Summary
Coenzyme Q is a redox active lipid essential for aerobic respiration. The Coq4 polypeptide is required for Q biosynthesis and growth on non-fermentable carbon sources, however its exact function in this pathway is not known. Here we probe the functional roles of Coq4p in a yeast Q biosynthetic polypeptide complex. A yeast coq4-1 mutant harboring an E226K substitution is unable to grow on nonfermentable carbon sources. The coq4-1 yeast mutant retains significant Coq3p O-methyltransferase activity, and mitochondria isolated from coq4-1 and coq4-2 (E121K) yeast point mutants contain normal steady state levels of Coq polypeptides, unlike the decreased levels of Coq polypeptides generally found in strains harboring coq gene deletions. Digitonin-solubilized mitochondrial extracts prepared from yeast coq4 point mutants show that Coq3p and Coq4 polypeptides no longer co-migrate as high molecular mass complexes by one- and two-dimensional Blue Native-PAGE. Similarly, gel filtration chromatography confirms that O-methyltransferase activity, Coq3p, Coq4p, and Coq7p migration are disorganized in the coq4-1 mutant mitochondria. The data suggest that Coq4p plays an essential role in organizing a Coq enzyme complex required for Q biosynthesis.
Keywords: Saccharomyces cerevisiae, Ubiquinone, Coenzyme Q, mitochondria, respiratory electron transport, Coq4
1. Introduction
Coenzyme Q, ubiquinone, (or Q), is a polyisoprenylated quinone lipid, necessary for respiratory electron transport. The length of the polyisoprenyl “tail” of Q is species dependent [1]. Q10 (the human Q isoform) is a popular dietary supplement and there is great interest in its use in treating mitochondrial, neurodegenerative and cardiovascular diseases [2–4]. However, de novo biosynthesis of Q is necessary to generate appropriate cellular quantities [5].
The Q biosynthetic pathway is best understood in several model organisms, including Escherichia coli, Saccharomyces cerevisiae, and Schizosaccharomyces pombe [6,7]. Human homologues of the yeast genes have been shown to restore Q biosynthesis in yeast mutants (reviewed in [8]). A group of respiratory deficient yeast coq mutants (coq1–coq9) were defined as Q-deficient because addition of Q to isolated mitochondria restored both NADH- and succinate-oxidation [9]. Although Q is present in other organellar compartments, its biosynthesis in S. cerevisiae takes place on the matrix side of the inner mitochondrial membrane, where all known Coq polypeptides reside [8].
Many lines of genetic evidence suggest that Q biosynthesis relies on one or more multi-subunit complexes of the Coq polypeptides. Yeast mutants harboring deletions in coq3, coq4, coq5, coq6, coq7, coq8, or coq9 genes accumulate only the first lipid Q-intermediate, 3-hexaprenyl-4-hydroxy-benzoic acid, detected after metabolic labeling with a dedicated precursor [U-14C]-4-hydroxybenzoic acid [10–12]. Coq3 O-methyltransferase activity is decreased in each of the coq null mutants relative to respiratory deficient control mutants (atp2, cor1), while COQ3 RNA levels remain constant, suggesting post-transcriptional regulation [13]. Furthermore, steady state levels of the Coq3, Coq4, Coq6, Coq7 and Coq9 polypeptides are profoundly decreased in mutants harboring deletions in any one of the COQ genes [14, 15]. The decrease in the Coq polypeptide levels observed in the coq mutants is hypothesized to be due to their degradation, secondary to destabilized Coq multi-subunit complex(es).
Recent evidence indicates direct physical interaction between the Coq polypeptides and the involvement of Q and Q-intermediates in the multi-subunit Coq polypeptide complex. Several of the Coq polypeptides in digitonin extracts of yeast mitochondria co-migrate at high molecular mass by gel filtration and BN-PAGE [15–17]. Coq3 O-methyltransferase activity is preserved under these detergent extraction conditions, and capture of a biotinylated form of Coq3p co-precipitates Coq4p [16]; recovery of HA-epitope-tagged Coq9 co-precipitates Coq4, Coq5, Coq6 and Coq7 polypeptides [15]. Demethoxy-Q, the penultimate intermediate in Q biosynthesis, co-migrates with the high molecular mass Q biosynthetic complex [16], and Q itself may be responsible for preserving high steady state levels of the Coq3 and Coq4 polypeptides [8, 17].
The Coq4 polypeptide is functionally conserved from yeast to humans [18], is essential for Coq3 O-methyltransferase activity [13], and production of Q [19]. However, the role Coq4p plays in the Q biosynthetic pathway is not known. In this work we have shown that Coq4p is necessary for the organization of the Coq polypeptide complex at high molecular mass. This suggests that the Coq4-mediated organization of Coq polypeptides in this multi-subunit complex is vital for the biosynthesis of Q.
2. Materials and Methods
2.1 Strains and growth media
Media were prepared as described [20]. Yeast strains (Table 1) were grown at 30μC in YPGal media (2% peptone, 1% yeast extract, 2% galactose, 0.1% dextrose), or YPG media (2% peptone, 1% yeast extract, 3% glycerol). Plate media contained 2% agar.
Table 1.
Genotypes and sources of S. cerevisiae strains
| Strain | Genotype | Source or Reference |
|---|---|---|
| C9E1 | MATa ade2-1 trp1-1 ura3-1 coq4-1 | [19] |
| C9E1Δ4 | MATa ade2-1 trp1-1 ura3-1 coq4::TRP1 | [19] |
| E745 | MATαmet6 coq4-2 | A. Tzagoloff* and this work |
| E718 | MATαmet6 coq4-2 | A. Tzagoloff* and this work |
| N83 | MATαmet6 coq4-3 | A. Tzagoloff* and this work |
| W303-1B | MATαade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein** |
| W303-1A | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein** |
| CC303 | W303-1B, coq3::LEU2 | [35] |
| W303-1AΔCOQ4 | W303-1A, coq4::TRP1 | [13] |
| W303-1AΔATP2 | W303-1A, atp2::LEU2 | [36] |
| NM101 | MATa leu 2-3,112 ura 3-52 coq7-1 | [37] |
Department of Biological Sciences, Columbia University, New York, USA.
