Abstract
Electroporation is a valuable tool for nucleic acid delivery because it can be used for a wide variety of cell types. Many scientists are shifting toward the use of cell types that are more relevant to in vivo applications, including primary cells, which are considered difficult to transfect. The ability to electroporate these cell types with nucleic acid molecules of interest at a relatively high efficiency while maintaining cell viability is essential for elucidating the pathway(s) in which a gene product is involved. We present data demonstrating that by optimizing electroporation parameters, nucleic acid molecules can be delivered in a highly efficient manner. We display transfection results for primary and difficult-to-transfect cell types including human primary fibroblasts, human umbilical vein endothelial cells, Jurkat cells, and two neuroblastoma cell lines [SK-N-SH (human) and Neuro-2A (mouse)] with plasmid DNAs and siRNAs. Our data demonstrate that by determining proper electroporation conditions, glyceraldehyde phosphate dehydrogenase mRNA was silenced in Jurkat cells when compared with negative control siRNA electroporations as early as 4 h post-transfection. Other experiments demonstrated that optimized electroporation conditions using a fluorescently labeled transfection control siRNA resulted in 75% transfection efficiency for Neuro-2A, 93% for human primary fibroblasts, and 94% for HUVEC cells, as analyzed by flow cytometry.
Keywords: electroporation, human primary fibroblasts, HUVEC, Jurkat, mammalian cells, MXcell, Neuro-2a, neuroblastoma, primary cells transfection, SK-N-SH
INTRODUCTION
The ability to modulate gene expression within primary cells is essential for functional genomics, pathway analysis, and medical applications. As these areas of research progress, the ability to study key interactions in cells more closely resembling in vivo conditions will be increasingly important. For many gene expression studies, the delivery of nucleic acid molecules into cells by transfection enables the downstream analysis of these key interactions. In this paper we describe electroporation—a simple and effective method of delivering DNA and siRNA into primary and other difficult-to-transfect cells. By rapidly optimizing electroporation conditions before proceeding with an experiment, it is possible to efficiently and consistently deliver nucleic acids into virtually any cell while maintaining cell viability.
Electroporation is a physical method of gene delivery. For this reason it is widely applicable to a variety of cell types, including animal, plant, and microbial.1 During electroporation, cells are exposed to a high-voltage pulse in the presence of exogenous nucleic acid. The high voltage causes the cellular membrane to be transiently permeabilized, allowing the foreign nucleic acids to enter the cell.2–4 Every cell type requires slightly different electroporation conditions that must be determined experimentally.
A few considerations need attention for optimal delivery of nucleic acids into cells by electroporation. Electric field strength and pulse duration are key parameters to maximize transfection efficiency and maintain cell viability.5 The pulse applied to the cells can be generated as two distinct wave forms: square and exponential decay. Square wave forms rely on a constant charge being applied to the cells for a set time. The use of square wave forms allows for the application of multiple pulses. During exponential decay wave forms, an initial voltage is set, and the duration of the decay (time constant) is the product of the capacitance setting and the resistance of the sample. Since the sample resistance results mainly from the ionic strength of the electroporation buffer, such that resistance is constant, one can empirically determine the effect of changing the capacitance setting on the pulse. The buffer components also influence transfection efficiency and cell viability. Traditionally, a buffer with high ionic strength (low resistance) such as phosphate buffered saline (PBS) or serum-free growth media is used in electroporation of mammalian cells at high capacitance. In this paper, we use a novel electroporation buffer designed to mimic the intracellular ionic strength to promote transfection efficiency and cell viability throughout the electroporation process. In addition to the electric parameters and buffer composition, electroporation is influenced by cell health and density and nucleic acid concentration and type. These parameters have been reviewed in detail.6
To optimize electroporation conditions we used an open-platform, plate-based electroporation system, which allowed us to vary several of the parameters detailed above in parallel. Here we describe the process of optimizing electroporation conditions, and the successful electroporation of plasmid DNA and siRNA into primary cells such as human primary fibroblasts (HPF), and four other cell lines which are typically difficult to transfect: human umbilical vein endothelial cells (HUVEC), Jurkat, and the neuroblastoma cell lines Neuro-2a (murine) and SK-N-SH (human).
