Abstract
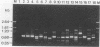
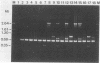
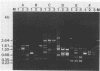
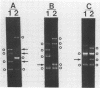
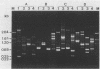
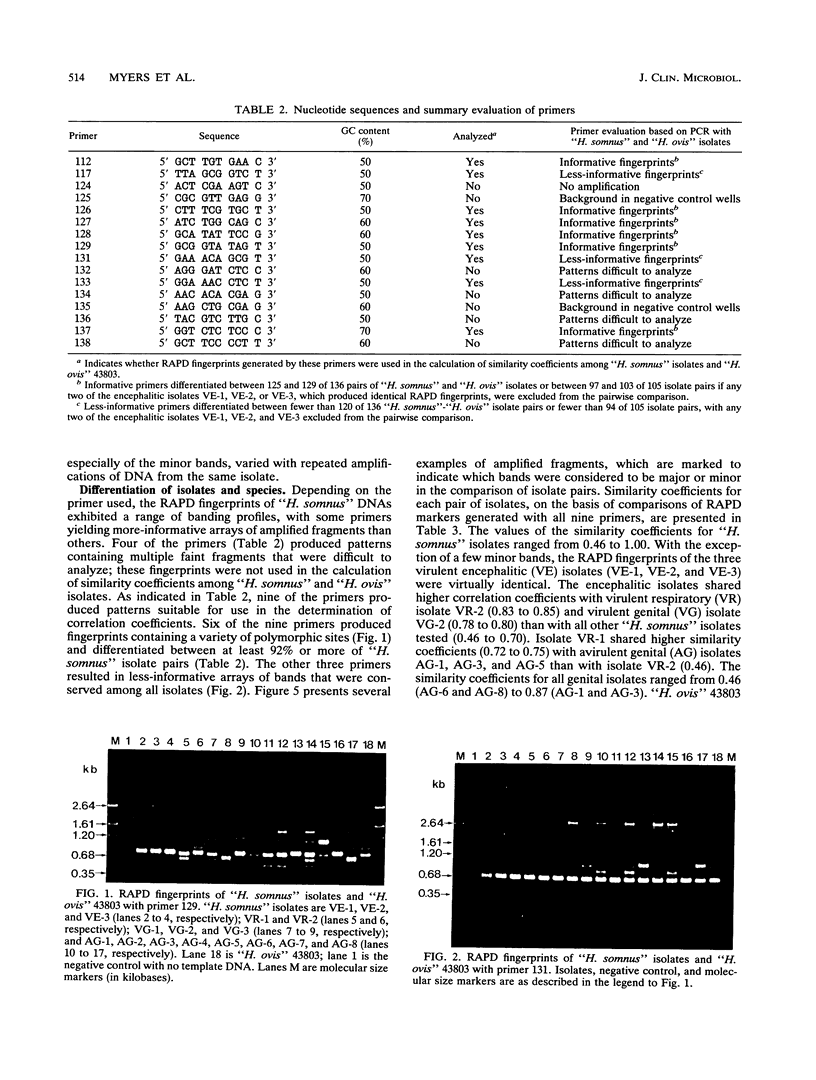
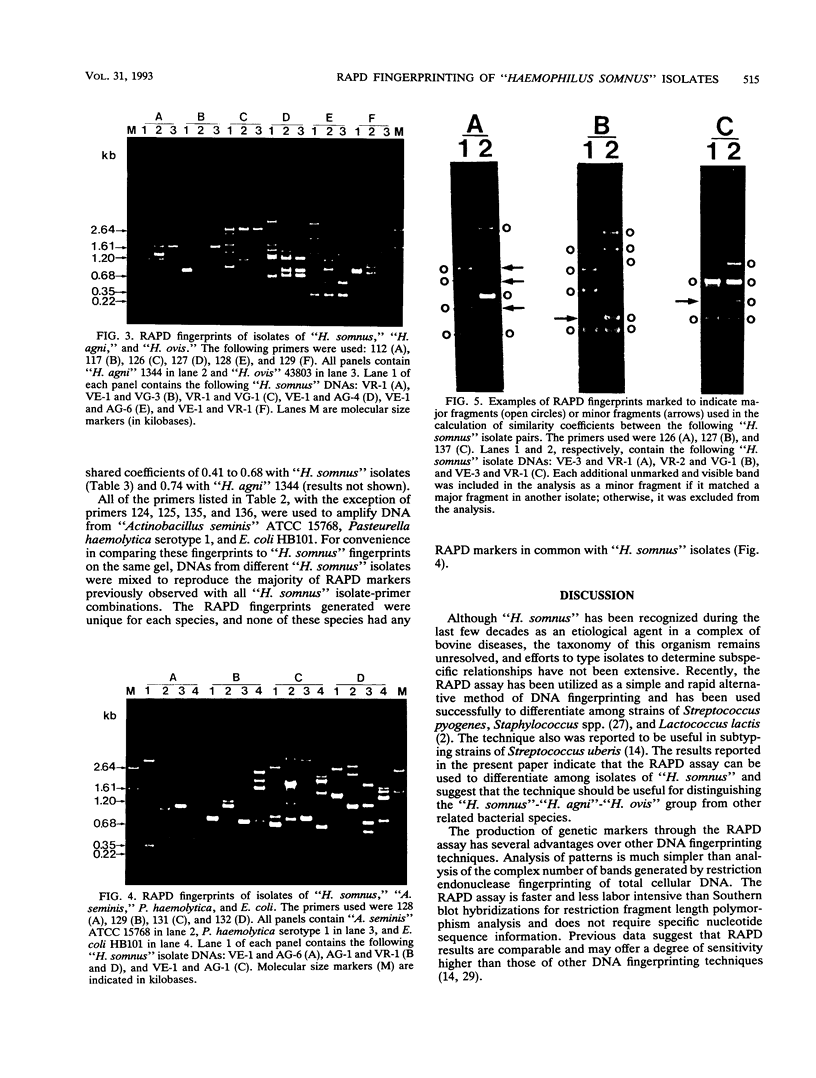
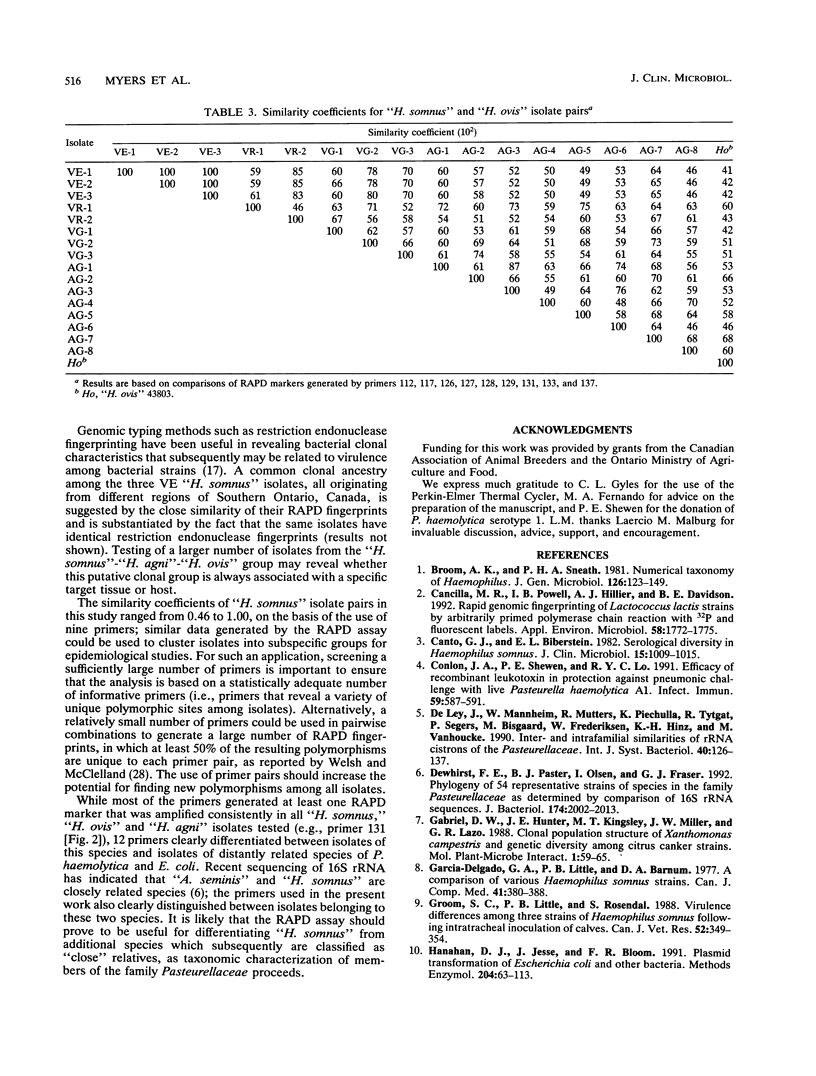
The random-amplified polymorphic DNA (RAPD) assay was used to generate DNA fingerprints for 16 isolates of "Haemophilus somnus," and one isolate each of "Haemophilus agni," "Histophilus ovis," "Actinobacillus seminis," Pasteurella haemolytica, and Escherichia coli. The RAPD assay differentiated among "H. somnus" isolates, which shared similarity coefficients of 0.46 to 1.00 on the basis of pairwise comparisons of RAPD markers produced with nine random decamer primers. Three virulent encephalitic "H. somnus" isolates exhibited identical banding patterns, suggesting a common clonal ancestry. The RAPD assay clearly distinguished between the "H. somnus"-"H. agni"-"H. ovis" group and the other bacterial species tested. The results of the present study suggest that DNA fingerprinting of "H. somnus" isolates by the RAPD assay could be valuable in revealing subspecific divisions within this largely unexplored species.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Broom A., Sneath P. H. Numerical taxonomy of Haemophilus. J Gen Microbiol. 1981 Sep;126(1):123–149. doi: 10.1099/00221287-126-1-123. [DOI] [PubMed] [Google Scholar]
- Cancilla M. R., Powell I. B., Hillier A. J., Davidson B. E. Rapid genomic fingerprinting of Lactococcus lactis strains by arbitrarily primed polymerase chain reaction with 32P and fluorescent labels. Appl Environ Microbiol. 1992 May;58(5):1772–1775. doi: 10.1128/aem.58.5.1772-1775.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto G. J., Biberstein E. L. Serological diversity in Haemophilus somnus. J Clin Microbiol. 