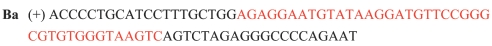Abstract
Here we describe a new DNA capture element (DCE) sensing system, based on the quenching and dequenching of a double-stranded aptamer. This system shows very good sensitivity and thermal stability. While quenching, dequenching, and separating the DCE systems made from different aptamers (all selected by SELEX), an alternative method to rapidly select aptamers was developed—the Aptamer Selection Express (ASExp). This process has been used to select aptamers against different types of targets (Bacillus anthracis spores, Bacillus thuringiensis spores, MS-2 bacteriophage, ovalbumin, and botulinum neurotoxin). The DCE systems made from botulinum neurotoxin aptamers selected by ASExp have been investigated. The results of this investigation indicate that ASExp can be used to rapidly select aptamers for the DCE sensing system.
Keywords: aptamer, ASExp, DCE, SELEX
INTRODUCTION
Aptamers are small single-strand DNA (ss-DNA) or RNA oligonucleotides with very high affinity to their targets.1–3 They bind to the targets with high selectivity and specificity due to their specific three-dimensional folded conformations. Aptamers offer the possibility of overcoming the limitations of antibodies and could replace antibodies in detection and identification systems for biological threat agents. Aptamers have many obvious advantages. For example, there is no need to use animals or cell cultures and, unlike antibody proteins, they do not require any special storage facilities. Once selected, aptamers can be produced at any time in large amounts and with high reproducibility by chemical synthesis. Since they are selected for a multitude of different substances, the field of application is very wide-ranging.4 The use of aptamers for sensing and diagnostic applications has been extensively explored.5–9 In many cases, aptamers show similar or even better performance when compared with antibodies in the laboratory.6–10
Currently, aptamers are selected by the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) process.11 This process is characterized by the repetition of successive steps of target binding and removal of unbound oligonucleotides, followed by elution, amplification, and purification of the selected oligonucleotides.11–13 The separation of target bound and unbound oligonucleotides during the process is the crucial step for successful aptamer selection. A conventional separation method is affinity chromatography in which the targets are immobilized onto column material.14 Therefore, considerable amounts of target molecules are necessary in order to reach sufficiently high loading of the column. Another type of labor-intensive aptamer separation which requires large volumes of library material is selection on nitrocellulose filters.14
Developing a sensing system that can detect and identify biological warfare agents rapidly, with high sensitivity, good thermal stability, and specificity has very important significance not only in defense and national security but also in commercial applications such as medical diagnostics, food safety, environmental monitoring, and so forth. Our purpose is to develop such a system based on aptamer technology. This system is called a DNA capture element, or DCE. Different DCE systems made from aptamers of Bacillus anthracis (Ba), Shiga toxin, botulinum neurotoxin (BoNT), and Francisella tularensis bacteria (all selected by SELEX) have been investigated. Cross-reactions were observed when the DCE systems interacted with different targets. When interaction of an individual target with a mixture of DCE systems was investigated, the need for a new type of separation method led to the development of a process for rapid aptamer selection—the Aptamer Selection Express (ASExp).15 This process combines the interaction of a target with double-stranded (ds)-DNA and magnetic separation technology. The ds-DNA interacts with the target, denaturing the ds-DNA. The single-stranded (ss)-DNA/target-bound complex can then be separated by magnetic beads bound with random ss-DNA. This separation method has been studied by reselecting aptamers against different targets, Ba spores, Shiga toxin, and F. tularensis bacteria. In contrast to SELEX, the use of magnetic beads for ss-DNA/target separation requires a very small amount of target providing for convenient handling.
First, we demonstrated the preparation of a new sensing system (DCE) based on a Ba aptamer. Secondly, using the ASExp selection method, the Ba aptamer was reselected and tested for agent selectivity. Finally, we tested the DCE’s of botulinum neurotoxin aptamers (type A and B light chain) selected using the ASExp method for agent specificity.