Department of Human Genetics, Columbia University, New York, USA.
2.2 Characterization of yeast coq4 mutations
PCR sequencing of yeast genomic DNA was accomplished as described [21], except the final concentration of MgCl2 was 2 mM and the total reaction volume was 100 μl.
2.3 Mitochondrial isolation and solubilization, gel filtration and BN PAGE analyses
Yeast cultures were grown to an OD600nm of 2–4 and mitochondria were isolated and purified on nycodenz gradients as described [22], except that mitochondria were collected from the interface of a 14%/21% step gradient. Dodecyl maltoside was purchased from Anatrace, Inc., Maumee, Ohio and digitonin from Biosynth International, Switzerland. Essentially all procedures were as described [16, 23]. The void volume in the gel filtration analyses was established by the elution of a protein complex predicted to consist of 96 copies of the 100-kDa major vault protein [24, 25]. BN-PAGE was performed as described with 5–10% gradient gels [16, 23]. Proteins were transferred to PVDF membranes (Immobilon P, Millipore) and the dyes removed by serial immersion in 100% methanol prior to transfer to nonfat milk blocking buffer consisting of PBS (140 mM NaCl, 10 mM Phosphate, pH 7.4), 1% nonfat milk, and 0.1% Tween 20.
2.4 Immunoblot analyses
Equal aliquots (200 μl) of each gel filtration fraction were precipitated with trichloroacetic acid (final concentration 10%) and resuspended in 25 μl PBS and 25 μl 2X sample buffer. 10 μl of each fraction were loaded on a 12% Tris-Glycine SDS-PAGE gel and were subsequently transferred to PVDF membranes. Primary antibodies generated to yeast polypeptides were added to non-fat milk blocking buffer at the following initial concentrations: Coq3p, 1:1000; Coq4p, 1:1000; Coq6p, 1:250; Coq7p, 1:500; Coq9p, 1:1000; Rip1p, 1:10,000; Cytc1and Atp2 (F1β), 1:1000. Solutions of primary antibodies were reused multiple times. Goat anti-rabbit and goat anti-mouse secondary antibodies conjugated to horseradish peroxidase (Calbiochem) were each used at a 1:7,500 dilution. Proteins were detected with the ECL Super Signal West Pico® kit (Pierce) and luminescence detected by VersaDoc™ image processing software (Bio-Rad).
2.5 Coq3 O-methyltransferase assays
Assays of Coq3 O-methyltransferase activity were performed as described [13] with the following modifications. Each 250 μl reaction contained 50 mM sodium phosphate, pH 7.0, 1 mM ZnSO4, 1 mM NADH, 10 mM DTT, 50 μM 2-farnesyl-5-hydroxy-6-methoxy-3-methyl-1,4 benzoquinone [16], and 6.9 μCi S-adenosyl–L-[methyl-3H]-methionine (55–81.5 Ci/mmol, Perkin Elmer). Coq3 O-methyltransferase assays contained aliquots of intact mitochondria (80–200 μg) or gel filtration fractions (200 μl), and were incubated at 37°C for 30 minutes. To stop the reaction, each tube was transferred to dry ice and samples were extracted twice with 1 ml of heptane. Extracts were dried under N2 gas and stored at −20°C. The 3H-labeled product, Q3, was separated by reverse phase HPLC chromatography (Betabasic C18, 150 × 4.6 mm, 5 μ, Thermo Electron Corporation). Initial conditions were: Solvent A (2 mM borate) 30% and Solvent B (95% acetonitrile, 5% 2 mM borate) 70%. After injection the buffer composition and flow rate were linearly changed (time min/%B/flow rate (ml/min): 0/70/1, 2/70/1, 5/100/1.4). After 14 minutes the mobile phase and flow rate reverted to the initial state. HPLC fractions (1 min) were collected, added to 10 ml of Safety Solvent (Research Products International), and subjected to scintillation counting. Coq3 O-methyltransferase assays of gel filtration fractions were performed on the same day the gel filtration fractions were collected.
2.6 Analysis of 14C-labeled Q6
Overnight cultures of wild-type and coq4 mutant yeast were used to inoculate 0.5 l of YPGal media containing 1 μCi [U-14C]4-hydroxybenzoic acid (450 Ci/mol; American Radiolabeled Chemicals, Inc., St. Louis, MO). The cultures were incubated at 30°C with shaking (200 rpm) for 22 h and yeast cell pellets were collected and extracted for crude lipids using a modification of the Radin method as described {Marbois, 2005 #275}. Crude lipid extracts were dried under N2 gas and stored at −20°C until analysis. Crude lipids were separated by reverse phase HPLC on a 5 micron, 4.6 × 100 mm, Beta Basic C18 column (Thermo-Finigan), with a water - acetonitrile gradient at 1ml/minute (minutes/% acetonitrile: 0/90; 3/90; 30/100). Fractions (1 ml) were collected and analyzed for radioactivity by scintillation counting as described [19]. Q6 was identified by co-migration with an authentic standard detected by UV at 274 nm.
3. Results
3.1 A putative Zinc ligand motif is conserved among Coq4 homologues
The amino acid sequence of Coq4p defines a distinct polypeptide family (COG 5031.1- pfam05019), and has only very limited secondary sequence homology to other protein families. A homologue of Coq4p is present in each of the represented eukaryotic genomes and is also present in certain species of alpha, beta, delta and gamma proteobacteria, and in cyanobacteria (Figure 1). Some of these prokaryotic species are not recognized as containing Q [26].
Fig. 1.
The Coq4p sequence contains a catalytic zinc ligand motif conserved in prokaryotic species. Current database searching of prokaryotic databases reveals Coq4p homologs among alpha-, beta-, delta-, and gamma-proteobacterial classes, as well as representatives within cyanobacteria. These homologues are identified as members of COG 5031.1 (pfam05019). The Gene bank identifier (gi) numbers for the protein sequences shown, in order top to bottom are: 24212654, 75701042, 85375819, 72122094, 108762091, and 119459668. The asterisks indicate the G120E, E121K, and E226K substitutions present in the coq4-3, coq4-2, and coq4-1 alleles, respectively. The boxed residues designate the location of a putative catalytic zinc ligand motif.
