Abstract
BACKGROUND:
HCG is produced by syncytiotrophoblast of placenta. It delays the apoptosis of corpus luteum and functions in implantation. Its possible role in male reproduction has been raised. HCG beta subunit is encoded by CGB, CGB5, CGB7 and CGB8 genes located at 19q13.3 in a common genome cluster with beta subunit non-coding CGB1 and CGB2. We conducted a sensitive quantification and comparison of CGB gene expression in human trophoblastic (blastocysts, n=6; normal/failed pregnancy, n=51) and non-malignant non-trophoblastic tissues (15 different tissue types, samples n=241).
METHODS:
Real-time RT-PCR.
RESULTS:
We showed a wide transcriptional window of CGB genes in normal pregnancy, a significant reduction in recurrent miscarriages, and a high expression (especially CGB1/CGB2) in ectopic and molar pregnancies. Expression was several orders of magnitude lower in the non-placental tissues, with the highest CGB levels being seen in testis, prostate, thymus, skeletal muscle and lung samples. The contribution of CGB1/CGB2 to the summarized expression of six CGB genes was not proportional to their gene dosage: 1/1000 to 1/10000. An interesting exception was the testis exhibiting a much higher CGB1/CGB2 to total CGB mRNA ratio of ∼1/3, corresponding to gene dosage.
CONCLUSIONS:
The expressional profile of CGB genes, activated already in blastocyst stage, is associated with the status of pregnancy. The presence of CGB transcripts in testes, and in particular CGB1/CGB2 transcripts, may indicate a role in male reproductive tract.
Keywords: Chorionic gonadotropin beta genes, normal and failed pregnancy, trophoblastic and non-trophoblastic CGB expression, CGB in male reproduction
Introduction
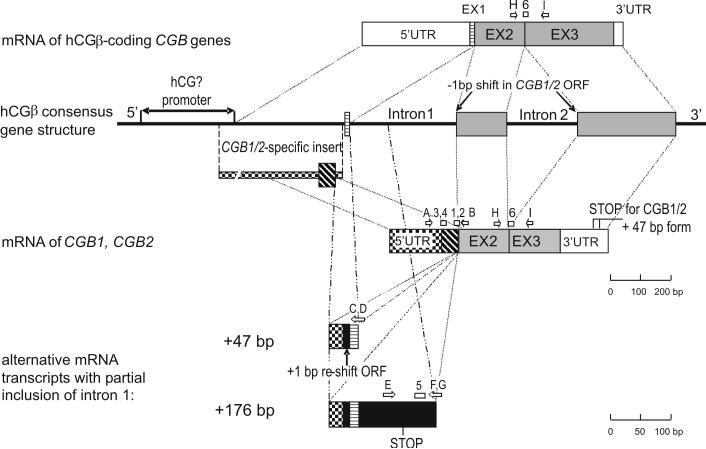
Human chorionic gonadotropin (HCG) is an evolutionary young hormone arisen in primate lineage (Talmadge et al., 1984; Maston and Ruvolo, 2002). HCG like other glycoproteins (thyroid stimulating hormone - TSH, follicle stimulating hormone – FSH, luteinizing hormone – LH) consists of two subunits. The alpha subunit is shared by all four glycoproteins, and the beta subunit is specific for each hormone. Unlike the other glycoproteins that are synthesized in the anterior lobe of pituitary gland, HCG is mainly produced by syncytiotrophoblast of placenta during pregnancy (Pierce and Parsons, 1981). The structure of anthropoid primate-specific hemochorial placenta enables the transport of the large glycoprotein into the maternal circulation, where it exerts effects for fetal benefit (King, 1993). In addition to its endocrine function of delaying the apoptosis of the corpus luteum during the first trimester of pregnancy, HCG has several paracrine effects essential in the process of implantation (Srisuparp et al., 2001, Cameo et al., 2004), angiogenesis and placentation (Toth et al., 2001; Zygmunt et al., 2002; Herr et al, 2007) and development of maternal immunotolerance (Kaylisli et al., 2003). In addition to its function in pregnancy, elevated levels of HCG or its free beta subunit in serum is found to be as a sensitive tumor marker for trophoblastic tumors and testicular germ cell tumors (Bates and Longo, 1985; Seppälä et al, 1986; Braunstein 1990; Duffy 2007). HCG and/or CGB proteins are present in a subset of cases with other tumor types, but their association is sporadic and the proteins have not been established as having functional, diagnostic or case management relevance in these other settings. HCG beta subunit is encoded by four genes (CGB, CGB5, CGB7, CGB8) that reside in a common genome cluster with the evolutionarily ancestral LHB and two recently duplicated genes CGB1 and CGB2 at chromosome 19q13.3 (Talmadge et al., 1984; Maston and Ruvolo, 2002; Hallast et al., 2005; 2007). Unlike the HCG beta coding genes that each produce a single mRNA transcript, CGB1 and CGB2 have four splice variants as originally described by Bo and Boime (1992). The major transcript of CGB1/CGB2 (marked hereinafter as a transcript of CGB1/CGB2) codes for a unique 5′UTR and exon 1, and one basepair shifted open reading frame (ORF) for exon 2 and exon 3 (Fig.1). This predicts a completely different protein from HCG. The alternative mRNAs (marked as CGB1/CGB2+47 bp, +166 bp, +176 bp, respectively) contain additional 47 bp, 166 bp or 176 bp of DNA sequence from intron 1 (Fig.1) that alter the amino acid sequence of transcript.
Figure 1.
Schematic representation of the structure of the CGB genes and transcripts as well as localization of primers and Taqman probes used in the study. The coding segments of all CGB genes are marked on the consensus gene structure with grey boxes, except for the exon I for hCGβ-coding genes (horizontal stripes) and for CGB1/CGB2 (diagonal stripes). The 5′UTR of mRNA transcribed from hCGβ-coding genes (white box) differs from the 5′UTR for CGB1/CGB2 (chequered box); 3′UTR of the two groups of gene is of variable length (white box). Alternative +47 bp CGB1/CGB2 mRNA forms contain additional sequence from CGB1/CGB2-specific intron I (22 bp, black box), including the fragment corresponding to the hCGβ 5′UTR (10 bp, chequered box) and exon I (15 bp, horizontal stripes), resulting in a re-shift the CGB1/CGB2 ORF to the ORF of hCGβ-coding transcripts. Due to sequence divergence in 3′UTR of CGB1/CGB2, the predicted STOP codons for +47 bp CGB1 and +47 bp CGB2 differ by 7 aminoacids. Alternative transcripts +166 bp (not shown in the figure) and + 176 bp contain additional 119 bp/129 bp sequence identical to the intronic part of all CGB genes (black box). The predicted STOP codon for +166 bp/+176 bp forms is located at position 355 from transcription start (hypothetical polypeptide 60 amino acids). Primer and probes use for real-time RT-PCR experiments are shown with white arrows or boxes and marked with letters or numbers, respectively. The oligonucleotide sequences are shown in Table I.