MATERIALS AND METHODS
Cell Culture
All cell lines were obtained from the American Type Culture Collection (ATTC). HUVEC cells (ATTC, CRL-1730) were cultured in EBM-2 medium (Lonza, Allen-dale, NJ). Jurkat cells (ATTC, TIB-152) were cultured in RPMI-1640 (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum. The murine Neuro-2a cells (ATTC, CCL-131) were cultured in Dulbeccoís Modified Eagleís Medium supplemented with 10% fetal bovine serum. Human neuroblastoma cell line SK-N-SH (ATCC, HTB-11) was grown in RPMI-1640 medium containing 25 mM HEPES and L-glutamine (Invitrogen) supplemented with 12.5% fetal bovine serum and 1% penicillin-streptomycin. Human fibroblast cells (ATTC, CRL-2703) were cultured in Iscove’s modified Dulbecco’s medium (Invitrogen) with 10% fetal bovine serum.
Cells were passaged 1 day prior to electroporation so that they were actively growing on the day of electroporation.
Electroporation
Prior to electroporation, the cells were washed with PBS, counted, and resuspended in Gene Pulser electroporation buffer (Bio-Rad, Hercules, CA) to a cell density of 1 × 106 cells/mL unless otherwise indicated, and mixed with nucleic acid.
The following plasmids were used: a luciferase expression plasmid (pCMVI-Luc) and a green fluorescent protein (GFP) expression plasmid (pEGFP-actin; Clontech Mountain View, CA). siLentMer Dicer-substrate siRNA for GAPDH, β-actin, and a nonsilencing negative control (NC) (Bio-Rad) were electroporated for the gene silencing studies. A fluorescently labeled transfection control (TC) siLentMer siRNA (Bio-Rad) was used to assess electroporation efficiency by flow cytometry. The plasmids were used at a concentration of 20 μg/mL, and siLentMer siRNAs were used at 100 nM.
The cells and nucleic acid in Gene Pulser electroporation buffer (Bio-Rad) were gently mixed and then transferred into electroporation plates (Bio-Rad). One hundred fifty microliters of the mix was used per well in a 96-well electroporation plate. The plate was electroporated using preset or manually entered protocols on the Gene Pulser MXcell electroporation system (Bio-Rad). One hundred microliters of the electroporated cells was transferred into 24-well tissue culture plates containing 500 μL of the appropriate growth media for each cell type and incubated at 37°C for 24 h unless otherwise indicated.
Analysis of Transfection
Prior to harvesting the cells, cell confluency was assessed by comparing the percentage of attached cells between different conditions. Suspension cell viability was determined by propidium iodide staining using the BD FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). Cells electroporated with pCMVI-Luc plasmid were washed and assayed for luciferase activity 24 h post-electroporation using a luminometer (MLX Microtiter Plate Luminometer; Dynex Technologies, Chantilly, VA) and a flash assay. Results are reported in relative light units. Cells electroporated with GFP plasmid or fluorescent TC siRNA were washed, detached with trypsin, and suspended in PBS for analysis by flow cytometry or fixed with 1.85% formaldehyde in PBS for analysis by fluorescent microscopy.
Delivery of siLentMer siRNA was also assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Total RNA was extracted from cells using the Aurum total RNA mini kit, and cDNA was synthesized using the iScript cDNA synthesis kit. Gene-specific primers were used to amplify the relevant message with iQ SYBR Green supermix on the iQ5 real-time PCR detection system (in at least triplicates) (all from Bio-Rad).
RESULTS
We report the optimal starting electroporation conditions as the parameters that resulted in the highest transfection efficiency or gene expression activity, paired with the highest cell viability (survival rate). Initial experiments resulted in quick identification of the best electroporation waveform conditions for each cell type transfected with either plasmid DNA or siRNA. This was followed by more thorough identification of the electroporation parameters which are considered to be waveform-dependent. In the end, experiments resulted in better transfection efficiency in conjunction with effective gene silencing or gene expression and high cell viability.