MATERIALS AND METHODS
Chemicals and Instruments
1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide HCl (EDC) was purchased from Pierce (Rockford, IL). Quantum Dots were purchased from Evident Technologies (Troy, NY). QXL 520 (quencher) and Hilyte Fluor 488 amine (fluorophore) were obtained from AnaSpec (San Jose, CA). ATP was purchased from Sigma-Aldrich (St. Louis, MO). Library of ss-DNA and all aptamers used were obtained from Sigma-Genosys (Woodlands, TX). Polybeads and magnetic beads were obtained from Poly-Sciences (Warrington, PA) and Spherotech (Lake Forest, IL), respectively. T4 polynucleotide kinase (OptiKinase) and imidazole were purchased from USB (Cleveland, OH) and Sigma-Aldrich, respectively. Phosphate buffered saline containing Mg+2 and Ca+2 was obtained from GIBCO (Carlsbad, CA). Shiga toxin and botulinum neurotoxins (A and B, light and heavy chain) were obtained from LIST Biological Laboratories (San Jose, CA). Bacillus anthracis spores (Sterne strain) were made from anthrax spore vaccine, Mobay Corporation (Pittsburgh, Pennsylvania). Tularemia bacteria were grown from the F. tularensis antigen (Becton-Dickson, Franklin Lakes, NJ). Sequencer was Applied Biosystems 3100 Genetic Analyzer (Foster City, CA), Microscope was Nikon E800 (Melville, NY), and plate reader was BioTek Synergy HT (Winooski, VT).
Concept of DCE System
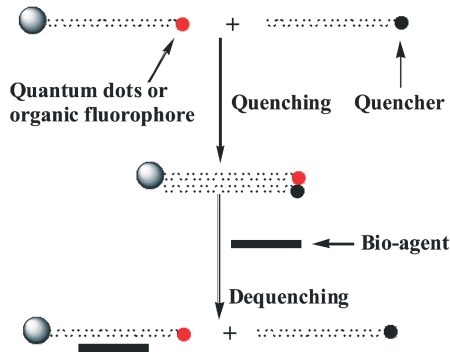
In this system, the primary strand aptamer has a quantum dot (or fluorophore) covalently linked to the 3′end and a bead covalently linked to the 5′end. The complementary strand aptamer has a quencher covalently linked to the 5 ′end. The two strands of DNA are annealed together and the quencher quenches the fluorescence of the quantum dot (or fluorophore). When a target agent is introduced to the DCE system, the double strands denature, enabling them to fluoresce brightly (Scheme 1).
SCHEME 1.
Quenching and dequenching the DNA capture element system. The fluorescence is determined by plate reader.
Preparation Procedures of DCE System
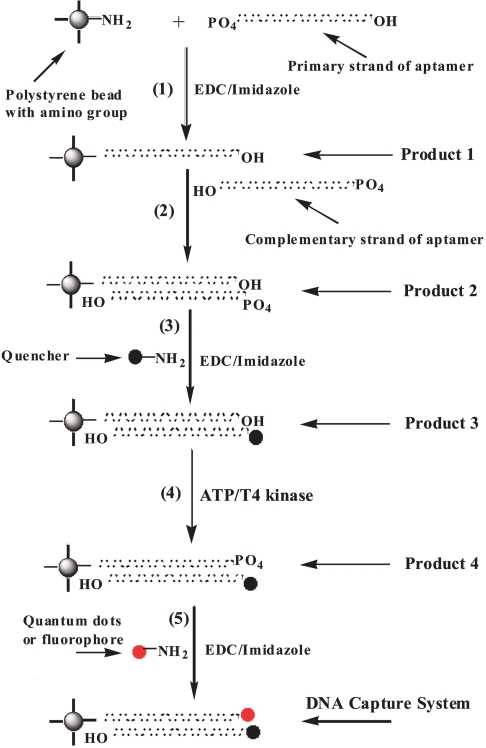
In Scheme 2, Step 1, primary DNA aptamer is added to amino polybeads in 0.1 M MOPS buffer. EDC and imidazole are then added. (Average number of primary strand DNA/bead = 5.5.) The reaction mixture is stirred at room temperature overnight. Product 1 is separated by centrifugation and washed with H2O, then resuspended in MOPS buffer.
