Abstract
The Purkinje cell degeneration (PCD) mutant mouse is characterized by a degeneration of cerebellar Purkinje cells and progressive ataxia. To identify the molecular mechanisms that lead to the death of Purkinje neurons in PCD mice, we used Affymetrix microarray technology to compare cerebellar gene expression profiles in pcd3J mutant mice 14 days of age (prior to Purkinje cell loss) to unaffected littermates. Microarray analysis, Ingenuity Pathway Analysis (IPA) and Expression Analysis Systematic Explorer (EASE) software were used to identify biological and molecular pathways implicated in the progression of Purkinje cell degeneration. IPA analysis indicated that mutant pcd3J mice showed dysregulation of specific processes that may lead to Purkinje cell death, including several molecules known to control neuronal apoptosis such as Bad, CDK5 and PTEN. These findings demonstrate the usefulness of these powerful microarray analysis tools and have important implications for understanding the mechanisms of selective neuronal death and for developing therapeutic strategies to treat neurodegenerative disorders.
Keywords: apoptosis, kinase, microarray, neurodegeneration, Purkinje cell
Introduction
The Purkinje Cell Degeneration (PCD) mutant mouse is characterized by the complete loss of the cerebellar Purkinje cells between second and sixth weeks following birth [1-3]. The PCD phenotype manifests as a true Mendelian autosomal recessive trait, with homozygous mutant mice accounting for one fourth of progeny from a heterozygous cross and exhibiting the Purkinje cell degeneration and subsequent ataxia. Mice homozygous for the mutation display an almost complete loss of Purkinje cells during adulthood. Heterozygous mutant mice account for half the progeny of a heterozygous cross and display only a delayed moderate (∼18%) degeneration of Purkinje cells and no ataxia [4].
The reason for the degeneration of Purkinje cells in PCD mutant mice has not been elucidated. Recently Nna1 (also referred to as agtpbp1; NM_023328 and NM_001048008), a gene related to neuronal regeneration [5], has been identified by classic genetic mapping methodologies as the gene harboring a genetic lesion in multiple allelic strains of PCD mutant mice [6]. To date eight independent allelic strains of PCD mouse mutants have been identified, conventionally referred to as Agtpbp1pcd, Agtpbp1pcd-2J, Agtpbp1pcd-3J etc [7]. We will refer to these as pcd1J, pcd2J, pcd3J, etc.
A recent study used Affymetrix microarray technology to examine gene expression profiles between cerebella of 4-month old pcd3J mice, a time where most Purkinje cells had died [8]. In that study, the goal was to identify candidate markers of cerebellar Purkinje neurons. The aforementioned study highlighted the need for a novel approach with new tools to answer the question of how the Purkinje neurons degenerate in the PCD mouse mutants. Microarray analysis is becoming an increasingly utilized and powerful tool for identifying relevant differences between the expression profiles of normal and diseased tissues. New computer analysis tools are constantly emerging to assist with mining of the massive amounts of raw data generated in microarray experiments. In the present study, we employed Affymetrix microarray analysis along with Ingenuity Pathway Analysis (IPA) software to compare the expression profiles of cerebella from P14 wild type and homozygous pcd3J/pcd3J mutant mice. We chose to use the pcd3J strain because the molecular lesion in the Nna1 gene of this PCD strain is known and pcd3J is established to be a loss-of-function allele [6]. Additionally, no marked degeneration of Purkinje neurons is observed even at P22 in this strain [9]. Therefore, we propose that the genes identified in this study may be involved in the initiation of neuronal death in the PCD mutants. These findings could give significant insight into mechanisms associated with chronic neurodegenerative disorders.
Materials and Methods
Animals
Pcd3J mice were purchased from Jackson Laboratories (Bar Harbor, ME; strain: BLBB/cByJ- Agtpbp1pcd-3J/J; stock# 003237). Homozygous pcd3J animals and their wild-type littermates were generated by crossing mice heterozygous for the pcd3J allele. Animals were genotyped as neonates by PCR as previously described [8]. Male wild-type and pcd3J heterozygous, null mutant mice (n=3/genotype) were euthanized at 14 days of age, immediately prior to Purkinje cell degeneration, and their cerebella carefully dissected and homogenized in TRIzol Reagent (Life Technologies, Rockville, MD, USA). All of the experimental procedures conformed to the humane treatment of animals as prescribed by the Institute for Laboratory Animal Research (Guide for the Care and Use of laboratory Animals) and were approved by the Morehouse School of Medicine Institutional Care and use Committee.
