Abstract
Apo E plays an important role in chylomicron and VLDL remnant processing, uptake or conversion to LDL. The type of lipoprotein that isolates in the LDL density of E2/2 subjects was investigated and the effect of the apo E isoforms on LDL mass was determined in all genotypes in a large group of Type 1 diabetics. Analysis of the LDL composition of E2/2 homozygotes (n = 6) compared to subjects with the common E3/3 isoform (n = 6) demonstrated an enrichment in apo E, unesterified cholesterol, phospholipid and triglyceride relative to apo B in E2/2 subjects, more typical of a dense IDL remnant than of LDL. Although diabetics were studied, these findings are considered to reflect those of the general population. Comparison of the lipoprotein distribution of homozygous and heterozygous subjects revealed that, as genotype changed from E4/4 (n = 22) to E3/4 (n = 262), E3/3 (n = 710) = E2/4 (n = 30), E2/3 (n = 151), E2/2 (n = 6), LDL cholesterol decreased significantly in a stepwise manner. The decrease was not in a specific subgroup of LDL. In conclusion, for E2/2 subjects, lipoproteins isolated in the LDL density range appear to be composed mainly of dense IDL remnants and some Lp(a). The apo E isoform also has a significant effect on LDL concentration in both homozygotes and heterozygotes.
Keywords: Apo E genotype, E2/2, LDL, VLDL remnants, LDL composition
1. Introduction
Apo E is an apolipoprotein found associated with chylomicrons, VLDL, their remnants and HDL in plasma. One of the most important roles for apo E in lipoprotein metabolism is the hepatic processing and uptake of chylomicron and VLDL remnants [1] mediated by apo E binding. Processing of the remnant is thought to lead to hepatic removal from plasma or to the maturation of the VLDL remnant (IDL) to LDL. Apo E, in humans, is present in three isoforms, apo E2, E3 and E4. These isoforms are expressed by alleles from a single gene, which are inherited co-dominantly, generating six possible phenotypes. Apo E3 is the most common isoform [2]. Apo E2 differs from E3 in that a cysteine is substituted for an arginine at position 158 whereas apo E4 differs from E3 due to an arginine being substituted for a cysteine at position 112. These substitutions result in a progressive impairment of binding to the apo B/E receptor with apo E4 binding most avidly and apo E2 having the weakest binding.
An example of the role of apo E in remnant uptake and maturation is seen in individuals homozygous for apo E2/2 who demonstrate an accumulation of cholesteryl ester-enriched VLDL and chylomicron remnants in the blood. Some of these E2 homozygotes exhibit a substantial increase in remnants in plasma, are hyperlipidemic (type III dyslipidemia or remnant removal disease) and have an increased risk of coronary heart disease. However, most individuals homozygous for apo E2 do not demonstrate excessive remnant accumulation [3] indicating that a secondary factor is required which causes the system to exceed the capacity to process the remnants formed [4,5]. It has thus been suggested that a change of arginine at position 158 to a cysteine on apo E2 compared to apo E3 reduces binding significantly but does not eliminate it completely since the majority of E2/2 subjects do not demonstrate excessive remnant accumulation and the associated hyperlipidemia.
Apo E2 homozygotes also demonstrate very low levels of LDL cholesterol [3]. The cause of the low LDL is controversial and several hypotheses have been put forth. One of the popular hypotheses is that the hepatic LDL receptor is up regulated due to (1) decreased delivery of cholesterol to the liver as a result of defective binding of apo E2-containing lipoproteins [1,6] and/or (2) a reduced competition between the apo E2-containing lipoproteins and apo B-100 containing LDL for the LDL receptor [1,7]. Both mechanisms would lead to more rapid uptake of LDL and thus maintenance of a low LDL level in the presence of normal conversion of VLDL to LDL in plasma. However, some lipoprotein tracer studies indicate that the presence of arginine at position 158 appears to be optimal [8] or required [9,10] for the maturation of VLDL to LDL. The study by Demant et al. [8] reported that the conversion of VLDL to LDL was substantially reduced with increased direct catabolism of IDL, but the plasma LDL was normal in composition other than there being a difference in percent composition of +1.9% for triglyceride and -3.2% for unesterified cholesterol. They conclude that it was the decreased conversion that accounted for the decrease in plasma LDL. Other tracer studies [9,10] concluded that very little endogenous VLDL of hyperlipidemic E2/E2 subjects is converted to LDL since the specific activity curve of LDL did not transect the decay curves of either VLDL or IDL and only a small fraction of the apo B radioactivity from VLDL appeared in LDL (less than 1%). It has also been have suggested that remnants are present in all apo B containing fractions including LDL in E2/2 subjects [11]. Thus, we were interested in determining the type of lipoprotein that isolates in the LDL density range using a density gradient ultracentrifugation technique that emphasizes the LDL density region and we postulate that, while conversion to LDL from VLDL may be reduced, the majority of the lipoprotein that isolate at an LDL density in E2 homozygotes may actually be dense IDL that cannot be converted to mature LDL. This is an important point to clarify in order to further our understanding of the importance of the role of apo E in LDL formation.
Therefore, the primary focus of the study was to investigate the type of lipoprotein particle that is isolated in the LDL density range in E2/2 subjects in order to address the controversy as to whether the particles are mature plasma LDL, or are dense IDL remnants that cannot be converted to LDL due to impaired binding of apo E2, indicative of a vital role for apo E in LDL formation. We had available a large subject population from which we were able to obtain plasma from E2/2 subjects and matched E3/3 subjects for comparison of LDL composition and thus used these Type 1 diabetics to investigate this question. Since these subjects with Type 1 diabetes are healthy and have low to normal lipids, the results are considered to reflect that of the normal population.
The secondary goal of the study was to determine whether the apo E genotype affects LDL cholesterol mass and density distribution in this large population of Type 1 diabetics in a manner similar to that reported in several comprehensive studies of the non-diabetic population [12-18]. These studies have demonstrated that the e4 allele is associated with higher LDL cholesterol and the e2 allele with lower LDL cholesterol. Three studies in Type 1 diabetics [19-21] have been carried out and found similar relationships but were substantially smaller in size than the present study and did not compare all genotypes. Also, none of the studies examined the LDL density distribution across the genotypes.