SCHEME 2.
Preparation procedure for DNA capture element sensing system. quantum dots were used here for the DNA capture element systems of Bacillus anthracis, Shiga, and tularemia aptamers (selected by the SELEX process). All of these agents were in turn used for aptamer re-selection. Sequences of these aptamers are shown in Figure 1. EDC, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide HCl.
In Step 2, excess complementary DNA is added to Product 1 in 0.1 M MOPS buffer. The mixture is stirred (or shaken) for one day at 60ºC, and then slowly cooled down to room temperature and kept for several days at room temperature. Product 2 is separated by centrifugation and washed with H2O.
In Step 3, Product 2 is added to a saturated solution of QXL 610 with EDC and imidazole (excess quencher is used). This reaction mixture is stirred overnight. Product 3 is separated by centrifuge and washed with H2O.
In Step 4, Product 3 is resuspended in 10 mL of reaction buffer (350 μL 2 M Tris-HCl, 100 μL of 1 M MgCl2, and 7.7 mg of dithiothreitol + H2O). Then 5 mL of ATP (27.6 mg ATP + H2O) and 2 μL of T4 polynucleotide kinase are added. The reaction mixture is shaken for 1–2 h at 37ºC. Product 4 is separated by centrifugation and washed with H2O.
In Step 5, Product 4 is added to a solution of quantum dots with EDC and imidazole in MOPS buffer (excess Q-Dots are used). The mixture is stirred overnight. Product 5 is separated by centrifugation and washed with H2O.
The sequences of Ba, Shiga, and tularemia aptamers used for making the DCEs are shown in Figure 1.
FIGURE 1.
Sequences of Bacillus anthracis (Ba), Shiga, and tularemia aptamers used for making DNA capture elements. All of these aptamers were selected by conventional SELEX. Sequences of the aptamers are red.
Re-selection of Ba Aptamers
The procedures of the aptamer re-selection are shown in Scheme 3. Approximately 0.5 μL of Ba spores (1.87 × 106 CFU/μL) were added to the mixture of Ba, Shiga, and tularemia DCE systems (DCE systems of all three aptamers were made based on Scheme 2, using 3 × 1013 aptamers for each batch). The interaction mixture was stirred at room temperature for 0.5 h. Then, 30 μL of magnetic beads with random ss-DNA attached (~7.485 × 106 bead, ~1.16 × 1012 ss-DNA) were added and stirred for another 0.5 h. The complex of aptamer/target/magnetic bead was separated by magnet, washed thoroughly with water, and re-suspended in Tris EDTA buffer, pH 7.4. The ss-DNA (aptamer) of the aptamer/target/magnetic bead complex was then amplified by polymerase chain reaction (PCR), cloned, and sequenced.
SCHEME 3.
procedures for Bacillus anthracis (Ba) aptamer re-selection.
PCR/Sequencing
The sample was first divided into 5-μL aliquots for amplification by PCR. Master Mix (per 5 μL template sample) was as follows: 5 μL buffer, 2 μL MgCl2, 2.5 μL DMSO, 1 μL betaine, 1 μL each dNTP, 2.5 μL F primer, 2.5 μL R primer, 0.54 μL taq polymerase, and 25.1 μL H2O. Betaine and increased DMSO were added to the master mix to eliminate polymerase jumping during PCR amplification.10 Using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA), 1 μL of amplified DNA was ligated into the provided vector (pCRII-TOPO; Invitrogen). To each ligation, 250 μL SOC Media was added and incubated at 37°C with shaking (225 rpm) for 1 h. Transformations were streaked to Xgal/IPTG LB/Amp plates (made in house) and incubated overnight at 37°C. Selected white and/or light blue colonies were isolated and grown O/N in 2 mL LB media with 2.5 mg/mL ampicillian additive. DNA was then isolated via standard alkaline lysis mini-prep protocol. Five microliters from miniprep (precipitate in 30 μL Tris EDTA buffer) was used as PCR template. Two colonies from each X-gal/IPTG LB/Amp plate underwent boil-prep with colony lysis buffer (1% Tween 20 in Tris-EDTA). Fifteen microliters was used as template in PCR. PCR products were precipitated with 5 M ammonium acetate and 100% isopropyl alcohol at −20°C overnight. PCR precipitates (1 μL each) were sequenced with BigDye Terminator v3.1 Ready Reaction Sequencing Standard Kit (Applied Biosystems, Part No. 4336935) and run on an Applied Biosystems 3100 automated sequencer.