1982 Jun;15(6):1009–1015. doi: 10.1128/jcm.15.6.1009-1015.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon J. A., Shewen P. E., Lo R. Y. Efficacy of recombinant leukotoxin in protection against pneumonic challenge with live Pasteurella haemolytica A1. Infect Immun. 1991 Feb;59(2):587–591. doi: 10.1128/iai.59.2.587-591.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ley J., Mannheim W., Mutters R., Piechulla K., Tytgat R., Segers P., Bisgaard M., Frederiksen W., Hinz K. H., Vanhoucke M. Inter- and intrafamilial similarities of rRNA cistrons of the Pasteurellaceae. Int J Syst Bacteriol. 1990 Apr;40(2):126–137. doi: 10.1099/00207713-40-2-126. [DOI] [PubMed] [Google Scholar]
- Dewhirst F. E., Paster B. J., Olsen I., Fraser G. J. Phylogeny of 54 representative strains of species in the family Pasteurellaceae as determined by comparison of 16S rRNA sequences. J Bacteriol. 1992 Mar;174(6):2002–2013. doi: 10.1128/jb.174.6.2002-2013.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Delgado G. A., Little P. B., Barnum D. A. A comparison of various Haemophilus somnus strains. Can J Comp Med. 1977 Oct;41(4):380–388. [PMC free article] [PubMed] [Google Scholar]
- Groom S. C., Little P. B., Rosendal S. Virulence differences among three strains of Haemophilus somnus following intratracheal inoculation of calves. Can J Vet Res. 1988 Jul;52(3):349–354. [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Jessee J., Bloom F. R. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- Harris F. W., Janzen E. D. The Haemophilus somnus disease complex (Hemophilosis): A review. Can Vet J. 1989 Oct;30(10):816–822. [PMC free article] [PubMed] [Google Scholar]
