Abstract
To standardize the amount of biological material between samples (e.g., number of cells or amount of tissue) for quantitative real-time reverse transcriptase PCR (qRT-PCR), the cycle of the target gene at which expression is detected (the cycle threshold, or Ct) is divided by the Ct of a gene either thought to be unaffected by experimental conditions or similarly expressed among donors. Genes that maintain cellular structure or homeostasis, referred to as housekeeping genes, or 18S ribosomal RNA are often used for this purpose. Although unstable or inconsistent housekeeping gene expression will misrepresent experimental effects on target gene expression, housekeeping genes are often chosen arbitrarily rather than systematically. We designed a simple and systematic approach towards selection of housekeeping genes based on Ct variance (as reflected by the standard deviation) and normality of distribution. We validated this approach by comparing stability and consistency of expression of 11 housekeeping genes across different types of cells, experimental treatments, and human donors. Finally, we demonstrated the consequences of inconsistent housekeeping gene expression on the calculation of target gene expression, and conclude that validation of stability of housekeeping gene expression by considering both distribution normality and standard deviation is straightforward and critical for proper experimental design.
Keywords: CD4+ T cells, housekeeping genes, quantitative real time reverse transcriptase PCR (RT-CR), cycle threshold
INTRODUCTION
Quantitative real time reverse transcriptase PCR (RT-PCR) instruments quantitatively measure fluorescence generated after each PCR cycle such that they detect replication of an amplicon more sensitively than standard PCR combined with gel electrophoresis and staining. Gene expression is often quantified in terms of cycle threshold (Ct), which is the PCR cycle at which the fluorescence measured between each cycle exceeds a threshold determined by background fluorescence at baseline.
Housekeeping genes (HKG) serve as a common denominator to which target gene expression is normalized, making the identification of stable HKG for real time RT-PCR a critical requirement for accurate and biologically meaningful analysis of gene expression. Fluctuations in HKG expression will misrepresent actual differences in target gene expression such that an increase in HKG expression reduces the apparent increase in target gene expression, and vice versa. Indeed, instability of HKG expression across experimental conditions as measured by ribonuclease protection assays,1 and across donors and human tissue types measured with microarray chips2 and real time qRT-PCR primer/probe sets,3 demonstrates that selection of a stably expressed HKG is critical for both semiquantitative and quantitative analysis of gene expression.4,5 Unfortunately, HKG chosen for this purpose are often selected arbitrarily rather than systematically.
For purposes of normalization, an ideal HKG is insensitive to experimental conditions such that fluctuations in HKG expression cluster symmetrically and tightly around a mean (i.e., normally distributed with a low standard deviation, or SD). Here we propose and validate a simple system to determine the appropriate choice of HKG to normalize target gene expression by cell lines and primary human cells. We demonstrate that the ideal HKG in this context is one that is normally distributed with a low SD independent of cell treatment or donor. By illustrating the consequences of systematic vs. arbitrary selection of HKG on the interpretation of experimental results, we demonstrate that HKG validation is relatively simple, straightforward, and critical for interpretation of experiments that use real time RT-PCR to measure gene expression.
METHODS AND MATERIALS
Cell Culture
A549 human respiratory epithelial cells were purchased from American Type Culture Collection (ATCC, Manassas, VA). Human lymphocytes and monocytes were purified from whole blood obtained from healthy donors by centrifugal countercurrent elutriation at the Department of Transfusion Medicine at the Clinical Center of the National Institutes of Health (clinical protocol number 99-CC-0168). Magnetic bead separation was used to purify CD4+ T cells from the lymphocytes, and to further purify monocytes from the elutriated preparation (Miltenyi Biotec, Auburn, CA).
A549 cells were grown in F12 media (Invitrogen, Carlsbad, CA) with 10% FCS (Hyclone, Logan UT) supplemented with 2 mM L-glutamine (Invitrogen), 100 U/mL penicillin and 100 μg/mL streptomycin (Biofluids, Biosource International, Rockville, MD). CD4+ T cells and monocytes were cultured in RPMI medium (Invitrogen) supplemented with 10% FCS, 2 mM L-glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin.