2. Methods
2.1. Subject selection
All subjects investigated were healthy individuals with Type 1 Diabetes Control and Complications Trial (DCCT) [22,23] and its follow-up the Epidemiology of Diabetes Interventions and Complications (EDIC) study [24]. This study was approved by the Human Subjects Review Committees of each site of the DCCT/EDIC studies, including the University of Washington. Of the 1347 subjects that underwent extensive investigation with respect to lipids and metabolic parameters, there were 1299 Caucasians, 48 African Americans, 12 Hispanics, 6 Asians and 1 Native American/Alaskan. Females comprised 47% of the total number. Procedures were followed in accordance with institutional guidelines. After a 12-14 h fast, blood was collected in 0.1% EDTA and immediately placed on ice. Plasma was obtained and immediately frozen at -70 °C for lipid measurements and lipoprotein separation by density gradient ultracentrifugation (DGUC). For comparison of the lipoprotein distributions and cholesterol content of the LDL peak, as determined by DGUC, the samples used for the investigation were those obtained at the last visit of the DCCT (1992) and included most of the E3/3 subjects (n = 710 of 736) and E4/4 subjects (n = 22 of 23) and all E2/3 subjects (n = 151), E3/4 subjects (n = 262) and E2/4 subjects (n = 30). The E2/2 (n = 6 of 8) distribution was also obtained using these samples. For analysis of the LDL composition and comparison to that of matched E3/3 subjects, a second sample from the last visit of the DCCT was unavailable, thus samples from year 4 to 6 of the EDIC study (1997-2000) were used for analysis of the lipid and apoprotein composition of the LDL fractions and of the lipoprotein distribution for comparison between the E2/2 and E3/3 subjects. For this comparison six E2/2 subjects were matched with six E3/3 subjects for age, sex and BMI. The E3/3 subjects were also selected to have triglyceride (TG), total cholesterol (TC), HDL cholesterol and LDL cholesterol within one standard deviation of those of the E2/2 subjects. Although they were not matched for HbA1c levels, there was no significant difference in HbA1c nor were there differences in age, BMI, TC, TG, HDL cholesterol and LDL cholesterol which confirms the successful selection of the subjects for these criteria.
2.2. Analysis of plasma lipids and apoproteins
Total cholesterol (TC), LDL cholesterol, cholesteryl ester (CE), unesterified cholesterol (UC), phospholipid (PL) and triglyceride (TG), for the investigation of LDL composition, were determined by standardized methodologies at the Northwest Lipid Research Laboratories [25]. Apo A-I, apo B and apo E were determined by nephelometry. Lp(a) was measured by an ELISA method [26]. To overcome the effects of apo(a) size heterogeneity on the measurement of Lp(a) levels, the ELISA utilizes a monoclonal antibody specific to a unique epitope present on Kringle 4 type 9 of apo(a) [27]. Thus, the assay provides an accurate determination of Lp(a) levels independent of the size of the apo(a) molecules. The assay has been extensively validated in a large number of individuals [26].
2.3. Density gradient ultracentrifugation
For determination of the lipoprotein cholesterol distribution, cholesterol content of the LDL peak and LDL composition, a discontinuous salt density gradient was created in an ultracentrifuge tube [28]. Briefly, 1 ml of plasma was combined with 1.5 ml of a d 1.006 g/ml NaCl solution and 1.5 ml of a d 1.21 g/ml solution and mixed, resulting in a final density of 1.0825 g/ml. A solution (8.5 ml) of d 1.006 g/ml NaCl was placed in a Beckman ultracentrifuge tube and underlayered with the 4 ml of the d 1.0825 g/ml solution, containing the plasma. Samples were centrifuged at 65,000 rpm for 70 min at 4 °C in a Beckman VTi 65.1 vertical rotor. Thirty-eight 0.34 ml fractions were collected from the bottom of the tube and the cholesterol concentration of each fraction was determined. Previously we have determined that VLDL is isolated in fractions 28-38, IDL in fractions 17-30 tailing into 31-34, LDL in fractions 7-18 and HDL in fractions 1-7. Fractions corresponding to: (1) dense IDL/very buoyant LDL, (2) buoyant LDL, and (3) dense LDL were pooled and analyzed for lipid and apoprotein composition for the comparison of E2/2 with E3/3. The same fractions were used for the E2/2 subject and the E3/3 subject paired comparison. The average fractions numbers were 17-20 for dense IDL/very buoyant LDL, 13-16 for buoyant LDL and 9-12 for dense LDL (includes the peak fraction of LDL in most cases). To obtain the cholesterol content of LDL, for the comparison of the LDL mass of the homozygotes and heterozygotes, the cholesterol concentrations in fractions 9-14 were summed which are the fractions that contain most of the LDL mass. Values reported in the tables and figures, other than those in Table 1, are those obtained from the density gradient and are not plasma values. We have investigated the effect of prolonged freezing (>10 years) at -70 °C on the cholesterol or apo B content of the lipoprotein profile obtained by DGUC and have not observed a significant change with time. This is dependent on the samples being immediately frozen and maintained at this temperature without thawing which is the case for the samples used in the present study.
Table 1.
Demographic characteristics and plasma lipids of E2/2 subjects and matched E3/3 subjects
| Age (yr) | Sex | BMI (kg/m2) | TC (mmol/l) | TG (mmol/l) | HDL (mmol/l) | LDL (mmol/l) | HbA1c (%) | |
|---|---|---|---|---|---|---|---|---|
| E2/2 subjects, n=6 | ||||||||
| 1. | 35 | F | 24.1 | 4.26 | 1.14 | 2.51 | 1.27 | 6.3 |
| 2. | 46 | F | 27.3 | 2.97 | 0.75 | 1.01 | 1.63 | 5.1 |
| 3. | 32 | M | 24.1 | 3.0 | 1.61 | 1.34 | 0.96 | 7.6 |
| 4.a | 31 | M | 27.3 | 4.88 | 2.81 | 1.11 | 2.52 | 8.4 |
| 5. | 43 | F | 22.2 | 4.55 | 1.24 | 1.76 | 2.25 | 8.1 |
| 6. | 45 | F | 25.3 | 3.7 | 1.01 | 2.2 | 1.06 | 9.0 |
| Mean | 38.7 | 25.1 | 3.89 | 1.43 | 1.65 | 1.6 | 7.4 | |
| S.D. | 6.8 | 2.0 | 0.8 | 0.73 | 0.61 | 0.65 | 1.5 | |
| E3/3 subjects, n=6 | ||||||||
| 1. | 31 | F | 22.9 | 4.19 | 0.64 | 1.65 | 2.25 | 7.0 |
| 2. | 42 | F | 27.3 | 4.03 | 0.47 | 1.71 | 2.12 | 6.7 |
| 3. | 29 | M | 22.4 | 4.32 | 1.02 | 1.32 | 2.56 | 9.2 |
| 4. | 33 | M | 26.9 | 4.63 | 1.25 | 1.01 | 3.07 | 11.6 |
| 5. | 44 | F | 22.2 | 4.11 | 1.13 | 1.6 | 2.02 | 7.1 |
| 6. | 41 | F | 25.2 | 4.37 | 1.01 | 1.89 | 2.04 | 7.4 |
| Mean | 36.7 | 24.5 | 4.27 | 0.92 | 1.53 | 2.34 | 8.2 | |
| S.D. | 6.4 | 2.3 | 0.21 | 0.30 | 0.31 | 0.41 | 1.9 |
There were no significant differences between the variables in the E2/2 subjects compared to the E3/3 subjects (p > 0.05).
See Section 3 for data on subject E2/2 #4.
2.4. Genotyping
Apo E genotyping was performed in all subjects according to a previously described method [29]. The 244 bp PCR amplicon containing the 112 and 158 restriction sites in exon 4 of the APO E gene was digested with the restriction enzyme HhaI and the fragments were electrophoretically separated on a 10% polyacrylamide gel and visualized with ethidium bromide staining. Negative and positive replicate controls were genotyped along with samples to ensure reproducibility between genotyping experiments and 5% of samples were repeated with 99-100% replicate agreement. Allele calls were independently made by two observers.
