Abstract
Establishment and maintenance of the blood system relies on self-renewing hematopoietic stem cells (HSCs) that normally reside in small numbers in the bone marrow niche of adult mammals. This Review describes the developmental origins of HSCs and the molecular mechanisms that regulate lineage-specific differentiation. Studies of hematopoiesis provide critical insights of general relevance to other areas of stem cell biology including the role of cellular interactions in development and tissue homeostasis, lineage programming and reprogramming by transcription factors, and stage- and age-specific differences in cellular phenotypes.
Introduction
The blood system serves as a paradigm for understanding tissue stem cells, their biology, and involvement in aging, disease, and oncogenesis. Because mature blood cells are predominantly short lived, stem cells are required throughout life to replenish multilineage progenitors and the precursors committed to individual hematopoietic lineages. Hematopoietic stem cells (HSCs) reside as rare cells in the bone marrow in adult mammals and sit atop a hierarchy of progenitors that become progressively restricted to several or single lineages (Orkin, 2000). These progenitors yield blood precursors devoted to unilineage differentiation and production of mature blood cells, including red blood cells, megakaryocytes, myeloid cells (monocyte/macrophage and neutrophil), and lymphocytes. As with all other stem cells, HSCs are capable of self-renewal—the production of additional HSCs—and differentiation, specifically to all blood cell lineages.
HSCs are defined operationally by their capacity to reconstitute the entire blood system of a recipient. In general, preparation of patients for transplantation with donor bone marrow containing HSCs entails destruction of host bone marrow by irradiation or by treatment with high-dose cytotoxic drugs, in part to provide ‘‘space’’ for donor HSCs within the marrow microenvironment (the niche) of the recipient. HSCs can be prospectively identified by monoclonal antibodies directed to surface markers, by dye efflux, or on the basis of their metabolic properties; HSCs can be separated from more-committed progenitors and other marrow cells by fluorescence-activated cell sorting (FACS). With contemporary methods, HSCs may be highly purified such that as few as one cell may provide long-term (>4 months) hematopoietic reconstitution in a recipient. Technical considerations regarding the assays for quantitation of HSCs and evaluation of their function have recently been reviewed (Purton and Scadden, 2007). Because no ex vivo assays can replace in vivo transplantation for measuring biological activity of HSCs, characterizing cell populations based on the expression of cell-surface markers cannot be considered synonymous with determining their function. During stress or other manipulations (such as in mutant animals), the surface marker profile of HSCs and their progenitors may be distorted.
Here, we discuss the developmental origins of the hematopoietic system and the molecular control of self-renewal and lineage determination. The process of hematopoiesis is generally conserved throughout vertebrate evolution. Manipulation of animal models, such as the mouse and zebrafish, has complemented and greatly extended studies of human hematopoiesis. Although not an entirely ideal experimental system, partial reconstitution of the blood system of immunodeficient mice (such as NOD/SCID strains) has been commonly employed to study human hematopoiesis. The remarkable regenerative properties of human HSCs arebest illustrated by the success of marrow transplantation in human patients, a current mainstay of therapy for a variety of genetic disorders, acquired states of bone marrow failure, and cancers.
Emergence of HSCs
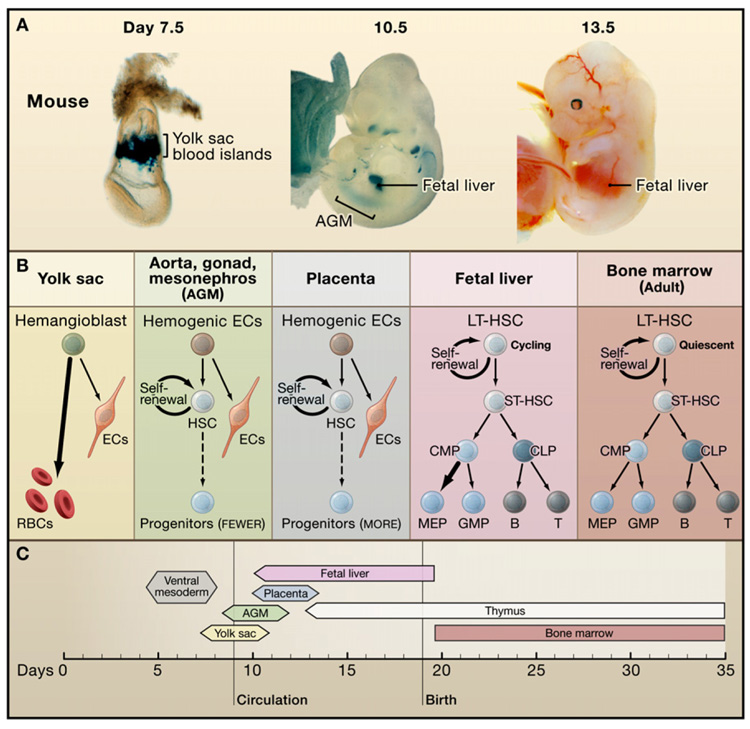
In vertebrates, the production of blood stem cells is accomplished by the allocation and specification of distinct embryonic cells in a variety of sites that change during development (Galloway and Zon, 2003) (Figure 1 and Figure 2). In mammals, the sequential sites of hematopoiesis include the yolk sac, an area surrounding the dorsal aorta termed the aorta-gonad mesonephros (AGM) region, the fetal liver, and finally the bone marrow (Figure 1). Recently, the placenta has been recognized as an additional site that participates during the AGM to fetal liver period. The properties of HSCs in each site differ, presumably reflecting diverse niches that support HSC expansion and/or differentiation and intrinsic characteristics of HSCs at each stage. For instance, HSCs present in the fetal liver are in cycle, whereas adult bone marrow HSCs are largely quiescent.
Figure 1. Developmental Regulation of Hematopoiesis in the Mouse.
(A) Hematopoiesis occurs first in the yolk sac (YS) blood islands and later at the aorta-gonad mesonephros (AGM) region, placenta, and fetal liver (FL). YS blood islands are visualized by LacZ staining of transgenic embryo expression GATA-1- driven LacZ. AGM and FL are stained by LacZ in Runx1-LacZ knockin mice. (Photos courtesy of Y. Fujiwara and T. North.).
(B) Hematopoiesis in each location favors the production of specific blood lineages. Abbreviations: ECs, endothelial cells; RBCs, red blood cells; LTHSC, long-term hematopoietic stem cell; ST-HSC, short-term hematopoietic stem cell; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; MEP, megakaryocyte/erythroid progenitor; GMP, granulocyte/macrophage progenitor.
(C) Developmental timewindows for shifting sites of hematopoiesis.
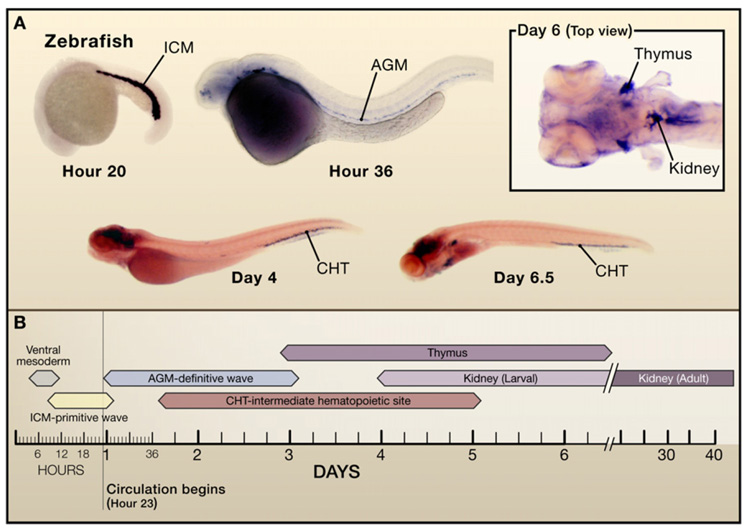
Figure 2. Hematopoietic Development in the Zebrafish.
(A) Hematopoiesis occurs first in the intermediate cell mass (ICM) and subsequently in the aorta-go-nad mesonephros (AGM) region and caudal hematopoietic tissue (CHT). Later hematopoietic cells are found in the kidney as well as in the thymus. In situ hybridization for GATA-1 at 30 hr (ICM), for c-myb at 36 hr (AGM), for SCL/tal1 at days 4 and 6.5 (CHT), and for c-myb at day 6 (top view) to demonstrate expression in the kidney marrow and thymus. (Photos courtesy of X. Bai and T. Bowman.)
(B) Developmental time windows for hematopoietic sites in the zebrafish.
Although there is little dispute regarding where HSCs are found during development, few topics have polarized investigators as much as the origin of HSCs. HSCs are derived from ventral mesoderm (see Review by C.E. Murry and G. Keller, page 661 of this issue). The contribution of each hematopoietic site (such as the yolk sac and fetal liver) to circulating blood in the fetus or adult was seemingly answered more than 25 years ago. Recent studies in mice and zebrafish, however, challenge the field with divergent views.
Multiple Waves of Hematopoiesis during Development
The initial wave of blood production in the mammalian yolk sac is termed ‘‘primitive.’’ The primary function for primitive hematopoiesis is production of red blood cells that facilitate tissue oxygenation as the embryo undergoes rapid growth. The hallmark of primitive erythroid cells is expression of embryonic globin proteins. The primitive hematopoietic system is transient and rapidly replaced by adult-type hematopoiesis that is termed “definitive.”
In mammals, the next site of hematopoietic potential is the AGM region. Hematopoietic cells were first detected in the aorta of the developing pig more than 80 years ago. Subsequently, studies of chick-quail chimeras and diploid-triploid Xenopus embryos demonstrated analogous AGM-like regions. Morphological examination revealed that a sheet of lateral mesoderm migrates medially, touches endoderm, and then forms a single aorta tube. Clusters of hematopoietic cells subsequently appear in the ventral wall. Similarly, an intraembryonic source of adult HSCs in mice capable of long-term reconstitution of irradiated hosts resides in the AGM region (Muller et al., 1994). At embryonic day 10.5, little HSC activity is detectable, whereas by day 11 engrafting activity is present.