We have previously studied the expression profile of individual CGB genes during normal and complicated pregnancy using a semi-quantitative Gene Scan Fragment Analysis approach (Rull and Laan, 2005). In this study we have addressed the fine-scale quantification of CGB genes in trophoblastic tissues (samples n=51) including normal pregnancy from the blastocyst (n=6) stage and different scenarios of pregnancy failure (recurrent miscarriages, ectopic and molar pregnancies). We also studied the transcriptional profile of CGBs in non-malignant non-trophoblastic human tissues (tissues n=15, samples n=39 for individual cDNAs and n=202 for tissue-specific pooled cDNAs). We enhanced the sensitivity of the transcript detection by using the real-time RT-PCR method, which enabled us to address separately the expressional profile of CGB1/CGB2. The screen for CGB expression in normal, non-malignant non-trophoblastic tissues was done to investigate whether these genes might have relevance outside of the blastocyst and pregnancy.
Materials and Methods
Experimental Subjects
The study was approved by the Ethics Committee of the University of Tartu, Estonia (protocols no. 117/9, 16.06.03, and 126/14, 26.04.2004), and a written informed consent was obtained from every patient. Blastocysts were kindly donated by six couples undergoing in vitro fertilization procedure at Nova Vita Clinic, Centre for Infertility Treatment and Medical Genetics (Dr. Peeter Karits). Chorionic villi/placental samples were obtained from females who underwent elective therapeutic abortion during first trimester of pregnancy (4-12 weeks of gestational age, n=10); therapeutic abortion during second trimester due to medical risks of pregnancy, where no fetal anomalies were detected (17-21 weeks of gestational age, n=8); normal delivery at term resulting from uncomplicated pregnancy (38-42 weeks of gestational age, n=12); surgical treatment of ectopic pregnancy (6-14 weeks of gestational age, n=8); cervical dilatation and uterine curettage because of recurrent (patients had ≥2 spontaneous abortions before the case) incomplete or missed abortion (6-17 weeks of gestational age, n=11); and because of molar pregnancy (9-10 weeks of gestational weeks, n=2). The autopsy material of selected tissues was obtained from Research Tissue Bank of Tartu University Hospital (cerebral cortex, liver, skeletal muscle, kidney, pancreas, spleen, small intestine, testes). Biopsy samples obtained during surgical therapeutic procedures upon informed consent were provided by Dr. Tõnu Vooder, Lung Clinic of Tartu University Hospital (lung and thymus) and by Dr. Margus Punab, Tartu University Hospital, Andrology Unit (testes). Tissue samples were snap-frozen in liquid nitrogen and stored at −80 °C or placed immediately into RNAlater solution (Ambion Inc, Austin TX) and kept at −20 °C until RNA isolation. For all biopsy/autopsy samples histological examination was carried out to confirm the non-malignancy of the tissues. In addition to clinical samples the cDNA from Human Multiple Tissue cDNA Panel I and II (Clontech Laboratories Inc, Mountain View, CA) was used to broaden the range of studied tissues.
RNA extraction and cDNA synthesis
Total RNA from 30-1000 mg of tissue was extracted and quantified as described previously (Rull and Laan, 2005). The integrity of RNA was checked by 2100 Agilent Bioanalyzer using RNA 600 Nano LabChips (Agilent Technologies, Santa Clara, CA). RNA was reverse transcribed to cDNA using random hexamers and SuperScript™ III Reverse Transcriptase according to manufacturer's protocol (Invitrogen Life Technologies, Carlsbad, CA). Briefly, a 1 μg aliquot of DNAse-treated total RNA was incubated with 50 ng random hexamer primers and 1 μl of a cocktail containing 10 mM of each dNTP at 70 °C for 5 min. The reaction was cooled on ice and remaining reagents were added (2 μl of 10 × reaction buffer, 100 nmol MgCl2,, 0.2 μmol dithiothreitol (DTT), 40 U RNAseOUT™, 200 U SuperScript™ III Reverse Transcriptase) and the reaction proceeded at 25 °C for 10 min and 50 °C for 50 min. The reverse transcription was inactivated by a 5 min incubation at 85 °C and cDNA was treated with 2 U of E.coli RNAse H at 37 °C for 25 min to remove the RNA template from the cDNA:RNA hybrid molecule. The minus reverse transcription controls were treated as described above except that the reactions lacked SuperScript™ III Reverse Transcriptase.
Primer and Taqman probe design, and real-time RT-PCR
All primers and probes (Table I) for real-time RT-PCR were designed using Primer Express version 2.0 (Applied Biosystems, Foster City, CA) taking into account polymorphic positions (SNP) in CGB genes identified by the resequencing study (Hallast et al., 2005). As the only splice-form discriminatory CGB1 Taqman probe included a polymorphic position (A/G; exon II, pos. 233 bp from transcription start) we designed two alternative detection oligos specific to either allele (Table I, Fig.1 probe 1,2). In subsequent calculations we averaged the results obtained from two differentially labeled probes. The quantification of mRNA transcript CGB1/CGB2+166 bp was omitted because of an unfavorable sequence (ACCCCCA) on the form-specific exon I/exon II boundary for positioning either primers or a probe. The primer-probe mix of the reference genes (GAPDH, Hs99999905_m1, amplicon length 122 bp; HPRT1, Hs99999909_m1, 100 bp) were purchased from Applied Biosystems (Foster City, CA). GAPDH is expressed at approximately the same level with hCG beta subunit coding genes and HPRT is transcribed in the similar activity as CGB1/CGB2 genes.
Table I.