Microarray sample preparation and hybridization
Total RNA was extracted with TRIzol and purified using an RNAqueous Kit (Ambion, Austin, TX, USA). RNA integrity was determined using an Agilent Bioanalyzer. RNA was converted to double-stranded cDNA (Invitrogen, Superscript Choice System, Carlsbad CA, USA) using a T7-(dT)24 primer. Cleanup of double-stranded cDNA used Phase Lock Gels (Eppendorf, Westbury, NY, USA) and Phenol/Chloroform/Isoamyl Alcohol (Sigma, St. Louis, MO, USA). cRNA was synthesized using a RNA transcript labeling kit (Enzo Diagnostics, Farmingdale, NY, USA). Biotin labeled cRNA was cleaned up using a GeneChip Sample Cleanup Module (Affymetrix Inc, Santa Clara, CA USA) and then quantified using a spectrophotometer. Twenty micrograms of the in vitro transcription product were fragmented by placing at 94°C for 35 minutes in Fragmentation buffer. Following fragmentation, 15 μg of the biotinylated cRNA was hybridized to an Affymetrix Murine Genome Array U74Av2 GeneChip. Samples were hybridized at 45°C for 16 hours, and then washed, stained with streptavidin-phycoerythrin, and scanned according to manufacturer's guidelines. RNA from each mouse was hybridized to a separate chip.
Microarray data processing
In these experiments, we used the mouse genome MG_U74Av2 Affymetrix GeneChip that has 12743 probe sets (genes). Replicate experiments were compared for each experimental group. GraphPad Prism analysis generated Person Correlation values and p-values for each replicate comparison. The correlation coefficient measured the strength of a linear relationship between two variables. Values of +1.0 to +0.7 represent a strong positive association with p<0.05. Using these values, we determined that the replicates were comparable.
Data analysis was performed using Affymetrix GCOS software. Single array analysis was used to build the databases of gene expression profiles. Three chips were used for each group: wild-type mice (WT; +/+), pcd3J heterozygous mice (HET; pcd/+), and pcd3J mutant mice (MUT; pcd/pcd). Affymetrix GCOS software was used to normalize and analyze data for each chip. Detection p-value (set at p<0.5) was used to statistically determine whether a transcript is expressed on a chip. The software generated a present (P), marginal (M) or absent (A) call for each transcript based on the p-value. Absolute calls (present, marginal, and absent) and the average difference (RNA abundance) for each gene on each chip were calculated using GCOS. One way ANOVA with Dunnett's post tests were performed on gene sets using GraphpadPrism version 4.0 for Windows (GraphPad software, San Diego, California, USA, www.graphpad.com) to measure correlation coefficients among replicates to determine reproducibility.
Data were then imported into Genespring software (Silicon Genetics, Redwood City, CA) for further analysis. Data analysis was performed on all replicate samples based on expression patterns. Genes that changed in expression levels by two-fold or more uniquely in the pcd/pcd compared to wild type litter mates were selected for further analysis. For selected genes, Ingenuity Pathway Analysis (IPA) version 2.0 (Ingenuity® Systems, www.ingenuity.com) was used to search for biological pathways and interrelationships between network genes in the subsets of candidate genes regulated in the different PCD genotypes. Generated data sets containing gene identifiers and corresponding expression values were uploaded into the application. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. A 2.0 fold change cutoff was set to identify genes whose expression was significantly differentially regulated. These genes, called Focus Genes, were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. Networks of these Focus Genes were then algorithmically generated based on their connectivity. To start building networks, the application queries the Ingenuity Pathways Knowledge Base for interactions between focus genes and all other gene objects stored in the knowledge base, then generates a set of networks with a network size of approximately 35 genes or proteins. Ingenuity Pathways Analysis then computes a score for each network according to the fit of the user's set of significant genes. The score is derived from a p-value and indicates the likelihood of the focus genes in a network being found together because of random chance. A score of 2 indicates a 1 in 100 chance that the focus genes are together in a network because of random chance. Therefore, scores of 2 or higher have at least a 99% confidence level of not being generated by random chance alone. Biological functions are then calculated and assigned to each network.