2.5. Statistics
To assess differences in the LDL composition of the six E2/2 subjects compared to the six matched E3/3 subjects a Wilcoxon Signed Rank test was used. This non-parametric test was chosen because the subjects were matched and it was not possible to determine if the values were normally distributed due the low number of subjects. This test reports differences in the median but, for clarity and comparison with other studies, the values in the tables are listed as means. The Wilcoxon Signed Rank test was also used to assess differences in age, BMI, lipids and HbA1c between the E2/2 subjects and the matched E3/3 subjects. To assess differences in the sum of the cholesterol content of the LDL peak between the various genotypes, a Kruskal-Wallis One Way Analysis of Variance on Ranks was carried out. To determine which specific genotypes differed from each other, a Wilcoxon Rank Sum test was used. This method of analysis was also used to determine differences between the cholesterol content of the individual fractions in the LDL peak among the various genotypes.
3. Results
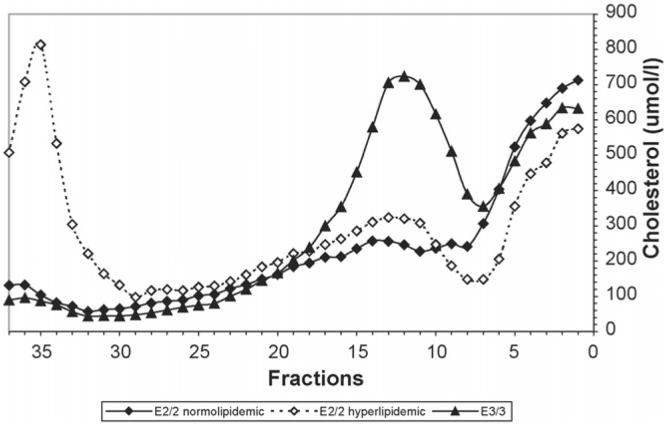
The lipoprotein distribution and LDL composition of the E2/2 subjects (n = 6) were compared to that of the E3/3 subjects (n = 6), matched for age, gender and BMI. Demographic characteristics are outlined in Table 1. There was no significant difference between age, BMI, cholesterol, triglyceride, LDL cholesterol, HDL cholesterol or HbA1c. Upon investigation of the plasma lipoprotein distribution, determined by DGUC, of the E2/2 subjects and comparing the distribution to that of the E3/3 subjects, marked differences in the profiles were noted (Fig. 1). Given that one of the E2/2 subjects was hyperlipidemic (subject #4, Table 1), this subject's distribution is shown separately from the average of the five normolipidemic E2/2 subjects. Compared to the E3/3 subjects, the dyslipidemic E2/2 subject demonstrated an increase in VLDL (in fractions 28-37) particularly in the dense VLDL (VLDL remnants) (in fractions 31-35) and in IDL (in fractions 17-30). In the absence of this dyslipidemic subject, the VLDL and IDL cholesterol distribution of the remaining normolipidemic E2/2 subjects were similar to, but appeared to be slightly higher, than that of the E3/3 subjects (Fig. 1). In both the normolipidemic as well as the hyperlipidemic E2/2 subjects, the LDL was extremely low compared to the E3/3 subjects, being the lowest in the normolipidemic individuals. HDL cholesterol concentrations appeared to be slightly increased in the normolipidemic E2/2 subjects but reduced in the hyperlipidemic subject compared to the E3/3 subjects. It is interesting to note that the dyslipidemic subject had normal lipids (TC = 3.41 mmol/l, TG 0.75 mmol/l) on enrollment in the DCCT at a time when the subject's BMI was normal (BMI 22 kg/m2). However, after intensive diabetes therapy which, in this individual, was associated with a substantial weight gain (BMI 26 kg/m2), hyperlipidemia became manifest (TC = 5.84 mmol/l, TG 4.62 mmol/l) at the end of the DCCT.
Fig. 1.
Lipoprotein distribution of apo E2/2 subjects compared to matched E3/3 subjects. Plasma lipoproteins from the subjects were separated by density gradient ultracentrifugation. The mean cholesterol content of each fraction is listed. VLDL isolates in fractions 28-37, IDL in fractions 17-30 with tailing into fractions 31-34, LDL isolates in fractions 7-18, with the majority of the mass in fractions 9-14, and HDL in fractions 1-7. The distribution of the hyperlipidemic apo E2/2 subject (n =1)(◊) is shown separately from that of the normolipidemic E2/2 subjects (n =5)(♦). The distribution of the E3/3 subjects (n =6)(▲) represents those matched with the E2/2 subjects.
In an attempt to address the controversy regarding the etiology of the low LDL cholesterol levels in E2/2 subjects, the composition of the LDL fractions as well as the dense IDL fractions was analyzed and compared to that of the apo E3/3 subjects. The fractions from the LDL and dense IDL density range were pooled into 3 groups corresponding to dense IDL/very buoyant LDL (pool 1, fractions 17-20), buoyant LDL (pool 2, fractions 13-16) and dense LDL (pool 3, fractions 9-12). The three pools were analyzed for lipid and apoprotein composition (Tables 2A and 2B). There was significantly less CE, UC, PL and apo B mass in the buoyant (pool 2) and dense LDL (pool 3) of the E2/2 subjects as compared to that of the E3/3 subjects, in accordance with the reduced mass in this region (fractions 13-16 and fractions 9-12 in Fig. 1). Relative to the apo B content, the dense IDL/very buoyant LDL (pool 1), buoyant LDL (pool 2) and dense LDL (pool 3) were substantially enriched in TG in the E2/2 subjects compared to the E3/3 subjects (TG/apo B, Table 2B). The TG/apo B ratios for buoyant LDL (pool 2) and dense LDL (pool 3) of the E2/2 subjects were similar to that of the dense IDL (pool 1) of the E3/3 subjects. There was not only an increase in TG relative to apo B in the buoyant (pool 2) and dense LDL (pool 3) of the E2/2 subjects, but also enrichment in apo E, PL and UC relative to apo B (apo E/apo B, PL/apo B, UC/apo B, Tables 2A and 2B) more similar to the ratios of dense IDL (pool 1) than of buoyant or dense LDL of the E3/3 subjects. The apo E/apo B ratios of the LDL fractions (pools 1-3, Table 2A) of the E2/2 subjects were 5-10 times greater than that of the E3/3 subjects. The ratio of TG/CE was also significantly higher in buoyant LDL (pool 2) and dense LDL (pool 3) in the E2/2 subjects compared to the E3/3 subjects, again closer to that of dense IDL (pool 1) rather than to buoyant or dense LDL of the E3/3 subjects. The enrichment in TG relative to CE was not due solely to the elevation in plasma TG of the hyperlipidemic E2/2 subject as the TG/CE ratio was at least two to three times higher in all E2/2 subjects compared to their E3/3 matches. In the case of Lp(a), the E2/2 subjects did not demonstrate an increase in any of the pools but there was significantly more Lp(a) relative to apo B in the buoyant and dense LDL of the E2/2 subjects than the E3/3 subjects. To further demonstrate that the composition of the lipoproteins isolated in the LDL density range in the E2/2 subjects was more like that of a dense IDL remnant than plasma LDL, we compared the lipid/apo B ratios to those of plasma IDL and LDL (Table 2B) calculated from reported lipid and apo B values [30,31]. We found that for the E3/3 subjects all lipid/apo B ratios and TG/CE values for buoyant LDL (pool 2) and dense LDL (pool 3) were similar to those of plasma LDL (Table 2B). In comparison, for the E2/2 subjects, the ratios for buoyant LDL (pool 2) and dense LDL (pool 3), though lower than plasma IDL, were substantially higher than plasma LDL for TG/apo B and PL/apo B, TG/CE was much closer to plasma IDL than LDL, and UC/apo B was the same as plasma IDL (Table 2B).
