Abstract
Type B gelatin-based engineered nanocarrier systems (GENS) have been used over the last several years as a non-condensing systemic and oral DNA delivery system. In this study, we have modified the surface of GENS with epidermal growth factor receptor (EGFR)-targeting peptide for gene delivery and transfection in pancreatic cancer cell lines. GENS were prepared by the solvent displacement method and the EGFR-targeting peptide was grafted on the surface using a hetero-bifunctional poly(ethylene glycol) (PEG) spacer. Plasmid DNA, encoding for enhanced green fluorescent protein (GFP), was efficiently encapsulated and protected from degrading enzymes in the control and surface-modified GENS. Upon incubation with EGFR over-expressing Panc-1 human pancreatic adenocarcinoma cells, the peptide-modified nanoparticles were found to be internalized efficiently by receptor-mediated endocytosis. Both quantitative and qualitative transgene expression efficiencies were significantly enhanced when plasmid DNA was administered with EGFR-targeted GENS relative to the control-unmodified gelatin or PEG-modified gelatin nanoparticle systems. Based on these preliminary results, EGFR-targeted GENS show tremendous promise as a safe and effective gene delivery vector with the potential to treat pancreatic cancer.
Key words: EGFP-N1 plasmid DNA, EGFR-targeted delivery, Panc-1 human pancreatic adenocarcinoma cells, transgene expression, type B gelatin nanoparticles
INTRODUCTION
Pancreatic cancer is the fourth leading cause of cancer-related deaths in the adult US population affecting as many as 32,000 patients per year (1). The disease is especially aggressive with an average 5-year survival rate of less than 5% and a median duration of survival of only 6 months after initial diagnosis (2). The poorly understood etiology, lack of early diagnostic methods, and a severe deficiency in effective treatment options for advanced stage disease, which has disseminated to the vital organs of the peritoneal cavity, are some of the critical factors that contribute to the high mortality rate associated with pancreatic cancer.
Cancer gene therapy comprises transfer of genetic constructs intended to alter the neoplastic phenotype of tumor cells and/or to provide additional protective effects. There are currently four main strategies for cancer gene therapy divided as follows: (1) enhancement of tumor suppressor function by introduction of wild-type p53 or other suppressor genes, (2) suicide gene therapy using DNA encoding for enzymes that activate prodrugs (e.g., herpes simplex virus thymidine kinase activation of ganciclovir), (3) augmentation of immune function using various cytokine and chemokine encoding genes, and (4) inhibition of tumor neovasculature by anti-angiogenic gene therapy (3–10). Pancreatic cancer can significantly benefit from gene therapy, especially when combined with cytotoxic and/or anti-angiogenic drugs to minimize metastatic potential (11,12).
The major challenge in systemic gene therapy is in the development of a suitable vector system that can deliver the gene of interest into the cell and allow for efficient and long-lived transfection (13). Viral and non-viral vectors are being examined for systemic gene delivery to target tissues and cells of interest. Despite their high transfection efficiency, viral vectors are plagued by significant toxicity such as fatal immunogenic responses and integration with host chromosome (14–16). As an alternative, non-viral vectors are attractive due to their flexibility in terms of the size and type of the nucleic acid construct that can be delivered. An additional important advantage of non-viral delivery systems is the promise of safety, especially when designed with biocompatible lipids and polymers. However, non-viral vectors in general do not demonstrate the same transfection efficiencies as their viral counterparts after systemic administration (17,18). Certain cationic lipids and polymeric delivery systems for plasmid DNA are also prone to significant toxicity issues (17,19).
Over the last several years, our group has investigated the potential of non-condensing type B gelatin-based engineered nanocarrier systems (GENS) for systemic and oral gene therapy (20–29). Type B gelatin, isolated from bovine or porcine collagen and hydrolyzed with a base (e.g., calcium hydroxide), has an isoelectric point of 4.5–5.5. As such, at pH 7.0, the negatively charged biopolymer can physically encapsulate reporter and therapeutic nucleic acid constructs as opposed to positively charged lipids and polymer that electrostatically condense DNA. The physically encapsulated plasmid DNA in a hydrogel-type matrix is protected in the systemic circulation and upon cellular transport. Additionally, the released plasmid DNA having a supercoiled structure at the nuclear membrane is critical for efficient uptake and transfection, especially in non-dividing cells (30). When poly(ethylene glycol) (PEG)-modified GENS were used for in vitro and in vivo delivery of reporter plasmid DNA (i.e., green fluorescent protein and beta-galactosidase expressing), there was over 60% transfection efficiency in vitro in NIH-3T3 murine fibroblast cells after 96 h of transgene administration and high levels of gene products in the tumor mass following intravenous and intratumoral delivery in Lewis lung carcinoma-bearing C57BL6/J mice (24). Moreover, we have examined the delivery of plasmid DNA encoding for the soluble form of human vascular endothelial growth factor receptor-1 (sVEGFR-1 or sFlt-1) in PEG-modified thiolated gelatin nanoparticles in an orthotopic MDA-MB-435 human breast adenocarcinoma xenograft established model in female Nu/Nu mice (26–29). Similar to the results with reporter plasmid, the sFlt-1 encoding plasmid DNA also was shown to transfect in the tumor mass upon intravenous administration. The expressed sFlt-1 was therapeutically active in suppressing tumor growth due to inhibition of neo-angiogenesis for up to 28 days following a single 60 μg plasmid DNA dose when administered with PEG-modified thiolated gelatin nanoparticles (29). In both of these gene delivery strategies, we have utilized passive tumor targeting with PEG-modified GENS based on the leaky tumor vascular and enhanced permeability and retention (EPR) effect.
Since pancreatic adenocarcinoma is known to have a high stromal content and is not as well vascularized as other solid tumors (e.g., breast adenocarcinoma), we hypothesize that a combination of passive and active targeted DNA delivery using epidermal growth factor receptor (EGFR)-targeted peptide surface modification of GENS will lead to enhanced transgene expression. More than 90% of human pancreatic cancers are known to over-express human epidermal growth factor receptor (HER) family. The HER family includes EGFR (or erbB-1), HER2 (or erbB-2), HER3 (or erbB-3), and HER4 (or erbB-4). EGFR expression in pancreatic cancer has been implicated in disease aggressiveness based on enhanced proliferation, metastasis, and resistance to chemo- and radiotherapies (31). Also, the increased level of EGFR expression in pancreatic cancer has been associated with advanced tumor stage, poor prognosis, and significantly shorter patient survival (32).
Specific cellular uptake of target-specific nanocarriers occurs through receptor-mediated endocytosis, where binding with the cell surface receptor leads to internalization of the entire nanocarrier–receptor complex and vesicular transport through the endosomal/lysosomal pathway (18). Based on the pioneering work of Rouslahti’s group, a number of peptide-based targeting ligands have been identified by screening phage display libraries (33). Peptide ligands, which have the advantage of high avidity of interaction with the target receptor through multiple points of contact, low immunogenicity, and easier surface modification of the nanocarrier systems, are being pursued by a number of groups as targeting moiety for cell-specific delivery (34). These peptide ligands have been conjugated to nanoparticle surface for targeted delivery to the endothelial cells on tumor neovasculature (e.g., RGD) or directly to tumor cells (35).