Results
Microarray analysis
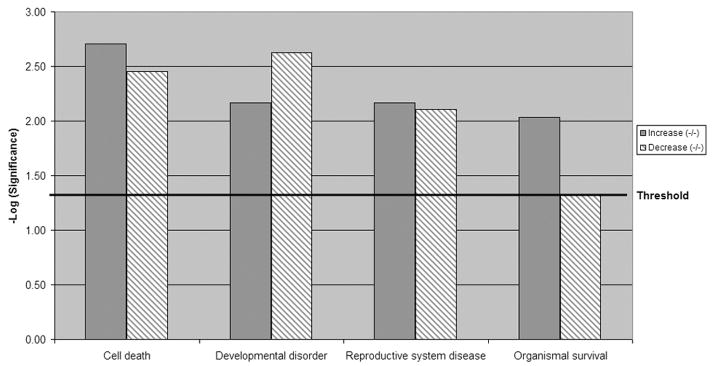
To understand how these genes interact or are interrelated, we employed the Ingenuity Pathway Analysis (IPA) tool. To determine the unique gene expression changes in pcd3J mice we compared the gene expression profiles of wild type (+/+) to the pcd/+ and pcd/pcd mice. There were 646 genes that increase in the pcd/pcd mice by 2.0 fold or more and 454 of those were unique to the pcd/pcd mice when genes found in the heterozygous mice were subtracted. There were 665 genes that decrease in the pcd/pcd mice (496 unique to the pcd/pcd mice) by 2 fold or more. The unique genes were then mapped to genetic networks by IPA. These networks were associated with known biological pathways. There are 11 networks that increased and 22 networks that decreased in the pcd/pcd mice (Table 1). Networks found to be highly significantly increased in the pcd/pcd mice were associated with cell adhesion, cell signaling, cell cycle, inflammatory disease and reproductive disease. Network categories that decreased included gene expression, immune function and cell death. Figure 1 shows graphically that the pcd/pcd mice uniquely upregulated genes are primarily associated with cell death/survival and developmental disorders.
Table 1.
| Top 5 networks generated from IPA for transcripts that increase 2-fold in homozygous mutant animals compared to wild type.
| ||||
|---|---|---|---|---|
| Network ID | Genes in Network | score | # Focus genes | Top Categories |
| 1 | AKT2, BRD8, CACNA1A, CDH1, CDK5, CHRNA2, CHRNA6, CHRNA7, CHRNA9
CHRNA10, CHRNB3, CHRND, DAB1, FHL1, GCN5L2, HGF, HRB2, INSM1, ITGB5, MAPK1 MPZ, NEF3, NEFH, NOTCH1, NOV, PHOX2A, PPP1R1A, PRKCA, PSMD3, RET, SLC1A1 SOX10, STX1A, TACC3, TH |
23 | 18 | Cardiovascular Disease, Cell-To-Cell Signaling and Interaction, Nervous System Development |
| 2 | AXL, BAD, BRRN1, C14ORF126, CALCR, CDC2, CDK5RAP3, EBI3, FLT3, GAS6, GPX3
GRB2, IFNGR1, IGHE, IL13, IL13RA1, IL13RA2, IL27RA, IL4R, MARCKS, NDUFA13 PHKA2, PIM2, PRKCQ, PTPRE, QSCN6, REG3A, REPS2, SERPINB3, SERPINB4 SLC6A4, SNCA, SOCS5, STAT3, UCHL1 |
21 | 17 | Cell Death, Cellular Growth and Proliferation Hematological Disease |
| 3 | ADFP, ANGPTL4, APBA1, APOA5, ARF6, ARFGAP1, CBX2, CCS, CPXM2, EGR1, FDPS
FTL, GGA1, GGA2, GGA3, GH1, HBB, IDH1, ITPR2, LGALS9, LIN7B, LNPEP, LPL LRRC8C, M6PR, PMM1, PPARG, RPL29, SIM1, SLC2A4, SST, SSTR1, SSTR4 SSTR5, XRCC1 |
19 | 16 | Cell Morphology, Cell Cycle, Reproductive System Development and Function |
| 4 | ABCC3, ARFGAP3, CASP12, CKMT2, CREB3, FABP1, FCGR1A, FCGR2B, FCGR3A
FGR, FOXO1A, KIF20A, KLHDC2, KLRC1, KLRD1, KLRK1, LAMA3, LAMA5, LAMB3 LAMC2, LIPE, MAP3K5, MAPK13, MLANA, MTTP, P4HB, RPS6KA5, SLK, STAC, STMN1 TEK, TIE1, TNF, TNNC1, TXN |
18 | 15 | Inflammatory Disease, Skeletal and Muscular Disorders, Lipid Metabolism |
| 5 | CBX7, CCNG1, CDKN2A, CRABP1, EZH2, FEN1, FOS, HAPLN1, IDE, IGF1, IGF2, IGH-3
IGHM, IL6, IL19, IL4R, KIF11, KIFC1, LIG1, MMP10, PCSK4, PRIM1, PSME1, PSME2, RAG1 |
18 | 15 | Cancer, Cell Cycle, Reproductive System Disease |
|
Significant networks generated from IPA for transcripts that decrease 2-fold in homozygous mutant animals compared to wild type.