Reagents
Ionomycin, phorbol myristate acetate (PMA), and lipopolysaccharide were purchased from EMD Chemicals (San Diego, CA), Sigma (St. Louis, MO), and Calbiochem (San Diego, CA), respectively. Recombinant human interferon-α10 (rhIFN-α10) was purchased from PBL Biomedical Labs Inc. (Piscataway, NJ).
Real Time PCR
Total RNA was extracted from cellular lysates using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RNA concentrations ranged from ~150–300 ng/μL as determined by an ND1000 spectrophotometer and NanoDrop 3.0.1 software (NanoDrop Technologies, Wilmington, DE); 260/280 ratios ranged from 1.85 to 2.04. First-strand cDNA was generated by reverse transcription of 1 μg total RNA per sample with random hexamers using SuperScript III Supermix (Invitrogen) according to the manufacturer’s instructions in a final reaction volume of 20 μL.
For RT-PCR, 1 μL of cDNA was added to TaqMan Fast Universal PCR Master Mix and TaqMan Gene Expression Assay primer/probe mixes (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions to achieve a final reaction volume of 20 μL. RT-PCR was performed using an Applied Biosystems 7900HT Fast Real-Time PCR System. The PCR protocol consisted of: initiation at 1 cycle at 50°C for 2 min and 1 cycle at 95°C for 10 min, followed by amplification for 40 cycles at 95°C for 15 sec and 60°C for 1 min. Ct data were collected via Sequence Detection Systems 2.3 software (Applied Biosystems). Each cell sample was assayed for each gene a minimum of two separate times.
Target Gene Expression Analysis
Raw fold changes in target gene expression (ΔCt) were calculated by transforming the difference in Ct values of treated vs. untreated cells: 2−(treated Ct − untreated Ct). Fold changes in target gene expression were then normalized to HKG via the published comparative 2−Δ ΔCt method using the formula:
Statistics
JMP Version 7 (SAS Institute, Cary, NC) was used to generate distribution curves, means and SD of the Ct values.
Analysis and Ranking of HKG in CD4+ T Cells
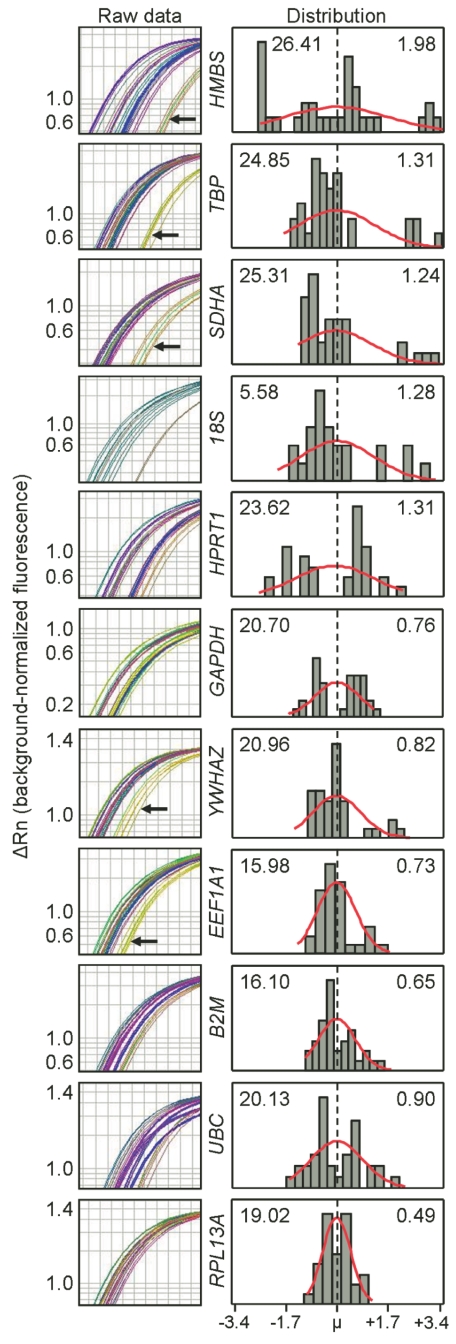
CD4+ T cells from 6 human donors were incubated with PMA and ionomycin or left unstimulated for 4 h, after which the RNA was harvested, reverse transcribed, and expression of 11 HKG (Table 1) was measured. The left column of Figure 1 compares the consistency of expression among HKG, as shown by the variable spread of amplification curves for each gene. Variation in expression as manifested by this spread was due to the effect of PMA and ionomycin on a specific HKG, differences in basal HKG expression among donors, or both. For example, a single donor consistently showed delayed amplification of HMBS, TBP, SDHA, YWHAZ, and EEF1A1 (Figure 1, black arrows) reflecting lower apparent levels of gene expression compared with other donors. In either case, the rightward shifts in amplification of these HKG, if used as normalization factors for a target gene, would erroneously lead to increased apparent expression of the target gene.
