- 34.Szczesna D, Guzman G, Miller T, Zhao J, Farokhi K, Ellemberger H, et al. The role of the four Ca2+ binding sites of troponin C in the regulation of skeletal muscle contraction. J Biol Chem. 1996;271:8381–8386. doi: 10.1074/jbc.271.14.8381. [DOI] [PubMed] [Google Scholar]
- 35.Sheng ZL, Francois JM, Hitchcock-DeGregori SE, Potter JD. Effects of mutations in the central helix of troponin C on its biological activity. J Biol Chem. 1991;266:5711–5715. [PubMed] [Google Scholar]
- 36.Dobrowolski Z, Xu GQ, Hitchcock-DeGregori SE. Modified calcium-dependent regulatory function of troponin C central helix mutants. J Biol Chem. 1991;266:5703–5710. [PubMed] [Google Scholar]
- 37.Fuchs F, Liou YM, Grabarek Z. The reactivity of sulfhydryl groups of bovine cardiac troponin C. J Biol Chem. 1989;264:20344–20349. [PubMed] [Google Scholar]
- 38.Regnier M, Rivera AJ, Wang CK, Bates MA, Chase PB, Gordon AM. Thin filament near-neighbour regulatory unit interactions affect rabbit skeletal muscle steady-state force-Ca2+ relations. J Physiol (Lond) 2002;540:485–497. doi: 10.1113/jphysiol.2001.013179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kreutziger KL, Gillis TE, Davis JP, Tikunova SB, Regnier M. Influence of enhanced troponin C Ca2+-binding affinity on cooperative thin filament activation in rabbit skeletal muscle. J Physiol. 2007;583:337–350. doi: 10.1113/jphysiol.2007.135426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swartz DR, Moss RL. Influence of a strong-binding myosin analogue on calcium-sensitive mechanical properties of skinned skeletal muscle fibers. J Biol Chem. 1992;267:20497–20506. [PubMed] [Google Scholar]
- 41.Guth K, Potter JD. Effect of rigor and cycling cross-bridges on the structure of troponin C and on the Ca2+ affinity of the Ca2+-specific regulatory sites in skinned rabbit psoas fibers. J Biol Chem. 1987;262:13627–13635. [PubMed] [Google Scholar]
- 42.Maron BJ. Hypertrophic cardiomyopathy: a systematic review. Jama. 2002;287:1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 43.Medin M, Hermida-Prieto M, Monserrat L, Laredo R, Rodriguez-Rey JC, Fernandez X, et al. Mutational screening of phospholamban gene in hypertrophic and idiopathic dilated cardiomyopathy and functional study of the PLN −42 C>G mutation. Eur J Heart Fail. 2007;9:37–43. doi: 10.1016/j.ejheart.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Schmitt JP, Kamisago M, Asahi M, Li GH, Ahmad F, Mende U, et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299:1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 45.Minamisawa S, Sato Y, Tatsuguchi Y, Fujino T, Imamura S, Uetsuka Y, et al. Mutation of the phospholamban promoter associated with hypertrophic cardiomyopathy. Biochem Biophys Res Commun. 2003;304:1–4. doi: 10.1016/s0006-291x(03)00526-6. [DOI] [PubMed] [Google Scholar]
- 46.Landstrom AP, Weisleder N, Batalden KB, Bos JM, Tester DJ, Ommen SR, et al. Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J Mol Cell Cardiol. 2007;42:1026–1035. doi: 10.1016/j.yjmcc.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]