Additional hematopoietic activity in the mouse embryo was detected subsequently in other sites, including the umbilical arteries and the allantois in which hematopoietic and endothelial cells are colocalized (Inman and Downs, 2007). Umbilical veins lack hematopoietic potential, suggesting that a hierarchy exists during definitive hematopoiesis in which HSCs arise predominantly during artery specification. In addition, significant numbers of HSCs are found in the mouse placenta (Gekas et al., 2005; Ottersbach and Dzierzak, 2005), nearly coincident with the appearance of HSCs in the AGM region and for several days thereafter. Placental HSCs could arise through de novo generation or colonization upon circulation, or both. The relative contribution of each of the above sites to the final pool of adult HSCs remains largely unknown.
Subsequent definitive hematopoiesis involves the colonization of the fetal liver, thymus, spleen, and ultimately the bone marrow. It is believed that none of these sites is accompanied by de novo HSC generation. Rather, their niches support expansion of populations of HSCs that migrate to these new sites. However, until very recently (as discussed below), there has been no evidence by fate mapping or direct visualization that HSCs from one site colonize subsequent sites.
Hemangioblasts and Hemogenic Endothelium
A common origin for blood and vascular cells, the “hemangio-blast,” was hypothesized a century ago, based largely on the intimate association of these lineages in the blood islands of the developing yolk sac. Sharing of markers between blood and blood vessel cells, and the impairment of both tissues in mutants, such as the mouse flk1 knockout (Shalaby et al., 1997) and zebrafish cloche (Stainier et al., 1995), are consistent with a common origin. Clonal studies using in vitro differentiating mouse embryonic stem (ES) cells provide the strongest evidence in favor of the existence of hemangioblasts (Choi et al., 1998). Furthermore, hemangioblast activity has been detected at the mid-streak stage of gastrulation and during the neural plate stage but is extremely transient in vivo (Huber et al., 2004). Despite these findings, formal proof of the hemangioblast hypothesis requires direct demonstration that a single cell divides asymmetrically to form blood and vascular derivatives in vivo.
Clonal analysis in mouse chimeras, however, presents contradictory evidence regarding the existence of the hemangioblast (Ueno and Weissman, 2006). Three different, stably marked ES cells were mixed and coinjected into host blastocysts. According to the hemangioblast hypothesis, each blood island of the yolk sac should be clonally derived. However, in these experiments more than a single ES cell often contributed to each blood island of the chimeric mice. The existence of the hemangioblast has also been addressed in zebrafish. A primitive wave of hematopoiesis occurs in a region called the intermediate cell mass that contains erythroid cells surrounded by venous endothelial cells (see Figure 2). Hematopoietic and endothelial markers segregate between the 3- to 10-somite period of development. By this time, there are few, if any, cells that might be considered hemangioblasts based on overlapping blood and blood vessel gene expression. Alternatively, hemangioblasts could appear before the 3-somite stage and also exhibit wider developmental potential than solely blood and blood vessels. Ventral mesoderm-mal cells are dedicated specifically to hematopoietic and endothelial fates. Fate-mapping studies have been performed in which a caged fluorescent dye is injected into the zebrafish embryo at the one-cell stage, and then at a later time the fluorescent dye is uncaged in single cells using a laser. Individual cells appear dedicated to hematopoietic and endothelial lineages at the 0- to 3-somite stage (Vogeli et al., 2006). However, other cell fates may also be present at this early time. Similarly, smooth muscle cells can be derived from populations of in vitro differentiated mouse ES cells exhibiting blood and blood vessel fates (Ema et al., 2003; Ema and Rossant, 2003). These studies support the existence of hemangioblasts, although it may be necessary to redefine the potential of these cells to include additional lineages (such as smooth muscle).
Principally based on morphology it has been proposed that as the AGM forms, “hemogenic endothelial” cells in the ventral wall of the aorta, rather than hemangioblasts, bud off HSCs. The program of hemogenic endothelial cell development may be regulated differently from that of presumptive hemangioblasts, given that the transcription factor requirements differ. For example, the transcription factor Runx1 is necessary for blood formation from hemogenic endothelium but not from yolk sac hemangioblasts (North et al., 1999, 2002). The potential to generate hematopoietic, endothelial, and smooth muscle cells has been attributed to another cell type, termed the mesoangioblast, present in the aorta (Cossu and Bianco, 2003). Perhaps, the presumptive mesoangioblast might be a precursor of the hemogenic endothelial cell.
Other work has indicated that mesenchymal cell populations in the subaortic region poke through the aorta and bud off HSCs (Bertrand et al., 2005). As this occurs, mesenchymal cells express endothelial-specific genes and ultimately express HSC-associated markers. These observations suggest an alternative model in which subaortic mesenchymal cells, which may also have smooth muscle potential, rather than hemogenic endothelial cells, are the source of future definitive HSCs.
Developmental Relationships between the Yolk Sac and the AGM
As with mesodermal derivatives, all blood cells in embryonic, fetal, and adult animals might arise from a small set of cells during development. Evidence for and against this notion is present in the literature. Fate mapping in the pre-gastrula Xenopus embryo with fluorescent dye injected into individual blastomeres of the 32-cell embryo demonstrated that different blastomeres contribute to primitive hematopoiesis and definitive HSC production (Ciau-Uitz et al., 2000). This finding contradicts the conclusion derived from diploid-triploid chimeric frogs that ventral mesoderm is the common origin of both primitive and definitive populations (Turpen et al., 1997). Technical aspects of fate mapping of the 32-cell embryo have been challenged (Lane and Sheets, 2002).
In situ hybridization and chimera studies in amphibians and birds suggest that the yolk sac and the AGM are derived independently and arise at different times in development (Turpen et al., 1997). With short-term culture and subsequent transplantation, mouse AGM tissue (isolated one day prior to the appearance of HSCs in vivo) generates cells with the capacity for long-term engraftment, whereas mouse yolk sac tissue does not (Cumano et al., 1996; Medvinsky and Dzierzak, 1996). The origin of HSCs in the AGM can be traced by Runx1 expression in the embryonic day 8.5 (E8.5) mouse embryo, just before the onset of circulation. Because functional activity of stem cells as determined by transplantation into irradiated adults occurs much later (at day 11), it is possible that cells of the yolk sac colonize the AGM through the circulation. In fact, HSC-like activity of yolk sac cells (as defined by a neonatal transplantation assay) (Palis et al., 2001) is detected as early as day 9, although circulation has started by that time. Conclusive resolution of the developmental relationship between cells of the yolk sac and AGM requires direct visualization of the migration event. Furthermore, the specific assay used to determine stem cell activity for one population of cells (such as immune reconstitution following irradiation of adult animals) may not be appropriate for a different stem cell population. Distinct host requirements, such as the use of neonatal recipients for cells of the yolk sac, may be necessary. Some of the intrinsic differences between cell populations, such as developmental stage, ease of access, the local niche, and whether they are dividing, may preclude a host transplant assay from detecting engraftment and multilineage reconstitution. Such questions will plague studies of other tissue stem cells, as these stem cells are defined by functional and biological readouts.
Does the Yolk Sac Contain HSCs?
Based on cell fate mapping and transplantation experiments in avian and amphibian species, the AGM has been widely viewed as the principal site for HSC production during vertebrate development. Accordingly, the yolk sac has often been relegated to a subservient position, despite older experiments suggesting that the yolk sac might be the source of adult hematopoiesis. Metcalf and Moore cultured E7.5 mouse embryos from which the yolk sac had been removed (Moore and Metcalf, 1970). Given that no hematopoietic cells appeared in the fetal liver following several days in culture, they concluded that the yolk sac was the major site of adult blood formation for the embryo. Although hematopoiesis in the yolk sac is largely primitive in character, progenitors within the yolk sac do give rise to definitive type cells in hematopoietic colony assays, an observation consistent with a yolk sac origin for definitive cells. This view was supported by other experiments in which specific donor-derived T cell populations appeared following transplantation of cells of the yolk sac into fetuses (Weissman et al., 1978).
In more recent work, Nishikawa and colleagues have also challenged the dogma that the yolk sac lacks definitive hematopoietic stem cells (Samokhvalov et al., 2007). The fate of early embryonic tissues was traced in transgenic mice in which Runx1 regulatory elements drive expression of hormonally activated Cre recombinase. Administration of tamoxifen to pregnant female mice at a particular developmental window permits the fate of cells expressing Runx1 (visualized by activation of a Flox-LacZ allele) to be assessed. Treatment of embryos at E7.5 led to prominent marking of fetal liver cells and adult hematopoietic cells. As the yolk sac is the only hematopoietic site at E7.5 and the only tissue known to express Runx1 at E7.5, these findings suggest that the yolk sac contains definitive HSCs (or cells that may give rise to HSCs). These experiments were interpreted to support the yolk sac as a site of HSC formation prior to the AGM, although consensus in the field is far from unanimous (DeWitt, 2007). Diploid-triploid transplants in frogs reveal that ~20% of adult blood in some animals is derived from the ventral blood island (the equivalent to the yolk sac), providing independent evidence that adult hematopoiesis may arise from the yolk sac region. Despite this finding, it is also clear that the analogous AGM region in Xenopus is the predominant contributor to adult hematopoiesis. The precise origin of HSCs in the adult remains a topic for further debate and study.
In Vivo Fate Mapping of Migrating Cells
Presumptive HSCs in the zebrafish express the transcription factors c-myb and Runx1 (see Figure 2). Caged fluorescein dye fate mapping of AGM cells has revealed a new hematopoietic region, the caudal hematopoietic tissue. Laser uncaging is targeted to a region of cells in which transgenic expression of green fluorescent protein (GFP) driven by an HSC-specific promoter marks HSCs in the AGM (Ferkowicz et al., 2003). This approach ensures that laser uncaging occurs specifically within HSCs. Multiple cells are uncaged and their fate is followed (Jin et al., 2007; Murayama et al., 2006). Uncaged cells of the AGM region that express CD41 (a surface marker of early HSCs) or c-myb appear later as fluorescent cell populations in the caudal hematopoietic tissue. The larval and adult site of hematopoiesis in the zebrafish is the kidney. Later on in the fate map experiments, the larval kidney becomes fluorescent, demonstrating that cells of the caudal hematopoietic tissue colonize the kidney. In addition, fluorescence is detected in the thymus. Recent evidence suggests direct population of thymic primordia through tissue planes, a finding consistent with earlier experiments in birds showing migration of progenitors to the thymus along the thoracic duct. Thus, population of the thymus may occur through circulation and direct migration through tissues. Alternatively, the caudal hematopoietic tissue may represent a site similar to the placenta or fetal liver prior to the onset of definitive hematopoiesis in the kidney.