Process of Rapid Aptamer Selection: Selection, Amplifying, Cloning, and Sequencing of Botulinum Neurotoxin Aptamers
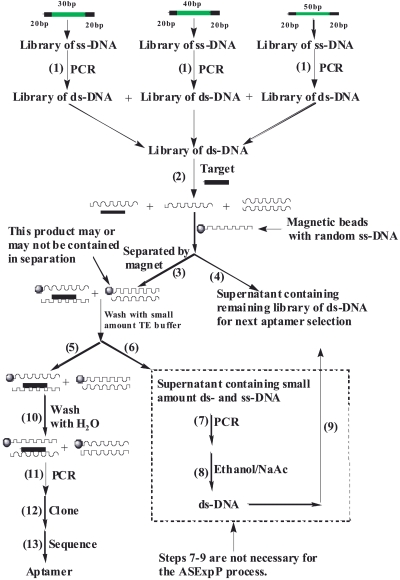
The procedure for rapid aptamer selection (ASExp) is shown in Scheme 4. The starting libraries consist of a multitude of ss-DNA fragments (~1015 molecules) with a central random region of 30, 40, and 50 nucleotides, flanked by 20-nucleotide constant regions that function as primer binding sites for PCR. Before selection, each library of ss-DNA was amplified in parallel PCR reactions to produce a library of ds-DNA. After ethanol precipitation, each ds-DNA library was resuspended in phosphate buffered saline (1×, with Mg+2 and Ca+2) and mixed together. A small amount of target was then added to the mixture of ds-DNA libraries and stirred for 0.5 h (~0.75 ng botulinum neurotoxin type A or B; light chain was used). The target denatured the ds-DNA and bound to the ss-DNA (aptamer). Then, magnetic beads bound with random ss-DNA (60mer) were added and interacted with the aptamer/target complex. A complex of aptamer/target/magnetic beads is formed and separated by magnet [a complex of complementary strand aptamer (attacked by target) and magnetic bead may also be formed and separated]. The supernatant containing the remaining library of ds-DNA was separated from the reaction mixture for reuse. (Steps 6–9 in Scheme 4 are not related to the aptamer selection, just for reusing the ds-DNA library.) The aptamer/target/magnetic bead complex was washed thoroughly with water. Lastly, the aptamer of the aptamer/target/magnetic bead complex was amplified by PCR, cloned, and sequenced. Sequences of BoNT aptamers (three aptamers for type A light chain, and 2 aptamers for type B light chain) were obtained (see Figure 7 below).
SCHEME 4.
Rapid aptamer selection (ASExp) for generating DNA aptamers for specific targets: botulinum neurotoxin; type A and B light chain. TE, Tris/EDTA buffer.
FIGURE 7.
Sequences of botulinum neurotoxin aptamers selected by ASExp. *Three base pairs are not correctly sequenced and no sequence of complementary strand was obtained. **No sequence of complementary strand was obtained.
Fluorescence Measurements
Fluorescence changes in the DCEs with and without agent were measured at a wavelength of 590 nm by a Bio-TEK Synergy HT Plate Reader (Winooski, VT). The DCEs (100 μL per well) were added to a 96-well plate. The fluorescence of each well was measured as a baseline before introducing the agent. The same amount of DCE without agent was used as a control. The RFU values were calculated as follows:
Fluorescence change = [Fluorescence of DCE and Agent – fluorescence of DCE (control)] – [(Fluorescence of DCE – fluorescence of DCE (control)].