Table 2A.
LDL protein composition of E2/2 subjects compared to E3/3 subjects
| Apo B (nmol/l) |
Apo E (nmol/l) |
Lp(a) (nmol/l) |
Apo E/Apo B |
Lp(a)/Apo B |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pool | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 |
| E2/2 n=6 | |||||||||||||||
| Mean | 67 | 100a | 81a | 39 | 53 | 86 | 0.1 | 1.4 | 2.2 | 0.63 | 0.54a | 1.12a | 0.002 | 0.02a | 0.03a |
| S.D. | 23 | 32 | 22 | 23 | 21 | 31 | 0.2 | 1.9 | 2.8 | 0.36 | 0.2 | 0.49 | 0.003 | 0.02 | 0.04 |
| E3/3 n=6 | |||||||||||||||
| Mean | 85 | 225 | 326 | 19 | 24 | 48 | 0.4 | 0.9 | 1.2 | 0.24 | 0.11 | 0.14 | 0.004 | 0.004 | 0.004 |
| S.D. | 23 | 34 | 92 | 15 | 12 | 23 | 0.6 | 0.4 | 0.8 | 0.2 | 0.06 | 0.03 | 0.006 | 0.002 | 0.004 |
Pool 1 = dense IDL/very buoyant LDL, fractions 17-20; Pool 2 = buoyant LDL, fractions 13-16; Pool 3 = dense LDL, fractions 9-12.
p < 0.05, E2/2 vs. E3/3.
Table 2B.
LDL lipid composition of E2/2 subjects compared to E3/3 subjects
| TG (μmol/l) |
CE (μmol/l) |
UC (μmol/l) |
PL (μmol/l) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pool | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 |
| E2/2, n=6 | ||||||||||||
| Mean | 58 | 52 | 42 | 101 | 140a | 120a | 89 | 105a | 96a | 89 | 123a | 136a |
| S.D. | 12 | 17 | 13 | 38 | 48 | 32 | 27 | 24 | 29 | 24 | 32 | 36 |
| E3/3, n=6 | ||||||||||||
| Mean | 51 | 62 | 60 | 106 | 331 | 469 | 92 | 181 | 230 | 90 | 204 | 292 |
| S.D. | 24 | 23 | 18 | 26 | 66 | 158 | 34 | 25 | 35 | 23 | 24 | 36 |
| TG/Apo B | CE/Apo B | PL/Apo B | UC/Apo B | TG/CE | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma IDL | 2582 | 1593 | 2253 | 1154 | 0.8 | ||||||||||
| Plasma LDL | 118 | 1440 | 800 | 517 | 0.08 | ||||||||||
| TG/Apo B |
CE/Apo B |
PL/Apo B |
UC/Apo B |
TG/CE |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | |
| E2/2, n=6 | |||||||||||||||
| Mean | 924a | 529a | 532a | 1477 | 1390 | 1499 | 1376 | 1264a | 1791a | 1389 | 1092a | 1264a | 0.69 | 0.39a | 0.35a |
| S.D. | 237 | 150 | 207 | 354 | 168 | 223 | 161 | 202 | 812 | 286 | 210 | 577 | 0.38 | 0.11 | 0.09 |
| E3/3, n=6 | |||||||||||||||
| Mean | 570 | 277 | 206 | 1264 | 1476 | 1422 | 1075 | 917 | 931 | 1069 | 806 | 730 | 0.46 | 0.2 | 0.15 |
| S.D. | 112 | 97 | 112 | 99 | 167 | 99 | 97 | 97 | 171 | 99 | 56 | 123 | 0.12 | 0.09 | 0.09 |
Pool 1 = dense IDL/very buoyant LDL, fractions 17-20; Pool 2 = buoyant LDL, fractions 13-16; Pool 3 = dense LDL, fraction 9-12. TG = triglyceride; CE = cholesteryl ester; UC = unesterified cholesterol; PL = phospholipid. Plasma IDL and LDL values were obtained from Refs. [30, 31].
p < 0.05, E2/2 vs. E3/3.
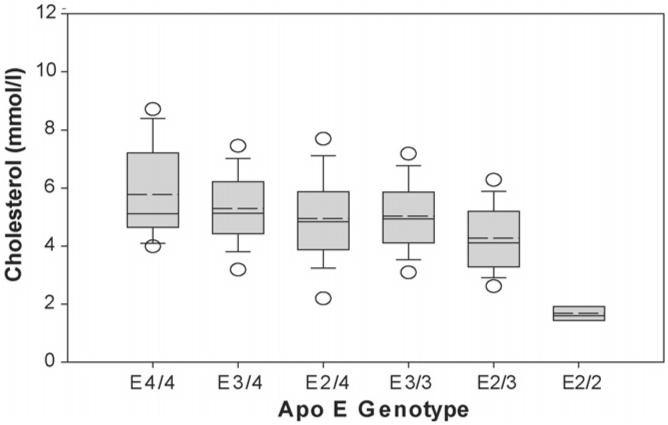
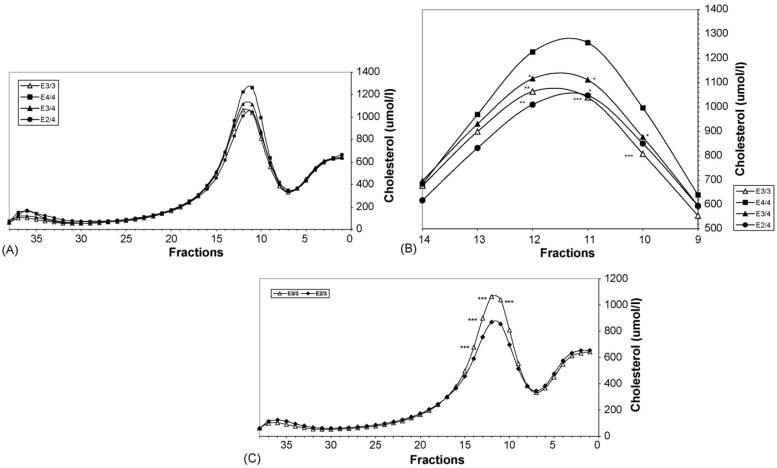
It is not only the apo E2/2 genotype that affects the LDL cholesterol mass, as differences were observed when the other genotypes were compared (Figs. 2 and 3A-C, Table 3). The effect of the apo e allele on the cholesterol content of the LDL peak appears to occur in a dose dependent manner with subjects homozygous for the e4 allele (n = 22) having the highest LDL cholesterol (sum of fractions 9-14), the E3 homozygotes (n = 710) an intermediate LDL level, and E2 homozygotes (n = 6) the lowest LDL cholesterol (Fig. 2). The presence of at least one e4 allele also affected LDL cholesterol levels in that the E3/4 subjects (n = 262) had more LDL cholesterol compared to the E3/3 subjects (n = 710) (p = 0.002). Interestingly, the presence of one e2 allele also impacted the lipoprotein distribution as the E2/3 subjects (n = 151) had less LDL cholesterol than the E3/3 subjects (p < 0.001). Conversely, the presence of apo E3 in the E2/3 subjects (n = 151) resulted in an increase in LDL cholesterol (p < 0.001), the appearance of a typical LDL peak, and an absence of remnant accumulation as compared to the E2/2 subjects (n = 6). It is also notable that subjects with one apo e4 and one apo e2 allele had a LDL cholesterol level equivalent to subjects with e3/3 alleles (Fig. 2). Thus, there was a significant difference in LDL peak mass between E2/2 and all other genotypes, between heterozygotes E3/4 and E3/3, 2/3, between E2/4 and E2/3, between E4/4 and E2/3, between E3/3 and E2/3, with a trend for a difference between E3/3 and E4/4 (p = 0.06) (Fig. 2, Table 3). When differences in the mass of cholesterol in the individual LDL fractions were analyzed rather than the summed LDL fractions (Fig. 3A-C), there were significant differences between all genotypes except for that of E3/3 with E2/4 and E2/4 with E3/4. Fig. 3A demonstrates the total lipoprotein cholesterol distribution of E4/4, E3/4, E3/3 and E2/4 subjects separated by DGUC. Fig. 3B is a magnification of the LDL region of Fig. 3A that includes fractions 9-14 and indicates significant differences between the cholesterol concentration