- Jayarao B. M., Bassam B. J., Caetano-Anollés G., Gresshoff P. M., Oliver S. P. Subtyping of Streptococcus uberis by DNA amplification fingerprinting. J Clin Microbiol. 1992 May;30(5):1347–1350. doi: 10.1128/jcm.30.5.1347-1350.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen B. E., Bjorvatn B., Lund V., Lindqvist B., Holten E. Differentiation of B15 strains of Neisseria meningitidis by DNA restriction endonuclease fingerprinting. J Infect Dis. 1984 Nov;150(5):672–677. doi: 10.1093/infdis/150.5.672. [DOI] [PubMed] [Google Scholar]
- Kwiecien J. M., Little P. B. Haemophilus somnus and reproductive disease in the cow: A review. Can Vet J. 1991 Oct;32(10):595–601. [PMC free article] [PubMed] [Google Scholar]
- Kwiecien J. M., Little P. B. Isolation of pathogenic strains of Haemophilus somnus from the female bovine reproductive tract. Can J Vet Res. 1992 Apr;56(2):127–134. [PMC free article] [PubMed] [Google Scholar]
- McGillivery D. J., Webber J. J., Dean H. F. Characterisation of Histophilus ovis and related organisms by restriction endonuclease analysis. Aust Vet J. 1986 Dec;63(12):389–393. [PubMed] [Google Scholar]
- Stephens L. R., Aukema R., Murray L. J. Antigenic heterogeneity of Haemophilus somnus. Aust Vet J. 1987 Apr;64(4):113–113. doi: 10.1111/j.1751-0813.1987.tb09643.x. [DOI] [PubMed] [Google Scholar]
- Stephens L. R., Humphrey J. D., Little P. B., Barnum D. A. Morphological, biochemical, antigenic, and cytochemical relationships among Haemophilus somnus, Haemophilus agni, Haemophilus haemoglobinophilus, Histophilus ovis, and Actinobacillus seminis. J Clin Microbiol. 1983 May;17(5):728–737. doi: 10.1128/jcm.17.5.728-737.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Genomic fingerprinting using arbitrarily primed PCR and a matrix of pairwise combinations of primers. Nucleic Acids Res. 1991 Oct 11;19(19):5275–5279. doi: 10.1093/nar/19.19.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]