In the present study, we have fabricated EGFR-targeted peptide-modified type B GENS for encapsulation of reporter plasmid DNA and have examined the potential for delivery and transfection in human pancreatic cancer cells. EGFR-specific peptide was conjugated to the GENS using a PEG spacer for long circulation time in the systemic circulation, passive targeted delivery by the EPR effect, and to allow for maximum flexibility of the peptide binding with the receptor on cell surface. Plasmid DNA encoding for enhanced green fluorescent protein (EGFP-N1) was encapsulated in the control and peptide-modified GENS and the quantitative and qualitative transfection efficiencies were measured in Panc-1 human pancreatic adenocarcinoma cells.
MATERIALS AND METHODS
Materials
Type B gelatin (2 bloom strength) with an average molecular weight of 40,000–50,000 Da, 100–115 mmol of free carboxylic acid per 100 g of protein, and an isoelectric point of 4.8–5.0 was purchased from Sigma-Aldrich Chemical Company (St. Louis, MO, USA). Monomethoxy-poly(ethylene glycol)-succinimidyl carboxymethyl (mPEG-SCM, MW 2,000 Da) and malemide-PEG-succinimidyl carboxymethyl (MAL-PEG-SCM, MW 2,000 Da) were purchased from Laysan Bio, Inc. (Arab, AL, USA). Four sequences of HER-targeting peptides, either with fluorescein isothiocyanate (FITC) groups or with C-G-G- spacer at the amino terminus were custom synthesized at Tufts University’s Peptide Synthesis Core Facility (Boston, MA, USA). EGFP-N1 reporter plasmid DNA (4.7 kB, Clontech), expressing enhanced green florescent protein (GFP), was purified by Elim Biopharmaceuticals, Inc. (Hayward, CA, USA). Lipofectin®, a cationic lipid transfection reagent, was purchased from Invitrogen (Carlsbad, CA, USA). Absolute ethyl alcohol (200 proof, 99.5% ACS reagent) was obtained from Acros Chemicals (Pittsburgh, PA, USA). Glyoxal 40% (w/v) and glycine were purchased from Fisher Scientific (Fair Lawn, NJ, USA). CellTiter 96® AQueous one solution cell proliferation assay was obtained from Promega Corporation (Madison, WI, USA). Fluorescence-free mounting medium, FluoromountG®, was obtained from Southern Biotech Associates (Birmingham, AL, USA). The human pancreatic and ovarian adenocarcinoma (i.e., Panc-1, Capan-1, and SKOV3) and murine fibroblast (i.e., NIH-3T3) cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA).
Preparation of Plasmid DNA-Encapsulated Nanoparticles
EGFP-N1 plasmid DNA was encapsulated in type B gelatin nanoparticles by the solvent displacement method under controlled temperature, pH, and stirring conditions as previously described (26). The final DNA concentration in gelatin solution was maintained constant at 0.5% (w/w). Following slow addition of dehydrated ethanol, precipitation, and formation of DNA-containing gelatin nanoparticles in the hydroalcoholic solvent system, 0.1 mL of 40% (v/v) glyoxal was added to crosslink the particles and the excess aldehyde groups were quenched with 0.2 M glycine. The formed crosslinked nanoparticles were isolated by centrifugation at 30,000 rpm for 45 min using a Beckman-Coulter ultracentrifuge equipped with a Ti70 rotor. The supernatant was discarded and the nanoparticle pellet was washed and freeze-dried.
Nanoparticle Surface Modification with PEG and Peptide
The surface of control and plasmid DNA-encapsulated type B gelatin nanoparticles was modified with mono-functional mPEG-SCM or hetero-bifunctional MAL-PEG-SCM. For mPEG grafting on gelatin nanoparticles, the pellet or freeze-dried powder was re-suspended in pH 7.4 phosphate buffer and incubated with different concentrations of mPEG-SCM under stirring conditions at room temperature for up to 2 h. To covalently attach EGFR-targeting peptide on the nanoparticle surface through a PEG spacer, MAL-PEG-SCM was reacted with gelatin nanoparticles and the peptide sequence was conjugated through the sulfhydryl group of the terminal cysteine residue in the C-G-G- spacer with maleimide group of PEG derivative. In order to achieve successful surface modification with PEG and peptide, two different methods of grafting were considered: (1) addition of the peptide with MAL-PEG-SCM and one-step PEG surface modification and peptide attachment to the nanoparticles or (2) sequential reaction of MAL-PEG-SCM to gelatin nanoparticles, centrifugation and washing of excess PEG derivative, followed by peptide conjugation. The final concentration of EGFR peptide for conjugation to the nanoparticle surface varied from 1 mg of peptide to 10 mg of peptide per 50 mg of nanoparticles. Following mPEG and EGFR-targeted peptide conjugation to GENS, the surface-modified nanoparticles were harvested by centrifugation and washing steps and freeze-dried.
Characterization of the Control and Surface-Functionalized Nanoparticles
Particle Size Analysis
The mean particle size and size distribution of control and plasmid DNA-encapsulated unmodified gelatin nanoparticles, PEG-modified gelatin nanoparticles, and EGFR peptide-conjugated gelatin nanoparticles were determined using ZetaPALS, 90Plus® (Brookhaven Instruments Corporation, Holtsville, NY, USA). This instrument uses light scattering principle to determine the effective hydrodynamic diameter of the nanoparticles in aqueous dispersion. The suspension of the nanoparticles was diluted with deionized distilled water and the particle size analysis was carried out at a scattering angle of 90° and at 25°C.
Surface Charge Measurements
The surface charge (zeta potential (ξ)) measurements of the control and plasmid DNA-encapsulated unmodified gelatin nanoparticles, PEG-modified gelatin nanoparticles, and EGFR peptide-conjugated gelatin nanoparticles in suspension of deionized water were performed using a Brookhaven Instrument’s ZetaPALS (Holtsville, NY, USA). The nanoparticles were dispersed in deionized water and the zeta potential values were measured at the default parameters of dielectric constant, refractive index, and viscosity of water. The zeta potential values were determined based on the electrophoretic mobility using the Smoluchowski–Helmholtz equation.
Scanning Electron Microscopy
Freeze-dried samples of unmodified gelatin nanoparticles, PEG-modified gelatin nanoparticles, and EGFR peptide-conjugated gelatin nanoparticles were mounted on an aluminum sample mount and sputter-coated with gold–palladium to enhance conductivity, minimize the buildup of surface charge, and improve backscatter signal. The samples were then observed with a Hitachi S-4800 (Pleasanton, CA, USA) field emission scanning electron microscope at the accelerating voltage of 3.0 kV and working distance of 8 mm.
Determination of Plasmid DNA Encapsulation Efficiency
In order to determine the encapsulation efficiency of plasmid DNA in unmodified, PEG-modified, and EGFR peptide-modified gelatin nanoparticles, the freeze-dried nanoparticle sample was dispersed in phosphate-buffered saline (PBS, pH 7.4) with 0.2 mg/mL protease and incubated at 37°C for up to 30 min. Following complete digestion of the gelatin matrix, the released plasmid DNA was assayed using PicoGreen® fluorescent reagent (Invitrogen) for double-stranded DNA quantitation according to the manufacturer’s instructions. It is important to note that PicoGreen® fluorescence emission does not correlate with functionality of plasmid DNA.