It is generally stated that HSCs of the fetal liver circulate to the adult bone marrow and, hence, are the source of adult hematopoiesis in birds and mammals (see Review by D.J. Laird et al., page 612 of this issue). In contrast, developmental studies reveal that the fetal liver and marrow are seeded at similar times during development (Delassus and Cumano, 1996). Direct tracking of cellular migration is required to distinguish these possibilities.
Pathways Involved in the Emergence of HSCs
The AGM has been characterized largely by morphology and functional assays, but the pathways involved in HSC generation remain incompletely defined. Studies of chick embryos demonstrate that endoderm has a prominent role and secretes inducing factors. Somitic mesoderm also contributes to the dorsal aspect of the aorta, and the addition of factors—such as VEGF, TGF-β, and FGF—to the somitic mesoderm leads to induction of hematopoietic tissue. In contrast, TGF-α and EGF suppressed formation of hematopoietic cells (Pardanaud and Dieterlen-Lievre, 1999).
Signaling pathways that regulate the induction of the AGM have recently been uncovered in mouse and zebrafish. Notch 1 is required for artery identity and aortic HSC production (Kumano et al., 2003). In the zebrafish mutant mindbomb that lacks Notch signaling, Runx1 overexpression rescues HSC production (Burns et al., 2005). Similarly, a Notch1 mutant is rescued by Runx1 overexpression, suggesting that Runx1 lies downstream or parallel to Notch signaling. Other pathways participate in the process including CoupTF-II (Pereira et al., 1999), as well as the CDX-HOX pathway (Davidson et al., 2003).
The Wnt/β -catenin and Notch-Delta signaling pathways influence the function of adult HSCs. Treatment of purified HSCs with Wnt3a protein leads to a modest increase in engrafting cells (Reya et al., 2003). Whereas a pulse of Wnt signaling appears to induce HSCs, constitutive Wnt activation by stabilized β-catenin leads to anemia, possibly by stem cell exhaustion as a consequence of prolonged Wnt signaling (Kirstetter et al., 2006; Scheller et al., 2006). Wnt signaling may be dispensable for adult HSC homeostasis, given that conditional knockout of β- or γ-catenin in hematopoietic cells fails to affect HSC number or engraftment potential (Cobas et al., 2004; Scheller et al., 2006). Stimulation of the Notch pathway also increases HSC activity and appears to be required for the increased self-renewal upon Wnt activation (Duncan et al., 2005). In addition to the Wnt and Notch pathways, new growth factors such as angio-poietin-like proteins appear capable of supporting ex vivo expansion of HSCs (Zhang et al., 2006).
A chemical genetic screen has recently revealed a role for the prostaglandin pathway in the production of HSCs in the zebra-fish. Treatment of embryos with prostaglandin E2 (PGE2) augments stem cell production (North et al., 2007), most likely through the EP4 receptor, a G-coupled receptor specifically expressed in the aorta region and activated by PGE2 (Villablanca et al., 2007). Prostaglandins also affect the homeostasis of definitive adult hematopoiesis, as shown by irradiation recovery assays, 5-fluorouracil stimulation assays, and long-term hematopoietic reconstitution. Thus, the emergence of HSCs in the aorta involves the prostaglandin pathway and the Notch-Runx pathways, which appear to be independent based on genetic relationships.
The hematopoietic system of the Drosophila embryo generates myeloid-like cells critical for tissue remodeling and engulfment and phagocytosis of dead cells. The emergence of sites of hematopoiesis during embryogenesis is remarkably similar to that of vertebrates. Drosophila progenitors are also formed adjacent to the circulatory system. Hematopoietic progenitors bud off from head mesoderm. These myeloid cells are transient and ultimately replaced by cells that bud off near the heart region and the great vessel. Vascular endothelial growth factor (VEGF) ligands are required for derivation of adult hematopoietic cells, as well as for attracting myeloid cells at specific sites (Cho et al., 2002). Genetic analysis demonstrates that specific signaling pathways, such as Notch, are required for the derivation of the lymph gland and a hemangioblast-like cell population (Mandal et al., 2004). Recent studies demonstrate that the lymph gland of the third instar larva of the fruit fly is patterned and contains a signaling center that expresses Hedgehog ligand (Krzemien et al., 2007; Mandal et al., 2007). Hedgehog cooperates with Notch ligands expressed in these regions to form a stem cell niche and regulates the cycling of hematopoietic progenitors. The search is underway for a similar signaling center in vertebrates. Hedgehog is also required for AGM hematopoiesis in the zebrafish (Gering and Patient, 2005). More recently, studies of human embryonic stem cells have indicated that factors such as hedgehog and bone morphogenetic protein (BMP) promote blood production during in vitro differentiation.
Niches
Stem cells depend on their microenvironment, the niche, for regulation of self-renewal and differentiation. Studies of Drosophila testes and ovarian stem cells have led to formulation of concepts that may be applicable to the niche in other tissues (Decotto and Spradling, 2005; see also the Review by S.J. Morrison and A.C. Spradling, page 598 of this issue). For instance, in the ovary, a hub cell directly binds to a stem cell and regulates its self-renewal and differentiation, in part though BMP signaling (see Minireview by R.M. Cinalli et al., page 559 of this issue). In the testis, an apical hub cell expresses the ligand Upd, an activator of the JAK-STAT signaling pathway in adjacent germ cells to control their self-renewal. By analogy to the Drosophila reproductive organs, investigators have sought an equivalent of the hub cell for the HSC.
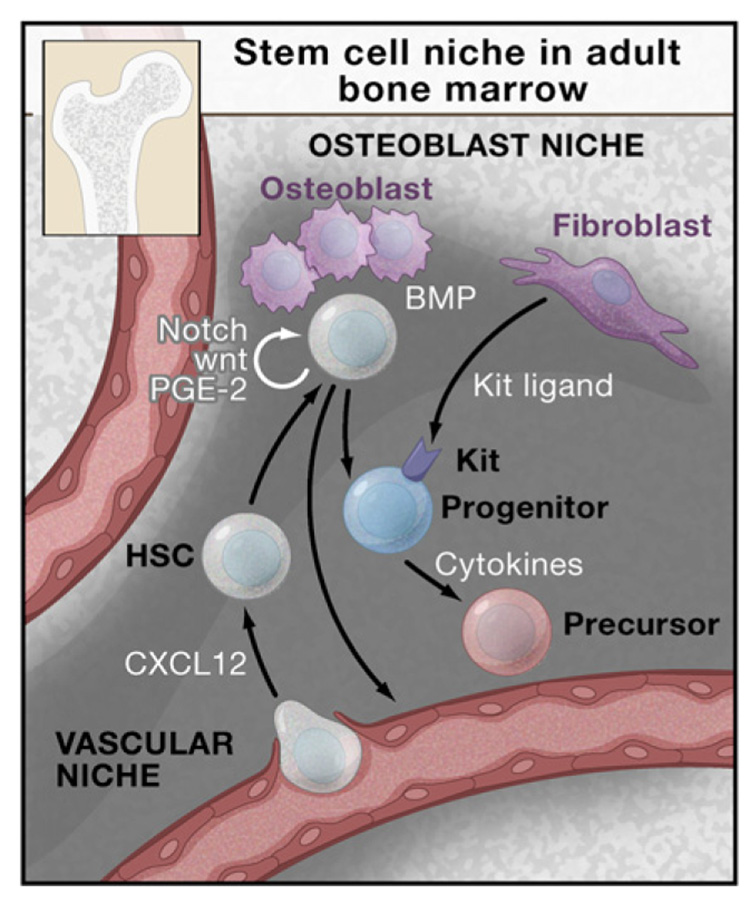
As the site of hematopoiesis changes during vertebrate development, the nature of the stem cell niche must also change. The adult bone marrow niche (depicted in Figure 3) has received most attention. Mutant mice in which the BMP pathway is disrupted have increased numbers of osteoblasts and HSCs (Calvi et al., 2003; Zhang et al., 2003). These findings suggest that osteoblasts may represent a critical component of the bone marrow niche for HSCs. As assessed by intravital microscopy, HSCs appear to reside in the periosteal region of calverium marrow (Sipkins et al., 2005). Transplanted GFP-marked or LacZ-marked HSCs appear to lodge adjacent to osteoblasts. Many factors, including ligands for Notch receptors and N-cadherin, are liberated by osteoblasts, although the contribution of these to adult hematopoiesis remains to be established. The role of N-cadherin as a mediator of interactions with osteoblasts (Zhang et al., 2003), as well as the prominence of osteoblasts for HSC adherence, have been challenged (Kiel et al., 2007). Recent findings suggest that HSCs are maintained in a quiescent state through interaction with thrombopoietin-producing osteoblasts (Yoshihara et al., 2007). The association of HSCs with osteo-blasts is countered by other studies that place HSCs adjacent to vascular cells. The chemokine CXCL12 regulates the migration of HSCs to the vascular cells (now called the vascular niche) (Kiel and Morrison, 2006). Taken together, these findings suggest that HSCs reside in various sites within the marrow and that their function might depend on their precise localization. Much of the existing debate may be semantic, however, if the osteoblastic and vascular niches are intertwined and not physically separate. Alternatively, HSCs may truly reside in distinct subregions, which may endow them with different activities. Cellular dynamics within the niche are relevant to clinical marrow transplantation. For example, recent findings suggest that antibody-mediated clearance of host HSCs facilitates occupancy of the niche and transplantation by exogenous HSCs (Czechowicz et al., 2007).
Figure 3. Stem Cell Niche in the Adult Bone Marrow.
HSCs are found adjacent to osteoblasts that are under the regulation of bone morphogenetic protein (BMP) (the osteobast niche). HSCs are also found adjacent to blood vessels (the vascular niche). The chemokine CXCL12 regulates the migration of HSCs from the circulation to the bone marrow. The osteoblast and vascular niches in vivo lie in close proximity or may be interdigitated. The marrow space also contains stromal cells that support hematopoiesis including the production of cytokines, such as c-Kit ligand, that stimulate stem cells and progenitors. Cytokines, including interleukins, thrombopoietin (Tpo), and erythropoietin (Epo), also influence progenitor function and survival.