We have re-examined amino acid sequence alignments of yeast Coq4p with prokaryotic homologues and identified a highly conserved putative zinc ligand motif HDxxH-(x)11-E (Figure 1). This motif is reminiscent of that found in catalytic zinc ligand domains [27–29] with structure similar to the thermolysin family of proteases. Importantly the final glutamate residue of the putative zinc ligand domain (Figure 1, asterisk) is altered in the coq4-1 allele, (E226K) [19]. The coq4-1 mutant was previously described to maintain a stable cohort of Coq polypeptides (Coq3p, Coq5p and Coq7p) as well as to maintain near wild-type levels of Coq4p [19]. This is in direct contrast to the decreased stability observed for these polypeptides in coq null strains [13].
We determined the nucleotide sequence of the coq4 alleles in three other yeast mutant strains of the coq4 complementation group (E718, E745, and N83). Each contained the silent nucleotide changes T231C and T360A previously detected in the COQ4 gene of the parental strain D273-10B [19]. An additional missense mutation G361A was detected in both E718 and E745, converting a GAG codon to AAG (E121K; termed coq4-2). A G359A mutation was detected in yeast strain N83. This mutation, coupled with the previous silent T360A, converts the GGT codon to GAA (G120E; termed coq4-3). Both amino acid substitutions occur in a highly conserved region among Coq4 eukaryotic homologues [18, 19], but are not as highly conserved among the prokaryotic homologs (Figure 1).
3.2 Coq3p O-methyltransferase activity and Coq polypeptide levels are preserved in yeast coq4 point mutants
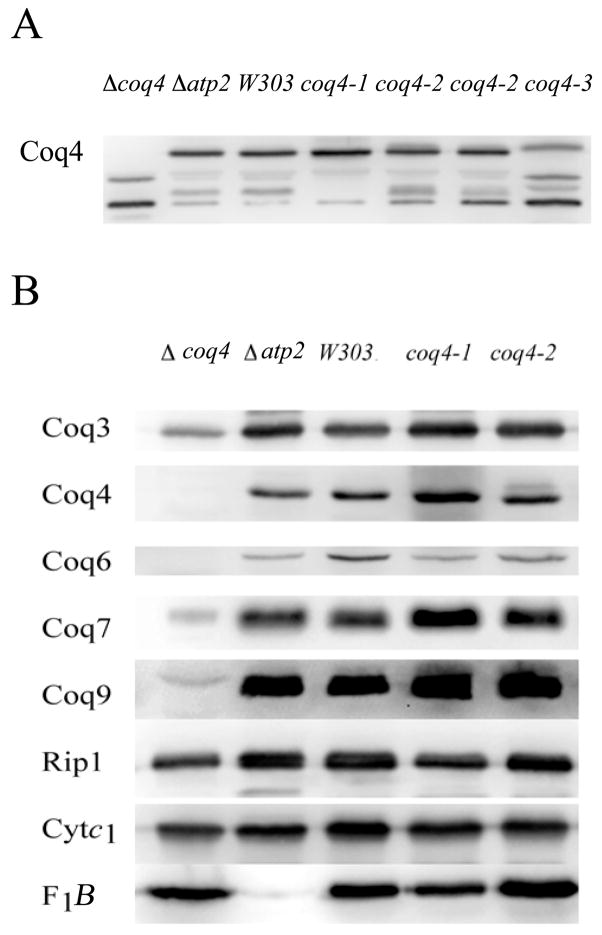
We have used the enzymatic assay for Coq3p as a bioassay for the presence of Q synthesis capability [16]. This assay revealed that mitochondria isolated from the coq4-1 mutant retain approximately 60% of the wild-type level of Coq3 O-methyltransferase activity (6.0 versus 9.8 pmol methyl groups/mg mito protein/h). In contrast, the Coq3 O-methyltransferase activity detected in mitochondria isolated from the coq4 null mutants is only 5–20% of wild-type levels [13]. Mitochondria isolated from the coq4 point mutant yeast strains were evaluated for Coq3 steady state polypeptide levels. The Coq4 polypeptide is readily detected in mitochondria isolated from the coq4-1, coq4-2, and coq4-3 point mutants (Figure 2A). At least two Coq4 products are observed; a large predominant polypeptide at 34 kDa, and a minor smaller product at 27 kDa, relative to non-specific bands identified in the coq4 null mitochondria. It is notable that initiation of Coq4 translation may occur at more than one site, as has been reported for yeast and human [18, 19]. The 27 kDa form is difficult to detect unless relatively large quantities of mitochondria are analyzed. For this reason, subsequent analyses focused on the predominant signal at 34 kDa. The coq4-1 and coq4-2 mutants also retain high steady state levels of Coq3p, Coq6p, Coq7p and Coq9p relative to the coq4 null mutant (Figure 2B). The steady state amounts of Rip1p, Cytc1, F1β (Atp2p) are not affected in the coq4 null mutant.
Fig. 2.
Coq3p, Coq4p and other Coq polypeptides are stable in mitochondria isolated from coq4 point mutants. Panel A; Mitochondria (75 μg protein) were analyzed by SDS-PAGE, and immunoblotted with anti-sera to Coq4p. Panel B, Purified mitochondria (10 μg protein) from the designated yeast strains were subjected to similar SDS-PAGE and blotting conditions. The gels were transferred to polyvinylidene fluoride membranes and standard immunodetection with the indicated antisera to Coq3, Coq4, Coq7 or Rip1 polypeptides was performed. Images were obtained via Bio-Rad Fx imager using VersaDoc™software.