Stability of Encapsulated Plasmid DNA by Agarose Gel Electrophoresis
The stability of the encapsulated plasmid DNA in the control and surface-modified nanoparticles was evaluated in the presence of protease (i.e., polymer matrix degrading enzyme) and DNAse, an endonuclease that catalyzes the degradation of both single- and double-stranded DNA, as described before (26). The control and functionalized nanoparticles were treated with 0.2 mg/mL protease and 0.2 U of DNAse, for 30 and 20 min, respectively, at 37°C. In some experiments, the nanoparticles were subjected to protease followed by either sequential or simultaneous DNAse treatment to show the degradation of released DNA or DNAse followed by protease treatment to show that the encapsulated plasmid DNA was not accessible to the endonucleases and remained intact. The activity of the DNAse was quenched with millimolar EDTA, which chelates the Ca+2 and Mg+2 ions that are required for the functioning of the enzyme. The resulting nanoparticulate solutions were diluted appropriately and mixed with fivefold diluted gel loading buffer. The samples were then loaded onto a 0.8% (w/v) pre-cast agarose gel (E-Gel®, Invitrogen) pre-stained with ethidium bromide at an equivalent DNA concentration of 100 ng/well in a 15-μL volume per well. The gels were run at 65 V for 15 min using a BioRad™ model 200/2 power supply, and the bands were visualized using a Kodak Gel-logic® 100 imaging system (Carestream Health, Rochester, NY, USA).
Baseline EGFR Expression in Cells
Two human pancreatic adenocarcinoma cell lines (Panc-1 and Capan-1) were analyzed by immunocytometry and Western blot techniques. Human ovarian adenocarcinoma (SKOV3) and murine fibroblast ((NIH-3T3) cells were used as positive and negative controls, respectively. Briefly, the cells were grown in fetal bovine serum-supplemented Dulbecco’s modified Eagle’s medium (DMEM; Panc-1, Capan-1, and NIH-3T3) or RPMI-1640 (SKOV3). After blocking non-specific binding, the fixed cells were incubated with 1:100 diluted primary rabbit EGFR antibody (Cell Signaling Technology, Danvers, MA, USA) for an hour at room temperature. The coverslips were then rinsed twice with washing buffer. Endogenous peroxidase activity was blocked and the cells were incubated with 1:200 horse-radish peroxidase-conjugated goat anti-rabbit secondary antibody (Cell Signaling Technology) and treated with diaminobenzidine (DAB) solution. In EGFR-positive cells, the peroxidase-catalyzed reaction results in the formation of dark-brown precipitates. The stained cells were then observed with a bright-field microscope at ×20 original magnification.
For Western blot analysis, the cell lysate was analyzed for total protein concentration using NanoOrange® (Invitrogen) protein quantitation kit. Twenty-five micrograms of total protein extract was run on a pre-cast sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) system at 150 V for 90 min. Subsequently, the protein bands on the gel were transferred onto a nitrocellulose membrane and blocked with 3% milk in Tween®-containing Tris buffer saline (TBS-t) for 1 h. Beta-actin from Panc-1 cells was used as a protein loading control. The membrane was incubated with 1:1,000 dilution of the primary rabbit EGFR antibody and primary rabbit beta-actin antibody overnight at 4°C. The membrane was then washed twice with TBS-t and incubated with 1:2,000 dilutions of the secondary anti-rabbit horse-radish peroxidase-conjugated IgG in TBS-t for 1 h at room temperature. After rinsing the excess secondary antibody with TBS-t and water, the ECL substrate (Pierce, Rockford, IL, USA) was added that is cleaved by peroxidase to give a chemiluminescent product. The chemiluminescent bands were then visualized using the Kodak Gel-logic® imaging system.
Selection of EGFR Peptide for Surface Modification
Four different peptide sequences were selected for comparison of their binding specificity to the human epidermal receptor family. The sequences of the selected peptides (from NH2 to COOH terminus) were: peptide-1 (i.e., G-G-R-S-V-G-L-G-R-L) as a negative control, peptide-2 (i.e., Y-C-D-G-F-Y-A-C-Y-M-D-V) (36), peptide-3 (i.e., A-A-E-E-I-Y-A-A-R-R-G) (37), and peptide-4 (i.e., Y-H-W-Y-G-Y-T-P-Q-N-V-I) (34). Peptides 2 and 3 are specific binders of HER2/neu receptor, while peptide 4 interacts specifically with EGFR. Panc-1 cells (3 × 105 cells per well) were grown in six-well plates in DMEM culture medium to achieve 80% confluency. Glass coverslips were added to some wells for florescence microscopy purposes. The cells were incubated with 10 μM concentrations of FITC-labeled peptides 1–4 in serum-free medium for 20 min. At the end of the incubation period, cells were rinsed twice with cold PBS, harvested, and fixed for flow cytometric analysis. Panc-1 cells on the coverslips were rinsed with cold PBS and fixed on glass slides with FluoromountG® for florescence microscopy.
Nanoparticle Cytotoxicity Profiles in Panc-1 Cells
The effect of unmodified, PEG-modified, and EGFR-targeted peptide (i.e., peptide-4 with full sequence C-G-G-Y-H-W-Y-G-Y-T-P-Q-N-V-I)-modified gelatin nanoparticles on the viability of Panc-1 human pancreatic adenocarcinoma cells was examined by incubating the cells with different nanoparticle concentrations ranging from 0 to 10 mg/mL. Untreated Panc-1 cells were used as negative control and poly(ethyleneimine) (MW 10,000 Da), a cytotoxic cationic polymer, was used as a positive control. Panc-1 cells were grown to 80% confluency in 96-well microplates and the regular cell culture medium was exchanged with serum-free media. The control and nanoparticle formulation were added and incubated for 4 h at 37°C. The cytotoxicity of the nanoparticle formulations, determined using the Promega’s (Madison, WI, USA) CellTiter 96® AQueous non-radioactive cell proliferation assay, is based on the principle that the mitochondrial dehydrogenase enzyme in metabolically active cells population converts the tetrazolium compound of MTS reagent to a purple colored formazan salt that is soluble in the culture medium. The absorbance of soluble formazan was measured at 490 nm with a Bio-Tek Synergy® HT plate reader (Winooski, VT, USA) and converted into percent cell viability relative to untreated control cells.
Cellular Uptake and Trafficking Studies
To evaluate the specificity of EGFR-mediated nanoparticle uptake and cellular distribution, the unmodified gelatin, PEG-modified gelatin, and EGFR-targeted peptide-modified gelatin nanoparticles were prepared as described above. However, instead of plasmid DNA, rhodamine-labeled dextran (MW 10,000 Da) was incorporated into the nanoparticles at 0.5% (w/w) concentration. Panc-1 human pancreatic adenocarcinoma cells were grown in culture on glass coverslips to 80% confluency. The fluorescently labeled nanoparticle formulations were incubated with Panc-1 cells for various time intervals ranging from 1 to 8 h at 37°C in serum-free media. To determine the specificity of EGFR peptide-modified nanoparticle interaction with Panc-1 cells, 10 μM soluble peptide was added 20 min prior to the addition of nanoparticles. The final concentrations of control and surface-modified gelatin nanoparticles (as adjusted by the loading efficiency) were 2 and 2.3 mg/mL respectively. Following the duration of incubation, excess nanoparticles in serum-free media were removed and the coverslip samples were washed three times with sterile PBS to remove any surface-bound nanoparticles. The coverslips were mounted on a clean glass slide with Fluoromount-G®. Differential interference contrast and epifluorescence microscopy was done using an Olympus 1X51 microscope system at ×40 original magnification.