of the individual fractions. Fig. 3C show the lipoprotein cholesterol distribution of E2/3 and E3/3 subjects. It is important to note that these figures not only demonstrate the decrease in the LDL cholesterol peak from genotype to genotype but also reveal that the decrease in the peak across the genotypes from E4/4 (Fig. 3A and B) through to E2/3 (Fig. 2C) is not due to a decrease in a particular LDL subgroup, i.e. light or dense LDL, but to a decrease in the overall LDL mass. In the case of the E2/2 subjects (Fig. 1) the peak is flattened and tends to be shifted towards the more buoyant fractions with a reduction in mass in the fractions corresponding to dense LDL. This is consistent with the presence of dense IDL rather than LDL, in concurrence with our findings upon analysis of the composition of the LDL of E2/2 subjects versus E3/3 subjects (Table 2A and B). Finally, it should be noted that the mean lipoprotein distribution of the 6 E3/3 subjects (from year 4 or 6 of EDIC) used for the comparison of the LDL composition (Fig. 1) was very similar to that of the total 710 E3/3 subjects (from the last visit of the DCCT) (Fig. 3A) indicating that the distribution profile is representative of individuals with the E3/3 genotype and the comparison using the 6 E3/3 subjects is a valid representation of the E3/3 population.
Fig. 2.
Peak LDL cholesterol concentration by apo E genotype. The cholesterol concentrations of the LDL peak (sum of cholesterol concentration in fractions 9-14 comprising the majority of the peak) of the E4/4 subjects (n = 22), E3/4 subjects (n = 262), E2/4 subjects (n = 30), E3/3 subjects (n = 710), E2/3 subjects (n = 151) and E2/2 (n = 6) were obtained from the lipoprotein distribution isolated by density gradient ultracentrifugation. The median (—) and the mean (- -) are indicated. The lower and upper end of the box indicates the 25th and 75th percentile, respectively. The lower and upper end of the error bars represent the 10th and 90th percentile, respectively. The lower and upper dots are the 5th and 95th percentile, respectively. See Table 3 for significance of differences between the cholesterol in the LDL peak between apo E genotypes.
Fig. 3.
(A) Plasma lipoprotein distribution of E4/4, E3/4, E3/3 and E2/4 genotypes obtained by density gradient ultracentrifugation. Plasma lipoproteins of E4/4 subjects (n = 22) (▀), E3/4 subjects (n = 262) (▴), E3/3 subjects (n = 710) (△) and E2/4 subjects (n = 30) (●) were isolated by density gradient ultracentrifugation and the cholesterol content of each fraction was analyzed. See Fig. 1 legend for location of VLDL, IDL, LDL and HDL. (B) Cholesterol distribution in LDL peak (Fractions 9-14). The LDL cholesterol distribution of fractions 9-14 of E4/4 subjects (n = 22) (▀), E3/4 subjects (n = 262) (▴), E3/3 subjects (n = 710) (△) and E2/4 subjects (n = 30) (●) was magnified from (A) and significance differences in the cholesterol content of the fractions relative to E4/4 are indicated (*p≤0.05, **p ≤ 0.01, ***p ≤ 0.001). Lipoproteins were isolated by density gradient ultracentrifugation and the cholesterol content of each fraction was analyzed. (C) Plasma lipoprotein distribution of E3/3 and E2/3 subjects obtained by density gradient ultracentrifugation. Plasma lipoproteins from E3/3 subjects (n = 710) (△) and E2/3 subjects (n = 151) (◆) were isolated by density gradient ultracentrifugation and the cholesterol content of each fraction was analyzed. Significance differences in the cholesterol content of the fractions of E3/3 compared to E2/3 are indicated (***p ≤ 0.001). See Fig. 1 legend for location of VLDL, IDL, LDL and HDL.
Table 3.
Significance of differences in LDL cholesterol (sum of fractions 9-14) between apo E genotypes
| E4/4 | E3/4 | E2/4 | E3/3 | E2/3 | E2/2 | |
|---|---|---|---|---|---|---|
| E4/4 | - | 0.32 | 0.11 | 0.06 | <0.001 | 0.002 |
| E3/4 | - | 0.195 | 0.002 | <0.001 | <0.001 | |
| E2/4 | - | 0.72 | 0.02 | 0.002 | ||
| E3/3 | - | <0.001 | <0.001 | |||
| E2/3 | - | <0.001 | ||||
| E2/2 | - |
Hyperglycemia did not affect the results as there was no significant difference between the genotype groups in HbA1c (E2/2 7.3% ± 1.7, E2/3 8.4% ± 1.4, E2/4 8% ± 1.2, 8.3% ± 1.5, E3/4 8.2% ± 1.4, E4/4 8.1% ± 1.4).
The study also confirms that, as is the case in nondiabetic subjects, most E2/2 subjects have low lipids, and that only with a second hit, such as the weight gain with intensive diabetes therapy [32], which occurred in the one hyperlipidemic E2/2 subject, that the effect of the e2 allele is manifested as hyperlipidemia and remnant removal disease. The study also demonstrates that the frequency of apo E2 homozygosity in these Type 1 diabetic subjects is similar to that of the normal population (0.7% versus 0.8%, respectively).