| ||||
| Network ID | Genes in Network | score | # Focus genes | Top Categories |
|
| ||||
| 1 | BST1, CCR2, CD40LG, CDX2, CMA1, COL17A1, CYR61, F10, GBP1, GDI2, GLA
HBB-B2, HLA-DRB1, IFNG, IL15, IL10RA, IL18R1, IL2RA, KLRK1, MAF, MMP9, NFE2L1 NOV, NR1H2, OAS1, OGN, PLP1, PRG1, PSAP, RSAD2, SERPINA5, SORL1, TFPI2 TIMP1, TYRP1 |
52 | 35 | Immunological Disease, Cellular Movement Inflammatory Disease |
| 2 | ABCF2, ACP5, BGLAP, CPA3, CTSB, DACH1, EDD1, EDG1, EYA3, FOS, GAST
HOXD10, HSD3B1, IBSP, INS, IRS2, IRS3, KIFAP3, KRT18, LYZ, NACA, NEK1, NRIP1 PC, PDPK1, PGAM2, PPARG, PSCD3, REN, RUNX1, SMAD3, SNW1, TSC1, VAV2 YWHAQ |
52 | 35 | Gene Expression, Carbohydrate Metabolism Cellular Development |
| 3 | ALDOB, ANXA1, BIRC1, BIRC4, CAPN2, CASP9, CDKN1B, CEP170, COX5A, COX6B2
COX7A1, CSND, CYP1B1, CYP3A2, DKK1, E2F3, ESR1, EXPI, FGB, GCG, GDF11 HNF4A, KIF5C, MAP3K7, MARK1, MCL1, PHLDB2, PLK1, PTEN, PTGDR, PTGS2, THBD WNK1, WNT11, YWHAG |
49 | 34 | Cancer, Cell Death, Reproductive System Disease |
| 4 | ADRA2A, APS, ATAD3A, CAPN2, CAPZB, CDC2L1, CTSB, CUL1, DDIT3, DDX5, DDX17
DDX50, DHX36, DUB2, FBL, FBXO15, HAS2, HNRPH1, HNRPU, HRAS, IGLL1, LGTN MGST3, MRPL12, MYBL1, MYC, PPID, RHOB, RPL32, SCEL, SLC25A5, SLC2A2 TNFRSF10B, VDAC2, XIST |
16 | 17 | Cell Death, Hepatic System Disease, Cancer |
| 5 | ARHGAP29, BRD4, CALM1, CD97, CISH, CSF3, CSF2RA, CSF2RB, CSF2RB2, CSF3R
CTLA4, EMR1, ES1, FOXD3, GADD45B, GADD45G, HDC, HOXC9, IL3, IL3RA, INPP5D KDR, LTA, MAPKAPK2, MID1, NCF1, PDE1A, PIM1, POU3F2, RPS14, SELL, STT3A TFDP1, TNFRSF1B, ZNF38 |
13 | 15 | Hematological System Development and Function Immune and Lymphatic System Development and Function, Tissue Morphology |
Figure 1.
Global functional category comparison of genes sets up-regulated or down-regulated in the homozygous mutant groups compared to wild type litter mates. Genes linked to cell death, developmental and reproductive dysfunctions were differentially expressed in the homozygous mutant group. Increasing value of −log (significance) indicated increased confidence for each category.