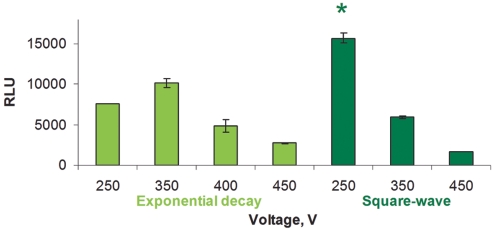
Electroporation of HUVEC cells resulted in high transfection efficiency using both plasmid (Figure 1) and siRNAs (Figure 2). To identify the best transfection conditions, the cells were electroporated with plasmid DNA (pCMVI-Luc), and manual protocols were entered on the Gene Pulser MXcell. Luciferase expression, expressed as relative light units, was measured in cell extracts 24 h after electroporation. The best square wave electroporation condition (250 V, 20 msec) for HUVEC cells resulted in 60% higher luciferase expression than the best exponential wave condition (350 V, 500 μF) (Figure 1, dark green). These protocols were used to deliver siRNAs to the HUVEC cells. First, cells were electroporated with a fluorescent TC siRNA in order to assess transfection efficiency which was determined to be about 94% by flow cytometry (Figure 2A). Then, siLentMer siRNAs targeting β-actin and GAPDH genes were electroporated into the cells. These experiments resulted in > 97% gene silencing as measured by RT-qPCR (Figure 2B), when compared with the NC siLentMer electroporations.
FIGURE 1.
Electroporation of HUVEC cells with pCMVI-Luc plasmid and Gene Pulser electroporation buffer, using exponential or square-wave conditions. Luciferase assay results are expressed in relative light units. The exponential wave electroporations varied the voltage and kept a constant capacitance (500 μF) and resistance (1000 Ω; light green). The square-wave electroporations varied the voltage and kept a constant pulse duration of 20 msec (dark green). *The condition providing the highest luciferase activity. RLU, relative light units.
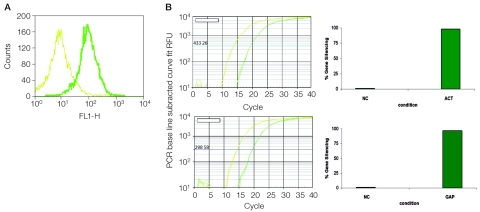
FIGURE 2.
Electroporation of HUVEC cells with siRNA. A: Overlaid histograms showing fluorescence of cells treated with electroporation buffer, but not electroporated (light green), and cells electroporated with fluorescent transfection control siRNA (dark green). B: RT-qPCR traces 24 h after electroporation using cDNA prepared from cells electroporated with β-actin siRNA (top panel, dark green) or negative control (top panel, light green) and GAPDH siRNA (bottom panel, dark green) or negative control (bottom panel,light green). ACT, β-actin siRNA; RFU, RFUGAQP; GAPDH siRNA; NC, negative control; RFU, relative light units. The corresponding qPCR results are expressed as percentage gene silencing in the right-most panels. Electroporation conditions used were A: exponential decay (350 V, 500 μF, 1000 Ω) and B: square-wave (250 V, 2000 μF, 1000 Ω, 20 msec).
The preset protocol Opt mini 96 wells/square exponential (which includes 3 square waves and 3 exponential-wave conditions) was used to identify the best waveform conditions for HPF cells. Exponential wave form protocols were more efficient than square wave protocols (data not shown). Using an exponential wave pulse of 250 V and 500 μF, 93% of the cells were successfully electroporated with the fluorescent TC siRNA (Figure 3A); 44% transfection efficiency was obtained with the pEGFP expression plasmid. The cell viability 24 h after siRNA or plasmid delivery was similar (data not shown). Gene silencing experiments using 100 nM of an siLentMer siRNA for the GAPDH gene showed GAPDH expression was reduced by 96.1% when compared with NC siLentMer siRNA electroporations (Figure 3B).
FIGURE 3.