4. Discussion
The present study has determined that, contrary to some of the current theories pertaining to the cause of low LDL in E2/2 subjects, lipoproteins that are isolated in the LDL density range are not simply low concentrations of typical plasma LDL but rather appear to be comprised mainly of dense IDLlike remnants and some Lp(a). This study has also confirmed the findings of previous studies in non-diabetics that the apo E isoform significantly impacts the LDL cholesterol content in both homozygotes as well as heterozygotes and, due to the large number of subjects available, was able to demonstrate an effect on LDL in all genotypes unlike other studies that examined this in subjects with Type 1 diabetes. These results were obtained using a density gradient ultracentrifugation technique that separates LDL into several fractions, allowing for examination of the LDL density distribution and cholesterol mass and for the separation of IDL/very buoyant LDL, buoyant LDL and dense LDL. This method of demonstrating differences in the LDL distribution of the apo E genotypes has never been previously employed.
The major goal of the study was to examine the nature of the particles that are isolated in the LDL density range in E2/2 subjects, based on their lipid and apoprotein composition in order to determine whether the lipoproteins were typical plasma LDL or were a different type of lipoprotein that varied in composition. In E2/2 subjects, in accordance with other studies, we found LDL to be substantially reduced, as indicated by the reduced amount of apo B, CE, UC and PL in both the buoyant and dense LDL fractions, but we also observed that there was no distinct LDL peak present as compared to all other homozygous or heterozygous apo E genotypes. On examination of the composition of the LDL of the E2/2 subjects and comparing it to that of E3/3 subjects, we found that the LDL particles were TG and apo E enriched. The fact that the fractions were also enriched in PL and UC relative to apo B and had a higher TG/CE ratio, which are all characteristic of remnants, suggests that the lipoproteins, though found in the LDL density range in the E2/2 subjects, may consist primarily of dense IDL remnant particles rather than true LDL. The composition of buoyant LDL and dense LDL in the E2/2 subjects were similar to that of the dense IDL fractions of the E3/3 subjects and the UC/apo B ratios were similar to that of plasma IDL, closer to plasma IDL than LDL for the TG/CE ratio and substantially higher than plasma LDL for TG/apo B and PL/apo B ratios. The presence of dense IDL remnants in the LDL region is likely due to the fact that the remnants cannot be converted to true LDL due to the lack of processing associated with apo E binding to the hepatocyte, and thus the remnants remain in plasma. Thus, conversion of VLDL to “LDL” may be reduced [8] but the particles that are present in the LDL density range appear to be mainly dense IDL. It is interesting to note that similar particles were formed in vitro [33] as a result of VLDL lipolysis by lipoprotein lipase (LPL), in the presence of cholesteryl ester transfer protein (CETP) and phospholipid transfer protein (PLTP) but in the absence of the liver and thus absence of apo E processing and hepatic lipase activity (Murdoch and Breckenridge, unpublished data). The particles formed under these conditions in vitro were isolated in the IDL, light LDL density range by ultracentrifugation and were enriched in apo E, TG, PL and UC, as were those of the E2/2 subjects observed in the present study. Although the enrichment in TG in the remnants which isolated in the LDL density range in the E2/2 subjects would have been expected to result in particles of decreased density relative to normal LDL, the increase in density of these particles may be accounted for by the enrichment with apo E and other apolipoproteins. Upon calculation of the density of the combined lipid and protein added per particle in the E2/2 subjects as compared to the E3/3 subjects, we found the resultant density of the additional lipid and protein to be 1.04 g/l, that of LDL, in concurrence with our findings. It is notable that, in the case of the E2/2 subject that became dyslipidemic, the increase in mass in the LDL density range appears to be due to an increase in these remnants and not an increase in LDL. Lp(a) was also present in the LDL density range in the buoyant but particularly in the dense LDL fractions and there was an increased amount in all E2/2 subjects relative to apo B. Since there was a decrease in apo B mass in buoyant and dense LDL, this does not imply that Lp(a) is higher in E2/2 subjects but indicates that Lp(a) makes up a greater percentage of the total lipoprotein mass in the buoyant and dense LDL fractions compared to E3/3 subjects. The apo E content in the dense LDL of the E2/2 subjects was 10 times that of the E3/3 and exceeded a ratio of 1 relative to apo B, as expected for IDL remnants. The ratio in dense IDL/very buoyant LDL and buoyant LDL was five to six times that of the E3/3 subjects but was somewhat less than anticipated if at least one apo E per particle is expected for remnants. One reason for this may be due to the presence of Lp(a) in the buoyant and dense LDL fractions which would effectively reduce this ratio. It should also be noted that it is possible that some typical plasma LDL may be present. If so, and assuming the Lp(a) is of normal composition, then the remaining particles which we refer to as dense IDL remnants would be even further enriched in TG, UC, PL and apo E relative to apo B than reported here.
These results are in concurrence with the findings of Chait et al. [9,10] in terms of their observation of very low conversion of IDL to LDL occurring in the E2/2 subjects since the lipoproteins that were isolated in the LDL density range in the present study appear to be more characteristic of IDL remnants than of plasma LDL. Possibly remnants were not detected in the LDL range in the studies of Chait et al. due to their precipitation method of isolation. The results of the present study are in contrast to those of Demant et al. [8] with respect to the composition of the particles that isolated at an LDL density. They reported that the LDL was similar to plasma LDL other than being triglyceride enriched and slightly lower in unesterified cholesterol. The reason for this discrepancy may be that their LDL was isolated sequentially with prolonged ultracentrifugation spins at 23 °C, a temperature at which CETP and PLTP are active as well as any LPL present on plasma lipoproteins. This is in contrast to our method of isolation, which is a rapid one-step technique and is carried out at 4 °C with the samples being consistently maintained at this temperature. Thus, the lipoprotein composition of the LDL isolated by Demant et al. may have been altered with processing and the values may not reflect that of the native particle composition. Furthermore they expressed the LDL composition as a percentage so that a change in the percentage composition of one parameter necessarily results in a change in that of the other parameters, whereas our measurements were reported on a mass basis and relative to apo B mass.
We also observed that it is not only the E2/2 genotype, associated with low LDL cholesterol, or the E4/4 genotype, associated with higher LDL cholesterol, that impact the LDL cholesterol concentration but that there appears to be a continuum of LDL cholesterol levels when comparing the various genotypes. As the genotype changes from E4/4 to E2/2, the LDL cholesterol concentration, determined by DGUC, decreases in a stepwise manner presumably because the apo e alleles are expressed co-dominantly. Thus the E2/4 subjects demonstrate LDL cholesterol similar to that of the E3/3 subjects. Furthermore, the separation of LDL by DGUC allowed for the analysis of the LDL density distribution which demonstrated that as LDL cholesterol decreased from E4/4 to E3/4, E3/3 = E2/4, E2/3, the decrease was in the total LDL peak and was not selective for a particular LDL subspecies such as buoyant or dense LDL. The only genotype that appeared to vary is that of the E2/2 subjects because there is no true LDL peak. Another important conclusion from our measurements of LDL mass across the genotypes is that subjects with only one e2 allele exhibited lower LDL cholesterol as compared to those with the e3 or e4 allele, indicating that it is not only the homozygotes that demonstrate an effect of the apo E allele on LDL cholesterol concentrations but heterozygotes are affected as well. Finally, it is noteworthy that E2/3 heterozygotes do not demonstrate remnant accumulation but have typical LDL indicating that the amount of apo E3 present is adequate for formation of LDL in these individuals. Our results pertaining to the impact of the e2 versus the apo e4 allele on LDL cholesterol are in agreement with studies of non-diabetics. Sing and Davignon [12] first published the observation that the presence of the e2 allele significantly reduced LDL cholesterol and the e4 allele increased it. This was later reported in several other studies [13-17] including an extensive analysis by Schaefer of a larger population stratified by sex and age [18]. There were no studies in Type 1 diabetics that investigated such a large population as that of the present study. Consequently, differences were determined in genotypes between groups based on the presence of E2 or E4 in most studies and the various genotypes were not individually compared [19-21,34]. In contrast to these studies, as well as those in non-diabetic subjects, we have demonstrated statistically significant differences between LDL cholesterol in almost all genotypes for the sum of the cholesterol in the LDL peak distribution, or between the concentrations of cholesterol in the individual LDL fractions.