Quantitative and Qualitative Transgene Expressions in Panc-1 Cells
Quantitative Expression Analysis
Panc-1 cells were grown in six-well cell culture plates containing to a density of 200,000 cells per well. EGFP-N1 plasmid DNA, encoding for enhanced green fluorescence protein, was administered in the unmodified, PEG-modified, and EGFR peptide-modified gelatin nanoparticles. Twenty micrograms of EGFP-N1 plasmid DNA complexed with 20 μL of Lipofectin®, a cationic lipid transfection reagent, was used as a positive control, and untreated cells were used as a negative control. After filtration, the DNA-containing nanoparticle suspension was added at a concentration equivalent to 20 μg of plasmid DNA per well and incubated with the cells at 37°C for a period of 4 h. Serum-free media was removed from each well and replaced with the standard growth media. Periodically after 24 h and up to 96 h post-administration, the Panc-1 cells were harvested and fixed with 4% formaldehyde solution. The fixed cell samples were placed in sheath fluid and analyzed for transfection on a FACScaliber flow cytometer (BD Biosciences, San Jose, CA, USA). GFP expression was detected using the FL-1 channel (530/30 emission). Data were analyzed using the Cell Quest Pro® software. The percent transfection efficiency was determined from the fluorescence emission of the gated GFP-positive cells relative to the total number of cells analyzed.
Additionally, after incubation with the control and surface-modified nanoparticles for different time intervals, the cells were lysed and the intracellular GFP concentrations were determined by an enzyme-linked immunoassay (ELISA). Using React-Bind® anti-GFP antibody-coated plates (Pierce, Rockford, IL, USA) and adding a GFP secondary antibody reactive to alkaline phosphatase, very low levels of GFP can be detected. Briefly, the cell supernatants were incubated with the anti-GFP coated plates for 24 h at 4°C, after washing with PBS–0.5% (w/v) Tween®-80, and GFP secondary antibody was added to each plate and was allowed to incubate for 2 h. After washing with PBS–Tween-80®, alkaline phosphatase was added and the product formed was measured with a Bio-Tek Synergy HT (Winooski, VT, USA) plate reader. NanoOrange® kit (Invitrogen) assay was again used to measure the total cellular protein and the expressed GFP concentration was reported as nanogram per milligram of total protein.
Qualitative Expression Analysis
Panc-1 cells were grown in six-well cell culture plates containing a glass coverslip, and EGFP-N1 plasmid DNA-encapsulated unmodified, PEG-modified, and EGFR peptide-modified gelatin nanoparticles were administered at a 20-μg DNA dose per well. Twenty micrograms of EGFP-N1 plasmid DNA complexed with 20 μL of Lipofectin®, a cationic lipid transfection reagent, was used as a positive control and untreated cells were used as a negative control. After 6 h, serum-free media was removed from each well and replaced with the standard growth media. After 24 to 96 h post-administration of plasmid DNA, the coverslips were washed three times with sterile PBS and mounted on a microscopic slide containing Fluoromount-G®. GFP expression in Panc-1 cells was observed by using an Olympus 1X51 fluorescence microscope at ×40 original magnification.
Data Analysis
All results were obtained from sample sizes of four or greater and were expressed as mean ± standard error of the mean. Statistical analyses were performed using a two-sample t test to measure statistical significance between pairs of results and p < 0.05 was considered to be significant.
RESULTS AND DISCUSSION
Preparation and Characterization of Control and Surface-Modified GENS
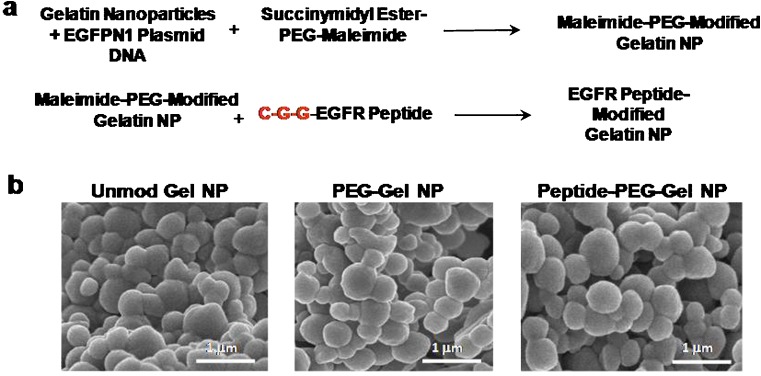
Using an optimized protocol based on pH- and temperature-controlled solvent exchange between water and ethanol, gelatin nanoparticles of less than 200 nm in diameter were reproducibly synthesized. Figure 1a shows the sequential two-step reaction for the preparation of EGFR peptide-modified GENS. In the first step, the reactive amines on the surface of the gelatin nanoparticles react with the succinimidyl-carboxymethyl (SCM) group of the heterobifunctional maleimide-PEG-SCM resulting in maleimide-PEG-modified GENS. In the second step, the maleimide group on the surface was conjugated with the sulfhydryl functionality of the terminal cysteine residue of the C-G-G- spacer in the peptide sequence. Further stabilization of the control and EGFR peptide-modified targeted nanoparticles was achieved by crosslinking the GENS with glyoxal. The SEM micrographs in Fig. 1b show spherical smooth nanoparticles of the controls and EGFR peptide-modified nanoparticles.
Fig. 1.
a Chemical reaction scheme illustrating surface modification of type B gelatin nanoparticles with epidermal growth factor receptor (EGFR) binding peptide through a poly(ethylene glycol) (PEG) spacer and b scanning electron microscopy of control, PEG-modified, and EGFR-targeted gelatin nanoparticles
Table I shows the hydrodynamic particle diameter, surface charge (zeta potential), and DNA encapsulation efficiency of the control and EGFR-targeted gelatin nanocarriers. The average hydrodynamic diameters of the control, PEG-modified, and EGFR peptide-modified GENS were between 150 and 180 nm with a narrow size distribution. Inclusion of plasmid DNA and surface modification with PEG and EGFR peptide did not significantly affect the particle size. Previous studies have shown that long-circulating PEG-modified nanoparticles of less than 200 nm can be efficiently delivered to tumor mass upon systemic administration due to the EPR effect. We anticipate that the EGFR peptide-modified nanoparticles will also extravasate out of the tumor neovasculature and achieve both passive and active targeted delivery when administered in vivo. Surface charge of the control and surface-modified DNA-encapsulated GENS was between −15 to −19 mV. These results are consistent with our previous findings and confirm that the plasmid was physically encapsulated in the nanocarrier matrix and not adsorbed on the surface. The DNA loading efficiency of control, PEG-modified, and EGFR peptide-modified GENS was >97% when the plasmid was added at 0.5% (w/w) in gelatin solution. Surface modification with PEG and EGFR peptide did not affect the DNA loading efficiency.
Table I.