RESULTS AND DISCUSSION
Fluorescent Change Resulting From Interaction of Ba Spores with Ba DCE Sensing System
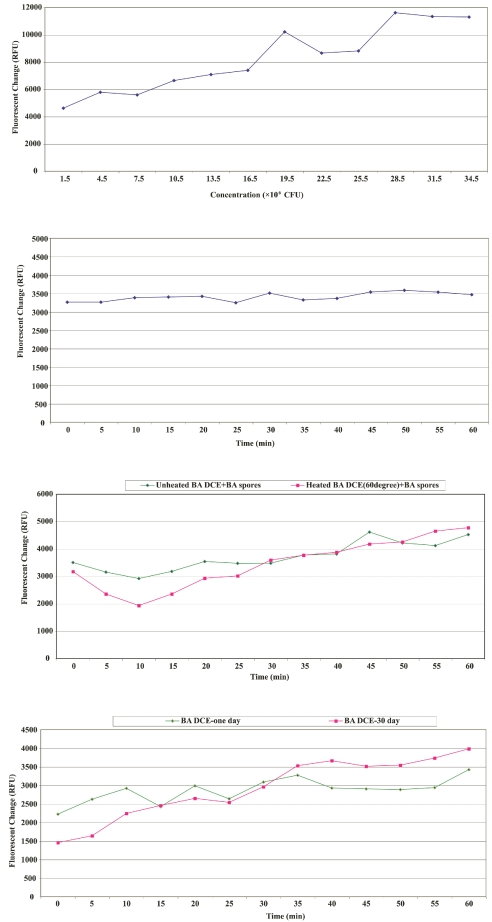
A new sensing system based on aptamer technology was studied. Figure 2A and B show the fluorescence change of the Ba DCE sensing system. Figure 2A does not show apparent fluorescence vs. time change because the plate reader was in shaking mode prior to the initial read out. Other fluorescence measurements did show this trend, however, such as Figure 2C and D and Figure 3. In Figure 4, microscopic images A–D confirmed that quenching and dequenching of the DCE sensing system occurs.16,17 Image B illustrated that the fluorescence of quantum dots was almost quenched by quencher, and image D showed strong fluorescence due to denaturing by Ba spore interaction. The quenched fluorescence returned because the Ba DCE was de-quenched by Ba spores. Image C showed Ba spores gathering around beads, indicating that the Ba spores bound to the aptamer. Figure 2C and D showed that the DCE sensing system possessed very good stability, even when heated at 60ºC for 4 h. Figure 3 showed that cross-reaction occurred when the Ba DCE system interacted with different targets: Ba spores, Shiga toxin, and tularemia bacteria. When Shiga toxin or tularemia DCE systems interacted with these three agents, the same results were obtained (data not shown). The reason(s) for the cross-reactivity within the three DCE systems tested are unknown at this time. The cause(s) are being investigated and actions are being taken to produce a more selective and specific DCE sensing system.
FIGURE 2.
A: Fluorescence change of Bacillus anthracis DNA capture element (Ba DCE) vs. concentration change of Ba spores. B: Fluorescence change of Ba DCE vs. time from interaction of Ba DCE with 1.2 × 106 CFu/μL of Ba spores. C: Thermal stability study of Ba DCE. Ten microliters of Ba spores (1.5 × 105 CFU/μL) were used for both Ba DCE experiments. The red line is from Ba DCEs that were heated for 4 h at 60°C. D: Stability study of Ba DCE. Ten microliters of Ba spores (1.5 × 105 CFU/μL) were used for both Ba DCEs experiments. The red line is from Ba DCEs that were kept for 30 d at room temperature.
FIGURE 3.
Selectivity study of Bacillus anthracis DNA capture elements (Ba DCE). Ten microliters of Ba spores (1.5 × 105 CFU/μL), 5 ng Shiga toxin, and 2 μL tularemia agent (5 × 105 CFU/μL) were used in each respective experiment.
FIGURE 4.