Electroporation of HPF cells with siRNA and plasmid DNA, using an exponential wave pulse of 250 V, 500 μF, 1000 Ω. A: Flow cytometry results 24 h post-electroporation of 100-nM fluorescent TC siRNA or 20 μg/mL pEGFP. The results show the percentage of transfected cells. B: RT-qPCR results 24 h post-electroporation of 100 nM GAPDH and negative control siLentMer siRNA. qPCR traces are shown in the middle, and results expressed as percentage of gene silencing are shown on the right. HPF, human primary fibroblasts; NC, negative control; RLU, relative light units.
An exponential waveform was previously identified as the most efficient for transfection of Jurkat cells (Gene Pulser Xcell electroporation system, Bio-Rad). We focused our efforts on the parameters specific to exponential decay pulses using plasmid DNA (pCMVI-Luc), first selecting the best voltage (Figure 4A), and then the best capacitance (Figure 4B). The optimal electroporation conditions for transfection of this plasmid were 250 V, 300 μF, and 1000 Ω (in Figure 4B). Using the same electroporation conditions, Jurkat cells were transfected with a GAPDH siLentMer siRNA to silence GAPDH gene expression. Four hours after electroporation, GAPDH expression was reduced by over 88% when compared with NC siLentMer electroporation, and expression continued to be reduced up to 48 h after electroporation (Figure 5). Longer time points were not tested in this experiment, although previous studies have shown that the silencing effects of these siRNAs can persist in cells up to 10 d.7 Mouse neuroblastoma cells (Neuro-2a) were electroporated with fluorescent TC siRNA or the GFP-expression plasmid at cell densities of 2 × 106 and 4 × 106 cells/mL. Electroporation efficiency for siRNA was monitored by flow cytometry, and for plasmid DNA, by fluorescent microscopy (Figure 6). Two exponential wave electroporation conditions provided equivalent results (200 V, 500 μF, 1000 Ω, and 250 V, 350 μF, 1000 Ω; data not shown). We observed about 75% transfection efficiency for siRNAs. We also examined electroporation of the human neuroblastoma cell line SK-N-SH. Combining luciferase activity and cell viability data (Figure 7), we determined that the best electroporation parameters were square wave, 200 V, 20 msec with either 1 × 106 cells/mL or 3 × 106 cells/mL. Additionally, we observed over 90% silencing of GAPDH using identical electroporation conditions and either 50 or 100 nM GAPDH siLentMer siRNA (data not shown).
FIGURE 4.
Luciferase activity 24 h after electroporation of Jurkat cells. An exponential waveform was used to determine the optimal conditions for electroporation. A: Optimization of voltage at 300 μF and 1000 Ω. B: Optimization of capacitance at 250 V and 1000 Ω. Results are expressed in RLU, relative light units. *, The condition providing the highest luciferase expression.
FIGURE 5.
Time course of GAPDH gene silencing in Jurkat cells. Jurkat cells were electroporated using an exponential wave pulse of 250 V, 300 μF, and 1000 Ω. Cells electroporated with GAPDH siRNA (dark green) showed an 88% reduction of the GAPDH transcript as compared with negative control cells. Values were normalized to negative control (light green bars at 0% gene silencing).
FIGURE 6.
Electroporation of Neuro-2a cells using an exponential waveform pulse of 250 V, 350 μF, and 1000 Ω. Cells were assayed 24 h after transfection. A: Cells analyzed by flow cytometry. Nonelectroporated negative control cells (top) and electroporated cells (bottom) were treated with fluorescent TC siRNA in electroporation buffer. We used 4 × 106 cells/mL in these experiments. B: Cells analyzed by microscopy. Cells were electroporated with pEGFP plasmid DNA. Top: Bright field image of Neuro-2A cells. Bottom: fluorescent image. We used 2 × 106 cells/mL in these electroporations.
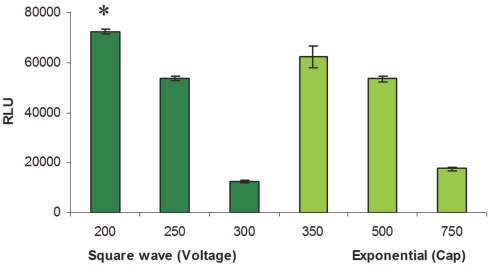
FIGURE 7.