Particle Size, Surface Charge, and Plasmid DNA Encapsulation Efficiency of Control and EGFR-Targeted Gelatin Nanoparticles
| Nanoparticle system | Hydrodynamic diameter (nm) | Zeta potential (mV) | Plasmid DNA encapsulation efficiency (%) |
|---|---|---|---|
| Blank gelatin nanoparticles | 124 ± 13.0a | −13.5 ± 0.8 | – |
| Plasmid DNA-encapsulated gelatin nanoparticles | 145 ± 16.5 | −14.7 ± 2.3 | 99.0 ± 0.82 |
| Plasmid DNA-encapsulated PEG-modified gelatin nanoparticles | 172 ± 9.0 | −15.3 ± 1.8 | 99.0 ± 0.40 |
| Plasmid DNA-encapsulated EGFR-targeted gelatin nanoparticles | 185 ± 8.0 | −19.0 ± 2.1 | 97.0 ± 0.65 |
aMean ± SE (n = 4)
Stability of the Encapsulated Plasmid DNA
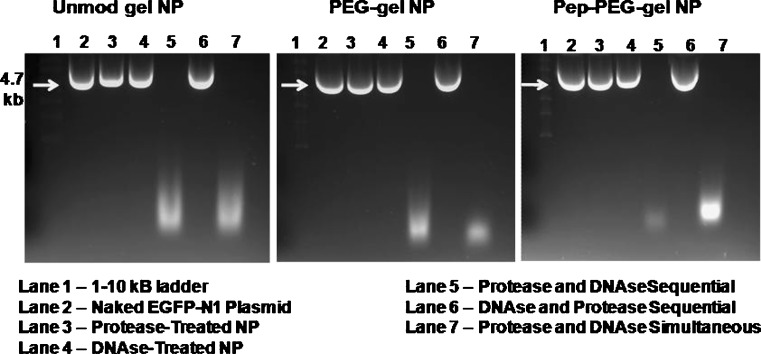
Agarose gel electrophoresis was used to examine the stability of encapsulated plasmid DNA due to process condition as well as after exposure to protease and DNAse. Figure 2 shows the electrophoretic mobility pattern of DNA from unmodified gelatin, PEG-modified gelatin, and peptide-modified gelatin nanoparticles. Intact supercoiled plasmid DNA was recovered from all of the nanoparticle formulations after treatment with protease as shown in lane 3. Similarly, when the nanoparticles were first treated with DNAse and then the plasmid was extracted by proteolytic digestion of the gelatin matrix, intact supercoiled DNA was again harvested from the system as shown in lane 4. On the other hand, when the nanoparticles were first treated with protease and the extracted DNA was treated with DNAse, as shown in lane 5, there was complete degradation of the plasmid. Lane 6 again shows protection of the encapsulated DNA when nanoparticles were exposed to DNAse and then to protease, and finally in lane 7, the plasmid is degraded when both protease and DNAse were added together with the nanoparticles. These results show that the nanoparticles can physically encapsulate and protect the plasmid from nucleases during systemic circulation and upon cellular transport.
Fig. 2.
Agarose gel electrophoresis to evaluate the stability of encapsulated plasmid DNA in control and epidermal growth factor receptor (EGFR)-targeted peptide modified type B gelatin-based engineered nanoparticles (GENS). The nanoparticle samples were digested with protease and treated with DNAse followed by running the extracted DNA on 1.2% agarose gels at 70 V for 30 min. Lane 1 has 1–10 kB DNA ladder, lane 2 had intact naked EGFP-N1 plasmid DNA, lane 3 had intact plasmid DNA extracted from GENS after protease digestion, lane 4 had plasmid DNA extracted from DNAse treated intact nanoparticles, lane 5 had plasmid DNA extracted from protease-treated GENS and then sequentially treated with DNAse, lane 6 had plasmid DNA extracted after DNAse treatment of intact nanoparticles and then sequentially treated with protease, and lane 7 had plasmid DNA extracted after protease and DNAse simultaneous treatment of GENS
Baseline EGFR Expression in Pancreatic Cancer Cells
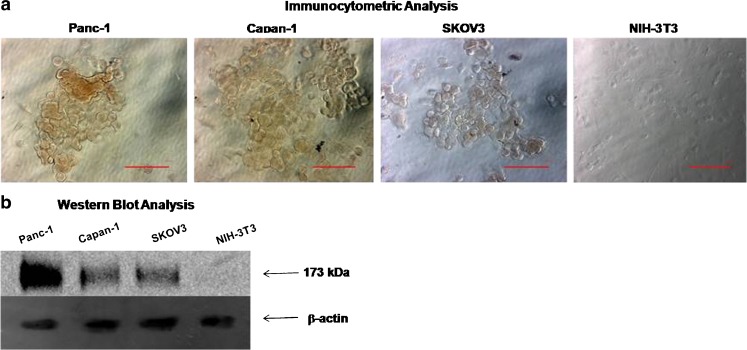
Human pancreatic adenocarcinoma cells, Panc-1 and Capan-1 cells, along with SKOV3 ovarian adenocarcinoma cells, and NIH-3T3 murine fibroblast cells (negative control) were evaluated for EGFR expression using immunocytometric and Western blot analyses. The results in Fig. 3 show that Panc-1 cells had the highest expression levels relative to Capan-1 and SKOV3 cells as shown by the DAB staining in immunocytometric analysis and the immunoblot density. NIH-3T3 fibroblast cells do not express EGFR and this was confirmed by both immunocytometric and Western blot analyses. Based on the highest level of EGFR expression, further studies were carried out with Panc-1 pancreatic adenocarcinoma cells.
Fig. 3.
Baseline epidermal growth factor receptor (EGFR, 175 kDa) expression analysis by a immunocytometry and b Western blot analysis in human pancreatic adenocarcinoma cells (Panc-1 and Capan-1). Additionally, EGFR expression in human ovarian adenocarcinoma (SKOV3) cells was evaluated as positive control and murine fibroblast (NIH-3T3) cells served as negative control. In Western blot analysis, beta-actin from Panc-1 cells was used as a loading control. Scale bar in the immunocytometric microscopy images in a represent a distance of 20 μm
Binding Specificity of EGFR Peptide to Panc-1 Cells
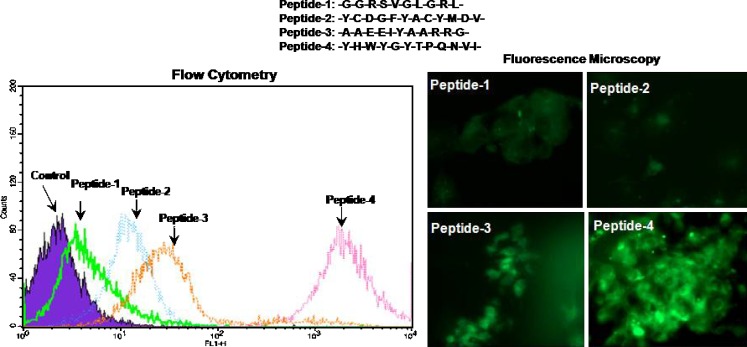
Four different peptide sequences known to possess binding specificity to the human epidermal receptor family members were chosen based on information in the scientific literature. Flow cytometry and fluorescence microscopy data in Fig. 4 show the binding of these FITC-labeled peptides to Panc-1 human pancreatic adenocarcinoma cells following 20 min with 10 μM concentration. Peptide-1, which is known to bind with EGFR, was found to have the lowest binding affinity followed by peptide-2 and peptide-3, which are known to interact with HER2 rather than EGFR. Peptide-4, another EGFR binding peptide, was found to have the highest affinity to Panc-1 cells based on flow cytometry and fluorescence data. Based on these results, the highly specific EGFR-targeting peptide sequence (i.e., peptide-4 with the amino acid sequence Y-H-W-Y-G-Y-T-P-Q-N-V-I) was chosen for surface modification of the nanoparticles to facilitate active targeted delivery in Panc-1 cells.
Fig. 4.