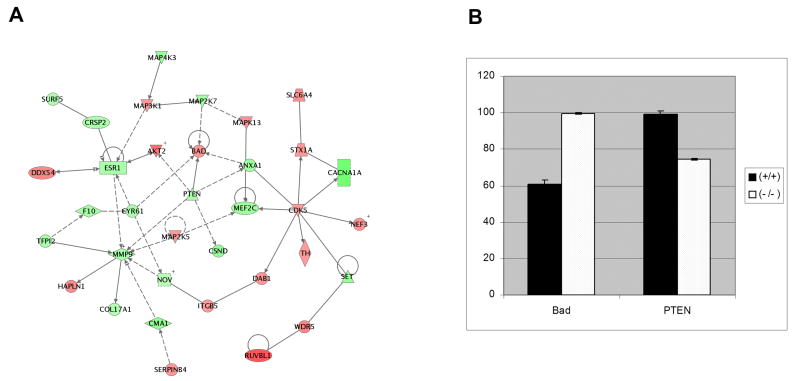
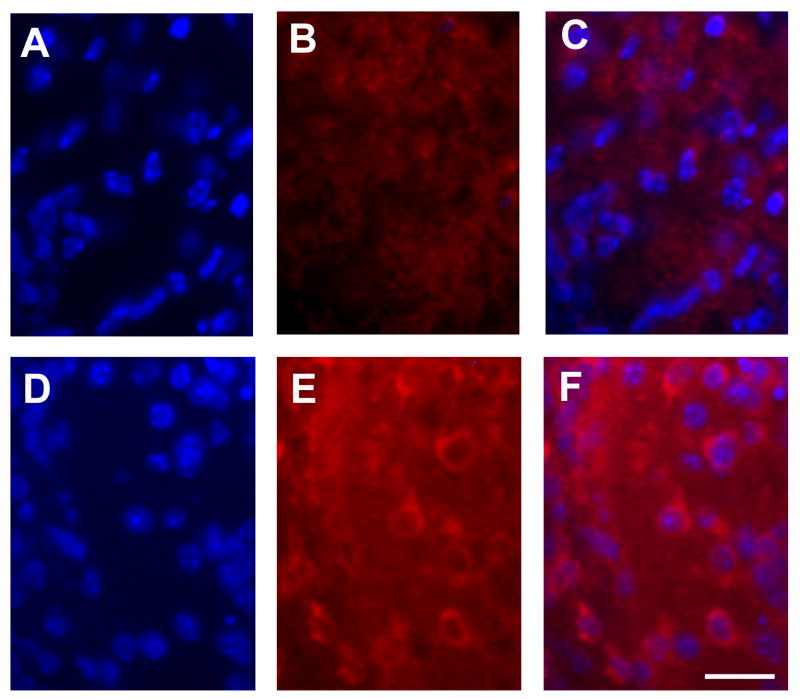
We further examined the most significant functional networks in pcd/pcd mice. Table 2 is a summary of the genes regulated in the most significant functional network in pcd/pcd mice. The network shown in figure 2A is a graphical representation of the molecular relationships between genes in the top pathway in pcd/pcd mice, respectively. Genes or gene products are represented as nodes (filled shapes), and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least 1 reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Pathways Knowledge Base. Red nodes are transcripts that increase in expression and green nodes represent transcripts that decrease in expression. IPA identified a major network in the pcd/pcd mice where the genes in the network focused around the following nodes: pro-apoptotic Bad, CDK5, phosphatase and tensin homologue deleted on chromosome 10 (PTEN) and matrix metalloproteinase 9 (MMP-9) (Fig. 2a). CDK5 and PTEN are capable of directly or indirectly regulating most the genes in this network while Bad and MMP-9 appear to serve as effector proteins. Real-time RT-PCR validated the expression patterns of selected genes highlighted in the microarray analysis (Fig. 2b). Consistent with the microarray results, Bad protein expression was absent in the Purkinje cell layer of Nna1 +/+ mice highly expressed in Purkinje cells of Nna1-/- mutants (Fig. 3).
Table 2.
| IPA Network analysis. Transcripts that increase or decrease by 2-fold or more in homozygous mutant compared to wild type.
| |||
|---|---|---|---|
| Name | Description | Fold Change | Family |
| RUVBL1 | RuvB-like 1 | 3.38 | Transcription regulator |
| AKT2 | V-akt murine thymoma viral oncogene homolog 2 | 2.84 | Kinase |
| WDR5 | WD repeat domain 5 | 2.44 | Other |
| DDX54 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 | 2.40 | Transcription regulator |
| HAPLN1 | Hyaluronan and proteoglycan link protein 1 | 2.30 | Other |
| SLC6A4 | Neurotransmitter transporter, serotonin | 2.26 | Transporter |
| MAPK13 | Mitogen-activated protein kinase 13 | 2.26 | Kinase |
| NEF3 | Neurofilament 3 | 2.16 | Other |
| BAD | BCL2-antagonist of cell death | 2.11 | Other |
| DAB1 | Disabled homolog 1 | 2.11 | Other |
| MAP3K1 | Mitogen-activated protein kinase kinase kinase 1 | 2.11 | Kinase |
| STX1A | Syntaxin 1A (brain) | 2.09 | Transporter |
| MAP2K5 | Mitogen-activated protein kinase kinase 5 | 2.09 | Kinase |
| SERPINB4 | Serpin peptidase inhibitor, clade B (ovalbumin) | 2.08 | Other |
| TH | Tyrosine hydroxylase | 2.05 | Enzyme |
| ITGB5 | Integrin, beta 5 | 2.03 | Other |
| CDK5 | Cyclin-dependent kinase 5 | 2.02 | Kinase |
| NOV | Nephroblastoma overexpressed gene | -2.02 | Growth factor |
| COL17A1 | Collagen, type XVII, alpha 1 | -2.11 | Other |
| SURF5 | Surfeit 5 | -2.24 | Other |
| CRSP2 | Cofactor required for Sp1 transcriptional activation | -2.34 | Transcription regulator |
| ESR1 | Estrogen receptor 1 | -2.35 | Nuclear receptor |
| CYR61 | Cysteine-rich, angiogenic inducer, 61 | -2.38 | Other |
| TFPI2 | Tissue factor pathway inhibitor 2 | -2.43 | Other |
| MMP9 | Matrix metallopeptidase 9 | -2.47 | Peptidase |
| PTEN | Phosphatase and tensin homolog | -2.48 | Phosphatase |
| F10 | Coagulation factor X | -2.53 | Peptidase |
| MAP2K7 | Mitogen-activated protein kinase kinase 7 | -2.54 | Kinase |
| ANXA1 | Annexin A1 | -2.62 | Other |
| MEF2C | MADS box transcription enhancer factor 2 | -2.70 | Transcription regulator |
| MAP4K3 | Mitogen-activated protein kinase kinase kinase kinase 3 | -2.88 | Kinase |
| SET | SET translocation (myeloid leukemia-associated) | -2.88 | Phosphatase |
| CSND | Casein alpha s2-like B | -2.92 | Other |
| CMA1 | Chymase 1, mast cell | -3.09 | Peptidase |
| CACNA1A | Calcium channel, voltage-dependent | -3.89 | Ion channel |
Figure 2.