Luciferase activity of SK-N-SH cells electroporated with pCMVI-Luc using the preset protocol Opt Mini 96/Sqr, Exponential. Three square-wave well sets were used to test differing voltages with a pulse duration of 20 msec (dark green), while three exponential well sets were used to test capacitances at constant voltage (250 V) and resistance (1000 Ω) (light green). Two cell densities were tested: 1 × 106 cells/mL and 3 × 106 cells/mL (only 3 × 106 cells/mL data are shown here). *, The condition providing the highest luciferase activity. Cap, capacitance; RLU, relative light units.
We have identified optimal electroporation starting conditions (Table 1) for the cell types mentioned here, as well as many others, by testing multiple electroporation parameters simultaneously.
TABLE 1.
Optimal Starting Conditions for Different Cell Types Using the Gene Pulser MXcell Electroporation System and Gene Pulser Electroporation Buffer
| Cell Line | Nucleic Acid | Starting Conditions |
|---|---|---|
| HUVEC | siRNA/plasmid | Sq: 250 V, 20 msec, 2000 μF, 1000 Ω* |
| Exp: 350 V, 500 μF, 1000 Ω | ||
| Jurkat | siRNA | Exp: 250 V, 300 μF, 1000 Ω |
| Neuro-2a | siRNA | Exp: 250 V, 350 μF, 1000 Ω |
| SK-N-SH | siRNA/plasmid | Sq: 200 V, 20 msec, 2000 μF, 1000 Ω |
| HPF | siRNA | Exp: 250 V, 500 μF, 1000 Ω |
| A549 | Sq: 300 V, 2000 μF, 1000 Ω, 20 msec | |
| CHO K-1 | siRNA | Sq: 250 V, 2000 μF, 1000 Ω, 20 msec |
| plasmid | Sq: 250 V, 2000 μF, 1000 Ω, 2 × 10 msec | |
| CHO DG44 | siRNA/plasmid | Sq: 300 V, 2000 μF, 1000 Ω, 20 msec* |
| Exp: 250 V, 500 μF, 1000 Ω | ||
| MCF-7 | siRNA | Exp: 250–300 V, 250 μF, 1000 Ω |
| HeLa | siRNA | Exp: 250 V, 350 μF OR Sq: 250 V, 20 msec* |
| plasmid | Exp: 250 V, 200 μF OR Exp: 250 V, 350 μF | |
| COS-7 | plasmid | Sq: 220 V, 2000 μF, 1000 Ω, 20 msec |
| K562 | plasmid | Exp: 300 V, 250 μF, 1000 Ω |
Conditions found to yield nearly indistinguishable results.
DISCUSSION
We show that electroporation can be a very efficient method for introducing nucleic acids into cells of interest, including those that are often considered difficult to transfect. High transfection efficiency for the cells described here was achieved by first identifying the most favorable electroporation waveform, and then by refining the parameters related to that waveform. For example, in a square wave protocol, we can modify the voltage and pulse duration, while in an exponential wave protocol, we can make adjustments to the voltage and capacitance settings. We assessed the results to confirm that our conditions maximized both transfection efficiency and cell viability. The best electroporation conditions for any cell type can be obtained by using an open plate-based system which allows adjustment of parameters including waveform, voltage, capacitance, and pulse duration.
We chose to work with two cell types that offer the advantage of being nontransformed (noncancerous), “native” cells. However, since these cells are not immortalized, they have a limited life span in culture and can be very difficult to transfect and maintain. HUVEC cells are often used as a model for endothelial cells from blood vessels8 and optimal electroporation rates of 40% have been reported.9 We were able to obtain almost 98% transfection efficiency by making adjustments in the electroporation parameters. It is interesting to note that several references in the literature report the use of exponential wave electroporation of HUVEC cells,8–10 while square wave electroporation was reported to be optimal in two papers11,12 and the work we present here.