How niches modulate self-renewal is a challenge for future studies. The generation of premalignant myeloproliferative syndromes in mice with abnormal niches underscores the need for precise control in vivo (Perry and Li, 2007). Remarkably, the site of hematopoiesis is not conserved in vertebrate evolution. For instance, the site of adult hematopoiesis is the kidney in fish. The frog forms adult blood in the liver, and birds and mammals form blood in the marrow. In the frog Rana temporaria, the site of hematopoiesis switches between the liver and bone marrow depending on the season (Maslova and Tavrovskaia, 1993).
Little is known regarding the nature of the niche for embryonic hematopoietic sites. Are diverse properties attributed to embryonic, fetal, and adult HSCs due to differences in their respective niches? To what extent are factors shared among different developmental or anatomical niches?
Transcription Factors in Hematopoietic Development
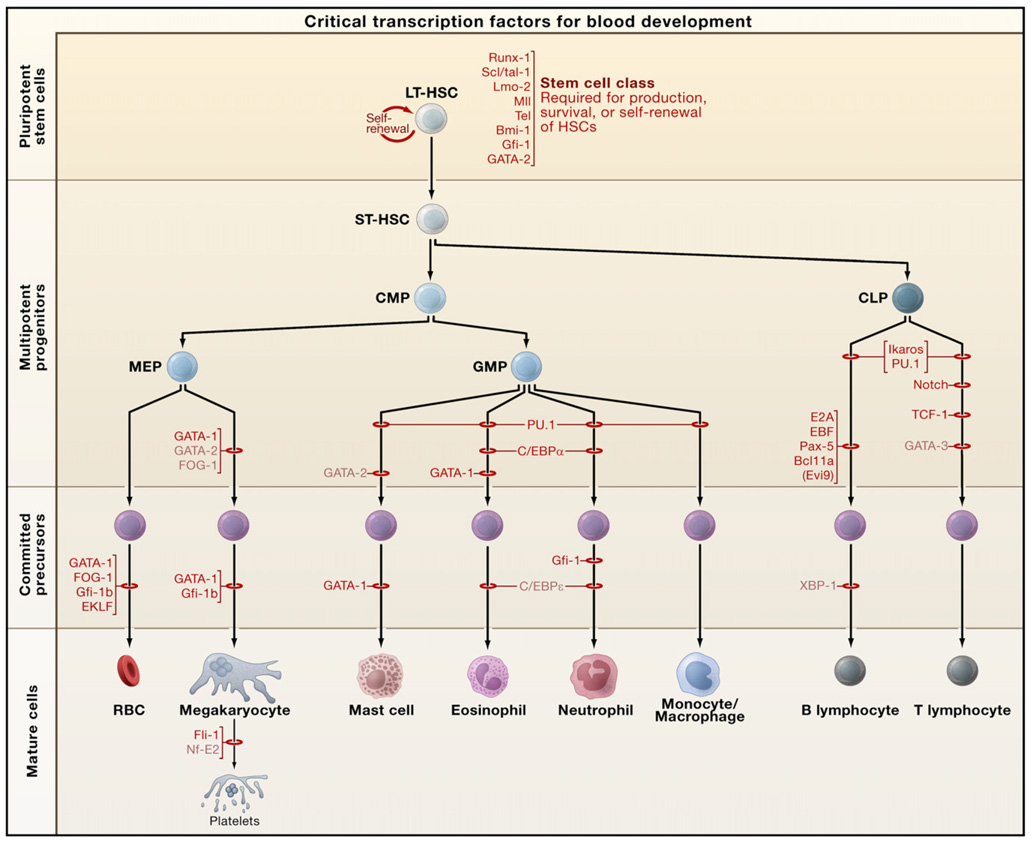
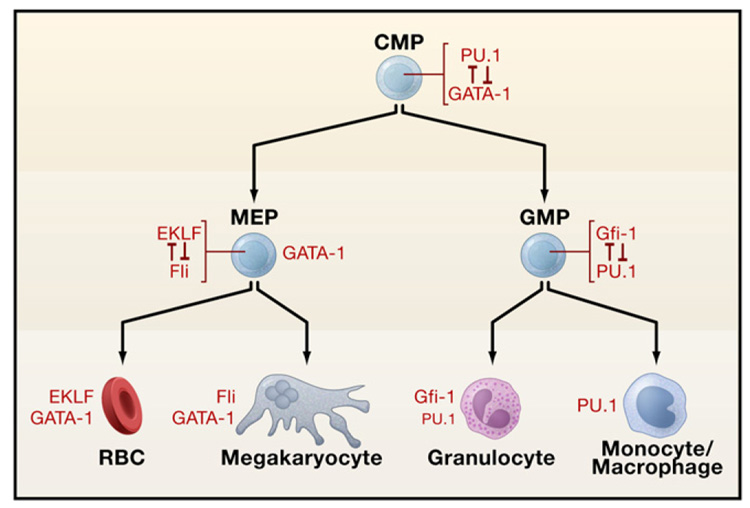
As intrinsic determinants of cellular phenotype, transcription factors provide an entry point for unraveling how HSCs develop during embryogenesis and how lineage-restricted differentiation is programmed (Orkin, 2000). Here we focus on principles and concepts that have emerged and consider how these may inform other organ/tissue systems. Recent reviews provide additional discussion of transcription factors in different hematopoietic lineages (Nutt and Kee, 2007; Iwasaki and Akashi, 2007; Kim and Bresnick, 2007; Rothenberg, 2007). Insights into the functions of the critical transcription factors have rested predominantly on findings from either conventional or conditional gene knockouts in mice and from forced expression experiments, all complemented by developmental studies in other model organisms (e.g., zebrafish, chicken, Drosophila, Xenopus). The transcription factors that are critical for hematopoiesis encompass virtually all classes of DNA-binding proteins, rather than favoring a specific family. A remarkable feature of transcription factors in the hematopoietic system is that the majority are involved in chromosomal translocations or with somatic mutations in human hematopoietic malignancies. Furthermore, experimental manipulation of the genes for such factors in mice often promotes malignancy. Hematopoietic cell fate is intertwined with the origins of leukemias. Requirements for several transcription factors, as established through conventional gene targeting, are summarized in Figure 4 (see also the SnapShot by S.H. Orkin and L.I. Zon, page 631 of this issue).
Figure 4. Requirements of Transcription Factors in Hematopoiesis.
The stages at which hematopoietic development is blocked in the absence of a given transcription factor, as determined through conventional gene knockouts, are indicated by red bars. The factors depicted in black have been associated with oncogenesis. Those factors in light font have not yet been found translocated or mutated in human/mouse hematologic malignancies. Abbreviations: LT-HSC, long-term hematopoietic stem cell; ST-HSC, short-term hematopoietic stem cell; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; MEP, megakaryocyte/erythroid progenitor; GMP, granulocyte/macrophage progenitor; RBCs, red blood cells.
For discussion purposes, one may distinguish between factors required for HSC formation or function and those employed in lineage-specific differentiation. Among the ‘‘HSC transcription factors’’ are MLL (for mixed lineage-leukemia gene), Runx1, TEL/ ETV6, SCL/tal1, and LMO2, whose genes account in toto for the majority of known leukemia-associated translocations in patients. In these instances, the translocations either deregulate expression of the locus, as in the case of SCL/tal1 and LMO2 in T cell acute leukemias, or generate chimeric fusion proteins, as in myeloid and lymphoid leukemias associated with MLL, Runx1, and TEL/ETV6. The above distinction is arbitrary in that all of these HSC transcription factors also serve roles later within differentiation of individual blood lineages, and conversely, factors that appear to have more lineage-restricted roles (such as PU.1, Gfi-1, C/EBPα) act within HSCs. The redeployment of transcription factors at different stages of blood cell development is reflected in part in the dynamic patterns of expression of key regulators and complicates analysis of in vivo requirements. Both temporal- and lineage-restricted conditional inactivation are often needed to reveal a meaningful phenotype. These circumstances reflect parsimony with regard to the repertoire of factors required to achieve complex gene regulation and differentiation.
Factors Essential for Formation of HSCs
The transcription factors required to program mesoderm toward a hematopoietic fate are of special interest. The basic-helix-loop- helix (bHLH) factor SCL/tal-1 and its associated protein partner, the Lim-domain containing LMO2, are individually essential for development of both the primitive and definitive (or adult) hematopoietic systems (Kim and Bresnick, 2007). In their absence, no blood cells are generated. Within the primitive system at the yolk sac stage, these factors are thought to function within the hemangioblast to specify a blood rather than a vascular fate. The genes encoding the SET-domain containing histone methyltransferase MLL and runt-domain Runx1 proteins are essential for generation of HSCs within the AGM (and possibly at other sites) (Orkin, 2000). In the absence of Runx1, no hematopoietic clusters (representing presumptive HSCs) form in the dorsal aorta in mice. As noted above, in zebrafish Runx1 lies downstream of Notch signaling, which is required for the induction of hematopoiesis. In addition, BMP signaling restricts hemato-vascular development of lateral mesoderm, possibly acting through a pathway involving LMO2 and presumably GATA-2 (Burns et al., 2005). MLL, like its Drosophila counterpart trithorax, functions in maintenance of, but not initiation of, HOX gene expression. MLL also appears to lie upstream of HOXB4 (and presumably other HOX genes) in HSC specification. Leukemias in which MLL function is perturbed due to chromosomal translocations may arise in part as a secondary consequence of changes in HOX gene expression. Recently, it has been demonstrated that expression of an MLL fusion gene (MLL-AF9) in granulocyte/ macrophage progenitors (GMPs) induces a “HSC stem cell-like” signature that includes various HOX genes (Krivtsov et al., 2006). The acquisition of a stem cell signature by leukemic GMPs may contribute to self-renewal of leukemia stem cells.
Study of a zebrafish mutant defective for blood formation identified the caudal-related factor Cdx4 as an inducer of blood specification (Davidson et al., 2003). Indeed, Cdx4-deficient embryos are rescued by expression of various homeobox genes (e.g., HoxA9) but not by SCL/tal1 (Yan et al., 2006). Activation of Cdx4 expression in mouse ES cells alters the pattern of HOX gene expression, augments in vitro blood formation, and cooperates with HOXB4 in the generation of long-term reconstituting hematopoietic progenitors (Wang et al., 2005). Recent evidence indicates that the pathway to Cdx4 in the zebrafish is initiated by the TATA-box binding protein-related factor 3 (Trf3) (Hart et al., 2007).