3.3 The coq4 point mutant strains fail to grow on YPG, are respiratory defective and lack Q
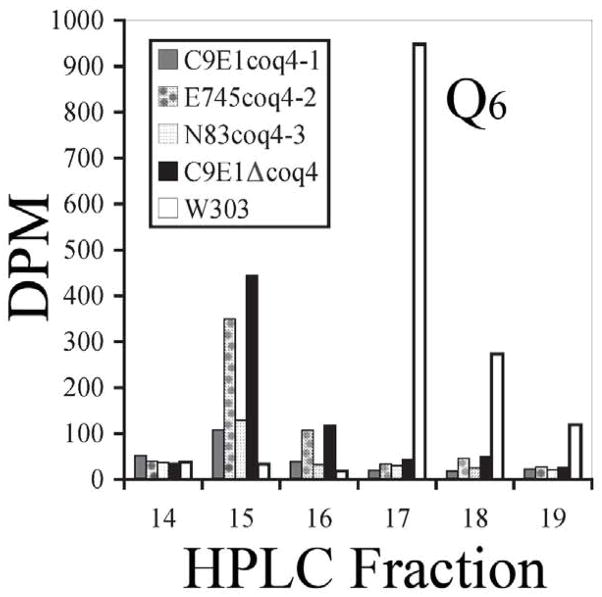
Previous analyses indicated the coq4-1 mutant had defects in respiration [19]. The coq4-1, coq4-2 and coq4-3 mutant strains are incapable of growth on YPG (data not shown). Lipid extracts of these mutant yeast strains were prepared after radiolabeling with the dedicated precursor [U-14C]-4-HB, and separated by reverse phase HPLC (Figure 3). The results show that these strains do not produce detectable quantities of Q6, and instead produce a more polar Q-intermediate, similar to that detected in the coq4 null mutant (fraction 15, Figure 3), identified previously as 3-hexaprenyl-4-hydroxybenzote [19].
Fig. 3.
Yeast coq4 point mutants have defects in Q biosynthesis. Lipid extracts prepared from the designated yeast strains labeled with [U-14C]4-hydroxybenzoic acid were separated by reverse phase HPLC, 1 ml fractions collected, and 14C-radioactivity was determined by scintillation counting. Values are plotted as the 14C-radioactivity in dpm. A coenzyme Q6 standard eluted in fraction 17.
3.4 The Coq3p/Coq4p complex is disorganized in the coq4-1 mutant
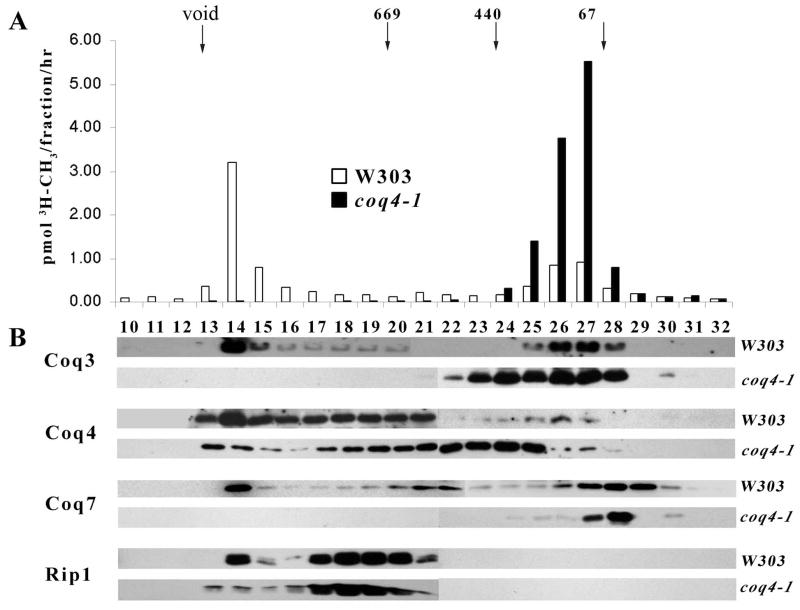
Gel filtration analyses were performed in order to study the oligomeric state of Coq3p and Coq4p within the putative Coq biosynthetic complexes of mitochondria from W303-1B wild-type yeast versus those from the coq4-1 mutant. Coq3p (EC2.1.1.114) is a key functional component of the complex [16], as it performs two O-methyltransferase steps in Q biosynthesis, and mediates the formation of Q itself [30]. The enzymatic assay of Coq3p O-methyltransferase, converts DMeQ3H2 (2-farnesyl-5-hydroxy-6-methoxy-3-methyl-1,4-benzoquinol) to Q3H2, and provides a surrogate for Q biosynthesis and complex activity. In W303-1B mitochondria (Figure 4A, open bars), Coq3 O-methyltransferase activity eluted predominantly in fraction 14, preceding the migration point of thyroglobulin (669 kDa) the highest molecular weight standard. Coq3 O-methyltransferase activity also eluted in fractions 26–27, corresponding to a size range of 50 to 150 kDa. SDS-PAGE and immunoblot analyses of the gel filtration fractions show that Coq3p is detected in two distinct peaks that correspond to the regions of Coq3 O-methyltransferase activity (Figure 4B). The elution profiles of a mitochondrial protein of known size (Rip1p/Isp2p) were also detected in the expected gel filtration fractions. A dramatic shift of Coq3p dependent O-methyltransferase activity (Figure 4A, black bars) and the Coq3p polypeptide are observed in the elution profiles of mitochondria from the coq4-1 mutant. The distribution of the Coq7p in the gel filtration fractions of W303-1B wild-type mitochondria elutes in similar fractions of high mass as found for Coq3p, however the lower mass fractions where Coq3p and Coq7p are abundant are distinct.
Fig. 4.
