Abstract
The c-myb proto-oncogene encodes an obligate hematopoietic cell transcription factor important for lineage commitment, proliferation, and differentiation. Given its critical functions, c-Myb regulatory factors are of great interest but remain incompletely defined. Herein we show that c-Myb expression is subject to posttranscriptional regulation by microRNA (miRNA)–15a. Using a luciferase reporter assay, we found that miR-15a directly binds the 3′-UTR of c-myb mRNA. By transfecting K562 myeloid leukemia cells with a miR-15a mimic, functionality of binding was shown. The mimic decreased c-Myb expression, and blocked the cells in the G1 phase of cell cycle. Exogenous expression of c-myb mRNA lacking the 3′-UTR partially rescued the miR-15a induced cell-cycle block. Of interest, the miR-15a promoter contained several potential c-Myb protein binding sites. Occupancy of one canonical c-Myb binding site was demonstrated by chromatin immunoprecipitation analysis and shown to be required for miR-15a expression in K562 cells. Finally, in studies using normal human CD34+ cells, we showed that c-Myb and miR-15a expression were inversely correlated in cells undergoing erythroid differentiation, and that overexpression of miR-15a blocked both erythroid and myeloid colony formation in vitro. In aggregate, these findings suggest the presence of a c-Myb–miR-15a autoregulatory feedback loop of potential importance in human hematopoiesis.
Introduction
The c-myb proto-oncogene is the founding member of a family of genes, A-myb (MYBL1), B-myb (MYBL2), and c-myb (MYB), which encode transcription factors that regulate genes important for lineage fate, cell proliferation, and maturation.1–9 In hematopoietic cells, c-Myb is required for normal cell development, as demonstrated by the fact that homozygous c-Myb null mice die in utero at approximately day 15 of severe anemia10 and silencing c-myb expression in vitro interferes with normal myeloid colony formation.11 c-Myb's role in lineage determination is illustrated by recently described transactivation domain mutations that are associated with marrow megakaryocyte hyperplasia and profound thrombocytosis.12 Other important functions are alluded to by the observation that disrupting c-Myb expression with dominant negative mutants interferes with cell-cycle progression and sensitizes leukemia cells to DNA damaging agents.13 It is becoming increasingly accepted that Myb function is cell type– and context-dependent.14
It is well known that c-myb expression is highest in primitive hematopoietic cells and that down-regulation of its expression is an essential prerequisite for terminal differentiation.15 It has been suggested that within this general framework, setting precise c-Myb levels is required for proper functioning during cell development.16–20 The very short half-life of c-myb's mRNA and protein is consistent with this suggestion, but mechanistic details regarding the cis- and trans-acting factors which regulate the protein's synthesis and turnover are not fully known. The 5′ promoter region of c-myb has binding sites for a large number of proteins that appear to regulate its expression both positively and negatively, including c-Ets-1, NF-κB, E2F, WT1, and c-Myb itself.21–25 In the murine system, a transcriptional pause site within the first intron of the gene may also play a role in varying c-Myb levels during hematopoietic cell development.26,27 That expression is responsive to cues from the external environment is demonstrated by the fact that interleukin-2 (IL-2) up-regulates c-myb expression in T lymphocytes via the PI3K, protein kinase B pathway.22 Other important elements of c-myb's regulatory network remain to be defined.
MicroRNAs (miRNAs) constitute a newly appreciated class of noncoding RNAs, 21 to 23 nucleotides in length, which play an important role in regulating gene expression at the posttranscriptional level. They are derived from endogenously expressed transcripts with characteristic hairpin structures. These are processed by 2 ribonucleases of the RNase III family, Drosha and Dicer, into short hairpins of the characteristic length.28 MiRNAs regulate mRNA usage by hybridizing to specific, but difficult to predict,29 sites within the 3′ UTR of candidate mRNAs. In this manner they inhibit translation of the mRNA by interfering with 80s ribosome assembly,30 association of eIF4E to the m7G cap,31 or alternatively, by directing physical destruction of the mRNA. miRNA genes constitute approximately 1% to 2% of the known genes in eukaryotes and it has been predicted that miRNAs may regulate protein production from as many as 10% to 30% of all human genes.32 Accumulating evidence indicates that miRNA function is critical during normal cellular development and homeostasis, and abnormalities in miRNA activity have also been found to contribute to the pathogenesis of several hematologic malignancies including CLL and acute leukemia.33 Nonetheless, validated miRNA targets remain limited. The growing association of microRNAs and leukemogenesis caused us to wonder if they might also regulate expression of c-Myb, a gene whose involvement in the pathogenesis of leukemia and lymphoma has long been hypothesized and recently directly demonstrated.34,35 Herein we report that c-Myb and miR-15a form an autoregulatory feedback loop, with functional significance in normal hematopoietic cells, and perhaps malignant human hematopoietic cells as well.