Temporal and Stage-Specific Requirements for Hematopoietic Regulators
Within the definitive hematopoietic system, fetal liver and bone marrow HSCs differ in many properties, including cell-surface markers, developmental potential, and cell-cycle status. Transplantation experiments in mice suggest that some characteristics that distinguish fetal liver and adult HSCs are intrinsically regulated (Bowie et al., 2007). Furthermore, HSCs in older age mice exhibit different self-renewal and gene expression patterns than those from younger animals (see Review by D. Rossi et al., page 681 of this issue). How the distinctive properties of HSCs at different developmental stages are programmed is of particular interest in correlating biological read-outs with molecular determinants. Recently, the HMG-box containing factor Sox17, which is also critical to endoderm specification, has been identified as critical for generation of fetal, but not adult, HSCs (Kim et al., 2007). “Geriatric” HSCs are less efficient at homing to and engrafting in the bone marrow, possibly linked to their increased cycling frequency. In addition, the differentiation potential of older HSCs is biased toward myeloid versus lymphoid lineages (Sudo et al., 2000). Several changes in gene expression of old versus young HSCs have been described, including increased expression of leukemia-associated genes and decreased expression of genes contributing to DNA damage repair, genomic integrity, and chromatin remodeling (Rossi et al., 2005; Nijnik et al., 2007). Some of these properties are posited to predispose older HSCs to myeloid leukemias (see Review by D. Rossi et al.).
Moreover, the transcription factors required for specification and formation of HSCs may not be required continuously for the subsequent survival or self-renewal of HSCs. Although SCL/tal1 is an obligate factor for hematopoietic fate specification during development, conditional inactivation in adult HSCs has surprisingly little consequence on maintenance or self-renewal of HSCs and multipotent progenitors (Mikkola et al., 2003). Under these circumstances, the role of this factor in maturation of erythroid and megakaryocytic cells is revealed. Similarly, inactivation of Runx1 in adult HSCs does not ablate HSC properties but instead perturbs differentiation of specific lineages (megakaryocytes, lymphocytes) (Ichikawa et al., 2004). Such observations point to differences in the transcription factor composition of emerging HSCs and adult HSCs and suggest that the phenotype of HSCs is quite stable.
Multilineage Gene Expression in HSCs
Generally, the expression of the lineage-affiliated transcription factors can be readily reconciled with the simple hierarchy diagrams of hematopoiesis (see Figure 1 and Figure 4). For instance, GATA-1 is highly expressed in megakaryocytic/erythroid progenitors (called MEPs) that give rise to megakaryocyte and red blood cell precursors, whereas a “myeloid factor,” such as C/EBPα, is present in GMPs. Indeed, in committed progenitors and precursors one can conveniently match cell-surface phenotypes (defined by monoclonal antibodies) and the subset of hematopoietic transcription factors expressed in these cells. However, this relationship breaks down at earlier stages in the hierarchy. A simple one-to-one correspondence of lineage-restricted transcription factors and progenitors is challenged by findings that earlier multipotential progenitors and HSCs express markers of disparate lineages even within single cells, albeit generally at low levels (Orkin, 2003). This phenomenon, termed lineage priming, suggests that the fate of these immature cells is not sealed and that lineage selection is largely a process in which alternative possibilities are extinguished rather than one in which new programs are imposed on an otherwise blank slate.
Lineage priming may be an efficient means by which chromatin invested in important hematopoietic programs is maintained in an available or open configuration in HSCs. Transient repression of alternative fates, followed by more permanent silencing, maintains the inherent plasticity of multipotential progenitors. Moreover, the coexistence of transcription factors representing different lineages within a common cell (the HSC or immature progenitor) offers the potential for immediate “crosstalk” between different fates at the molecular level (see below).
Recently, by FACS sorting of cells initiating expression GATA- 1 or PU.1, it has been demonstrated that short-term repopulating HSCs may be further subdivided into those committed to myeloerythroid and myelolymphoid lineages (Arinobu et al., 2007). As these findings illustrate, continued fractionation of HSC or progenitor populations reveals increasing diversity in the choice of lineage. Thus, the schematic lineage diagrams that are generally presented cannot be taken literally but rather as guides to the options available to progenitors. The extent to which HSCs exhibit developmental potential beyond hematopoiesis remains controversial (Graf, 2002). Experiments purportedly demonstrating “plasticity” through transplantation of marrow cells to recipient mice are plagued by possible cell fusion of differentiated hematopoietic cells with host cells and by inadequate characterization of input populations.
Mechanisms of Action for Principal Hematopoietic Regulators
The requirements and functions of the principal transcriptional regulators are context dependent (Orkin, 2000). The key lineage- restricted factors are endowed with the complementary tasks of promoting their own lineage differentiation while simultaneously acting against factors favoring other choices (Figure 5). Combining positive and antagonistic roles in the major regulators provides an efficient means for resolving and reinforcing lineage choices. Numerous examples of this principle of lineage programming have been described. GATA-1 and PU.1 promote erythroid/ megakaryocytic/eosinophil and myeloid differentiation, respectively. The proteins physically interact and antagonize each other’s actions. In vivo confirmation of the complementary roles of GATA-1 and PU.1 has been shown in zebrafish. Inhibition of GATA-1 expression by morpholinos shifts hematopoietic progenitors to a myeloid fate, whereas the converse occurs upon inhibition of PU.1 expression (Galloway et al., 2005; Rhodes et al., 2005). Other examples of direct antagonism by hematopoietic transcription factors include the relative relationships of C/ EBP and FOG1 with respect to eosinophil and multipotential cell fates (Querfurth et al., 2000), EKLF and Fli-1 for erythroid and megakaryocytic choice (Starck et al., 2003), GATA-3 and T-bet for TH1 and TH2 cells (Usui et al., 2006), and Gfi1 and PU.1 for neutrophil versus monocyte outcomes (Dahl et al., 2007). In the absence of Gfi1 in mice, neutrophil precurors fail to mature and also incompletely silence monocyte/macrophage gene expression (Hock et al., 2003).
Figure 5. Transcription Factor Antagonism in Lineage Determination.
Examples of antagonism are depicted in red. The transcription factors present in the mature precursors following choice of specific lineage are shown at the bottom in black. Abbreviations: CMP, common myeloid progenitor; MEP, megakaryocyte/erythroid progenitor; GMP, granulocyte/macrophage progenitor; RBCs, red blood cells.
The critical contribution of repression to lineage selection is illustrated by loss-of-function studies of Pax5. Proper B cell development requires Pax5 (Nutt and Kee, 2007). In its absence, proB cells assume a multipotential phenotype and differentiate (under appropriate growth factor conditions) to T-, NK-, or dendritic cells, macrophages, neutrophils, or erythroid precursors. Repression of critical growth factor receptors, for example macrophage-colony-stimulating factor (M-CSF) receptor, restricts lineage choice during normal development. In effect, Pax5 commits progenitors to a B cell fate, while other B cell transcription factors, such as E2A and EBP, specify lineage-appropriate gene activation.
In T-lymphoid development, Notch signaling serves a similar but more focused role as it functions as a commitment factor, principally by repressing factors, such as PU.1, associated with other outcomes. Although GATA-3 has been viewed as a T cell-specific transcription factor, recent work indicates that it functions only in the context of Notch signaling to promote T cell development (Rothenberg, 2007). As recently stated, lineage programming in the T cell lineage is more the consequence of “negotiation” rather than instruction.
Mechanisms of Lineage Programming
The context-dependent action and direct antagonism between key transcriptional regulators are best accommodated by models in which factors interact directly within protein complexes (Orkin, 2000). In this regard, the GATA factors are illustrative (Kim and Bresnick, 2007). A protein complex in erythroid cells comprised of GATA-1 (or its close relative, GATA-2), LMO2 and its partner Ldb1, and SCL/tal1 and its heterodimeric partner E2A recognizes a consensus GATA-E-box DNA motif. Knockout of either LMO2 or SCL/tal1 leads to the absence of any hematopoietic progenitors in the early embryo. The identical phenotypes are consistent with the action of these proteins within a single complex that is required prior to any commitment to erythroid differentiation. Indeed, forced expression of GATA-1/2, SCL, and LMO2 converts Xenopus mesoderm efficiently to a hematopoietic fate. Remarkably, although the GATA/SCL/ LMO2/Lbd1 complex recognizes a composite DNA-binding site, later studies demonstrated that the DNA-binding activity of SCL (in a heterodimer with E2A) is dispensable for hematopoietic specification but required for full erythroid and megakaryocytic cell maturation (Porcher et al., 1999). The recruitment of DNA-binding-defective SCL to the protein complex accounts for in vivo function despite its inability to bind to DNA.
GATA factors form alternative protein complexes with a specific cofactor known as FOG (for friend of GATA) (Kim and Bresnick, 2007). Targeted mutation in mice has demonstrated the essential role of the GATA/FOG interaction for erythroid and megakaryocytic development. The GATA-1 (or GATA-2)/FOG1 complex in both erythroid and megakaryocytic lineages is physically associated with the NuRD chromatin remodeling complex via an NuRD binding motif at the N terminus of FOG1. The interaction of FOG1 with GATA factors mediates transcription repression and also may facilitate accessibility of the GATA factor to its DNA-binding motif in chromatin. GATA/SCL/LMO2/Ldb1 and GATA/FOG1 complexes appear to be mutually exclusive.
Other connections between hematopoietic factors and chromatin-associated proteins amplify this theme. Ikaros proteins also associate with the NuRD complex and participate in repression and the formation of heterochromatin (Kim et al., 1999). The erythroid factor EKLF interacts with Brg1, a critical component of the Swi/Snf ATP-dependent chromatin remodeling complex (Brown et al., 2002). Consistent with the relevance of this relationship to biological function, a Brg1 mutant protein, which retains ATPase activity and assembles into the Swi/Snf complex, fails to generate DNase I hypersensitivity and leads to embryonic lethality due to failure to activate normal β-globin expression (Bultman et al., 2005). Furthermore, another erythroid factor NF-E2 associates with the MLL2 complex, which is responsible for H3K4 histone methylation (Demers et al., 2007). In addition, the zinc-finger repressor Gfi protein recruits a CoREST/lysine demethylase (LSD1) complex to target genes in erythroid, megakaryocytic, and myeloid cells to control maturation of these lineages (Saleque et al., 2007). Further elucidation of the ways in which hematopoietic transcription factors interact and function with chromatin-associated factors in HSCs and individual lineages is an on-going challenge for the field.