The electroporation conditions reported in the HUVEC studies used voltage settings between 200 and 400 V and 950 μF.8–10 Our “next best condition” in Figure 1 is an exponential wave pulse at 350 V and 500 μF. Although the electroporation buffers were not specified in these reports, we infer that it was PBS or serum-free media. In order to obtain a similar time constant when using the Gene Pulser electroporation buffer (with higher resistance than either PBS or serum-free media), we recommend the capacitance setting be reduced by about one third to one half in exponential wave electroporations.
Human primary fibroblasts are used as a model of “normal” epithelial cells. These cells are important in the study of epithelial cell cancers and other diseases, and can be derived from skin and foreskin. Efficient electroporation of HPF has been reported in which cells were stably transfected and results reported in colonies/μg plasmid.13 We obtained over 90% electroporation efficiency with our optimized conditions.
Jurkat cells are immortalized T-cells commonly used in the study of T-cell signaling and leukemias, and are considered difficult to transfect. In our studies, we electroporated GAPDH-specific siRNA and reported the gene silencing to be 88% as compared with control siRNA-electroporated cells. Electroporation of Jurkat cells with an siRNA for the multidrug resistance gene MRP1 resulted in 70% silencing.14
Neuroblastoma cell lines are used as model systems for the study of neural cell development, as stimulation by commonly used differentiation agents such as phorbol esters, hydroxyphenyl retinamide, and cytosine arabino-side result in different responses depending on the cell lines.15 The human neuroblastoma cell line SK-N-SH has been transfected by calcium phosphate,16 lipid,17 and electroporation.18 The mouse neuroblastoma cell line Neuro-2a has been transfected by calcium phosphate19 and also by electroporation.20
It is interesting to note that we identified different waveforms for the two different neuroblastoma cell lines studied here. Thus, even with preliminary information regarding electroporation conditions of a similar cell type, it is quite possible to identify an even more efficient condition. We have demonstrated that even with no prior electroporation experience with a particular cell line (HUVEC or HPF), we were able to find optimal electroporation conditions in a matter of one or two experiments. Also, when starting with information obtained from the literature, as in the case of Jurkat cells, we could further improve upon the results by testing several electroporation parameters simultaneously. The differences noted in electroporation protocols published in the literature bring us to the point that there may be minor differences in the growth conditions of these cells and the electroporation buffers used. All of these variables may lead to slight differences in optimal transfection conditions, thus indicating the need to confirm that the electroporation conditions employed are in fact the most favorable so that the best possible outcome can be obtained. The plate-based system we used allowed the simultaneous testing of exponential and square waveforms within the same experiment, as well as other electrical parameters (capacitance, resistance, and voltage) or biological parameters (type and concentration of nucleic acid and cell density).
ACKNOWLEDGMENT
We thank Jamie Wibbenmeyer for her critical review of this manuscript and for her tireless support.