The DNA sequences of the primers and probes for real-time RT-PCR experiments
| Expression of CGB gene transcripts | ||||
|---|---|---|---|---|
| Real-time primer |
Fig.1
Label |
Sequence with Fluorescent Dye | Tm (°C) |
Product size |
| CGB1/2F | A | 5′-AACACCCCTCACTCCCTGTCT-3′ | 58.1 | 139 bp |
| CGB1/2R | B | 5′-ATGCTCAGCAGCAGCAACA-3′ | 57.8 | |
| alt47C1majR | C | 5′-GCAGCAGCCTCTGGAACATCT-3′ | 58.6 | 167/ 165 bp |
| alt47C1min/C2R | D | 5′-AGCAGCCCCTGGAACATCTC-3′ | 58.3 | |
| alt176C1/C2intF | E | 5′-GCCATCACTGGCATGAGAAG-3′ | 57.8 | 96 bp |
| alt176C1majR | F | 5′-AGCAGCAGCCTTGAAGCTTACT-3′ | 58.6 | |
| alt176C1min/C2R | G | 5′-AGCAGCAGCCCTGAAGCTTAC-3′ | 59.8 | |
| CGBallF | H | 5′-TCACCGTCAACACCACCATCT-3′ | 59.8 | 121 bp |
| CGBtotalR | I | 5′-ATGGACTCGAAGCGCACATC-3′ | 59.8 | |
| Taqman probe | ||||
| CGB1A | 1 | 5′-FAM-ACATGTCAAAGAGGCTG-MGB-3′ | 69.2 | |
| CGB1G | 2 | 5′-VIC-CATGTCAAAGGGGCTG-MGB-3′ | 69.9 | |
| CGB1 † | 3 | 5′-FAM-CATGTCCACATTCCCAGTG-MGB-3′ | 68.2 | |
| CGB2† | 4 | 5′-FAM-CATGTCCACATCCCCAGT-MGB-3′ | 67.9 | |
| alt176C1/2 | 5 | 5′-FAM-CTCTTTCTGGAGGAGCGT-MGB-3′ | 68.3 | |
| CGBall | 6 | 5′-FAM-CCACCATGACCCGCGT-MGB-3′ | 68.6 | |
|
| ||||
| Primers for plasmid construction | ||||
|
| ||||
| CGB8F | 5′-AGCACCTTTCTCGGGTCAC-3′ | 60.2 | 791 bp | |
| CGB8R | 5′-GGCCTTTGAGGAAGAGGAGT-3′ | 59.5 | ||
| CGB1F | 5′-GAGGAAGGGGAACTGCATCT-3′ | 60.6 | 686/ 689 bp |
|
| CGB2F | 5′-AGGGAGGAAGGGGAACTGTA-3′ | 59.9 | ||
| CGB1, CGB2R | 5′-TGCGGATTGAGAAGCCTTTA-3′ | 60.8 | ||
| CGB1Fnested | 5′-GAAGGGGAACTGCATCTGAG-3′ | 59.8 | 406 bp* | |
| CGB1Rnested | 5′-GGTAGTTGCACACCACCTGA-3′ | 59 | ||
Tm, melting temperature; F, forward primer; R, reverse primer;
the probe was used to detect CGB1/CGB2 + 47 bp alternative mRNA form
the primers amplified CGB1 + 176 bp mRNA form, 581 bp and CGB1 + 166 bp mRNA alternative form, 571 bp.
The real-time RT-PCRs was performed using Applied Biosystems 7900HT Fast Real-time PCR system in 384 micro-well plates. The 10 μl PCR reactions consisted of 1 μl cDNA product, 5 μl TaqMan® Universal PCR Master Mix (2X) containing AmpliTaq Gold® DNA Polymerase and AmpErase® UNG (Applied Biosystems, Foster City, CA), as well as primers and a probe in following concentration: 500 nM forward and reverse primer, 100 nM probe for target genes; or 0.5 μl ready-to-use endogenous control gene primer-probe mix (20X) for GAPDH or HPRT1. The PCR protocol was identical for all runs: 50°C for 2 min and 95 °C for 10 min to initiate the reaction followed by 50 two-step amplification cycles (15″ 95 °C; 1′ 60 °C). Each sample was run in triplicate. Statistical analysis included the data only from the reactions where minus cDNA gave the negative result.
Gene Scan Fragment Analysis was conducted as described (Rull and Laan, 2005), except that no discrimination of the transcripts for each individual hCG beta coding gene and CGB1/CGB2 was carried out.
Construction of transcripts-specific and negative control plasmids
We cloned from normal first trimester placenta tissue RT-PCR amplified (primer sequences in Table I) and gel-puried cDNA fragments of CGB8 (representing four HCG beta subunit coding genes), CGB1 (=CGB2) and CGB1+176 bp splice-form, which were used for real-time RT-PCR optimization and preparation of standard curves for absolute quantification. All inserts were cloned into pTZ57R/T vector (InsTAclone™ PCR Cloning Kit, Fermentas Life Sciences, Burlington, Canada) and plasmid DNA was extracted with Nucleo Bond® PC 500 Plasmid Purification Kit (Macherey-Nagel GmbH&Co KG, Düren, Germany). As an additonal negative control for the oligo specificity to mRNA and to assure the presence of CGB1/CGB2 alternative forms (exclude genomic contamination) we used a cloned CGB1 genomic fragment, which contained the regions targeted by Taqman primers and probes.
Data analysis
For quantification of the target genes two methods were used: comparative Ct (cycle threshold, cycle number at which the PCR amplification crosses the background fluorescence) method and standard curve method. The calculations by Ct method was carried out by two formulas differing in taking into account PCR efficiency (Livak and Schmittgen, 2001; Pfaffl, 2001).The calculated relative expression level of CGB genes by two methods was strongly correlated (r2=0.95-0.99). Standard curve of serial dilutions of plasmid cDNA was run over eight logs covering 107-101 copies of target transcript per reaction. Copy number of CGB transcripts was quantified according to guidelines by Applied Biosystems (http://www.appliedbiosystems.com/support/apptech/#rt_pcr) and as described by Reimer et al. (2000).
For statistical analysis a Pearson correlation test and a linear regression model were used. All statistics were performed using R 2.4.1, a free software environment for statistical computing and graphics (http://www.r-project.org/)
Results
Real-time RT-PCR experiment validation
Reproducibility of the real-time RT-PCR experiments was validated by high amplification efficiency (100+/−10%) as well as low intra-assay (<1.9%) and inter-assay (<2%) variation coefficients. Although the expression of the two reference genes differed from each other (GAPDH > HPRT1) in trophoblastic samples (by average 6.8+/−0.7 cycles) as well as in non-trophoblastic tissues (from 2.0-2.7 cycles in testis to 8.6-9.0 cycles in skeletal muscle samples), the results were concordant using either of the endogenous controls.
Proportional expression of CGB transcripts
Consistent with previous reports the major sources of CGB mRNA transcription were HCG beta coding genes (CGB, CGB5, CGB7, CGB8). The contribution of CGB1 and CGB2 genes to the summarized expression of all CGB genes was not proportional to their gene dosage: in the first trimester of normal pregnancy these two of the total six CGB gene copies (1/3) provided only 1/1000 to 1/5000 of the CGB mRNA transcripts, during the second and third trimester the contribution was even lower (∼1/10 000; Table II). In comparison to normal implantation process the trophoblastic tissue from the cases of ectopic pregnancy was characterized by higher transcriptional activity of CGB1/CGB2 and/or lower expression level of HCG beta subunit coding genes: CGB1/CGB2 contributed 1/500 to 1/3000 of summarized CGB transcripts. This expressional peak of CGB1/CGB2 confirms the original finding by semi-quantitative analysis (Rull and Laan, 2005).
Table II.