Coq3, Coq4, and Coq7 are displaced from high molecular mass complex in coq4-1 mutant mitochondria as compared to wild-type. A, Digitonin-extracts (2:1, g/g, detergent/protein) of mitochondria (2 mg protein) from wild-type W303 (white bars) versus coq4-1 (black bars) were separated by Superose 6 gel filtration chromatography. An aliquot of each fraction (200 of 600 μl) was assayed for Coq3-dependent O-methyltransferase activity as described in Experimental Procedures, and is depicted as pmol methyl groups/fraction/h. B, Coq3, Coq4, Coq7 or Rip1 polypeptides were detected in eluate fractions by immunoblot analyses. The elution positions of Coq3p, Coq4p and Coq7p in wild-type mitochondria are as determined previously [17], and are shown for purposes of comparison. The column void volume was established by the elution position of the vault particles in fraction 13. Elution position of calibration standards are indicated: 669 kDa, thyroglobulin; 440 kDa, ferritin; and 67 kDa, bovine serum albumin.
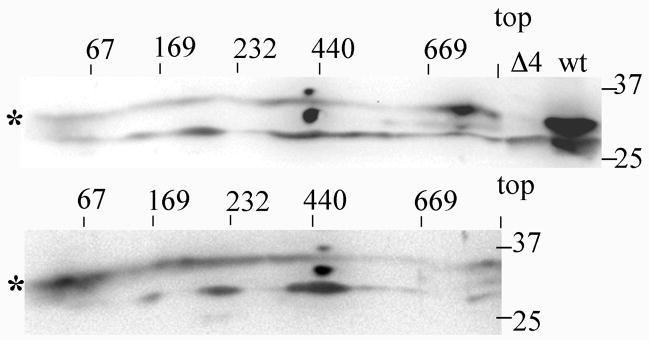
Because essentially all of Coq4p in the digitonin-solubilized mitochondria migrates as high mass complex by gel filtration, we also studied the migrations of Coq4p by BN-PAGE analyses. Analyses of Coq4p migration in the mitochondrial extracts from W303-1B wild-type and the coq4-1 mutant by two dimensional BN-PAGE (Figure 5) mimic the results of gel filtration (Figure 4B), although a broader distribution of the Coq4 polypeptide is observed in the 2D-BN-PAGE. Importantly both techniques show the most abundant signal of Coq4 is found at the highest mass location (above the 669 kDa marker). In the coq4-1 mitochondrial extracts this high mass location is disturbed and the Coq4 polypeptide appears more abundant at a lower mass (below the 67 kDa marker).
Fig. 5.
Coq4p is displaced from high molecular mass in the coq4-1 mutant strain. Upper panel: 200 μg of mitochondria were isolated from wild-type W303-1A, solubilized with digitonin as described in Figure 4, and subjected to BN-PAGE (5–13.5% gradient gel) in the first dimension and SDS-PAGE (10–13%) in the second dimension, followed by immunoblot analysis with antisera to Coq4. Aliquots of control mitochondria from a coq4 null mutant (Δ4) or from W303-1A (wt) were subjected to just the second dimension (SDS-PAGE) in lanes adjacent to the BN-gel separation (upper panel only). Lower panel: 200 μg of mitochondria were isolated from the C9E1 coq4-1 mutant. In both panels, the asterisks designate the Coq4p signal, (distinct from the non-specific signal identified in the coq4 null mutant – see upper panel). The separation of protein markers in the first dimension is indicated: 669kDa, thyroglobulin; 440 kDa, ferritin; 232 kDa, catalase; 140 kDa, lactate dehydrogenase, and 67 kDa, bovine serum albumin.
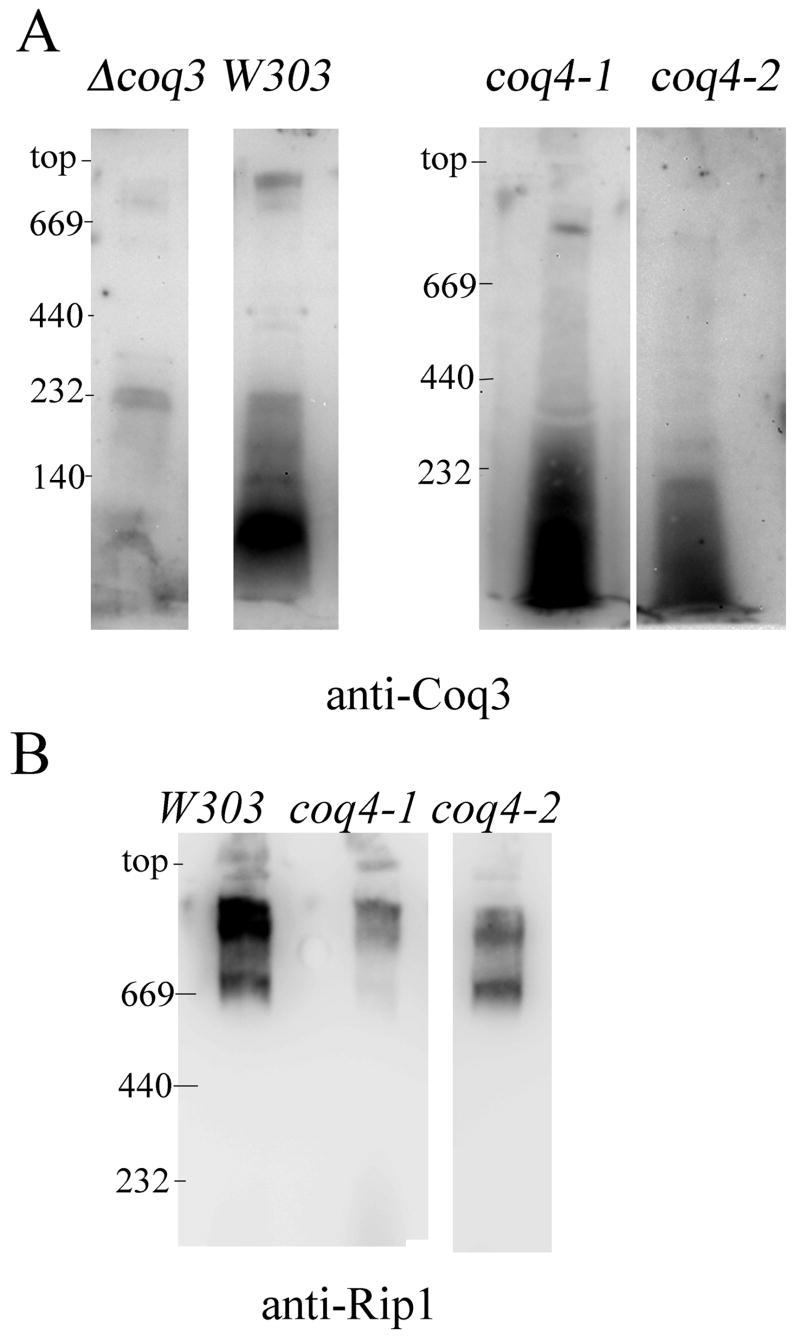
Figure 6A shows the one-dimensional BN-PAGE and subsequent immuno-detection of Coq3p in digitonin-solubilized extracts of mitochondria isolated from W303-1B versus the coq4-1 and coq4-2 mutants. A small amount of Coq3p is detected in coq4-1 mutant mitochondrial extracts at high mass in these immunoblots (shown with enhanced exposure relative to the wild-type sample). However, the amount of Coq3p at low mass in the coq4-1 mutant far exceeds that present in the corresponding wild-type sample. These analyses confirm and extend the gel filtration analyses, and indicate that the high mass location of Coq3p in W303-1B mitochondria has been disorganized in mitochondria of the coq4-1 and coq4-2 mutants to a lower molecular mass. High molecular mass complexes containing Rip1p, a subunit of the bc1 complex, are detected in the coq4-1 and coq4-2 mutants, but are altered in size and abundance (Figure 6B).