(A) Network analysis. This network from is IPA analysis of gene sets up- or down- regulated in the homozygous mutant compared to wild type litter mates The network is displayed as nodes (genes or gene products) and edges (lines; biological relationships between nodes). Solid lines denote direct interactions, while dotted lines represent indirect interactions between the genes. Genes colored green displayed two-fold or larger decreases, while genes colored red displayed two-fold or larger increases. Table 2 is a description on the genes represented in this network. (B) Real-time RT-PCR analysis of transcripts from pcd3J mice. The RNA expression of selected genes was assessed by real-time RT-PCR in pcd/+ and pcd/pcd mice and compared to wild-type mice.
Figure 3.
Bad protein expression in the Purkinje cell layer of the cerebellum of the P14 Nna1+/+ and Nna1-/- mutant mice. Cells in the Purkinje cell layer were counterstained with 4′,6-diamidino-2-phenylindole (DAPI), a nuclear label (A, D). Bad staining was absent in the Purkinje cell layer of Nna1 +/+ mice (B) but expression was detected in Purkinje cells of Nna1-/- mutants (E). Figures C and F are merged images indicating co-localization of DAPI and Bad in NNa1+/+ (C) and Nna1-/- mice (F). The scale bar is 20 μm.
Discussion
We examined the relative expression of genes in the cerebella of 14-day old pcd/pcd mice and clustered the differentially expressed genes into functional categories. Since the earliest cytological signs of Purkinje cell degeneration are not observed until after postnatal day 15 in PCD mice [1, 9], it is plausible that genes altered in these mice are related to the death of Purkinje cells. This study indicates that there are unique differences in gene expression between wild-type and pcd/pcd mice and suggests mechanisms that may be involved in the Purkinje cell death that characterize this mutant. Many previous microarray-based studies have simply reported large list of all of the individual genes that exhibit altered expression in a particular experimental paradigm. Although important information can be gathered from these lists, it usually remains unclear whether the genes identified are functionally relevant or merely indicators of the condition. Furthermore, in many cases, disease states may results in changes in particular classes of genes rather than the same individual gene in every circumstance [10, 11]. Therefore, we chose to also focus on functional classes of genes that may contribute to the development of Purkinje cell death in the PCD mutant mice.
To determine the functions that may lead to neuronal death in the pcd/pcd mice, we compared genes that were uniquely altered in pcd/pcd mice. IPA analysis of pcd/pcd mice indicated that genes and biological processes were regulated that promoted the death of Purkinje neurons. Bad is a well-known Bcl-2-associated pro-apoptotic protein [12]. Following an apoptotic stimulus, dephosphorylation of Bad allows its interaction with Bcl-XL and Bcl-2 thus facilitating neuronal apoptosis. Overexpression of Bad in cerebellar granule neurons results in cell death [12]. Bad was increased two-fold in the pcd/pcd mice. However, no change in Bad expression was seen in pcd/+, consistent with the lack of neuronal apoptosis in these mice.