Expression of CGB genes in human tissues determined by Gene Scan Fragment Analysis and Taqman Real-Time methods
| No of samples |
Summarized CGB |
CGB1/ CGB2
|
|||||
|---|---|---|---|---|---|---|---|
| cDNA panela |
Biopsies/ autopsies |
Gene Scanc | TaqMand | Gene Scanc | TaqMand | ||
| Trophoblastic tissue: | |||||||
| Calibrator, I trimb | − | 1 | + | 11873350 | + | 15000 | |
| Normal, I trim | − | 10 | + | 2308383 | + | 879 | |
| Ectopic pregnancy | − | 8 | + | 13163945 | + | 15067 | |
| Miscarriage, I trim | − | 11 | + | 194371 | + | 134 | |
| Placenta, II trim | − | 8 | + | 147295 | +/− | 15 | |
| Placenta, III trim | 8 | − | + | 161600 | nd | 28 | |
| Testis | 45 | 5 | + | 722 | + | 453 | |
| Prostate | 32 | − | +/− | 491 | +/− | 17 | |
| Thymus | 18 | 5 | +/− | 280 | + | 4 | |
| Skeletal muscle | 8 | 3 | nd | 254 | nd | nd | |
| Lung | 4 | 5 | +/− | 160 | + | nd | |
| Small intestine | 32 | 3 | nd | 86 | nd | nd | |
| Ovary | 14 | − | nd | 50 | nd | 3 | |
| Kidney | 5 | 3 | nd | 28 | nd | 5 | |
| Spleen | 3 | 3 | nd | 23 | +/− | nd | |
| Heart | 3 | − | nd | 20 | nd | nd | |
| Brain | 2 | 3 | nd | 20 | nd | nd | |
| Liver | 1 | 3 | nd | 28 | nd | nd | |
| Colon | 20 | 3 | nd | 9 | nd | 6 | |
| Pancreas | 15 | 3 | nd | nd | +/− | nd | |
| Peripheral blood leucocytes |
NA | − | nd | nd | nd | nd | |
NA – not available, trim – trimester, nd – not determined
pooled cDNAs from two tissue panels (see Materials and Methods),
the calibrator sample (first trimester normal pregnancy) selected based on the highest expression of the target gene in the group and used in relative quantification
detection of the target gene in at least in two runs of three (+); one run of three (+/−)
mRNA molecules of the target gene/50ng of total RNA used for cDNA synthesis
Interestingly, we detected approximately equal fractions of the originally described CGB1/CGB2 transcript coding for a hypothetical protein and of the alternatively spliced CGB1/CGB2 + 176 bp mRNA that contains a premature stop-codon (Fig.1-2). This observation contributes to the uncertainty of the function and role of CGB1/CGB2 in human. The other reported alternative splice-form (+47 bp; Fig.1) was transcribed at the borderline (in trophoblastic samples) or even below (in non-trophoblastic tissues) the reliable quantitation limit of real-time RT-PCR. The transcription initiation of different splice forms of CGB1/CGB2 correlated within a sample (r2=0.82-0.89).
Figure 2.
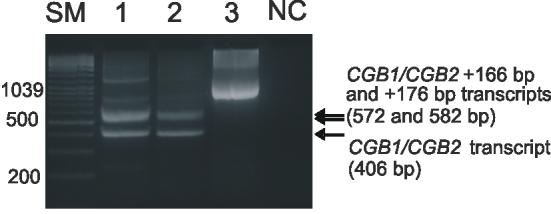
Detection of CGB1/CGB2 mRNA in trophoblastic tissue from the first trimester of a normal intrauterine pregnancy (Lane 1) and an ectopic pregnancy (Lane 2). Alternatively spliced transcripts CGB1/CGB2, CGB1/CGB2 + 166bp and CGB1/CGB2 + 176 bp are represented by the RT-PCR amplified fragments sized 406 bp, 572 bp and 582 bp, respectively. The CGB1CGB/2 + 166bp and the CGB1/CGB2 + 176 bp transcripts are indistinguishable from each other at the 3% agarose gel. The detection sensitivity of EtBr stained DNA separated on agarose gel is not enough for the visualization low expressed CGB1/CGB2 + 47 bp splice-variant. The amplified genomic DNA (Lane 3) is 1040 bp. NC, negative control with water as a template; SM, size marker (Gene Ruler™ 100 bp DNA Ladder Plus, Fermentas Life Sciences).
Transcription of CGB genes in trophoblastic tissue during the normal and complicated pregnancy
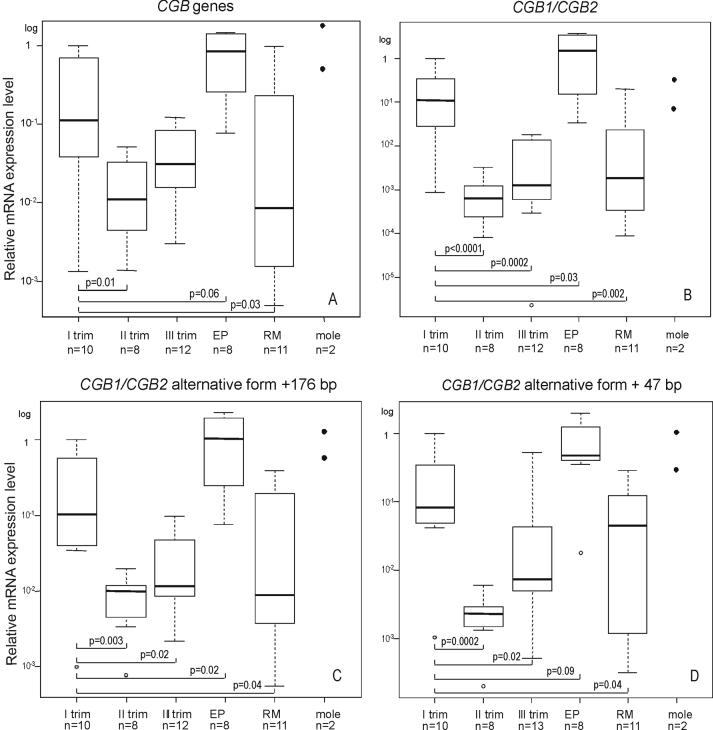
As expected, during normal pregnancy the highest summarized expression of all CGB genes was detected for the first trimester (p=0.01 and p=0.11 compared to the second and third trimester, respectively) (Fig. 3A). In cases of recurrent miscarriages (RM) the summarized transcriptional activity of all CGB genes was significantly reduced compared to normal pregnancy at same gestational age (p=0.03; Fig.3A). Inversely, in cases of ectopic pregnancy (EP), the total CGB mRNA level tended to be higher than the distribution described for normal intrauterine gestation (p=0.06). The expression of the three alternative CGB1/CGB2 transcripts followed the same pattern: the strongest expression during first trimester compared to second and third trimester of normal pregnancy (p<0.004; p<0.04, respectively), a reduced transcriptional activity in cases of RM (p<0.05) and more abundant CGB1/CGB2 mRNA transcripts in trophoblastic tissues from EP compared to normal pregnancy of same gestational age (0.02≤p≤0.09, Fig.3B-D).
Figure 3.