TABLE 1.
Housekeeping Genes Evaluated for This Study
| Gene Symbol | Gene Name | RefSeq Accession # |
|---|---|---|
| B2M | Beta-2-microglobulin | NM_004048.2 |
| EEF1A1 | Eukaryotic translation elongation factor 1 alpha 1 | NM_001402.5 |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase | NM_002046.3 |
| HMBS | Hydroxymethylbilane synthase | NM_000190.3 |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | NM_000194.1 |
| RPL13A | Ribosomal protein L13a | NM_012423.2 |
| SDHA | Succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | NM_004168.2 |
| TBP | TATA box binding protein | NM_003194.3 |
| UBC | Ubiquitin C | NM_021009.4 |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activa- | NM_145690.1, |
| tion protein, zeta polypeptide | NM_003406.2 | |
| 18S | Eukaryotic 18S rRNA | X03205.1 * |
GenBank Accession Number
FIGURE 1.
Distribution of HKG expression by CD4+ T cells. CD4+ T cells were incubated with PMA and ionomycin or left unstimulated for four hours. The left column shows raw RT-PCR amplification. Black arrows indicate replicates from a single donor. The right panel shows distribution of Ct values for each gene, with the Ct mean shown in the left corner and the standard deviation in the right corner. The abscissa denotes the distribution midpoint (broken line) and range of Ct data; N ≥ 23 per HKG.
The right column of Figure 1 shows the distribution, mean, and SD of each gene’s Ct values. Ct values of HMBS appeared to be least normally distributed and had the highest SD. By contrast, the Ct values of RPL13A appeared normally distributed and had the lowest SD, indicating that RPL13A is expressed consistently among donors despite stimulation of the CD4+ T cells with PMA and ionomycin. Importantly, Ct values may appear normally distributed with low SD (RPL13A) or high SD (e.g., UBC), or the distribution is not normal with either low SD (B2M) or high SD (HMBS). Together, these examples suggest that distribution and variance are independent criteria to be used together to determine the optimal HKG.
Similar analyses of multiple experiments using the A549 human respiratory epithelial cell line also demonstrated significant variation of expression of HKG (data not shown), albeit less than the peripheral blood CD4+ T cells (HKG maximum spread of 2.4 cycles for A549 cells vs. 6.8 cycles for CD4+ T cells. However, the most consistently expressed HKG in A549 were UBC, HMBS and GAPDH, contrasting with the best candidate HKG in CD4 T cells, RPL13A, thus demonstrating cell specificity of HKG stability.
Differential HKG Consistency Between Treatment Groups and Among Donors
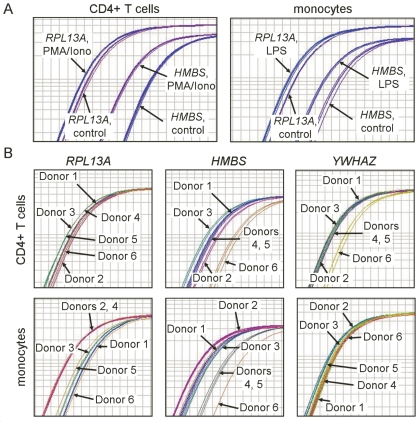
We next asked whether HKG that were stable between treatment conditions were also consistently expressed among donors. CD4+ T cells were treated with PMA and ionomycin, monocytes were treated with lipopolysaccharide, and expression of each of the eleven HKG was measured and compared to unstimulated control. Expression of the 11 HKG in response to stimulation varied widely, ranging from nonresponsiveness to substantial up-regulation. In response to stimulation, RPL13A expression by CD4+ T cells and monocytes shifted only slightly. In contrast, HMBS is a poor choice for both cell types as stimulation dramatically increased its expression (Figure 2A).
FIGURE 2.