Evaluation of four different epidermal growth factor receptor (EGFR) binding peptide specificity and affinity to Panc-1 human pancreatic adenocarcinoma cells. Panc-1 cells were exposed to fluorescein isothiocyanate-labeled peptide-1 to peptide-4 at 10 μM final concentration for 20 min followed by washing with sterile phosphate-buffered saline (pH 7.4) and fixation in 4% paraformaldehyde. Flow cytometric analysis of peptide binding and fluorescence microscopy images
Cytotoxicity of Control and Surface-Modified GENS
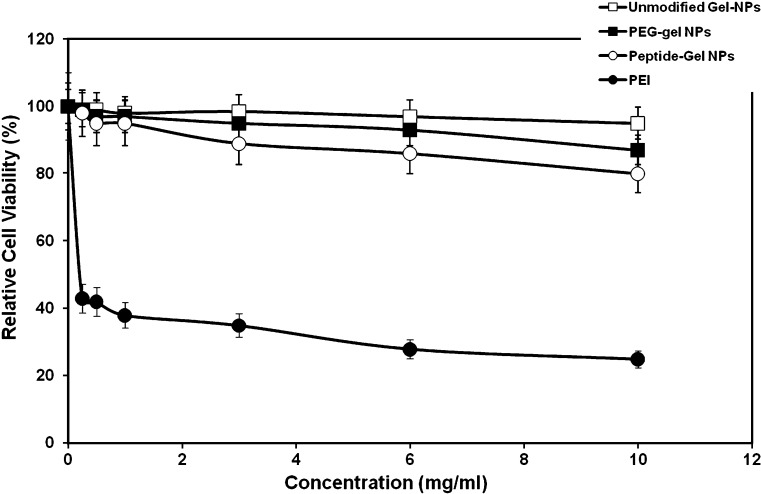
Cytotoxicity of control and surface-modified GENS was evaluated by incubating the nanoparticles at different concentrations with Panc-1 cells. Cell viability was determined using the MTS reagent assay, which is based on tetrazolium salt conversion to soluble formazan chromogen by dehydrogenase enzyme found in metabolically active cells. The percent cell viability relative to untreated control cells was determined as a function of the concentration unmodified, PEG-modified, and peptide-modified GENS. The results shown in Fig. 5 indicate that unmodified gelatin and PEG-modified gelatin nanoparticles were relatively non-toxic, as previously shown, over most of the concentration range examined. The slight decrease in cell viability at 10 mg/mL with unmodified gelatin and PEG-modified gelatin nanoparticles, however, was attributed to the high viscosity of gelatin in the cell culture medium rather than specific toxicity of the biopolymer to the cells. This observation was confirmed when free gelatin at the same concentration was added to the cell culture medium. The EGFR peptide-modified GENS showed a slight decrease in cell viability with increasing concentrations. For instance, at the highest concentration of 10 mg/mL, the relative cell viability with EGFR peptide-modified GENS was 85%. Based on the observation of cellular morphology, the decrease in cell viability with EGFR-targeted nanoparticles was probably due to inhibition of EGFR-mediated cell proliferation rather than direct cell kill. In comparison, poly(ethyleneimine), the positive control, had a relative cell viability of 40% at 0.5 mg/mL concentration and decreased even further to 22% at 10 mg/mL concentration. Based on these results, EGFR peptide-modified GENS are relatively biocompatible and could potentially be useful in in vivo gene delivery applications.
Fig. 5.
Cytotoxicity profiles of control (unmodified) type B gelatin nanoparticles, poly(ethylene glycol)-modified gelatin nanoparticles (PEG-Gel NP), and epidermal growth factor receptor (EGFR)-targeted gelatin nanoparticles (Peptide-Gel NP) as a function of concentration in Panc-1 human pancreatic adenocarcinoma cells. Polyethylenimine (PEI), a known cytotoxic cationic polymer, was used as positive control. Panc-1 cells were treated with nanoparticle concentrations ranging from 0 to 10 mg/ml for a period of 4 h. Following the incubation period, percent cell viability relative to untreated control was evaluated using the MTS (formazan) reagent
Receptor-Mediated Cellular Uptake in Panc-1 Cells
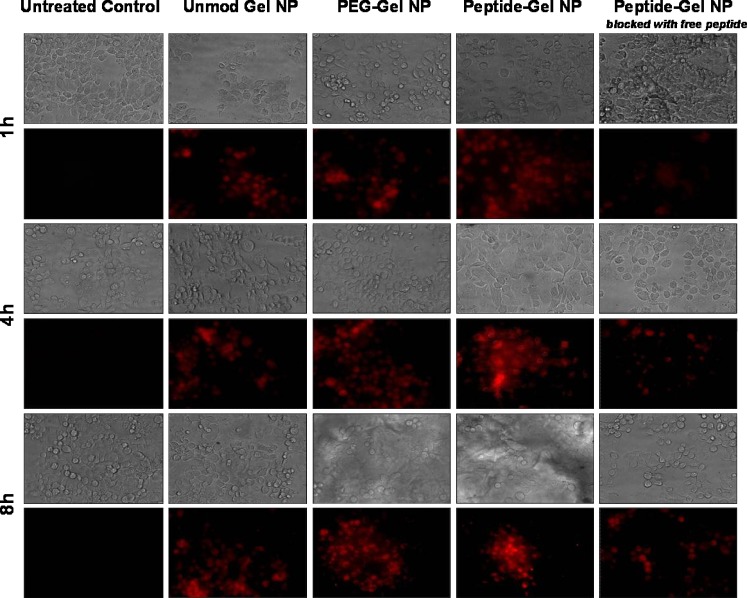
To confirm surface accessibility of EGFR-targeting peptide and receptor-mediated endocytotic uptake of nanoparticles, rhodamine-labeled dextran encapsulated control and surface-modified GENS were incubated with Panc-1 cells. Periodically, differential interference contrast and fluorescent images were acquired to visualize nanoparticle uptake and distribution in Panc-1 cells. In Fig. 6, the fluorescent micrographs show that unmodified gelatin and PEG-modified gelatin nanoparticles were internalized in Panc-1 cells after 4 h of incubation. On the other hand, EGFR peptide-modified GENS were rapidly internalized within the first hour of incubation and higher nanoparticle density was observed at longer time points. However, with increasing duration up to 8 h, the peptide-modified nanoparticle uptake was found to be reduced probably due to saturation of the available receptors on cell surface. In the last panel on the right in Fig. 7, the cells were first treated with 10 μM free EGFR peptide for 20 min and then incubated with the rhodamine dextran-encapsulated peptide-modified nanoparticles. Based on these results, which show a significant decrease in nanoparticle uptake, the specificity of EGFR peptide-modified nanoparticles to bind with its receptor on Panc-1 cells was confirmed.
Fig. 6.
Differential interference contrast and epifluorescence microscopic analysis of cellular uptake and trafficking of rhodamine-labeled dextran encapsulated in control (unmodified) type B gelatin nanoparticles, poly(ethylene glycol)-modified gelatin nanoparticles (PEG-Gel NP), and epidermal growth factor receptor (EGFR)-targeted gelatin nanoparticles (Peptide-Gel NP) in Panc-1 human pancreatic adenocarcinoma cells at 1, 4, and 8 h post-incubation. The specificity of EGFR-targeted nanoparticle uptake and internalization in Panc-1 cells was confirmed by incubation of the cells with free peptide 20 min before the addition of peptide-modified nanoparticles
Fig. 7.