Microscopic images of quenched and de-quenched Bacillus anthracis DNA capture elements (DCE). A–D are of the same sample, taken before and after adding Ba spores, respectively. A and C are visible light, while B and D are fluorescence. in B, most of the quantum dot fluorescence is quenched. C shows Ba spores binding to the aptamer around the beads.
Results of Re-selection of Ba Aptamer
The process of Ba aptamer re-selection is shown in Scheme 3. Figures 5 and 6 and Table 1 show the results of an aptamer re-selection by Ba spores interacting with a mixture of DCEs. This may suggest that the Ba spore has a stronger affinity to the Ba aptamer than to Shiga toxin and tularemia bacteria. Only sequences of Shiga toxin and tularemia aptamers were obtained separately when Shiga toxin and tularemia bacteria were used as targets to interact with a mixture of Ba, Shiga toxin, and tularemia DCEs individually. All of the three aptamer re-selections were repeated and the same results were obtained (data not shown). These results show that we can select aptamers for the DCE sensing system based on the aptamer reselection process.
FIGURE 5.
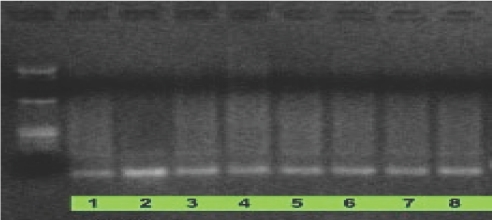
Electophoresis image after polymerase chain reaction of the product separated from reaction mixture shown in Scheme 3. (Agarose gel 2.5%; standard used in Lane 1 is Fermentas ZipRuler DNA Ladder 2200–20,000 bp.) original re-selection sample was aliquoted into 8 samples, all wells are identical.
FIGURE 6.
Sequence of aptamer selected from the interaction of Bacillus anthracis (Ba) spores with a mixture of Ba, Shiga, and tularemia DCEs (Scheme 3). Sequence of aptamer selected from Scheme 3 re-selection is same as that of Ba aptamer selected by SELEX. No sequences of Shiga and tularemia aptamers were obtained.
TABLE 1.
Fluorescence Data of Aptamer/Ba Spore/Magnetic Bead Complex Separated From Interaction of Ba Spores with the DCE Mixture of Ba, Shiga, and Tularemiaa, b
| Magnetic Beads
|
|||
|---|---|---|---|
| With Random DNA: Control
|
Plus Sample: After Washing
|
||
| Time (min) | D6 | D7 | Diff. |
| 0 | 959 | 2588 | 1629 |
| 2 | 1008 | 2757 | 1749 |
| 4 | 955 | 2880 | 1925 |
| 6 | 988 | 2929 | 1941 |
| 8 | 982 | 2977 | 1995 |
| 10 | 955 | 2998 | 2043 |
See Scheme 3.
Same amount of magnetic beads were used as control. Fluorescence of aptamer/Ba spore/magnetic bead complex was measured after washing thoroughly. Ba, Bacillus anthracis.
Results of BoNT Aptamers Selected by ASExp
Procedures of selection, separation, cloning, and sequencing of BoNT aptamers (type A and B light chain) by ASExp are described in Scheme 4 and sequences of aptamers selected are shown in Figure 7.
Fluorescence Test Results From the Interaction of Botulinum Neurotoxin DCE Systems (Made From Botulinum Neurotoxin Aptamers) with Botulinum Neurotoxin
Four DCE systems were made, the sequences of which are shown in Figure 8. The preparation procedures are described in Scheme 4. DCE-1 and DCE-2 were made from aptamers selected by ASExp against BoNT type A and B light chain, respectively. DCE-3 and DCE-4 were made from aptamers selected by the SELEX process against BoNT type A light chain and BoNT holotoxin, respectively. The organic fluorophore, Hylite Fluor 488 amine, was used in BoNT DCEs 1–4.
FIGURE 8.
Sequences of aptamers for making botulinum neurotoxin DNA capture elements (DCE) 1–4. Sequences of aptamers are red and primers are black.