The involvement of multiprotein complexes in the action of he-matopoietic transcription factors predicts that relative concentrations of particular factors should influence the choice of lineage. Protein complexes constitute a convenient platform for direct competition, as well as for cooperative action. Considerable data argue for concentration-dependent effects in lineage choice and differentiation. Experiments in zebrafish cited above in which the pattern of erythroid versus myeloid development is affected by inhibition of expression of GATA-1 or PU.1 (Galloway et al., 2005; Rhodes et al., 2005) are consistent with this model. The relative action of GATA-1 and PU.1 fits a simple quantitative model that predicts a metastable undifferentiated progenitor state when both proteins are present at low levels and differentiation to one of two alternatives when one of the proteins is present at higher levels (Roeder and Glauche, 2006). Taking advantage of PU.1-null fetal liver progenitors, Singh and colleagues have proposed that high-level PU.1 expression directs macrophage differentiation, whereas low-level expression promotes B cell formation (DeKoter and Singh, 2000). Subsequently, they proceeded to show that low-level PU.1-expressing bi-phenotypic (macrophage/neutrophil) cells are relatively stable, and increased PU.1 levels promote macrophage differentiation by influencing a circuit of counterantagonistic repressors, Egr-1/2/Nab-2 and Gfi1 (Laslo et al., 2006). A mathematical model, similar to that described above for GATA-1 and PU.1, has been derived to account for these observed effects in the two myeloid lineages.
MicroRNAs (miRNAs) provide an additional level of control beyond the transcription factors (Shivdasani, 2006) (see Minireview by B. Stadler and H. Ruohola-Baker, page 563 of this issue). Ongoing studies of the involvement of miRNAs in hematopoiesis will reveal their roles in lineage decisions, stem cell to progenitor transitions, niche control, and cell function. The transcription of blood cell-specific miRNAs is likely to be driven by the complexes discussed above, providing a complex regulatory network. Several miRNAs are highly expressed in specific hematopoietic lineages and manipulation of their levels has been correlated with changes in cellular properties or differentiation (Chen et al., 2004). For example, miR-150 impinges on B cell differentiation by targeting c-myb mRNA (Xiao et al., 2007), whereas miR-155 is required for T helper cell generation and germinal center activity (Rodriguez et al., 2007; Thai et al., 2007). Moreover, conditional inactivation of the gene encoding Dicer, an essential component in the processing of pre-miRNAs to miRNAs, leads to multiple defects in T-lineage lymphopoiesis (Muljo et al., 2005; Cobb et al., 2006). Unpublished experiments indicate that Dicer is also required in the setting of bone marrow transplantation for radioprotection of lethally irradiated recipient mice (cited in Martinez and Busslinger, 2007).
Lineage Reprogramming
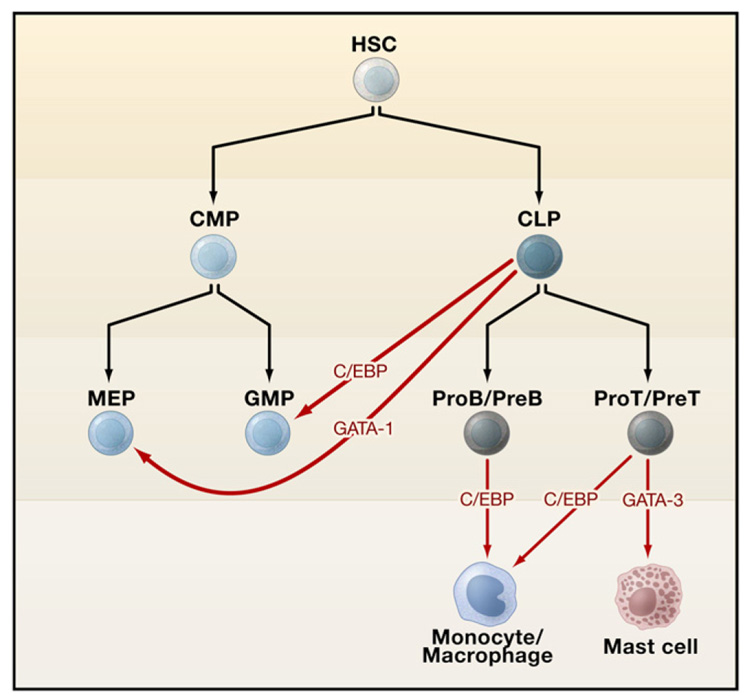
Cellular differentiation was once considered unidirectional, that is, once progenitors have committed to a particular linear pathway their fate is sealed. Accumulating evidence in the hematopoietic system and in other systems (see Review by R. Jaenisch and R. Young, page 567 of this issue) dispels this notion and provides a strong foundation for cellular reprogramming. Indeed, cells of one hematopoietic lineage can be converted to another through the forced expression of carefully chosen transcription factors (Figure 6). Knowledge of the rules governing how transcription factors that direct cellular lineages act informs directed attempts to reprogram one lineage to another. In an early example based on such logic, Querfurth et al. (2000) queried the significance of the lack of FOG-1 in eosinophils, despite their dependence on GATA-1 for differentiation. Forced expression of FOG-1 in avian eosinophils downregulated expression and function of C/EBP β, an essential eosinophil factor, leading to acquisition of a multipotent phenotype. Conversely, downregulation of FOG-1 by C/EBP β is a critical step in eosinophil lineage commitment. In other experiments, forced expression of GATA-1 was shown to drive early myeloid avian progenitors to erythroid, eosinophilic, or thromboblastic (megakaryocytic) precursors (Kulessa et al., 1995). Moreover, introduction of GATA-1 into GMPs and common lymphoid progenitors redirects their commitment to megakaryocytic/erythroid progenitors or to erythroid cells/mast cells/basophils (Iwasaki and Akashi, 2007). Committed B- and T-lymphoid cells can be reprogrammed to functional macrophages through expression of C/EBPα (Laiosa et al., 2006; Xie et al., 2004). Furthermore, preT cells can be reprogrammed to myeloid dendritic cells through PU.1 expression. Cells that are reprogrammed transit through an intermediate state in which markers of both myeloid and lymphoid lineages are expressed, indicative of the stepwise nature of the process. Resolution of the intermediate state leads to stable unilineage differentiation. Notch and GATA-3 appear to counteract reprogramming of preT cells to macrophages or dendritic cells. Recently, GATA- 3, traditionally viewed as a T cell-restricted factor, has been shown to direct mast cell reprogramming from proT cells (Taghon et al., 2007). In addition, mid-stage thymocytes (at the DN2 stage) can be converted to mast cells through growth in culture medium containing interleukin (IL)-3 and stem cell factor (Kit-ligand) in the absence of Notch.
Figure 6. Reprogramming of Hematopoietic Lineages.
The orange arrows depict lineage reprogramming upon expression of the transcription factors GATA-1, C/EBP, or GATA-3. Abbreviations: HSC, hematopoietic stem cell; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; MEP, megakaryocyte/erythroid progenitor; GMP, granulocyte/ macrophage progenitor.
Cancer: A Perturbation of the Hematopoietic Transcriptional Network
Of the more than two dozen regulators designated “hematopoietic transcription factors,” nearly all are intimately associated with hematopoietic malignancy (Figure 4). Indeed, the majority of genes encoding these factors were discovered either through analysis of chromosomal translocations found in human leukemias or study of cooperating leukemia genes during insertional mutagenesis in the mouse. Disturbance of the homeostatic balance of the critical transcriptional regulators is a defining feature of leukemias. In the setting of chromosomal translocations that lead to misexpression of a factor, such as SCL/tal1 or LMO2, target genes may be inappropriately activated or repressed in early lymphoid progenitors. Chimeric transcription factors generated through chromosomal translocations exert multiple downstream effects, including improper target gene activation or repression, inhibition of function of other critical factors, and recruitment of alternative chromatin-modifying enzymes to target loci (Rosenbauer and Tenen, 2007). Leukemia is not the consequence of nonspecific transcriptional effects but rather the end result of attacks at vulnerable points in a network. This observation is best exemplified by somatic mutations in GATA-1 in Down Syndrome- associated megakaryocytic leukemia (Wechsler et al., 2002), PU.1 and C/EBPα in myeloid leukemias (Mueller et al., 2002; Pabst et al., 2001), and Pax5 and other B cell factors (for example, E2A and EBF) in B-lymphoid leukemias (Mullighan et al., 2007). In addition to somatic mutation of the principal hematopoietic transcription factors, lesions in more broadly utilized signaling pathways that control specific lineage differentiation may underlie hematopoietic malignancy. This circumstance is illustrated by consistent somatic mutation of Notch in T cell leukemias (Weng et al., 2004). As an essential component of the regulatory network for T cell commitment and development, Notch is a preferred target for somatic mutation in T cell leukemia.
Messages for the Wider Stem Cell Field
As arguably the most “mature” organ system under study, the hematopoietic system constitutes a model for other subfields of stem cell biology. The hematopoietic system continues to evolve as a model, however, as numerous critical issues remain to be addressed. It is likely that many of the concepts derived from work in the blood field will be revisited in other organ systems. Nonetheless, it is also apparent that nature has explored alternative pathways to tissue organization and development. Hence, differences should be anticipated, particularly with respect to the extent to which true stem cells are required for the maintenance of different tissues or cell populations within an organ. Adult stem cell populations are generated during embryogenesis. For other organs, distinct stage-specific programs regulate stem cell homeostasis and tissue differentiation. As illustrated by umbilical cord blood stem cells, transient populations may have therapeutic value. Reflection on the history and recent advances in the hematopoietic field leads to several “lessons” for the stem cell field:
Precise characterization of the cells within the hematopoietic hierarchy has been instrumental in providing an adequate framework for biological and molecular studies. The prospective isolation of subsets of cells, coupled with in vitro colony-forming assays and quantitative in vivo transplantation methods, has greatly facilitated molecular studies of both normal and malignant hematopoiesis. HSCs differ in their properties depending on their location (fetal liver, bone marrow, placenta) and on the age of the organism. Hence, thorough cell biological studies are fundamental to approaching mechanisms that regulate stem cell function.