Fig. 6.
Coq3p has a high mass location in wild-type mitochondria and is displaced in mitochondria from coq3, atp2, coq4-1, and coq4-2 mutant yeast strains. One-dimensional BN-PAGE analyses (5–13.5% gradient gel) of digitonin-solubilized mitochondria from coq3 null (Δcoq3), wild-type (W303), coq4-1 or coq4-2 yeast strains. A, immuno-detection of Coq3p. B, the blot in A was stripped and re-probed with antisera to Rip1p.
4. Discussion
The data presented support a hypothesis that the presence of a multi-subunit Coq polypeptide complex at high molecular mass is necessary for Q biosynthesis in yeast. Here we characterize two yeast coq4 point mutants that lack Q, yet retain high steady state levels of the Coq3, Coq4, Coq6, Coq7, and Coq9 polypeptides and Coq3 O-methyltransferase activity. In contrast, levels of these Coq polypeptides and O-methyltransferase activity in the coq4 null mutant are drastically decreased. Typically Coq3 O-methyltransferase activity is profoundly decreased in yeast mutants harboring mutations in any of the other yeast COQ genes. What is the nature of the defect in Q biosynthesis in the coq4 point mutants? Each is shown to lack a high molecular mass complex of Coq polypeptides. The Coq3, and Coq7 polypeptides, and O-methyltransferase activity are no longer organized in a high molecular mass complex, and instead are detected only at low molecular mass by gel filtration and BN-PAGE separation techniques. Thus the formation of Q associated with the Coq3 O-methyltransferase in vitro assay does not, in fact, reflect de novo Q biosynthesis per se. Our interpretation is that a cohort of Coq polypeptides must be organized in a high molecular mass complex, and that this complex is essential for Q biosynthesis.
It is intriguing that high steady state levels of the Coq3 and Coq7 polypeptides are present in the coq4 point mutant yeast, given that neither resides in the high molecular mass complex in the coq4-1 mutant. Similarly, it is intriguing that the Coq4-1 polypeptide persists in high molecular weight fractions (Fig. 4). We surmise that in vivo a weak complex (or partial complex) is formed, protecting Coq3p and Coq7p from proteolysis. However these complexes are less stable in the coq4-1 mutant and thus only sub-complexes are observed upon in vitro solubilization and size separation; both Coq3p and Coq7p elute from the gel filtration column at higher than monomeric mass. Recent work indicates that the phosphorylation state of Coq3 impacts its association with large Q-biosynthetic polypeptide complex, and that Coq8/Abc1 is required for Coq3 phosphorylation [31]. Unlike Coq3p, Coq9p was shown to persist in the large Q-biosynthetic complex in the coq8/abc1 mutant [31]. Thus, variations in the phosphorylation state of Coq3, perhaps resulting from phosphatase activity in vitro, may affect the proportion of Coq3 retained in the Q biosynthetic complex, but allow other Coq polypeptide interactions to persist.
The large Q-biosynthetic complex containing Coq3 is present in cor1 and crd1 null yeast mutants, and is thus distinct from and independent of the complex III/IV supercomplexes [16, 31]. Conversely, as visualized by Rip1, the altered pattern of the bc1 complexes in the BN-PAGE analysis indicates that the respiratory super-complexes are affected in the coq4-1 and coq4-2 mutants. However, this phenotype cannot be attributed to the lack of Q per se, because the pattern of Rip1 is not noticeably affected in a coq7 null mutant [17], yet is profoundly diminished in a coq3 null mutant [16]. Because none of these coq mutants produce Q, and each accumulates the same predominant early Q-intermediate, the differences might be due to functional interactions of certain Coq polypeptides with respiratory super-complexes, or may simply reflect effects of different genetic background of the yeast strains examined.
The yeast coq4 point and coq4 null mutants lack Q and accumulate the typical prenylated upstream lipid Q-intermediate 3-hexaprenyl-4-hydroxybenzoate [19], indicating that Coq4p participates in one or more ring modifications in Q biosynthesis. Invariant amino acid residues present in both eukaryotic and prokaryotic Coq4 homologues, HDxxH-(x)11-E (Figure 1) are consistent with a putative catalytic zinc ligand motif. A very similar motif is present in the thermolysin family members of zinc endoproteases [32]. The E226K amino acid substitution in the coq4-1 mutant is predicted to disrupt the invariant glutamate ligand of this motif. Although it is possible that Coq4 serves a proteolytic processing function, there is no evidence that the other Coq polypeptides migrate differently in the coq-4 mutant relative to wild-type (see Figure 2). It is possible that the putative zinc ligand of Coq4 is sensitive to the redox state of mitochondria, as there is a growing body of literature on the role of redox regulation of protein liganded zinc [33]. We speculate that the observed disorganization of Coq3p in the coq4-1 mutant may derive from a defective coordination of zinc.