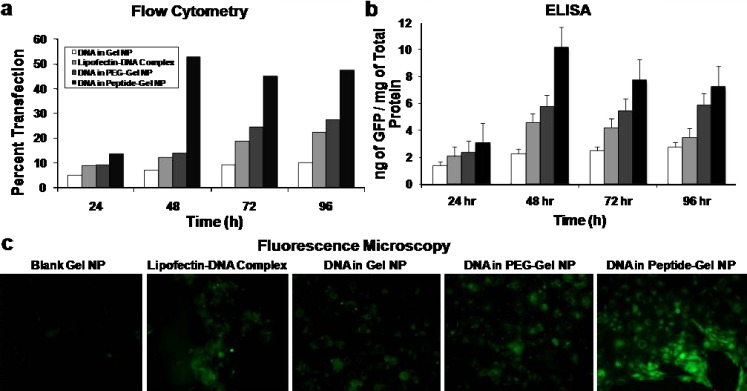
Quantitative and qualitative in vitro enhanced green fluorescence protein (GFP) transgene expression efficiency studies in control (unmodified) type B gelatin nanoparticles, poly(ethylene glycol)-modified gelatin nanoparticles (PEG-Gel NP), and epidermal growth factor receptor (EGFR)-targeted gelatin nanoparticles (Peptide-Gel NP) in Panc-1 human pancreatic adenocarcinoma cells. Quantitative analysis was performed by a flow cytometry and b enzyme-linked immunoassay for GFP at different time points from 24 to 96 h post-transfection. c Qualitative analysis of GFP transfection was performed by epifluorescence microscopy after 48 h. The Panc-1 cells were treated with the plasmid DNA dose of 20 μg per 200,000 cells for a period of 4 h, followed by washing with sterile phosphate-buffered saline (pH 7.4) and replacement of regular cell culture medium. Epifluorescence microscopy images were obtained at ×40 original magnification. Cells treated with blank gelatin nanoparticles served as a negative control, while the commercial cationic lipid-based DNA transfection reagent, Lipofectin®, was used as a positive control
Quantitative and Qualitative In Vitro GFP Transfection in Panc-1 Cells
Flow cytometry and ELISA were used to measure quantitative GFP transfection efficiency in Panc-1 cells upon administration of unmodified, PEG-modified, and EGFR peptide-modified GENS. As shown in Fig. 7a, flow cytometry analysis showed an increasing percent transfection of Panc-1 cells with increasing duration from 24 to 96 h post-administration. For instance, plasmid DNA delivered using EGFR peptide-targeted GENS showed a transfection efficiency of approximately 12% at 24 h post-administration, which increased to 48% after 96 h post-administration. In comparison, the transfection efficiencies with DNA encapsulated in PEG–gelatin nanoparticles or DNA complexed with Lipofectin®, a cationic lipid transfection reagent, were significantly lower. Lipofectin® was also found to induce significant toxicity in Panc-1 cells, especially at longer time intervals. In addition to the flow cytometry results, ELISA was performed on the cell lysates of the transfected Panc-1 cells to quantify the levels of expressed GFP as a function of total protein. The ELISA results in Fig. 7b showed the highest GFP production in cells transfected with the EGFR-targeted nanoparticles, similar to the transfection efficiency pattern seen with flow cytometry analysis. For instance, at 24 h post-administration, there was approximately 3 ng of expressed GFP per milligram of total protein. The expressed GFP concentration increased to 10 ng/mg after 48 h post-administration and 7.2 ng/mg at 96 h post-administration when EGFP-N1 plasmid DNA was administered in EGFR peptide-modified GENS. The slight decrease in GFP expression levels at 72 h and beyond was probably due to cell replication that dilutes the transfected GFP.
The quantitative transgene expression results are further corroborated by the qualitative fluorescence microscopic images of Panc-1 cells acquired 48 h after administration of EGFP-N1 plasmid DNA in control and EGFR peptide-modified GENS. As seen from Fig. 7c, a large population of the cells in the microscope’s field of view was positive for GFP expression when the plasmid was administered in EGFR peptide-modified nanoparticles. Plasmid DNA administered in unmodified gelatin and PEG-modified gelatin nanoparticle-treated Panc-1 also showed GFP expression albeit at a lower level.
CONCLUSIONS
In this study, we have formulated EGFR-targeted gelatin-based nanoparticulate gene delivery system and examined the uptake and transfection efficiency in EGFR-expressing Panc-1 human pancreatic adenocarcinoma cells. Control and surface-modified GENS efficiently encapsulated plasmid DNA (EGFP-N1) and protected the payload against process condition- as well as DNAse-induced degradation. The cytotoxicity profile of EGFR peptide-modified GENS was slightly higher than the unmodified or PEG-modified gelatin nanoparticles at higher concentrations. This may be due to EGFR’s role in cell proliferation, which would be inhibited by the binding of targeted nanoparticles. EGFR peptide-modified GENS were able to interact with Panc-1 cells through receptor-specific interaction leading to higher cellular internalization capacity relative to other controls.
Quantitative and qualitative GFP transfection results demonstrated the superior transfection efficiency of EGFR-targeted GENS in comparison with the cationic lipid transfection reagent Lipofectin®, unmodified gelatin nanoparticles, and PEG-modified gelatin nanoparticles. The enhanced cellular uptake and transfection properties of EGFR-targeted peptide-modified GENS were attributed to the presence of surface-accessible peptide bound through PEG spacer that facilitates receptor-mediated cellular internalization in Panc-1 cells. Non-condensing type B gelatin with lower cytotoxicity profile protects the payload during vesicular transport in cells and allows for the release of supercoiled plasmid DNA at the nuclear membrane, which can be efficiently imported into the nucleus leading to enhanced transgene expression efficiency. Overall, EGFR peptide-modified GENS show superior DNA delivery properties in human pancreatic cancer cells in vitro. These results are highly significant in the development of safe and effective in vivo gene delivery vector for the treatment of pancreatic cancer.
Acknowledgments
Padmaja Magadala is a Fellow in the Nanomedicine Science and Technology Interdisciplinary Graduate Education and Research Training (IGERT) program. This pre-doctoral program is supported by the National Cancer Institute (NCI) and the National Science Foundation (NSF). Scanning electron microscopy was performed at Northeastern University’s Electron Microscopy Center. We deeply appreciate the assistance of Mr. Luis Brito with the flow cytometry analysis. Flow cytometry studies were performed in Professor Vladimir Torchilin’s Lab at Northeastern University.