REFERENCES
- 1.Shigekawa K, Dower WJ. Electroporation of eukaryotes and prokaryotes: A general approach to the introduction of macromolecules into cells. Biotechniques. 1988;6:742–751. [PubMed] [Google Scholar]
- 2.Tsong TY. Electroporation of cell membranes. Biophys J. 1991;60:297–306. doi: 10.1016/S0006-3495(91)82054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rols M-P, Golzio M, Delteil C, Teissie J. In vitro delivery of drugs and other molecules to cells. In: Jaroszeski MJ, Heller R, Gilbert R, editors. Methods in Molecular Medicine. Vol. 37. Totowa, NJ: Humana Press; 2000. pp. 83–97. [DOI] [PubMed] [Google Scholar]
- 4.Rols MP. Electropermeabilization: A physical method for the delivery of therapeutic molecules into cells. Biochim Biophys Acta. 2006;1758:423–428. doi: 10.1016/j.bbamem.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Rols M-P, Teissié J. Electropermeabilization of mammalian cells to macromolecules: Control by pulse duration. Biophys J. 1998;75:1415–1423. doi: 10.1016/S0006-3495(98)74060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heiser WC. Transformation of Mammalian Cells. In: Tymms MJ, editor. Methods in Molecular Biology. Vol. 130. Totowa, NJ: Humana Press; 2000. pp. 117–134. Available as a reprint from Bio-Rad Laboratories literature no. RP0010. [DOI] [PubMed] [Google Scholar]
- 7.Kim DH, Behlke MA, Rose SD, et al. Synthetic dsRNA dicer substrated enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 8.van Leeuwen EBM, van der Veen AY, Hoekstra D, et al. Transfection of Small Numbers of Human Endothelial Cells by Electroporation and Synthetic Amphiphiles. Eur J Vasc Endovasc Surg. 1999;17:9–14. doi: 10.1053/ejvs.1998.0677. [DOI] [PubMed] [Google Scholar]
- 9.Ear T, Giguére P, Fleury A, et al. High efficiency transient transfection of genes in human umbilical vein endothelial cells by electroporation. J Immunol Methods. 2001;257:41–49. doi: 10.1016/s0022-1759(01)00445-8. [DOI] [PubMed] [Google Scholar]
- 10.Yamauchi K, Nishimura Y, Shigematsu S, et al. Vascular endothelial cell growth factor attenuates actions of transforming growth factor-β in human endothelial cells. J Biol Chem. 2004;279:55104–55108. doi: 10.1074/jbc.M407423200. [DOI] [PubMed] [Google Scholar]
- 11.Ovcharenko D, Jarvis R, Hunicke-Smith S, Kelnar K, Brown D. High-throughput RNAi screening in vitro: From cell lines to primary cells. RNA. 2005;11:985–993. doi: 10.1261/rna.7288405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Boeuf F, Houle F, Huot J. Regulation of vascular endothelial growth factor receptor 2-mediated phosphorylation of foal adhesion kinase by heat shock protein 90 and Src kinase activities. J Biol Chem. 2004;279:39175–39185. doi: 10.1074/jbc.M405493200. [DOI] [PubMed] [Google Scholar]
- 13.Fountain JW, Lockwood WK, Collins FS. Transfection of primary human skin fibroblasts by electroporation. Gene. 1988;68:167–172. doi: 10.1016/0378-1119(88)90610-5. [DOI] [PubMed] [Google Scholar]
- 14.Hammond CL, Marchan R, Krance SM, Ballatori N. Glutathione export during apoptosis requires functional multidrug resistance-associated proteins. J Biol Chem. 2007;282:14337–14347. doi: 10.1074/jbc.M611019200. [DOI] [PubMed] [Google Scholar]
- 15.Thiele CJ. Neuroblastoma. In: Masters J, editor. Human Cell Culture. Vol. 1. Lancaster UK: Kluwer Academic Publishers; 1988. pp. 21–19853. [Google Scholar]
- 16.Ghosh C, Song W, Lahiri DK. Efficient DNA transfection in neuronal and astrocytic cell lines. Mol Biol Rep. 2000;27:113–121. doi: 10.1023/a:1007173906990. [DOI] [PubMed] [Google Scholar]
- 17.Tang F, Hughes JA. Introduction of a disulfide bond into a cationic lipid enhances transgene expression of plasmid DNA. Biochem Biophys Res Commun. 1998;242:141–145. doi: 10.1006/bbrc.1997.7923. [DOI] [PubMed] [Google Scholar]
- 18.Spagnol G, Doneda P, Cavanna B, et al. Expression of glycosylated recombinant human myelin-associated glycoprotein on a neuroblastoma cell line and its reactivity with HNK-1 but not human anti-MAG antibodies. Neurosci Lett. 1998;246:157–160. doi: 10.1016/s0304-3940(98)00261-4. [DOI] [PubMed] [Google Scholar]
- 19.Chen CA, Okayama H. Calcium phosphate-mediated gene transfer: A highly efficient transfection system for stably transforming cells with plasmid DNA. Biotechniques. 1988;6:632–638. [PubMed] [Google Scholar]
- 20.Lawman MJP, Lawman PD. Antigen modified cancer cell vaccines for cancer therapy. United States Patent 7348015, 2008.