Notched box plot for the distribution of the relative expression level of CGB transcripts in placenta during the first, second and trimester of normal pregnancy, in case of recurrent miscarriage (RM), ectopic pregnancy (EP) and molar pregnancy. The boxes represent the 25th and 75th percentiles. The median is denoted as the line that bisects the boxes. The whiskers are lines extending from each end of the box covering the extent of the data on 1.5 X interquartile range. Circles represent the outlier values. P-values reflecting the differences between groups are shown.
Relative mRNA quantization with real-time RT-PCR comparative Ct method for A, summarized CGB genes transcription; B, CGB1/CGB2 transcripts; C, D CGB1/CGB2 +176 bp and CGB1/CGB2 +47 bp alternative forms. The trophoblastic sample with highest summarized transcription activity of CGB genes from first trimester normal pregnancy is used as calibrator sample.
Expression of CGB genes in blastocysts and molar pregnancy
We explored the expression of CGB genes in trophoblastic tissues obtained from molar pregnancies (n=2) and from blastocysts (n=6). The transcription level of all CGB transcripts in molar pregnancy was at the upper range of the distribution obtained from normal first trimester pregnancy (n=10) (Fig.3A-D). In mRNA extracted from pooled blastocysts material the summarized level of CGB transcripts was as high as the estimated median (Fig. 3A) for trophoblastic samples from the first trimester of normal pregnancy (10-fold lower compared to the calibrator) using relative quantification. However, low yield of the total mRNA extracted from blastocysts prevented the exact calculation of the amount of mRNA entering the cDNA synthesis and thus, no absolute quantification of CGB mRNA molecules was possible. This supports the suggestions that the full activation of CGB genes occurs in early preimplantation stage and is essential for normal implantation (Bonduelle et al., 1988; Jurisicova et al., 1999). Interestingly, despite the low general expression level of CGB1/CGB2 and their alternative forms, their transcripts were also detected already in blastocyst material both by real-time RT-PCR (data not shown) as well as even by semi-quantitative analysis (Fig.4).
Figure 4.
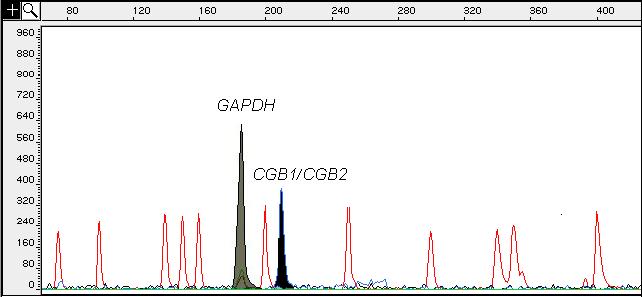
Gene Scan Fragment Analysis electrophoretogram showing the amplified fluorescent-labeled products of CGB1 and CGB2 (6-FAM; 207 bp, black peak) gene and GAPDH (HEX; 196 bp, grey peak) from cDNA of a single blastocyst material. The x-axis shows the size of the detected fragments in base pairs (bp), and the y-axis represents the relative intensity of fluorescence (RFU). The empty peaks mark the GeneScan-500 TAMRA internal size standard.
Expression of CGB genes in non-trophoblastic tissues
To avoid contamination and false-positive results facilitated by low expression level, we addressed the transcriptional profile of CGB genes in non-trophoblastic tissues by two alternative methods: a semi-quantitative RT-PCR combined with Gene Scan Fragment analysis (Bellet et al., 1997; Miller-Lindholm et al., 1997; Rull and Laan, 2005) and quantification with real-time RT-PCR using TaqMan method (Reimer et al., 2000). Gene Scan Fragment analysis was used to screened CGB transcripts from commercial human tissue cDNA panels (15 pooled cDNAs from 1-45 subjects). For real-time RT-PCR experiments we used identical cDNA panels as well as analyzed additional RNAs (n=3-5) prepared from various human non-malignant tissues (Table II). The results obtained from tissue panel and analyzed selection of clinical samples and autopsies were concordant. As expected, real-time RT-PCR method proved to be more sensitive resulting in detection of CGB transcripts in 13 of the studied 15 non-trophoblastic tissues. The reliable expression of CGB genes was detected in testis, prostate, thymus, skeletal muscle and lung samples (160-722 cDNA copies per PCR reaction; Table II). However, the transcription level was still very low, from 1/2500 (testis) to 1/12500 (lung) of the CGB expression level in first trimester placenta. In several tissues (ovary, small intestine, kidney, spleen, liver, heart, brain and colon) CGB transcripts were present at the level of the borderline detection by the real-time RT-PCR method (9-86 copies per reaction; Table II). Finally, no CGB transcripts were detected in pancreas and peripheral blood leukocytes. Semi-quantitative Gene Scan Fragment analysis identified CGB transcripts only in four non-trophoblastic tissues (testis, prostate, thymus and lung). Thus, the detection limit of this method is >100 cDNA copies per PCR reaction.
Due to low expression CGB1/CGB2 mRNA was amplified by real-time RT-PCR only in a limited number of non-trophoblastic tissues: testis, prostate, thymus, ovary, kidney and colon. Interestingly, CGB1/CGB2 are ∼16 times more transcribed in testis (453 copies per reaction) than in the second (15 copies per reaction) and third trimester of pregnancy (28 copies per reaction; Table II). Also, in testis the proportional contribution of HCG-β coding genes and CGB1/CGB2 to the total CGB transcript pool is very different from placenta, ∼66%/33% (722 copies versus 453 copies per reaction; Table II).
Discussion
This study reports a detailed mRNA transcription profile of CGB genes in trophoblastic (normal/complicated pregnancy) and in non-malignant non-trophoblastic human tissues. We chose the real-time RT-PCR method allowing sensitive quantification of the CGB transcripts in extra-uterine tissues expressing these placental genes at low level. In addition, the enhanced sensitivity of the experiment enabled to provide more confident data about the expression of low-transcribed CGB1/CGB2 genes.
Both, semi-quantitative Gene Scan Fragment analysis (Miller-Lindholm et al., 1997; Rull and Laan, 2005) and quantitative real-time RT-PCR experiments indicated a wide range of distribution of the transcriptional level for the six CGB genes in normal intrauterine pregnancy and a significant correlation between CGB mRNA abundance in placenta with HCG concentration in the maternal serum. There is evidence that in cases of recurrent miscarriages low HCG levels in urine/serum (Gerhard and Runnenbaum, 1984; Buyalos et al., 1992; Letterie and Hibbert, 2000) may result as a direct consequence of a significant reduction in the expression of CGB genes. We are aware that ongoing biological processes to reject fetus from uterus during the miscarriage and induced therapeutic abortion may change the expression pattern of the genes in some extent, although decrease of hormone in serum after abortion considerably slow (Steier et al. 1984). Low transcriptional activation of CGB genes might result from an inadequate function of decidual cells and syncytiotrophoblasts producing the lactogenic hormones required for immunological adaptation of the embryo and trophoblast invasion (Lyall, 2002; Burton et al, 2007). However, the low CGB mRNA level compared to normal gestational age-matched placentas may indicate not only maternal susceptibility to a miscarriage, but also chromosomal aberrations of the conceptus. In case of trisomy 18 (Edwards syndrome), decreased concentration of maternal serum HCG during first trimester is the consequence of impairment in the transcription of HCG beta subunit genes (Brizot et al., 1996). The situation for trisomy 21 Down's pregnancies is more complicated. In vitro studies following the differentiation process of cultured cytotrophoblast cells showed a significant decrease in CGB mRNA transcription and consequently the secretion of HCG for trisomy 21 placentaes causing a defect and/or a delay formation of syncytiotrophoblast formation (Massin et al., 2001). But it is well known that Down's syndrome is associated with elevated, not reduced maternal serum concentrations of HCG as well as free HCG beta subunit (reviewed by Knöfler, 1999).