The present study has demonstrated that the presence of the apo e2 allele significantly impacts the lipoprotein distribution and composition and, as observed in non-diabetics, most E2/2 subjects have low lipids, with a predominance of remnants isolating in the LDL density range. It confirms that only with a second hit, such as the weight gain with intensive diabetes therapy [32] that occurred in the one hyperlipidemic E2/2 subject, that the effect of the e2 allele is manifested as hyperlipidemia and severe remnant removal disease.
This study was carried out in Type 1 diabetics who have low-normal lipids but are hyperglycemic in some cases. Although hyperglycemia may result is a shift in LDL from buoyant to dense [35] associated with insulin resistance and hypertriglyceridemia [36], it does not account for the differences in the comparisons between genotypes carried out in this study nor does it negate the extrapolation of these results to the normal non-diabetic population. In the comparison of the LDL composition of the E2/2 subjects with the E3/3 subjects there was no significant difference in HbA1c between the two groups, thus hyperglycemia did not account for the differences between the groups. Furthermore the effect of hyperglycemia on LDL density cannot account for the difference between the remnant-like composition of the LDL in E2/2 subjects and the composition typical of LDL observed in the E3/3 subjects when comparing the two groups or extrapolating the results to the normal population. Hyperglycemia cannot account for the differences in the LDL in the all genotype comparison in that there were no significant differences in HbA1c between the groups. Furthermore we did not observe a shift in the LDL density in the comparison of the E4/4 to E3/4, E3/3, E2/4, E2/3 subjects, but rather a progressive decrease in mass of the overall LDL peak. In the E2/2 subjects there is a flattening of the LDL peak with more mass in the buoyant fractions as well as the fractions that contain IDL and less in the dense. However this difference appears to be accounted for by the fact that there is a preponderance of remnants rather than LDL in this region.
In summary, upon investigation of the LDL composition of apo E2/2 subjects as compared to that of E3/3 subjects, we found that the small amount of apparent “LDL” in the E2/2 subjects appears to be largely made up of very dense IDL remnants and some Lp(a) and the results suggest that true plasma LDL may not be present as the major lipoprotein constituent. Finally, the fact that the apo E genotype has a marked effect on LDL cholesterol (but not density other than in the E2/2 subjects) in the heterozygotes as well as the homozygotes is an important observation with respect to the effect of apo E on LDL metabolism.
Acknowledgements
This study was supported by National Institute of Health Grants DK02456, EDIC-NO1 DK62204 Epidemiology of Diabetes Interventions and Complications, the Juvenile Diabetes Foundation, the General Clinical Research Centers Program (MO1 RR0000037), National Center for Research Resources, the Clinical Nutrition Research Unit (DK35816) and contracts with the Division of Diabetes, Endocrinology and Metabolic Diseases of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Health. We would like to thank the subjects of the DCCT/EDIC program and the investigators who participated in the study.
Abbreviations
- Apo E
apolipoprotein E
- DGUC
density gradient ultracentrifugation
References
- [1].Mahley RW, Huang Y, Rall SC., Jr Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia). Questions, quandaries, and paradoxes. J Lipid Res. 1999;40:1933–49. [PubMed] [Google Scholar]
- [2].Utermann G, Langenbeck U, Beisiegel U, Weber W. Genetics of the apolipoprotein E system in man. Am J Hum Genet. 1980;32:339–47. [PMC free article] [PubMed] [Google Scholar]
- [3].Rall SC, Jr, Weisgraber KH, Mahley RW. Isolation and characterization of apolipoprotein E. Methods Enzymol. 1986;128:273–87. doi: 10.1016/0076-6879(86)28073-8. [DOI] [PubMed] [Google Scholar]
- [4].Utermann G. Apolipoprotein E mutants, hyperlipidemia and arteriosclerosis. Adv Exp Med Biol. 1985;183:173–88. doi: 10.1007/978-1-4613-2459-1_14. [DOI] [PubMed] [Google Scholar]
- [5].Rall SC, Jr, Newhouse YM, Clarke HR, et al. Type III hyperlipoproteinemia associated with apolipoprotein E phenotype E3/3. Structure and genetics of an apolipoprotein E3 variant. J Clin Invest. 1989;83:1095–101. doi: 10.1172/JCI113988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Davignon J, Gregg RE, Sing CF. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis. 1988;8:1–21. doi: 10.1161/01.atv.8.1.1. [DOI] [PubMed] [Google Scholar]
- [7].Woollett LA, Osono Y, Herz J, Dietschy JM. Apolipoprotein E competitively inhibits receptor-dependent low density lipoprotein uptake by the liver but has no effect on cholesterol absorption or synthesis in the mouse. Proc Natl Acad Sci USA. 1995;92:12500–4. doi: 10.1073/pnas.92.26.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Demant T, Bedford D, Packard CJ, Shepherd J. Influence of apolipoprotein E polymorphism on apolipoprotein B-100 metabolism in normolipemic subjects. J Clin Invest. 1991;88:1490–501. doi: 10.1172/JCI115459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chait A, Brunzell JD, Albers JJ, Hazzard WR. Type-III hyperlipoproteinaemia (“remnant removal disease”). Insight into the pathogenetic mechanism. Lancet. 1977;1:1176–8. doi: 10.1016/s0140-6736(77)92717-9. [DOI] [PubMed] [Google Scholar]
- [10].Chait A, Hazzard WR, Albers JJ, Kushwaha RP, Brunzell JD. Impaired very low density lipoprotein and triglyceride removal in broad beta disease: comparison with endogenous hypertriglyceridemia. Metabolism. 1978;27:1055–66. doi: 10.1016/0026-0495(78)90151-8. [DOI] [PubMed] [Google Scholar]
- [11].Havel RJ, Kotite L, Vigne JL, et al. Radioimmunoassay of human arginine-rich apolipoprotein, apoprotein E. Concentration in blood plasma and lipoproteins as affected by apoprotein E-3 deficiency. J Clin Invest. 1980;66:1351–62. doi: 10.1172/JCI109988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sing CF, Davignon J. Role of the apolipoprotein E polymorphism in determining normal plasma lipid and lipoprotein variation. Am J Hum Genet. 1985;37:268–85. [PMC free article] [PubMed] [Google Scholar]