One of the signaling molecules specifically upregulated in the pcd/pcd mice was cdk5. Cdk5 is a neuronal-specific cyclin-dependent kinase (cdk) that is involved in neuronal migration, differentiation and development [13-16]. Dysregulation of cdks has been shown to play a key role in neurodegenerative diseases, including Alzheimer's disease, amyotrophic lateral sclerosis, and Niemann-Pick Type C. Cdk5 inhibitors were shown to reduce the rate of Purkinje neuron death in a mouse model of Niemann-Pick Type C [14]. Therefore, it is plausible that upregulation of cdk5 plays a direct role in accelerated Purkinje cell death in pcd/pcd mice. Conversely, one intriguing phosphatase, PTEN, was specifically downregulated in the pcd/pcd mice. In mouse brain, PTEN is expressed in neurons and is particularly abundant in cerebellar Purkinje neurons [17, 18]. Recent studies using PTEN conditional knockouts have indicated a key role for PTEN in regulating Purkinje neuron survival [19-21]. Using En2 promoter-driven Cre mice, PTEN was deleted at embryonic day 9.5 in both neurons and glia at the midbrain-hindbrain junction. The resulting mice were also ataxic and there was degeneration of cerebellar Purkinje cells. When Cre was driven by an L7 promoter, PTEN was deleted postnatally in Purkinje cells. These mice displayed Purkinje cell degeneration. Studies are underway to determine the cellular localization of the protein and to regulate the expression of identified genes by deletion and expression strategies.
As previously stated, Nna1 was identified as the gene harboring a genetic lesion in multiple strains of PCD mutant mice [6]. The presence of several consensus motifs, including, most notably, 2 distinct nuclear localization signals, an acidic rich domain reminiscent of a canonical transcriptional transactivating domain, an ATP/GTP binding site, and a metallocarboxypeptidase domain, within the Nna1 polypeptide sequence led to the hypothesis that Nna1 may be a transcriptional regulatory factor [5]. The observed partitioning of overexpressed Nna1 protein between the nucleus and cytoplasm of transfected neurons supports the notion of Nna1 as a transcriptional regulatory molecule [5]. These findings, taken together, support the hypothesis that mutation of Nna1 may result in changes in global transcriptional profiles as seen in our microarray studies.
In summary, we used gene expression profiling and powerful data analysis tools to gain insight into mechanisms associated with neuronal degeneration and neuroprotection. IPA is a powerful software application that facilitates the rapid biological interpretation of gene lists generated from high-throughput microarray data. The remarkable potential for these methods of analyzing large microarray data sets resides in the ability to convert the results of expression profiling studies from studying individual gene to biological themes or pathways that may be involved in the initiation and/or progression of disease processes. The network analysis could be a powerful tool for developing effective therapeutic targets since in identifies focus genes within a biological pathway that have the most potential to regulate other genes in that pathway. In addition, these pathway tools could be employed to compare data sets from different individual experiments and potentially compare findings observed in different species. We acknowledge that is not possible from this study to determine whether these genes or pathways are the primary cause of Purkinje cell death in the pcd3J mutant. However, studies are underway to modulate the function of pathways and individual genes identified in this study in different cerebellar cell types. Analysis of the identified pathways will yield important information concerning the regulation of gene expression during neuronal degeneration during development and in neuropathological disorders. Therefore, the results of this study could have significant clinical implications and may aid in developing treatments for neurodegenerative disorders by providing novel targets for therapeutic intervention.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mullen RJ, Eicher EM, Sidman RL. Purkinje cell degeneration, a new neurological mutation in the mouse. Proc Natl Acad Sci U S A. 1976;73:208–212. doi: 10.1073/pnas.73.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landis SC, Mullen RJ. The development and degeneration of Purkinje cells in pcd mutant mice. J Comp Neurol. 1978;177:125–143. doi: 10.1002/cne.901770109. [DOI] [PubMed] [Google Scholar]