Lack of consistency of housekeeping gene expression between treatment groups and among donors. A: Differences between treatment and untreated control in RT-PCR amplification replicates from a single donor’s CD4+ T cells and monocytes. CD4+ T cells were treated with PMA and ionomycin; monocytes were treated with lipopolysaccharide. B: Relative expression of RPL13A, HMBS, and YWHAZ in untreated CD4+ T cells or monocytes from 6 donors.
We next examined variation of HKG expression among 6 donors in untreated CD4+ T cells and monocytes and found that RPL13A was expressed with little variation by CD4+ T cells regardless of donor, but expression of HMBS and YWHAZ varied widely (Figure 2B, upper panels). For monocytes, in contrast, expression of both RPL13A and HMBS varied widely among the 6 donors while expression of YWHAZ remained highly consistent (Figure 2B, lower panels), reinforcing the discrepancies among stable HKG in different cell types.
Effect of HKG Selection on Analysis of Target Gene Expression
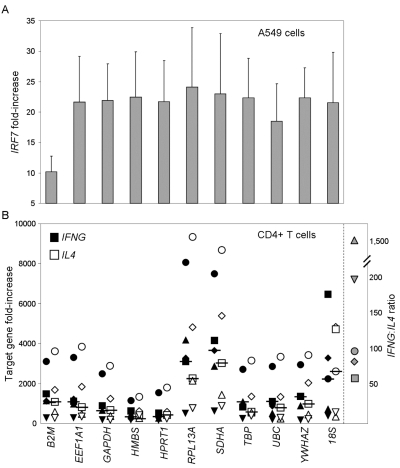
To demonstrate the consequences of normalizing to a HKG that is variably expressed, we treated A549 human respiratory epithelial cells with IFN-α10 and compared expression of the IFN-responsive gene IRF7 and the 11 HKG to unstimulated control cells. Figure 3A shows the increase in expression of IRF7 when normalized to each of the HKG using the ΔΔCt method. Because IFN-α10 increased expression of B2M, normalization to this HKG diminished the perceived IRF7 response. While normalization to the other 10 HKG generated reasonably consistent increases in mean IRF7 expression, there was wide variation between the HKG-normalized data such that SD ranged from 4.93 (for YWHAZ) to 9.91 (for SDHA). Of these HKG, GAPDH exhibited the best combination of normal distribution (data not shown) and relatively low deviation of normalized data (SD = 6.0). Therefore, normalization to GAPDH would most likely demonstrate statistical significance when there is an actual biological difference in target gene expression.
FIGURE 3.
Housekeeping gene selection affects interpretation of target gene expression. A: Mean and SD of IRF7 gene expression by A549 cells after treatment with IFN-10,, shown as fold-increase over unstimulated control and normalized to each housekeeping gene (indicated on abscissa) using the Δ ΔCt method; N ≥ 24 per housekeeping gene. B: IFNG (filled symbols) and IL4 (open symbols) gene expression by CD4+ T cells from 5 donors after treatment with phorbol myristate acetate and ionomycin, normalized to each HKG. The horizontal bars indicate the medians. Each symbol represents a different donor. The ordinate on the right side indicates the IFNG:IL4 ratio for each donor after treatment of CD4+ T cells with phorbol myristate acetate and ionomycin.
We then collected CD4+ T cells from 5 donors and tested expression of IFNG and IL4 in response to treatment with PMA and ionomycin, and used the ΔΔCt method to normalize expression of these target genes to the eleven HKG. Figure 3B demonstrates that the choice of HKG dramatically affected the perceived increase in expression of each cytokine. Furthermore, the magnitude of variation in expression values among the 5 donors was dependent upon the HKG. Some HKG exhibited large spread among donors (e.g., RPL13A and SDHA), while other HKG exhibited tight clustering among donors (e.g., HMBS and HPRT1). Figure 1 demonstrated that RPL13A is the best choice of HKG for CD4+ T cells, suggesting that the wide spread in IFNG and IL4 fold-increase over control after normalization to RPL13A (Figure 3B) most accurately represents the donor variation in target gene expression in response to PMA and ionomycin. Of note, RPL13A was a poor HKG choice in A549 cells (data not shown), again demonstrating the cell dependency of the choice of HKG.