In contrast to recurrent miscarriages, the expression of the CGB genes in extra-uterine pregnancy (EP) is up-regulated and has a narrower expressional window (Fig.3). The CGB transcriptional activity in molar pregnancy falls into the same range as in EP and is responsible for high serum HCG levels (this report; Feng et al., 2006). In cases of EP, the discrepancy between the HCG concentration in serum and CGB mRNA transcription level in trophoblastic tissues contrasts the trisomy 21 pregnancies: low hormone versus high transcriptional activity. In both physiological conditions, post-transcriptional processes like mRNA stability, translation initiation, posttranslational modifications, hormone assembly, activity and stability determine the fine regulation of HCG molecule biosynthesis and properties. Indeed, in Down syndrome the defective differentiation leads to the accumulation of cytotrophoblasts producing the structurally and functionally distinct hyperglycosylated hormone variant HCG-H instead of the regular HCG (reviewed by Cole, 2007). The abundance of HCG-H might be one of the reasons why the fetuses with trisomy 21 unlike most other chromosomal anomalies are not aborted during the gestation. In EP, the low hormone levels refer to the deficient hormone assembly and the high expressional activity of CGB genes might reflect the failure of a normal negative autoregulatory feedback. Hormonal effects on the stability of a specific mRNA can profoundly alter its steady-state concentration (e.g. reviewed by Staton et al., 2000; Ing, 2004).
In addition to the detection of summarized transcription of all CGB genes we analyzed isolated expression of the most recently duplicated, the African great ape specific CGB1/CGB2 and their alternative products both in trophoblastic and non-trophoblastic tissues. We have recently shown that the human and chimpanzee CGB1/CGB2 genes have no ORF disrupting mutations (Hallast et al., 2007), as well as there are signals that these novel genes evolve under positive selection (Hallast et al., unpublished data). The expressional activity of CGB1/CGB2 within a particular sample was correlated with the total CGB transcript level suggesting a concerted transcriptional activation of the gene cluster. The conserted manner of the expression might be explained by either the transcriptional activation of the genome cluster through simultaneous chromatin modifications (e.g. demethylation of the abundant CpG-island within the cluster) and/or by highly homologous DNA sequence (>95%) in the promoter regions of all CGB genes. The low level of the transcription of CGB1 and CGB2 could result from modified promoter region lacking functionally important binding sites for two Ets-2 transcription factors (Fig.1; Ghosh et al., 2003). The central role of Ets-2 in trophoblast differentiation and function has been found in several other studies (Roberts et al., 2003; Chakrabarty et al., 2007). It is still an open question whether the CGB1/CGB2 genes are pseudogenes producing mRNA transcripts as a consequence of general expressional activation of the CGB gene cluster or code for a functional protein. The pseudogene status is supported by extremely low transcription level compared to HCG beta genes (with the exception of testis) and no structural similarity of the predicted hypothetical protein to other known proteins. However, there is growing evidences that transcribed pseudogenes are not entirely without purpose. For example, they can act as potential regulators of gene expression and effectors of the function of known genes (Hirotsune et al., 2003, Korneev et al., 1999). As concluded by Khaitovich et al. (2006) these types of regulatory transcripts tend to be expressed at low levels, sometimes at or below detection limit and the transcripts are located close to annotated genes or even within genes, in introns or on the opposite DNA strand. The transcripts produced by the pseudogenes located between protein coding genes show conserved tissue-specific expression patterns concordant with the protein-coding transcripts of known genes. Still, we cannot exclude the possibility that CGB1 and CGB2 code for a functioning protein – similarly to the HCG beta peptides (163 amino acids, 12 Cys), the predicted hypothetical CGB1/CGB2 peptide is enriched in cysteins (132 amino acids, 5 Cys) relative to regular globular proteins. Thus, concordant to HCG synthesis, Cys-Cys bridges (forming a cystein-knot) may be critical for the 3D structure and function of the hypothetical protein involving CGB1/CGB2 peptide (Ceslovas Venclovas, personal communication).
Despite the historic reports of CGB expression by individual cases of many tumors, to our knowledge there are only two larger previous studies addressing the transcriptional profile of CGB genes in normal non-malignant non-trophoblastic tissues (Bellet et al., 1997: semi-quantitative detection; Reimer et al., 2000: real time quantification with TaqMan). Consistent with these reports our data show that the expression of CGB genes varied among tissues. In all three studies the strongest non-trophoblastic CGB transcriptional activity was detected in testis, prostate and skeletal muscle, which is also concordant with the available gene expression microarray data (http://expression.gnf.org, 1153_f_at; Affymetrix Annotation). There is overlap between the two real-time experiments confirming reliably the transcription of CGB genes in thymus, colon, lung and kidney. Among the tissues studied in literature, CGB expression was detected also in uterus, mammary, thyroid, adrenal and pituitary glands (Bellet et al., 1997, Reimer et al., 2000, Dirnhofer at al., 1996).
The question about the biological function of the CGB transcripts in testis (Berger et al., 1994) and prostate (Dirnhofer et al., 1998) was raised already more than 10 years ago when all the different mRNA variants (including three alternative splicing products of CGB1/ CGB2) were identified in the cDNA pool from these tissues. Interestingly, compared to the placental tissue, the proportion of CGB1/CGB2 transcripts was increased in testis (Table II). The presence of CGB transcripts in testes and prostate indicating a possible role male reproductive tract is in agreement with a recent study showing that HCG alpha and HCG beta free subunits, but much less the alpha/beta dimers (HCG) are produced in high amounts in the prostate and testis, and are subsequently observed in seminal plasma (Berger et al., 2007). In testicular Leydig cells exclusively HCG beta free subunits were identified. It has been suggested that HCG-derived molecules might act in auto-/paracrine or even transcrine ways in the prostate and the uterus.