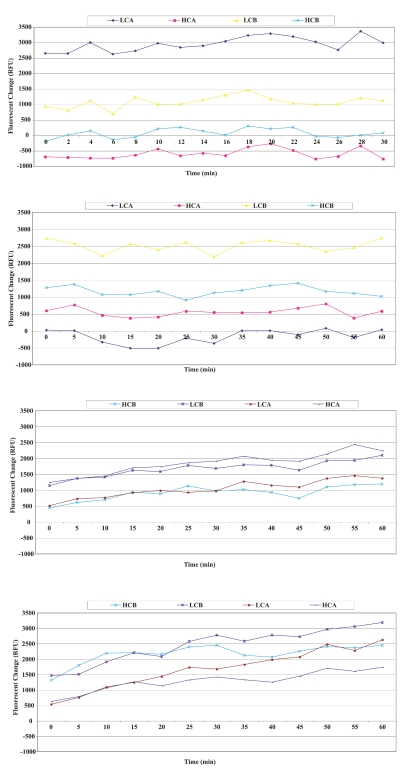
Four types of BoNTs (type A and B, heavy and light chain) were used to interact with each respective DCE system. The fluorescence changes resulting from the interactions of the four toxins (toxin type A light chain, toxin type A heavy chain, toxin type B light chain, and toxin type B heavy chain with DCEs 1–4 are shown in Figure 9A–D; 5 ng BoNTs were used for all fluorescent measurements). When BoNTs (type A and B light chain) interacted with a library of ds-DNA separately, as described in Scheme 4 for ASExp, sequences of three aptamers for type A light chain and two aptamers for type B light chain were obtained. Sequences of a complementary strand of #1 of type A light and #1 of type B light were also obtained. This suggests that some aptamers or their complementary strands may be lost during cloning and sequencing. Four BoNT DCE systems were made according to Scheme 2 and interacted with four different BoNTs. Figure 9A and B indicated that the BoNT DCE-1 can be denatured by type A (strongly) and B light chain (weakly) of BoNT toxins, but not by type A and B heavy chain. DCE-2 can be denatured by the type B light chain (strongly), type A and B heavy chain (weakly), and not by type A light chain of the toxin. It is not known if these results are related to the structure of the toxin or not. Ba, Shiga toxin, and tularemia DCE systems show cross-reaction between BoNT DCE-3, 4 and different types of toxins (Figure 9C and D). These results suggest that ASExp may be used as an alternative method to select aptamers for our DCE sensing system. ASExp possesses some advantages such as ease of operation and speed (10 h for selection process) as compared with SELEX. More efforts are being taken to study ASExp in order to improve the process and obtain aptamers with better selectivity or specificity.
FIGURE 9.
A: Fluorescence change resulting from the interactions of DCE-1 [made from aptamer against botulinum neurotoxin (BoNT), type A light chain by ASExp] with different types of BoNTs. B: Fluorescence change resulting from the interactions of DCE-2 (made from aptamer against BoNT, type B light chain by ASExp) with different types of BoNTs. C: Fluorescence change resulting from the interactions of DCE-3 (made from aptamer against BoNT, type A light chain by SELEX process) with different types of BoNTs. D: Fluorescence change resulting from the interactions of DCE-4 (made from aptamer against BoNT holotoxin by SELEX) with different types of BoNTs. HCA, toxin type A heavy chain; HCB, toxin type B heavy chain; LCA, toxin type A light chain; LCB, toxin type B light chain.
CONCLUSION
This study illustrates a new sensing system with very good sensitivity and thermal stability based on ds-aptamers. Results showed that ds-aptamers are denatured and de-quenched. In addition, an alternative method for rapid aptamer selection was demonstrated (ASExp) based on the use of a ds-DNA library and magnetic beads bound with random ss-DNA for fast separation. ASExp has been applied to select aptamers against different types of targets successfully, such as B. anthracis spores, B. thuringiensis spores, MS-2 bacteriophage, and ovalbumin (only BoNT aptamers are reported here). Both aptamer selection systems SELEX and ASExp seem to have cross-reaction problems. However, ASExp provides a much faster and easier selection due to the small amount of target required, resulting in low cost when compared with SELEX. We conclude that the BoNT aptamers selected by ASExp, and the study of their DCEs interactions, show that ASExp can be used as an alternative rapid selection method for apatmers.