The “classical” hierarchy diagram depicting progenitors arising in an orderly fashion from a prototypical HSC provides a seductive, but overly simplified view. HSCs may be described more accurately as groups of cells with varying developmental potentials based on intrinsic networks driven by transcription factors and inputs from the cellular niches in which they reside. Processes, such as lineage priming coupled with plastic decisions driven by transcription factor competition, confer great flexibility on the options of HSCs. The molecular events of self-renewal must be coordinated with these steps. Current views of HSCs must account for a spectrum of cells with varying engraftment potential and distinct biases toward subsequent myeloid or lymphoid lineage choices. These complex features of HSC biology are likely to be revisited in studies of other tissue-dedicated stem cells.
Parallel investigation in diverse species has accelerated an understanding of blood cell development. Although an important goal is application of fundamental knowledge of hematopoietic stem cell biology to the treatment of human disease, studies in mice, fish, and flies are mutually reinforcing. Although the anatomical features of blood formation in the latter organisms differ from that of mammals, critical growth factor signaling and transcriptional pathways are shared. Parallel investigation of different species takes advantage of the strengths of each and complements work with human hematopoietic cells.
Elucidation of the transcriptional network that controls lineage choice and differentiation sets the stage for directed reprogramming of cellular lineages. Knowing the principal factors that govern the cellular lineages identifies candidate regulators for functional testing. Further study of the sequence of molecular events accompanying lineage conversion is likely to provide important mechanistic clues regarding how complex cellular reprogramming occurs (Rossant, 2007).
The remarkable link between hematopoietic transcription factors and malignancy underscores how disturbance of a transcriptional network lies at the crux of oncogenesis. As most hematopoietic transcription factors were discovered through study of chromosomal translocations in leukemia, investigators should pursue the normal developmental functions of genes disrupted or brought into fusion products in various solid tumors, both in sarcomas and epithelial cancers, given the recent appreciation for the importance of translocations in these settings. More generally, the transcriptional regulators participating in cell-specific gene expression provide entry points into the transcriptional network controlling self-renewal and differentiation, the hallmarks of stem cells.
ACKNOWLEDGMENTS
We thank Steve Moskowitz for help with the figures.
REFERENCES
- Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, et al. Reciprocal activation of GATA-1 and PU.1marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Bertrand JY, Giroux S, Golub R, Klaine M, Jalil A, Boucontet L, Godin I, Cumano A. Characterization of purified intraembryonic hematopoietic stem cells as a tool to define their site of origin. Proc. Natl. Acad. Sci. USA. 2005;102:134–139. doi: 10.1073/pnas.0402270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie MB, Kent DG, Dykstra B, McKnight KD, McCaffrey L, Hoodless PA, Eaves CJ. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc. Natl. Acad. Sci. USA. 2007;104:5878–5882. doi: 10.1073/pnas.0700460104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Pattison S, van Ree J, Coghill E, Perkins A, Jane SM, Cunningham JM. Distinct domains of erythroid Kruppel-like factor modulate chromatin remodeling and transactivation at the endogenous beta-globin gene promoter. Mol. Cell. Biol. 2002;22:161–170. doi: 10.1128/MCB.22.1.161-170.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes Dev. 2005;19:2849–2861. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 2005;19:2331–2342. doi: 10.1101/gad.1337005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Cho NK, Keyes L, Johnson E, Heller J, Ryner L, Karim F, Krasnow MA. Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell. 2002;108:865–876. doi: 10.1016/s0092-8674(02)00676-1. [DOI] [PubMed] [Google Scholar]
- Choi K, Kennedy M, Kazarov A, Papadimitriou JC, Keller G. A common precursor for hematopoietic and endothelial cells. Development. 1998;125:725–732. doi: 10.1242/dev.125.4.725. [DOI] [PubMed] [Google Scholar]
- Ciau-Uitz A, Walmsley M, Patient R. Distinct origins of adult and embryonic blood in Xenopus. Cell. 2000;102:787–796. doi: 10.1016/s0092-8674(00)00067-2. [DOI] [PubMed] [Google Scholar]
- Cobas M, Wilson A, Ernst B, Mancini SJ, MacDonald HR, Kemler R, Radtke F. Beta-catenin is dispensable for hematopoiesis and lymphopoiesis. J. Exp. Med. 2004;199:221–229. doi: 10.1084/jem.20031615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, et al. A role for Dicer in immune regulation. J. Exp. Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossu G, Bianco P. Mesoangioblasts–vascular progenitors for extravascular mesodermal tissues. Curr. Opin. Genet. Dev. 2003;13:537–542. doi: 10.1016/j.gde.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Cumano A, Dieterlen-Lievre F, Godin I. Lymphoid potential, probed before circulation in mouse, is restricted to caudal intraembryonic splanchnopleura. Cell. 1996;86:907–916. doi: 10.1016/s0092-8674(00)80166-x. [DOI] [PubMed] [Google Scholar]
- Czechowicz A, Kraft D, Weissman IL, Bhatacharya D. Efficient transplantation via antibody-based clearance of hematopoietic stem cell niches. Science. 2007;318:1296–1299. doi: 10.1126/science.1149726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl R, Iyer SR, Owens KS, Cuylear DD, Simon MC. The transcriptional repressor GFI-1 antagonizes PU.1 activity through protein-protein interaction. J. Biol. Chem. 2007;282:6473–6483. doi: 10.1074/jbc.M607613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ, Ernst P, Wang Y, Dekens MP, Kingsley PD, Palis J, Korsmeyer SJ, Daley GQ, Zon LI. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature. 2003;425:300–306. doi: 10.1038/nature01973. [DOI] [PubMed] [Google Scholar]
- Decotto E, Spradling AC. The Drosophila ovarian and testis stem cell niches: similar somatic stem cells and signals. Dev. Cell. 2005;9:501–510. doi: 10.1016/j.devcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- Delassus S, Cumano A. Circulation of hematopoietic progenitors in the mouse embryo. Immunity. 1996;4:97–106. doi: 10.1016/s1074-7613(00)80302-7. [DOI] [PubMed] [Google Scholar]
- Demers C, Chaturvedi CP, Ranish JA, Juban G, Lai P, Morle F, Aebersold R, Dilworth FJ, Groudine M, Brand M. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol. Cell. 2007;27:573–584. doi: 10.1016/j.molcel.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt N. Rewriting in blood: blood stem cells may have a surprising origin. Nat. Rep. Stem Cells. 2007 Published online June 21, 2007. [Google Scholar]
- Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat. Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- Ema M, Rossant J. Cell fate decisions in early blood vessel formation. Trends Cardiovasc. Med. 2003;13:254–259. doi: 10.1016/s1050-1738(03)00105-1. [DOI] [PubMed] [Google Scholar]
- Ema M, Faloon P, Zhang WJ, Hirashima M, Reid T, Stanford WL, Orkin S, Choi K, Rossant J. Combinatorial effects of Flk1 and Tal1 on vascular and hematopoietic development in the mouse. Genes Dev. 2003;17:380–393. doi: 10.1101/gad.1049803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferkowicz MJ, Starr M, Xie X, Li W, Johnson SA, Shelley WC, Morrison PR, Yoder MC. CD41 expression defines the onset of primitive and definitive hematopoiesis in the murine embryo. Development. 2003;130:4393–4403. doi: 10.1242/dev.00632. [DOI] [PubMed] [Google Scholar]
- Galloway JL, Zon LI. Ontogeny of hematopoiesis: examining the emergence of hematopoietic cells in the vertebrate embryo. Curr. Top. Dev. Biol. 2003;53:139–158. doi: 10.1016/s0070-2153(03)53004-6. [DOI] [PubMed] [Google Scholar]
- Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev. Cell. 2005;8:109–116. doi: 10.1016/j.devcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Gekas C, Dieterlen-Lievre F, Orkin SH, Mikkola HK. The placenta is a niche for hematopoietic stem cells. Dev. Cell. 2005;8:365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Gering M, Patient R. Hedgehog signaling is required for adult blood stem cell formation in zebrafish embryos. Dev. Cell. 2005;8:389–400. doi: 10.1016/j.devcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Graf T. Differentiation plasticity of hematopoietic cells. Blood. 2002;99:3089–3101. doi: 10.1182/blood.v99.9.3089. [DOI] [PubMed] [Google Scholar]
- Hart DO, Raha T, Lawson ND, Green MR. Initiation of zebrafish haematopoiesis by the TAT-box-binding protein-related factor Trf3. Nature. 2007;450:1082–1085. doi: 10.1038/nature06349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S, Orkin SH. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003;18:109–120. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- Huber TL, Kouskoff V, Fehling HJ, Palis J, Keller G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature. 2004;432:625–630. doi: 10.1038/nature03122. [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Asai T, Saito T, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Ogawa S, Kurokawa M, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- Inman KE, Downs KM. The murine allantois: emerging paradigms in development of the mammalian umbilical cord and its relation to the fetus. Genesis. 2007;45:237–258. doi: 10.1002/dvg.20281. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26:726–740. doi: 10.1016/j.immuni.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Jin H, Xu J, Wen Z. Migratory path of definitive hematopoietic stem/progenitor cells during zebrafish development. Blood. 2007;109:5208–5214. doi: 10.1182/blood-2007-01-069005. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Morrison SJ. Maintaining hematopoietic stem cells in the vascular niche. Immunity. 2006;25:862–864. doi: 10.1016/j.immuni.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Radice GL, Morrison SJ. Lack of evidence that hematopoietic stem cells depend on N-cadherin-mediated adhesion to osteoblasts for their maintenance. Cell Stem Cell. 2007;1:204–217. doi: 10.1016/j.stem.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130:470–483. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Kim SI, Bresnick EH. Transcriptional control of erythropoiesis: emerging mechanisms and principles. Oncogene. 2007;26:6777–6794. doi: 10.1038/sj.onc.1210761. [DOI] [PubMed] [Google Scholar]
- Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat. Immunol. 2006;7:1048–1056. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Krzemien J, Dubois L, Makki R, Meister M, Vincent A, Crozatier M. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature. 2007;446:325–328. doi: 10.1038/nature05650. [DOI] [PubMed] [Google Scholar]
- Kulessa H, Frampton J, Graf T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 1995;9:1250–1262. doi: 10.1101/gad.9.10.1250. [DOI] [PubMed] [Google Scholar]
- Kumano K, Chiba S, Kunisato A, Sata M, Saito T, Nakagami-Yamaguchi E, Yamaguchi T, Masuda S, Shimizu K, Takahashi T, et al. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 2003;18:699–711. doi: 10.1016/s1074-7613(03)00117-1. [DOI] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Lane MC, Sheets MD. Primitive and definitive blood share a common origin in Xenopus: a comparison of lineage techniques used to construct fate maps. Dev. Biol. 2002;248:52–67. doi: 10.1006/dbio.2002.0717. [DOI] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Mandal L, Banerjee U, Hartenstein V. Evidence for a fruit fly hemangioblast and similarities between lymph-gland hematopoiesis in fruit fly and mammal aorta-gonadal-mesonephros mesoderm. Nat. Genet. 2004;36:1019–1023. doi: 10.1038/ng1404. [DOI] [PubMed] [Google Scholar]
- Mandal L, Martinez-Agosto JA, Evans CJ, Hartenstein V, Banerjee U. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature. 2007;446:320–324. doi: 10.1038/nature05585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Busslinger M. Life beyond cleavage: the case of Ago2 and hematopoiesis. Genes Dev. 2007;21:1983–1988. doi: 10.1101/gad.1591407. [DOI] [PubMed] [Google Scholar]
- Maslova MN, Tavrovskaia TV. Zh. Evol. Biokhim. Fiziol. 1993;29:211–214. [PubMed] [Google Scholar]
- Medvinsky A, Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- Mikkola HK, Klintman J, Yang H, Hock H, Schlaeger TM, Fujiwara Y, Orkin SH. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature. 2003;421:547–551. doi: 10.1038/nature01345. [DOI] [PubMed] [Google Scholar]
- Moore MA, Metcalf D. Ontogeny of the haemopoietic system: yolk sac origin of in vivo and in vitro colony forming cells in the developing mouse embryo. Br. J. Haematol. 1970;18:279–296. doi: 10.1111/j.1365-2141.1970.tb01443.x. [DOI] [PubMed] [Google Scholar]
- Mueller BU, Pabst T, Osato M, Asou N, Johansen LM, Minden MD, Behre G, Hiddemann W, Ito Y, Tenen DG. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood. 2002;100:998–1007. doi: 10.1182/blood.v100.3.998. [DOI] [PubMed] [Google Scholar]
- Muller AM, Medvinsky A, Strouboulis J, Grosveld F, Dzierzak E. Development of hematopoietic stem cell activity in the mouse embryo. Immunity. 1994;1:291–301. doi: 10.1016/1074-7613(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. Genomewide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin HF, Handin RI, Herbomel P. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25:963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck NA. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- North TE, de Bruijn MF, Stacy T, Talebian L, Lind E, Robin C, Binder M, Dzierzak E, Speck NA. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Orkin SH. Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet. 2000;1:57–64. doi: 10.1038/35049577. [DOI] [PubMed] [Google Scholar]
- Orkin SH. Priming the hematopoietic pump. Immunity. 2003;19:633–634. doi: 10.1016/s1074-7613(03)00302-9. [DOI] [PubMed] [Google Scholar]
- Ottersbach K, Dzierzak E. The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev. Cell. 2005;8:377–387. doi: 10.1016/j.devcel.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, Behre G, Hiddemann W, Tenen DG. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat. Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- Palis J, Chan RJ, Koniski A, Patel R, Starr M, Yoder MC. Spatial and temporal emergence of high proliferative potential hematopoietic precursors during murine embryogenesis. Proc. Natl. Acad. Sci. USA. 2001;98:4528–4533. doi: 10.1073/pnas.071002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardanaud L, Dieterlen-Lievre F. Manipulation of the angiopoietic/ hemangiopoietic commitment in the avian embryo. Development. 1999;126:617–627. doi: 10.1242/dev.126.4.617. [DOI] [PubMed] [Google Scholar]
- Pereira FA, Qiu Y, Zhou G, Tsai MJ, Tsai SY. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999;13:1037–1049. doi: 10.1101/gad.13.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JM, Li L. Disrupting the stem cell niche: good seeds in bad soil. Cell. 2007;129:1045–1047. doi: 10.1016/j.cell.2007.05.053. [DOI] [PubMed] [Google Scholar]
- Porcher C, Liao EC, Fujiwara Y, Zon LI, Orkin SH. Specification of hematopoietic and vascular development by the bHLH transcription factor SCL without direct DNA binding. Development. 1999;126:4603–4615. doi: 10.1242/dev.126.20.4603. [DOI] [PubMed] [Google Scholar]
- Purton LE, Scadden DT. Limiting factors in murine hematopoietic stem cell assays. Cell Stem Cell. 2007;1:263–270. doi: 10.1016/j.stem.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Querfurth E, Schuster M, Kulessa H, Crispino JD, Doderlein G, Orkin SH, Graf T, Nerlov C. Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev. 2000;14:2515–2525. doi: 10.1101/gad.177200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, Kanki JP. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev. Cell. 2005;8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder I, Glauche I. Towards an understanding of lineage specification in hematopoietic stem cells: a mathematical model for the interaction of transcription factors GATA-1 and PU.1. J. Theor. Biol. 2006;241:852–865. doi: 10.1016/j.jtbi.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- Rossant J. Stem cells: the magic brew. Nature. 2007;448:260–262. doi: 10.1038/448260a. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ, Weissman IL. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. USA. 2005;102:9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV. Negotiation of the T lineage fate decision by transcription-factor interplay and microenvironmental signals. Immunity. 2007;26:690–702. doi: 10.1016/j.immuni.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol. Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- Samokhvalov IM, Samokhvalova NI, Nishikawa S. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature. 2007;446:1056–1061. doi: 10.1038/nature05725. [DOI] [PubMed] [Google Scholar]
- Scheller M, Huelsken J, Rosenbauer F, Taketo MM, Birchmeier W, Tenen DG, Leutz A. Hematopoietic stem cell and multilineage defects generated by constitutive beta-catenin activation. Nat. Immunol. 2006;7:1037–1047. doi: 10.1038/ni1387. [DOI] [PubMed] [Google Scholar]
- Shalaby F, Ho J, Stanford WL, Fischer KD, Schuh AC, Schwartz L, Bernstein A, Rossant J. A requirement for Flk1 in primitive and definitive hematopoiesis and vasculogenesis. Cell. 1997;89:981–990. doi: 10.1016/s0092-8674(00)80283-4. [DOI] [PubMed] [Google Scholar]
- Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646–3653. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipkins DA, Wei X, Wu JW, Runnels JM, Cote D, Means TK, Luster AD, Scadden DT, Lin CP. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005;435:969–973. doi: 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier DY, Weinstein BM, Detrich HW, 3rd, Zon LI, Fishman MC. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121:3141–3150. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- Starck J, Cohet N, Gonnet C, Sarrazin S, Doubeikovskaia Z, Doubeikovski A, Verger A, Duterque-Coquillaud M, Morle F. Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol. Cell. Biol. 2003;23:1390–1402. doi: 10.1128/MCB.23.4.1390-1402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo K, Ema H, Morita Y, Nakauchi H. Age-associated characteristics of murine hematopoietic stem cells. J. Exp. Med. 2000;192:1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat. Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Turpen JB, Kelley CM, Mead PE, Zon LI. Bipotential primitive- definitive hematopoietic progenitors in the vertebrate embryo. Immunity. 1997;7:325–334. doi: 10.1016/s1074-7613(00)80354-4. [DOI] [PubMed] [Google Scholar]
- Ueno H, Weissman IL. Clonal analysis of mouse development reveals a polyclonal origin for yolk sac blood islands. Dev. Cell. 2006;11:519–533. doi: 10.1016/j.devcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Usui T, Preiss JC, Kanno Y, Yao ZJ, Bream JH, O’Shea JJ, Strober W. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J. Exp. Med. 2006;203:755–766. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villablanca EJ, Pistocchi A, Court FA, Cotelli F, Bordignon C, Allende ML, Traversari C, Russo V. Abrogation of prostaglandin E2/EP4 signaling impairs the development of rag1+ lymphoid precursors in the thymus of zebrafish embryos. J. Immunol. 2007;179:357–364. doi: 10.4049/jimmunol.179.1.357. [DOI] [PubMed] [Google Scholar]
- Vogeli KM, Jin SW, Martin GR, Stainier DY. A common progenitor for haematopoietic and endothelial lineages in the zebrafish gastrula. Nature. 2006;443:337–339. doi: 10.1038/nature05045. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yates F, Naveiras O, Ernst P, Daley GQ. Embryonic stem cell-derived hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 2005;102:19081–19086. doi: 10.1073/pnas.0506127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM, Crispino JD. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat. Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- Weissman IL, Papaioannou V, Gardner RL. Differentiation of Normal and Neoplastic Hematopoietic Cells. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1978. Fetal hematopoietic origins of the adult hematolymphoid system; pp. 33–47. [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JP, 4th, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Yan J, Chen YX, Desmond A, Silva A, Yang Y, Wang H, Hua X. Cdx4 and menin co-regulate Hoxa9 expression in hematopoietic cells. PLoS ONE. 2006;1:e47. doi: 10.1371/journal.pone.0000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara H, Arai F, Hosokawa K, Hagiwara T, Takubo K, Nakamura Y, Gomei Y, Iwasaki H, Matsuoka S, Miyamoto T, et al. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell. 2007;1:685–697. doi: 10.1016/j.stem.2007.10.020. [DOI] [PubMed] [Google Scholar]
- Zhang CC, Kaba M, Ge G, Xie K, Tong W, Hug C, Lodish HF. Angiopoietin-like proteins stimulate ex vivo expansion of hematopoietic stem cells. Nat. Med. 2006;12:240–245. doi: 10.1038/nm1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, Ross J, Haug J, Johnson T, Feng JQ, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]