Acknowledgements
We thank all the patients, who participated in the study; dr. Helle Karro for providing facilities for patient material collection at the Women's Clinic of Tartu University Hospital, Estonia; drs. Peeter Karits, (Nova Vita Clinic, Centre for Infertility Treatment and Medical Genetics), Tõnu Vooder (Lung Clinic of Tartu University Hospital) and Kristjan Välk (Department of Biotechnology, Institute of Molecular and Cell Biology, University of Tartu) for handling the clinical samples. We are thankful to the scientific and administrative staff at the Serono Reproductive Biology Institute, Rockland for providing bases for this collaboration study.
Funding
The study has been financed by Howard Hughes Medical Institute International Scholarship (grant #55005617) and International Senior Research Fellowship (grant no. 070191/Z/03/Z) in Biomedical Science in Central Europe for M.L. Additionally it has been by supported Estonian Science Foundation (grant no. 5796) and Estonian Ministry of Education and Science (core grants no. 0182721s06 and 0182641s04). K.R. is a recipient of scholarships from Kristjan Jaak Stipend Program and Estonian-American Andreas and Elmerice Traks Academic Fund. P.H. is a recipient of a stipend from the World Federation of Scientists' and L.U. is a recipient of stipend from the Estonian-Revelia Academic Fund.
References
- Bates SE, Longo DL. Tumor markers: the value and limitations in the management of cancer patients. Cancer Treat Rev. 1985;12:163–207. doi: 10.1016/0305-7372(85)90037-4. [DOI] [PubMed] [Google Scholar]
- Berger P, Kranewitter W, Madersbacher S, Gerth R, Geley S, Dirnhofer S. Eutopic production of human chorionic gonadotropin beta (hCG beta) and luteinizing hormone beta (hLH beta) in the human testis. FEBS Lett. 1994;343:229–233. doi: 10.1016/0014-5793(94)80561-x. [DOI] [PubMed] [Google Scholar]
- Berger P, Gruschwitz M, Spoettl G, Dirnhofer S, Madersbacher S, Gerth R, Merz WE, Plas E, Sampson N. Human chorionic gonadotropin (hCG) in the male reproductive tract. Mol Cell Endocrinol. 2007;260-262:190–196. doi: 10.1016/j.mce.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Bellet D, Lazar V, Bieche I, Paradis V, Giovangrandi Y, Paterlini P, Lidereau R, Bedossa P, Bidart JM, Vidaud M. Malignant transformation of nontrophoblastic cells is associated with the expression of chorionic gonadotropin beta genes normally transcribed in trophoblastic cells. Cancer Res. 1997;57:516–523. [PubMed] [Google Scholar]
- Bo M, Boime I. Identification of the transcriptionally active genes of the chorionic gonadotropin beta gene cluster in vivo. J Biol Chem. 1992;267:3179–3184. [PubMed] [Google Scholar]
- Bonduelle ML, Dodd R, Liebaers I, Van Steirteghem A, Williamson R, Akhurst R. Chorionic gonadotrophin-beta mRNA, a trophoblast marker, is expressed in human 8-cell embryos derived from tripronucleate zygotes. Hum Reprod. 1988;3:909–914. doi: 10.1093/oxfordjournals.humrep.a136808. [DOI] [PubMed] [Google Scholar]
- Braunstein GD. Placental proteins as tumor markers. Immunol Ser. 1990;53:673–701. [PubMed] [Google Scholar]
- Brizot ML, Jauniaux E, Mckie AT, Farzaneh F, Nicolaides KH. Placental mRNA expression of alpha and beta human chorionic gonadotrophin in early trisomy 18 pregnancies. Mol Hum Reprod. 1996;6:463–465. doi: 10.1093/molehr/2.6.463. [DOI] [PubMed] [Google Scholar]
- Burton GJ, Jauniaux E, Charnock-Jones DS. Human Early Placental Development: Potential Roles of the Endometrial Glands. Placenta Suppl A. 2007:S64–69. doi: 10.1016/j.placenta.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buyalos RP, Glassman LM, Rifka SM, Falk RJ, Macarthy PO, Tyson VJ, DiMattina M. Serum beta-human chorionic gonadotropin, estradiol and progesterone as early predictors of pathologic pregnancy. J Reprod Med. 1992;37:261–266. [PubMed] [Google Scholar]
- Cameo P, Srisuparp S, Strakova Z, Fazleabas AT. Chorionic gonadotropin and uterine dialogue in the primate. Reprod Biol Endocrinol. 2004;2:50–56. doi: 10.1186/1477-7827-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A, Roberts MR. Ets-2 and C/EBP-beta are important mediators of ovine trophoblast Kunitz domain protein-1 gene expression in trophoblast. BMC Mol Biol. 2007;8:14. doi: 10.1186/1471-2199-8-14. doi:10.1186/1471-2199-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirnhofer S, Berger C, Hermann M, Steiner G, Madersbacher S, Berger P. Coexpression of gonadotropic hormones and their corresponding FSH- and LH/CG-receptors in the human prostate. Prostate. 1998;35:212–220. doi: 10.1002/(sici)1097-0045(19980515)35:3<212::aid-pros7>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Dirnhofer S, Hermann M, Hittmair A, Hoermann R, Kapelari K, Berger P. Expression of the human chorionic gonadotropin-beta gene cluster in human pituitaries and alternate use of exon 1. J Clin Endocrinol. 1996;81:4212–4217. doi: 10.1210/jcem.81.12.8954017. [DOI] [PubMed] [Google Scholar]
- Duffy MJ. Role of tumor markers in patients with solid cancers: a critical review. Eur J Int Med. 2007;18:175–184. doi: 10.1016/j.ejim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Feng HC, Tsao SW, Ngan HYS, Kwan HS, Shih SM, Xue WC, Chiu PM, Chan KW, Cheung ANY. Differential Gene Expression Identified in Complete Hydatidiform Mole by Combining Suppression Subtractive Hybridization and cDNA Microarray. Placenta. 2006;27:521–526. doi: 10.1016/j.placenta.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Gerhard I, Runnebaum B. Predictive value of hormone determinations in the first half of pregnancy. Eur J Obstet Gynecol Reprod Bio. 1984;17:1–17. doi: 10.1016/0028-2243(84)90075-3. [DOI] [PubMed] [Google Scholar]
- Ghosh D, Ezashi T, Ostrowski MC, Roberts RM. A central role for Ets-2 in the transcriptional regulation and cyclic adenosine 5′-monophosphate responsiveness of the human chorionic gonadotropin-beta subunit gene. Mol Endocrinol. 2003;17:11–26. doi: 10.1210/me.2002-0223. [DOI] [PubMed] [Google Scholar]