- 3.Roffler-Tarlov S, Beart PM, O'Gorman S, Sidman RL. Neurochemical and morphological consequences of axon terminal degeneration in cerebellar deep nuclei of mice with inherited Purkinje cell degeneration. Brain Res. 1979;168:75–95. doi: 10.1016/0006-8993(79)90129-x. [DOI] [PubMed] [Google Scholar]
- 4.Doulazmi M, Hadj-Sahraoui N, Frederic F, Mariani J. Diminishing Purkinje cell populations in the cerebella of aging heterozygous Purkinje cell degeneration but not heterozygous nervous mice. J Neurogenet. 2002;16:111–123. doi: 10.1080/01677060213157. [DOI] [PubMed] [Google Scholar]
- 5.Harris A, Morgan JI, Pecot M, Soumare A, Osborne A, Soares HD. Regenerating motor neurons express Nna1, a novel ATP/GTP-binding protein related to zinc carboxypeptidases. Mol Cell Neurosci. 2000;16:578–596. doi: 10.1006/mcne.2000.0900. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Gonzalez A, La Spada AR, Treadaway J, Higdon JC, Harris BS, Sidman RL, Morgan JI, Zuo J. Purkinje cell degeneration (pcd) phenotypes caused by mutations in the axotomy-induced gene, Nna1. Science. 2002;295:1904–1906. doi: 10.1126/science.1068912. [DOI] [PubMed] [Google Scholar]
- 7.Wang T, Morgan JI. The Purkinje cell degeneration (pcd) mouse: an unexpected molecular link between neuronal degeneration and regeneration. Brain Res. 2007;1140:26–40. doi: 10.1016/j.brainres.2006.07.065. [DOI] [PubMed] [Google Scholar]
- 8.Rong Y, Wang T, Morgan JI. Identification of candidate Purkinje cell-specific markers by gene expression profiling in wild-type and pcd(3J) mice. Brain Res Mol Brain Res. 2004;132:128–145. doi: 10.1016/j.molbrainres.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Wang T, Parris J, Li L, Morgan JI. The carboxypeptidase-like substrate-binding site in Nna1 is essential for the rescue of the Purkinje cell degeneration (pcd) phenotype. Mol Cell Neurosci. 2006;33:200–213. doi: 10.1016/j.mcn.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 11.Nisenbaum LK. The ultimate chip shot: can microarray technology deliver for neuroscience? Genes Brain Behav. 2002;1:27–34. doi: 10.1046/j.1601-1848.2001.00013.x. [DOI] [PubMed] [Google Scholar]
- 12.Akhtar RS, Ness JM, Roth KA. Bcl-2 family regulation of neuronal development and neurodegeneration. Biochim Biophys Acta. 2004;1644:189–203. doi: 10.1016/j.bbamcr.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Jeong YG, Lee KY, Lee BC, Lee NS, Won MH, Fukui Y. Post-natal changes of cyclin-dependent kinase 5 activator expression in the developing rat cerebellum. Anat Histol Embryol. 2005;34:20–26. doi: 10.1111/j.1439-0264.2004.00555.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhang M, Li J, Chakrabarty P, Bu B, Vincent I. Cyclin-dependent kinase inhibitors attenuate protein hyperphosphorylation, cytoskeletal lesion formation, and motor defects in Niemann-Pick Type C mice. Am J Pathol. 2004;165:843–853. doi: 10.1016/S0002-9440(10)63347-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohshima T, Gilmore EC, Longenecker G, Jacobowitz DM, Brady RO, Herrup K, Kulkarni AB. Migration defects of cdk5(-/-) neurons in the developing cerebellum is cell autonomous. J Neurosci. 1999;19:6017–6026. doi: 10.1523/JNEUROSCI.19-14-06017.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe G, Pena P, Shambaugh GE, 3rd, Haines GK, 3rd, Pestell RG. Regulation of cyclin dependent kinase inhibitor proteins during neonatal cerebella development. Brain Res Dev Brain Res. 1998;108:77–87. doi: 10.1016/s0165-3806(98)00032-7. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Liu F, Ross AH. PTEN regulation of neural development and CNS stem cells. J Cell Biochem. 2003;88:24–28. doi: 10.1002/jcb.10312. [DOI] [PubMed] [Google Scholar]
- 18.Li L, Liu F, Salmonsen RA, Turner TK, Litofsky NS, Di Cristofano A, Pandolfi PP, Jones SN, Recht LD, Ross AH. PTEN in neural precursor cells: regulation of migration, apoptosis, and proliferation. Mol Cell Neurosci. 2002;20:21–29. doi: 10.1006/mcne.2002.1115. [DOI] [PubMed] [Google Scholar]
- 19.Backman SA, Stambolic V, Suzuki A, Haight J, Elia A, Pretorius J, Tsao MS, Shannon P, Bolon B, Ivy GO, Mak TW. Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat Genet. 2001;29:396–403. doi: 10.1038/ng782. [DOI] [PubMed] [Google Scholar]
- 20.Kwon CH, Zhu X, Zhang J, Knoop LL, Tharp R, Smeyne RJ, Eberhart CG, Burger PC, Baker SJ. Pten regulates neuronal soma size: a mouse model of Lhermitte-Duclos disease. Nat Genet. 2001;29:404–411. doi: 10.1038/ng781. [DOI] [PubMed] [Google Scholar]
- 21.Marino S, Krimpenfort P, Leung C, van der Korput HA, Trapman J, Camenisch I, Berns A, Brandner S. PTEN is essential for cell migration but not for fate determination and tumourigenesis in the cerebellum. Development. 2002;129:3513–3522. doi: 10.1242/dev.129.14.3513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.