ACKNOWLEDGMENTS
This work was sponsored by internal funding from the Air Force Research Laboratory, Human Effectiveness Directorate and National Institutes of Heath (SBIR 1R43AI058561-01).
REFERENCES
- 1.Famulok M, Mayer G, Blind M. Nucleic acid aptamers—from selection in vitro to applications in vivo. Acc Chem Res. 2000;33:591–599. doi: 10.1021/ar960167q. [DOI] [PubMed] [Google Scholar]
- 2.Gold L, Brody E, Heilig J, Singer B. One, two, infinity: Genomes filled with aptamers. Chem Biol. 2002;9:1259–1264. doi: 10.1016/s1074-5521(02)00286-7. [DOI] [PubMed] [Google Scholar]
- 3.Ellington AD, Corad R. Aptamers as potential nucleic acid pharmaceuticals. BiotechnolAnnu Rev. 1995;I:185–214. doi: 10.1016/s1387-2656(08)70052-8. [DOI] [PubMed] [Google Scholar]
- 4.Jayasena SD. Aptamer: An emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45(9):1628–1650. [PubMed] [Google Scholar]
- 5.Osborne SE, Matsumura I, Ellington AD. Aptamers as therapeutic and diagnostic reagents. Curr Opin Chem Biol. 1997;1:5–9. doi: 10.1016/s1367-5931(97)80102-0. [DOI] [PubMed] [Google Scholar]
- 6.Famulok M. Oligonucleotide aptamers that recognize small molecules. Curr Opin Struct Biol. 1999;9:324–329. doi: 10.1016/S0959-440X(99)80043-8. [DOI] [PubMed] [Google Scholar]
- 7.Hartig JS, Najafi-Shoushtari SH, Gruene I, et al. Protein-dependent ribozymes report molecular interactions in real time. Nat Biotechnol. 2002;20:717–722. doi: 10.1038/nbt0702-717. [DOI] [PubMed] [Google Scholar]
- 8.Nutiu R, Li Y. Structure-switching signaling aptamers. J Am Chem Soc. 2003;125:4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 9.Nutiu R, Li Y. Kinetics of the color change of the cocaine sensor at various concentrations of cocaine. Chem Eur J. 2004;10:1868. [Google Scholar]
- 10.Vivekananda J, Kiel JL. Anti-Francisella tularensis DNA aptamers detect tularemia antigen from different subspecies by Aptamer-Linked Immobilized Sorbent Assay. Lab Invest. 2006:1–9. doi: 10.1038/labinvest.3700417. [DOI] [PubMed] [Google Scholar]
- 11.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to backteriophage T4 DNA Polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 12.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 13.Bruno JG, Kiel JL. Use of magnetic beads in selection and detection of biotoxin aptamers by electrochemiluminescence and enzymatic methods. Biotechniques. 2002 Jan;32:178–183. doi: 10.2144/02321dd04. [DOI] [PubMed] [Google Scholar]
- 14.Tombelli S, Minnuni M, Mascini M. Analytical applications of aptamers. Biosens Bioelectron. 2005;20:2424–2434. doi: 10.1016/j.bios.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Kiel JL, Holwitt EA, Fan M, Roper SD. Methods and Compositions for a Process of Rapid Selection and Production of DNA. Patent Pending, Patent and Trademark Office; 2008. [Google Scholar]
- 16.Nutiu R, Li Y. Aptamers with fluorescence-signaling properties. Methods. 2005;37:16–25. doi: 10.1016/j.ymeth.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Bishop J, Wilson C, Chagovetz AM, Blair S. Competitive displacement of DNA during surface hybridization. Biophys Let. 2007 Jan 1;92(1):L10–L12. doi: 10.1529/biophysj.106.097121. [DOI] [PMC free article] [PubMed] [Google Scholar]