- [13].Ehnholm C, Lukka M, Kuusi T, Nikkila E, Utermann G. Apolipoprotein E polymorphism in the Finnish population: gene frequencies and relation to lipoprotein concentrations. J Lipid Res. 1986;27:227–35. [PubMed] [Google Scholar]
- [14].Boerwinkle E, Utermann G. Simultaneous effects of the apolipoprotein E polymorphism on apolipoprotein E, apolipoprotein B, and cholesterol metabolism. Am J Hum Genet. 1988;42:104–12. [PMC free article] [PubMed] [Google Scholar]
- [15].Eichner JE, Kuller LH, Ferrell RE, Meilahn EN, Kamboh MI. Phenotypic effects of apolipoprotein structural variation on lipid profiles. III. Contribution of apolipoprotein E phenotype to prediction of total cholesterol, apolipoprotein B, and low density lipoprotein cholesterol in the healthy women study. Arteriosclerosis. 1990;10:379–85. doi: 10.1161/01.atv.10.3.379. [DOI] [PubMed] [Google Scholar]
- [16].Hallman DM, Boerwinkle E, Saha N, et al. The apolipoprotein E polymorphism: a comparison of allele frequencies and effects in nine populations. Am J Hum Genet. 1991;49:338–49. [PMC free article] [PubMed] [Google Scholar]
- [17].Xhignesse M, Lussier-Cacan S, Sing CF, Kessling AM, Davignon J. Influences of common variants of apolipoprotein E on measures of lipid metabolism in a sample selected for health. Arterioscler Thromb. 1991;11:1100–10. doi: 10.1161/01.atv.11.4.1100. [DOI] [PubMed] [Google Scholar]
- [18].Schaefer EJ, Lamon-Fava S, Johnson S, et al. Effects of gender and menopausal status on the association of apolipoprotein E phenotype with plasma lipoprotein levels. Results from the Framingham Offspring Study. Arterioscler Thromb. 1994;14:1105–13. doi: 10.1161/01.atv.14.7.1105. [DOI] [PubMed] [Google Scholar]
- [19].Winocour PH, Tetlow L, Durrington PN, Ishola M, Hillier V, Anderson DC. Apolipoprotein E polymorphism and lipoproteins in insulin-treated diabetes mellitus. Atherosclerosis. 1989;75:167–73. doi: 10.1016/0021-9150(89)90173-1. [DOI] [PubMed] [Google Scholar]
- [20].Eichner JE, Ferrell RE, Kamboh MI, et al. The impact of the apolipoprotein E polymorphism on the lipoprotein profile in insulin-dependent diabetes: the Pittsburgh Epidemiology of Diabetes Complications Study IX. Metabolism. 1992;41:347–51. doi: 10.1016/0026-0495(92)90066-j. [DOI] [PubMed] [Google Scholar]
- [21].Blaauwwiekel EE, Beusekamp BJ, Sluiter WJ, Hoogenberg K, Dullaart RP. Apolipoprotein E genotype is a determinant of low-density lipoprotein cholesterol and of its response to a low-cholesterol diet in Type 1 diabetic patients with elevated urinary albumin excretion. Diabet Med. 1998;15:1031–5. doi: 10.1002/(SICI)1096-9136(1998120)15:12<1031::AID-DIA705>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- [22].The Diabetes Control and Complications Trial (DCCT) Design and methodologic considerations for the feasibility phase. The DCCT Research Group. Diabetes. 1986;35:530–45. [PubMed] [Google Scholar]
- [23].The Diabetes Control and Complications Trial Research Group The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med. 1993;329:977–86. doi: 10.1056/NEJM199309303291401. [DOI] [PubMed] [Google Scholar]
- [24].Epidemiology of Diabetes Interventions and Complications (EDIC) Design, implementation, and preliminary results of a long-term follow-up of the Diabetes Control and Complications Trial cohort. Diabetes Care. 1999;22:99–111. doi: 10.2337/diacare.22.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Warnick GR. Enzymatic methods for quantification of lipoprotein lipids. Methods Enzymol. 1986;129:101–23. doi: 10.1016/0076-6879(86)29064-3. [DOI] [PubMed] [Google Scholar]
- [26].Marcovina SM, Albers JJ, Wijsman E, Zhang Z, Chapman NH, Kennedy H. Differences in Lp[a] concentrations and apo[a] polymorphs between black and white Americans. J Lipid Res. 1996;37:2569–85. [PubMed] [Google Scholar]
- [27].Marcovina SM, Albers JJ, Gabel B, Koschinsky ML, Gaur VP. Effect of the number of apolipoprotein(a) kringle 4 domains on immunochemical measurements of lipoprotein(a) Clin Chem. 1995;41:246–55. [PubMed] [Google Scholar]
- [28].Hokanson JE, Austin MA, Brunzell JD. Measurement and clinical significance of low-density lipoprotein subclasses. In: Rifai N, Warnick GR, Dominiczak MH, editors. Handbook of Lipoprotein Testing. American Association of Clinical Chemistry Press; Washington: 1997. pp. 267–82. [Google Scholar]
- [29].Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31:545–8. [PubMed] [Google Scholar]
- [30].McFarlane C, Young IS, Hare L, Mahon G, McEneny J. A rapid methodology for the isolation of intermediate-density lipoprotein: characterization of lipid composition and apoprotein content. Clin Chim Acta. 2005;353:117–25. doi: 10.1016/j.cccn.2004.10.010. [DOI] [PubMed] [Google Scholar]
- [31].Eisenberg S. Plasma lipoprotein conversions: the origins of low-density and high-density lipoproteins. Ann N Y Acad Sci. 1980;348:30–47. doi: 10.1111/j.1749-6632.1980.tb21289.x. [DOI] [PubMed] [Google Scholar]
- [32].Purnell JQ, Hokanson JE, Marcovina SM, Steffes MW, Cleary PA, Brunzell JD. Effect of excessive weight gain with intensive therapy of type 1 diabetes on lipid levels and blood pressure: results from the DCCT. Diabetes Control and Complications Trial. Jama. 1998;280:140–6. doi: 10.1001/jama.280.2.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Murdoch SJ, Breckenridge WC. Effect of lipid transfer proteins on lipoprotein lipase induced transformation of VLDL and HDL. Biochim Biophys Acta. 1996;1303:222–32. doi: 10.1016/0005-2760(96)00105-1. [DOI] [PubMed] [Google Scholar]
- [34].Salo MK, Rantanen R, Huupponen T, Lehtimaki T, Jokela H. Apolipoprotein E phenotypes and plasma lipids in diabetic children and adolescents. Eur J Pediatr. 1993;152:564–8. doi: 10.1007/BF01954081. [DOI] [PubMed] [Google Scholar]
- [35].Purnell JQ, Marcovina SM, Hokanson JE, et al. Levels of lipoprotein(a), apolipoprotein B, and lipoprotein cholesterol distribution in IDDM. Results from follow-up in the Diabetes Control and Complications Trial. Diabetes. 1995;44:1218–26. doi: 10.2337/diab.44.10.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Carr MC, Brunzell JD. Abdominal obesity and dyslipidemia in the metabolic syndrome: importance of type 2 diabetes and familial combined hyperlipidemia in coronary artery disease risk. J Clin Endocrinol Metab. 2004;89:2601–7. doi: 10.1210/jc.2004-0432. [DOI] [PubMed] [Google Scholar]