The presence of Coq4p homologues in delta proteobacteria (e.g. Myxococcus xanthus) and cyanobacteria (e.g. Anabaena variabilis) is intriguing as these prokaryotes lack Q, but do produce other isoprenoid quinones [34]. M. xanthus produce menaquinone, a polyisoprenylated napthoquinone, and A. variabilis produce phylloquinone, plastoquinone, and perhaps also alpha-tocopherolquinone [26, 34, 35]. The conserved zinc ligand motif in COQ4 homologues of prokaryotic species that do not synthesize Q suggests a possible broader role for Coq4p than we previously suspected. It is tempting to speculate that Coq4 functions to organize the synthesis of one or more of the isoprenoid quinones. The M. xanthus genome contains a number of other COQ gene homologues, including COQ1/ispB (a polyprenyldiphosphate synthase that generates the tail precursor), COQ5/ubiE (a C-methyltransferase that functions in both Q and menaquinone biosynthesis) and COQ8 (a putative kinase required for Q biosynthesis). Thus it is tempting to speculate that Coq4 in M. xanthus may function in the synthesis of menaquinone, while A. variabilis Coq4 may function in producing phylloquinone or plastoquinone. Conversely, many prokaryotic species (e.g. E. coli) that do synthesize Q lack a COQ4 homolog.
Our previous work indicates that Coq4 and Coq3 polypeptides interact [16], and that Coq4 interacts with the Coq5, Coq6, Coq7, and Coq9 polypeptides [15]. In light of the data presented here, Coq4 appears to be involved in organizing a high molecular mass complex of Coq polypeptides, and this complex is required for Q biosynthesis in yeast.
Acknowledgments
We thank Dr. David Auld for his comments on the Coq4p zinc ligand motif. We thank Dr. Valerie Kickhoefer, and Dr. Leonard Rome who provided the vault polypeptide complex to define the void volume of the gel filtration analyses. We thank Drs. Bernard Trumpower and Lee McAlister-Henn for antisera to Rip1p and IDH, respectively. We also thank Drs. Carla Koehler and Steven Claypool for advice and support. This work was supported by the National Institutes of Health grant GM45952.
Footnotes
The abbreviations used are: BN-PAGE, Blue Native Polyacrylamide gel electrophoresis; PBS, phosphate buffered saline; Q, coenzyme Q or ubiquinone;
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crane FL. Distribution of Quinones. In: Morton RA, editor. Biochemistry of Quinones. Vol. 1. Academic Press; London: 1965. pp. 183–206. [Google Scholar]
- 2.Debray FG, Lambert M, Mitchell GA. Disorders of mitochondrial function. Curr Opin Pediatr. 2008;20:471–482. doi: 10.1097/MOP.0b013e328306ebb6. [DOI] [PubMed] [Google Scholar]
- 3.Galpern WR, Cudkowicz ME. Coenzyme Q treatment of neurodegenerative diseases of aging. Mitochondrion. 2007;7(Suppl):S146–153. doi: 10.1016/j.mito.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Pepe S, Marasco SF, Haas SJ, Sheeran FL, Krum H, Rosenfeldt FL. Coenzyme Q10 in cardiovascular disease. Mitochondrion. 2007;7(Suppl):S154–167. doi: 10.1016/j.mito.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Bhagavan HN, Chopra RK. Coenzyme Q10: absorption, tissue uptake, metabolism and pharmacokinetics. Free Radic Res. 2006;40:445–453. doi: 10.1080/10715760600617843. [DOI] [PubMed] [Google Scholar]
- 6.Meganathan R. Ubiquinone biosynthesis in microorganisms. FEMS Microbiol Lett. 2001;203:131–139. doi: 10.1111/j.1574-6968.2001.tb10831.x. [DOI] [PubMed] [Google Scholar]
- 7.Kawamukai M. Biosynthesis, bioproduction and novel roles of ubiquinone. Journal of Bioscience and Bioengineering. 2002;94:511–517. doi: 10.1016/s1389-1723(02)80188-8. [DOI] [PubMed] [Google Scholar]
- 8.Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 2007;7S:S62–S71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzagoloff A, Dieckmann CL. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poon WW, Marbois BN, Faull KF, Clarke CF. 3-Hexaprenyl-4-hydroxybenzoic acid forms a predominant intermediate pool in ubiquinone biosynthesis in Saccharomyces cerevisiae. Arch Biochem Biophys. 1995;320:305–314. doi: 10.1016/0003-9861(95)90014-4. [DOI] [PubMed] [Google Scholar]
- 11.Poon WW, Do TQ, Marbois BN, Clarke CF. Sensitivity to treatment with polyunsaturated fatty acids is a general characteristic of the ubiquinone-deficient yeast coq mutants. Molec Aspects Med. 1997;18:s121–s127. doi: 10.1016/s0098-2997(97)00004-6. [DOI] [PubMed] [Google Scholar]
- 12.Johnson A, Gin P, Marbois BN, Hsieh EJ, Wu M, Barros MH, Clarke CF, Tzagoloff A. COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae. J Biol Chem. 2005;280:31397–31404. doi: 10.1074/jbc.M503277200. [DOI] [PubMed] [Google Scholar]
- 13.Hsu AY, Do TQ, Lee PT, Clarke CF. Genetic evidence for a multi-subunit complex in the O-methyltransferase steps of coenzyme Q biosynthesis. Biochim Biophys Acta. 2000;1484:287–297. doi: 10.1016/s1388-1981(00)00019-6. [DOI] [PubMed] [Google Scholar]