References
- 1.Jemal A., Tiwari R. C., Murray T., Ghafoor A., Samuels A., Ward E., Feuer E. J., Thun M. J. Cancer statistics, 2004. CA Cancer J. Clin. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 2.Huber B. E. Gene therapy strategies for treating neoplastic disease. Ann. N. Y. Acad. Sci. 1994;716:6–11. doi: 10.1111/j.1749-6632.1994.tb21699.x. [DOI] [PubMed] [Google Scholar]
- 3.Roth J. A. Adenovirus p53 gene therapy. Expert. Opin. Biol. Ther. 2006;6:55–61. doi: 10.1517/14712598.6.1.55. [DOI] [PubMed] [Google Scholar]
- 4.Roth J. A. Gene replacement strategies for lung cancer. Curr. Opin. Oncol. 1998;10:127–132. doi: 10.1097/00001622-199803000-00008. [DOI] [PubMed] [Google Scholar]
- 5.Roth J. A. Gene replacement strategies for cancer. Isr. J. Med. Sci. 1996;32:89–94. [PubMed] [Google Scholar]
- 6.Roth J. A., Grammer S. F. Gene replacement therapy for non-small cell lung cancer: a review. Hematol. Oncol. Clin. North Am. 2004;18:215–229. doi: 10.1016/S0889-8588(03)00144-8. [DOI] [PubMed] [Google Scholar]
- 7.Roth J. A., Grammer S. F. Tumor suppressor gene therapy. Methods Mol. Biol. 2003;223:577–598. doi: 10.1385/1-59259-329-1:577. [DOI] [PubMed] [Google Scholar]
- 8.Roth J. A., Grammer S. F., Swisher S. G., Komaki R., Nemunaitis J., Merritt J., Meyn R. E. P53 gene replacement for cancer-interactions with DNA damaging agents. Acta. Oncol. 2001;40:739–744. doi: 10.1080/02841860152619160. [DOI] [PubMed] [Google Scholar]
- 9.Roth J. A., Swisher S. G., Meyn R. E. p53 tumor suppressor gene therapy for cancer. Oncology (Williston Park) 1999;13:148–154. [PubMed] [Google Scholar]
- 10.Wadhwa P. D., Zielske S. P., Roth J. C., Ballas C. B., Bowman J. E., Gerson S. L. Cancer gene therapy: scientific basis. Annu. Rev. Med. 2002;53:437–452. doi: 10.1146/annurev.med.53.082901.104039. [DOI] [PubMed] [Google Scholar]
- 11.Jacob D., Davis J. J., Zhang L., Zhu H., Teraishi F., Fang B. Suppression of pancreatic tumor growth in the liver by systemic administration of the TRAIL gene driven by the hTERT promoter. Cancer Gene Ther. 2005;12(2):109–115. doi: 10.1038/sj.cgt.7700773. [DOI] [PubMed] [Google Scholar]
- 12.Pearson A. S., Bouvet M., Evans D. B., Roth J. A. Gene therapy and pancreatic cancer. Front Biosci. 1998;3:E230–237. doi: 10.2741/a382. [DOI] [PubMed] [Google Scholar]
- 13.Blessing T., Remy J. S., Behr J. P. Monomolecular collapse of plasmid DNA into stable virus-like particles. Proc. Natl. Acad. Sci. U. S. A. 1998;95(4):1427–1431. doi: 10.1073/pnas.95.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marshall E. Gene therapy on trial. Science. 2000;288:951–957. doi: 10.1126/science.288.5468.951. [DOI] [PubMed] [Google Scholar]
- 15.Check E. Gene therapy: shining hopes dented—but not dashed. Nature. 2002;420:735. doi: 10.1038/420735b. [DOI] [PubMed] [Google Scholar]
- 16.Thomas C. E., Ehrhardt A., Kay M. A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003;4(5):346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 17.Lv H., Zhang S., Wang B., Cui S., Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release. 2006;114(1):100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 18.Panyam J., Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv. Drug Deliv. Rev. 2003;55:329–347. doi: 10.1016/S0169-409X(02)00228-4. [DOI] [PubMed] [Google Scholar]
- 19.Dellian M., Yuan F., Trubetskoy V. S., Torchilin V. P., Jain R. K. Vascular permeability in a human tumour xenograft: molecular charge dependence. Br. J. Cancer. 2000;82(9):1513–1518. doi: 10.1054/bjoc.1999.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhavsar M. D., Amiji M. M. Development of novel biodegradable polymeric nanoparticles-in-microsphere formulation for local plasmid DNA delivery in the gastrointestinal tract. AAPS PharmSciTech. 2008;9:288–294. doi: 10.1208/s12249-007-9021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhavsar M. D., Amiji M. M. Oral IL-10 gene delivery in a microsphere-based formulation for local transfection and therapeutic efficacy in inflammatory bowel disease. Gene. Ther. 2008;15(17):1200–1209. doi: 10.1038/gt.2008.67. [DOI] [PubMed] [Google Scholar]
- 22.Bhavsar M. D., Amiji M. M. Gastrointestinal distribution and in vivo gene transfection studies with nanoparticles-in-microsphere oral system (NiMOS) J. Control. Release. 2007;119:339–348. doi: 10.1016/j.jconrel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Bhavsar M. D., Tiwari S. B., Amiji M. M. Formulation optimization for the nanoparticles-in-microsphere hybrid oral delivery system using factorial design. J. Control. Release. 2006;110:422–430. doi: 10.1016/j.jconrel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Kaul G., Amiji M. Biodistribution and targeting potential of poly(ethylene glycol)-modified gelatin nanoparticles in subcutaneous murine tumor model. J. Drug Target. 2004;12:585–591. doi: 10.1080/10611860400013451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaul G., Amiji M. Tumor-targeted gene delivery using poly(ethylene glycol)-modified gelatin nanoparticles: in vitro and in vivo studies. Pharm. Res. 2005;22:951–961. doi: 10.1007/s11095-005-4590-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kommareddy S., Amiji M. Poly(ethylene glycol)-modified thiolated gelatin nanoparticles for glutathione-responsive intracellular DNA delivery. Nanomedicine. 2007;3:32–42. doi: 10.1016/j.nano.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kommareddy S., Amiji M. Biodistribution and pharmacokinetic analysis of long-circulating thiolated gelatin nanoparticles following systemic administration in breast cancer-bearing mice. J. Pharm. Sci. 2007;96:397–407. doi: 10.1002/jps.20813. [DOI] [PubMed] [Google Scholar]
- 28.Kommareddy S., Amiji M. Preparation and evaluation of thiol-modified gelatin nanoparticles for intracellular DNA delivery in response to glutathione. Bioconjug. Chem. 2005;16:1423–1432. doi: 10.1021/bc050146t. [DOI] [PubMed] [Google Scholar]
- 29.Kommareddy S., Amiji M. Antiangiogenic gene therapy with systemically administered sFlt-1 plasmid DNA in engineered gelatin-based nanovectors. Cancer Gene Ther. 2007;14:488–498. doi: 10.1038/sj.cgt.7701041. [DOI] [PubMed] [Google Scholar]
- 30.Neves C., Escriou V., Byk G., Scherman D., Wils P. Intracellular fate and nuclear targeting of plasmid DNA. Cell Boil. Toxicol. 1999;15:193–202. doi: 10.1023/A:1007693805849. [DOI] [PubMed] [Google Scholar]
- 31.Tobita K., Kijima H., Dowaki S., Kashiwagi H., Ohtani Y., Oida Y., Yamazaki H., Nakamura M., Ueyama Y., Tanaka M., Inokuchi S., Makuuchi H. Epidermal growth factor receptor expression in human pancreatic cancer: significance for liver metastasis. Int. J. Mol. Med. 2003;11:305–309. [PubMed] [Google Scholar]
- 32.Yamanaka Y., Friess H., Kobrin M. S., Buchler M., Beger H. G., Korc M. Coexpression of epidermal growth factor receptor and ligands in human pancreatic cancer is associated with enhanced tumor aggressiveness. Anticancer Res. 1993;13:565–569. [PubMed] [Google Scholar]
- 33.Jarvinen T. A., Ruoslahti E. Molecular changes in the vasculature of injured tissues. Am. J. Pathol. 2007;171:702–711. doi: 10.2353/ajpath.2007.061251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z., Zhao R., Wu X., Sun Y., Yao M., Li J., Xu Y., Gu J. Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics. FASEB J. 2005;19:1978–1985. doi: 10.1096/fj.05-4058com. [DOI] [PubMed] [Google Scholar]
- 35.Zitzmann S., Ehemann V., Schwab M. Arginine-glycine-aspartic acid (RGD)-peptide binds to both tumor and tumor-endothelial cells in vivo. Cancer Res. 2002;62:5139–5143. [PubMed] [Google Scholar]
- 36.Guillemard V., Nedev H., Berezov A., Murali R., Saragovi H. U. HER2-mediated internalization of a targeted prodrug cytotoxic conjugate is dependent on the valency of the targeting ligand. DNA Cell Biol. 2005;24(6):351–358. doi: 10.1089/dna.2005.24.351. [DOI] [PubMed] [Google Scholar]
- 37.Chan P., Nestler H. P., Miller W. T. Investigating the substrate specificity of the HER2/Neu tyrosine kinase using peptide libraries. Cancer Lett. 2000;160(2):159–169. doi: 10.1016/S0304-3835(00)00581-4. [DOI] [PubMed] [Google Scholar]