The similarity in the relative fold increase of expression of IFNG and IL4 reflects the relative disparity in basal levels of expression in the untreated control CD4+ T cells. When compared directly, the IFNG:IL4 ratio on the right side of Figure 3B demonstrates that consistent with our own experience7 and previous reports,8,9 expression of IFNG is ~50–1500 that of IL4 after treatment with PMA and ionomycin.
Normalization to HKG serves at least three critical functions. First, it insures that changes in target gene expression are selective and not reflective of a general transcriptional “ramp-up”; second, normalization to HKG allows for analysis of samples in which RNA amount is below levels of quantification; and third, normalization allows for combining data from multiple experiments. In addition, HKG analysis serves as a gross check for RNA/cDNA integrity and the quality of the PCR reaction when target gene expression is undetectable.
We present a straightforward protocol for identifying HKG that are stably expressed, either between experimental conditions or among multiple donors, based on the simple criteria of normal and tight (i.e., low SD) distribution of Ct values. Two Microsoft Excel-based programs, Bestkeeper10 and geNorm11 were written to facilitate HKG selection, but neither addresses the distribution of HKG expression values. In addition, both programs assume a stable relationship between different HKG within a cell in order to use algorithms that compare the variance of an individual HKG to all but the least consistent members of the set—an assumption that is not necessarily valid.
In addition to low variance, normal distribution not only supports the inference of HKG stability but provides insight into the technical quality and reproducibility of the experiment. Based on the criteria of normal distribution and low variance, the ideal HKG for the cell types we have studied are shown in Table 2. Given the simplicity and importance of systematically determining the proper HKG for normalization, we strongly suggest that this analysis be performed to best interpret data from studies that use quantitative RT-PCR.
TABLE 2.
Ideal Housekeeping Genes for Cell Types Used in This Study
| Cell Type | Ideal HKG |
|---|---|
| CD4+ T cells | B2M, RPL13A |
| Monocytes | HPRT1, SDHA |
| A549 cell line | GAPDH, UBC |
HKG, housekeeping genes.
ACKNOWLEDGMENTS
The findings and conclusions herein have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any Agency determination or policy. The authors wish to thank Mario Roederer for helpful discussions.
REFERENCES
- 1.Zhong H, Simons JW. Direct comparison of GAPDH, beta-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem Biophys Res Commun. 1999;259:523–526. doi: 10.1006/bbrc.1999.0815. [DOI] [PubMed] [Google Scholar]
- 2.Hsiao LL, Dangond F, Yoshida T, et al. A compendium of gene expression in normal human tissues. Physiol Genomics. 2001;7:97–104. doi: 10.1152/physiolgenomics.00040.2001. [DOI] [PubMed] [Google Scholar]
- 3.Tricarico C, Pinzani P, Bianchi S, et al. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309:293–300. doi: 10.1016/s0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- 4.Chang TJ, Juan CC, Yin PH, Chi CW, Tsay HJ. Up-regulation of beta-actin, cyclophilin and GAPDH in N1S1 rat hepatoma. Oncol Rep. 1998;5:469–471. doi: 10.3892/or.5.2.469. [DOI] [PubMed] [Google Scholar]
- 5.Bereta J, Bereta M. Stimulation of glyceraldehyde-3-phosphate dehydrogenase mRNA levels by endogenous nitric oxide in cytokine-activated endothelium. Biochem Biophys Res Commun. 1995;217:363–369. doi: 10.1006/bbrc.1995.2785. [DOI] [PubMed] [Google Scholar]
- 6.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Alston MA, Huang H, Rabin RL. Human T cell cytokine responses are dependent on multidrug resistance protein-1. Int Immunol. 2006;18:485–93. doi: 10.1093/intimm/dxh389. [DOI] [PubMed] [Google Scholar]
- 8.Wu CY, Demeure C, Kiniwa M, Gately M, Delespesse G. IL-12 induces the production of IFN-gamma by neonatal human CD4 T cells. J Immunol. 1993;151:1938–1949. [PubMed] [Google Scholar]
- 9.Picker LJ, Singh MK, Zdraveski Z, et al. Direct demonstration of cytokine synthesis heterogeneity among human memory/effector T cells by flow cytometry. Blood. 1995;86:1408–1419. [PubMed] [Google Scholar]
- 10.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 11.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]