- 14.Gin P, Clarke CF. Genetic Evidence for a Multi-subunit Complex in Coenzyme Q Biosynthesis in Yeast and the Role of the Coq1 Hexaprenyl Diphosphate Synthase. J Biol Chem. 2005;280:2676–2681. doi: 10.1074/jbc.M411527200. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh EJ, Gin P, Gulmezian M, Tran UC, Saiki R, Marbois BN, Clarke CF. Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch Biochem Biophys. 2007 doi: 10.1016/j.abb.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marbois B, Gin P, Faull KF, Poon WW, Lee PT, Strahan J, Shepherd JN, Clarke CF. Coq3 and Coq4 define a polypeptide complex in yeast mitochondria for the biosynthesis of coenzyme Q. J Biol Chem. 2005;280:20231–20238. doi: 10.1074/jbc.M501315200. [DOI] [PubMed] [Google Scholar]
- 17.Tran UC, Marbois B, Gin P, Gulmezian M, Jonassen T, Clarke CF. Complementation of Saccharomyces cerevisiae coq7 mutants by mitochondrial targeting of the Escherichia coli UbiF polypeptide: two functions of yeast Coq7 polypeptide in coenzyme Q biosynthesis. J Biol Chem. 2006;281:16401–16409. doi: 10.1074/jbc.M513267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casarin A, Jimenez-Ortega JC, Trevisson E, Pertegato V, Doimo M, Ferrero-Gomez ML, Abbadi S, Artuch R, Quinzii C, Hirano M, Basso G, Ocana CS, Navas P, Salviati L. Functional characterization of human COQ4, a gene required for Coenzyme Q10 biosynthesis. Biochem Biophys Res Commun. 2008;372:35–39. doi: 10.1016/j.bbrc.2008.04.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belogrudov GI, Lee PT, Jonassen T, Hsu AY, Gin P, Clarke CF. Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q synthesis. Arch Biochem Biophys. 2001;392:48–58. doi: 10.1006/abbi.2001.2448. [DOI] [PubMed] [Google Scholar]
- 20.Burke D, Dawson T. Stearns, Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Plainview, NY: 2000. [Google Scholar]
- 21.Akada R, Murakane T, Nishizawa Y. DNA extraction method for screening yeast clones by PCR. Biotechniques. 2000;28:668–670. 672, 674. doi: 10.2144/00284st02. [DOI] [PubMed] [Google Scholar]
- 22.Glick BS, Pon LA. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 23.Schagger H. Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 2001;65:231–244. doi: 10.1016/s0091-679x(01)65014-3. [DOI] [PubMed] [Google Scholar]
- 24.Kickhoefer VA, Garcia Y, Mikyas Y, Johansson E, Zhou JC, Raval-Fernandes S, Minoofar P, Zink JI, Dunn B, Stewart PL, Rome LH. Engineering of vault nanocapsules with enzymatic and fluorescent properties. Proc Natl Acad Sci U S A. 2005;102:4348–4352. doi: 10.1073/pnas.0500929102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephen AG, Raval-Fernandes S, Huynh T, Torres M, Kickhoefer VA, Rome LH. Assembly of vault-like particles in insect cells expressing only the major vault protein. J Biol Chem. 2001;276:23217–23220. doi: 10.1074/jbc.C100226200. [DOI] [PubMed] [Google Scholar]
- 26.Collins MD, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auld DS. Zinc coordination sphere in biochemical zinc sites. Biometals. 2001;14:271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- 28.Vallee BL, Auld DS. Short and long spacer sequences and other structural features of zinc binding sites in zinc enzymes. FEBS Lett. 1989;257:138–140. doi: 10.1016/0014-5793(89)81805-8. [DOI] [PubMed] [Google Scholar]
- 29.Vallee BL, Auld DS. Cocatalytic zinc motifs in enzyme catalysis. Proc Natl Acad Sci U S A. 1993;90:2715–2718. doi: 10.1073/pnas.90.7.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poon WW, Barkovich RJ, Hsu AY, Frankel A, Lee PT, Shepherd JN, Myles DC, Clarke CF. Yeast and rat Coq3 and Escherichia coli UbiG polypeptides catalyze both O-methyltransferase steps in coenzyme Q biosynthesis. J Biol Chem. 1999;274:21665–21672. doi: 10.1074/jbc.274.31.21665. [DOI] [PubMed] [Google Scholar]
- 31.Tauche A, Krause-Buchholz U, Rodel G. Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1/Coq8p. FEMS Yeast Res. 2008 doi: 10.1111/j.1567-1364.2008.00436.x. in press. [DOI] [PubMed] [Google Scholar]
- 32.Auld DS. Zinc Enzymes. In: King RB, editor. Encyclopedia of Inorganic Chemistry. Vol. 9. John Wiley & Sons; Chichester: 2005. pp. 5885–5927. [Google Scholar]
- 33.Maret W. Zinc Coordination Environments in Proteins as Redox Sensors and Signal Transducers. Antioxidants Redox Signal. 2006;8:1419–1441. doi: 10.1089/ars.2006.8.1419. [DOI] [PubMed] [Google Scholar]
- 34.Hiraishi A. Isoprenoid quinones as biomarkers of microbial populations in the environment. J Biosci Bioeng. 1999;88:449–460. doi: 10.1016/s1389-1723(00)87658-6. [DOI] [PubMed] [Google Scholar]
- 35.Hughes PE, Tove SB. Occurrence of alpha-tocopherolquinone and alpha-tocopherolquinol in microorganisms. J Bacteriol. 1982;151:1397–1402. doi: 10.1128/jb.151.3.1397-1402.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Do TQ, Schultz JR, Clarke CF. Enhanced sensitivity of ubiquinone deficient mutants of Saccharomyces cerevisiae to products of autooxidized polyunsaturated fatty acid. Proc Natl Acad Sci USA. 1996;93:7534–7539. doi: 10.1073/pnas.93.15.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marbois BN, Clarke CF. The COQ7 gene encodes a protein in Saccharomyces cerevisiae necessary for ubiquinone biosynthesis. J Biol Chem. 1996;271:2995–3004. doi: 10.1074/jbc.271.6.2995. [DOI] [PubMed] [Google Scholar]