- Hallast P, Nagirnaja L, Margus T, Laan M. Segmental Duplications and Gene Conversion: Human Luteinizing Hormone/ Chorionic Gonadotropin Beta Gene Cluster. Genome Res. 2005;15:1535–1546. doi: 10.1101/gr.4270505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallast P, Rull K, Laan M. The evolution and genomic landscape of CGB1 and CGB2 genes. Mol Cell Endocrinol. 2007;260-262:2–11. doi: 10.1016/j.mce.2005.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr F, Baal N, Reisinger K, Lorenz A, McKinnon T, Preissner KT, Zygmunt M. HCG in the regulation of placental angiogenesis. Results of an in vitro study. Placenta Suppl A. 2007:S85–93. doi: 10.1016/j.placenta.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Hirotsune S, Yoshida N, Chen A, Garrett L, Sugiyama F, Takahashi S, Yagami K, Wynshaw-Boris A, Yoshiki A. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature. 2003;423:91–96. doi: 10.1038/nature01535. [DOI] [PubMed] [Google Scholar]
- Ing NH. Steroid hormones regulate gene expression posttranscriptionally by altering the stabilities of Messenger RNAs. Biol Reprod. 2005;72:1290–1296. doi: 10.1095/biolreprod.105.040014. [DOI] [PubMed] [Google Scholar]
- Jurisicova A, Antenos M, Kapasi K, Meriano J, Casper RF. Variability in the expression of trophectodermal markers beta-human chorionic gonadotrophin, human leukocyte antigen-G and pregnancy specific beta-1 glycoprotein by the human blastocyst. Hum Reprod. 1999;14:1852–1858. doi: 10.1093/humrep/14.7.1852. [DOI] [PubMed] [Google Scholar]
- Kayisli UA, Selam B, Guzeloglu-Kayislim O, Demir R, Arici A. Human chorionic gonadotropin contributes to maternal immunotolerance and endometrial apoptosis by regulating Fas-Fas ligand system. J Immunol. 2003;171:2305–2313. doi: 10.4049/jimmunol.171.5.2305. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Kelso J, Franz H, Visagie J, Giger T, Joerchel S, Petzold E, Green RE, Lachmann M, Paabo S. Functionality of intergenic transcription: an evolutionary comparison. PLoS Genet. 2006;2:e171. doi: 10.1371/journal.pgen.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King BF. Development and structure of the placenta and fetal membranes of nonhuman primates. J Exp Zool. 1993;266:528–540. doi: 10.1002/jez.1402660605. [DOI] [PubMed] [Google Scholar]
- Knöfler M. Regulation of HCG during normal gestation and in pregnancies affected by Down's syndrome. Mol Hum Reprod. 1999;5:895–897. doi: 10.1093/molehr/5.10.895. [DOI] [PubMed] [Google Scholar]
- Korneev SA, Park JH, O'Shea M. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J Neurosci. 1999;19:7711–7720. doi: 10.1523/JNEUROSCI.19-18-07711.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letterie GS, Hibbert M. Serial serum human chorionic gonadotropin (hCG) levels in ectopic pregnancy and first trimester miscarriage. Arch Gynecol Obstet. 2000;263:168–169. doi: 10.1007/s004040050275. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;4:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lyall F. The human placental bed revisited. Placenta. 2002;23:555–562. doi: 10.1053/plac.2002.0850. [DOI] [PubMed] [Google Scholar]
- Massin N, Frendo JL, Guibourdenche J, Luton D, Giovangrandi Y, Muller F, Vidaud M, Evain-Brion D. Defect of syncytiotrophoblast formation and human chorionic gonadotropin expression in Down's syndrome. Placenta Suppl A. 2001:S93–97. doi: 10.1053/plac.2001.0658. [DOI] [PubMed] [Google Scholar]
- Maston GA, Ruvolo M. Chorionic gonadotropin has a recent origin within primates and an evolutionary history of selection. Mol Biol Evol. 2002;19:320–335. doi: 10.1093/oxfordjournals.molbev.a004085. [DOI] [PubMed] [Google Scholar]
- Miller-Lindholm AK, LaBenz CJ, Ramey J, Bedows E, Ruddon RW. Human chorionic gonadotropin-beta gene expression in first trimester placenta. Endocrinology. 1997;138:5459–5465. doi: 10.1210/endo.138.12.5618. [DOI] [PubMed] [Google Scholar]
- Pierce JG, Parsons TF. Glycoprotein hormones: structure and function. Annu Rev Biochem. 1981;50:465–495. doi: 10.1146/annurev.bi.50.070181.002341. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer T, Koczan D, Briese V, Friese K, Richter D, Thiesen HJ, Jeschke U. Absolute quantification of human chorionic gonadotropin-beta mRNA with TaqMan detection. Mol Biotechnol. 2000;14:47–57. doi: 10.1385/mb:14:1:47. [DOI] [PubMed] [Google Scholar]
- Roberts RM, Ezashi T, Rosenfeld CS, Ealy AD, Kubisch HM. Evolution of the interferon tau genes and their promoters, and maternal-trophoblast interactions in control of their expression. Reprod Suppl. 2003;61:239–251. [PubMed] [Google Scholar]
- Rull K, Laan M. Expression of β-subunit of human chorionic gonadotropin genes during the normal and failed pregnancy. Hum Reprod. 2005;20:3360–3368. doi: 10.1093/humrep/dei261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppälä M, Iino K, Rutanen EM. Placental proteins in oncology. Clin Obstet Gynaecol. 1986;13:593–610. [PubMed] [Google Scholar]
- Srisuparp S, Strakova Z, Fazleabas AT. The role of chorionic gonadotropin (CG) in blastocyst implantation. Arch Med Res. 2001;32:627–634. doi: 10.1016/s0188-4409(01)00330-7. [DOI] [PubMed] [Google Scholar]
- Staton JM, Thomson AM, Leedman PJ. Hormonal regulation of mRNA stability and RNA-protein interactions in the pituitary. J Mol Endocrinol. 2000;25:17–34. doi: 10.1677/jme.0.0250017. [DOI] [PubMed] [Google Scholar]
- Steier JA, Bergsjø P, Myking OL. Human chorionic gonadotropin in maternal plasma after induced abortion, spontaneous abortion, and removed ectopic pregnancy. Obstet Gynecol. 1984;64:391–394. [PubMed] [Google Scholar]
- Talmadge K, Vamvakopoulos NC, Fiddes JC. Evolution of the genes for the beta subunits of human chorionic gonadotropin and luteinizing hormone. Nature. 1984;307:37–40. doi: 10.1038/307037a0. [DOI] [PubMed] [Google Scholar]
- Toth P, Lukacs H, Gimes G, Sebestyen A, Pasztor N, Paulin F, Rao CV. Clinical importance of vascular LH/hCG receptors - a review. Reprod Biol. 2001;1:5–11. [PubMed] [Google Scholar]
- Zygmunt M, Herr F, Keller-Schoenwetter S, Kunzi-Rapp K, Munstedt K, Rao CV, Lang U, Preissner KT. Characterization of human chorionic gonadotropin as a novel angiogenic factor. J Clin Endocrinol Metab. 2002;87:5290–5296. doi: 10.1210/jc.2002-020642. [DOI] [PubMed] [Google Scholar]