Methods
Suspension cell cultures
Human embryonic kidney 293T (HEK293T) cells were maintained in Dulbecco modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (HyClone Laboratories, Logan, UT). K562 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS).
Human erythroid progenitor cells cultured from normal human hematopoietic cells (CD34+) were obtained from the University of Pennsylvania Stem Cell and Xenograft Core Facility. The University of Pennsylvania School of Medicine Institutional Review Board approved the acquisition and distribution of normal hematopoietic cells. These cells were then cultured in Iscove modified Dulbecco medium (IMDM; Invitrogen) supplemented with 20% of serum substitute BIT9500 (StemCell Technologies, Vancouver, BC), 10 U/mL erythropoietin (EPO; Amgen, Thousand Oaks, CA), and 10 ng/mL stem cell factor (SCF; R&D Systems, Minneapolis, MN) and harvested on days 1, 2, 3, 4, 6, 8, and 10 of culture, washed twice in Dulbecco phosphate-buffered saline (D-PBS; Invitrogen), and stored frozen at −80°C until thawed for the assays indicated in the text.
Colony-forming unit and burst-forming unit assays
Normal human bone marrow CD34+ cells (3.0 × 103) or ficoll-separated human bone marrow mononuclear cells (MNCs; 1.2 × 105), transfected with either a miR-15a mimic or control miR (Dharmacon, Lafayette, CO), were suspended in 900 μL IMDM containing 20% FBS and cytokines to stimulate colony-forming unit–erythroid (CFU-E) growth (EPO 10 U/mL; Amgen), colony-forming unit–granulocyte, macrophage (CFU-GM) growth (IL-3 20 ng/mL and GM-CSF 10 ng/mL; Amgen), or burst-forming unit–erythroid (BFU-E) growth (SCF 5 ng/mL and EPO 10 U/mL; Amgen). The suspended cells were added to 3.1 mL of methylcellulose and then transferred into 3 separate 35-mm Petri dishes (1.2 ml per dish). CFU-E, CFU-GM, and BFU-E were enumerated under an inverted microscope on day 14 of incubation.
Constructs
An 1191-bp fragment encompassing the entire human c-myb 3′-UTR was PCR-amplified with the following sense (5′-TGTCTAGAGACATTTCCAGAAAAGCAT-3′) and antisense (5′-GTTCTAGAAGGTAAAATAAGGGCACATCT-3′) primers using standard procedures and a proofreading polymerase (Platinum Pfu; Invitrogen). The cDNA clone (ATCC number 1415590), which contains the full length of c-myb 3′-UTR, was used as template. The PCR product was sub-cloned into the pCR 2.1 vector (Invitrogen). The c-myb 3′-UTR inserts were removed from the pCR 2.1 plasmid by XbaI digestion and ligated into a XbaI site located downstream of the firefly luciferase (f-luc) reporter gene (pGL3-Basic; Promega, Madison, WI) driven by a 900-bp 5′ flanking region from human Bub1 gene. The authenticity and orientation of the inserts relative to the luciferase gene were confirmed by sequencing. The resulting plasmids were designated pBub1/myb3U.
The expression plasmids pGL3/myb3U/ miR-15a1, pGL3/myb3U/miR-15a2, and pGL3/myb3U/miR-15a12, deleting the 2 miR-15a seed binding sites harbored in the c-myb 3′-UTR using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) or were simultaneously generated by using the pGL3/myb3U plasmid as template. The primers are available upon request.
Transfection
Partially double-stranded RNAs that mimic endogenous precursor miRNAs, hsa-miR-15a, hsa-miR-107, hsa-miR-150, and miRNA Negative Control no. 1, were obtained from Dharmacon (Lafayette, CO). In addition, anti-miR miRNA inhibitors (anti–miR-15a and anti-miR Negative Control #1), designed to inhibit endogenous miRNAs, were also obtained from Dharmacon. HEK293 T cells were transfected with the luciferase reporter constructs described above, pRL-CMV (Promega) and the appropriate miRNA precursor using Lipofectamine 2000 (Invitrogen). After 48 hours, f-luc and Renilla luciferase (r-luc) activities were determined using the dual-luciferase reporter assay system (Promega) and a luminometer. Alternatively, K562 cells were transfected with either specific miR mRNAs or anti-miR miRNA as described above with an Amaxa (Gaithersburg, MD) Nucleofector. Amaxa Nucleofector was also used to transfected CD34+ cells (106 cells/transfection) and mononuclear cells (107 cells/transfection) isolated from normal human bone marrow.
Quantitative real-time polymerase chain reaction for c-myb expression
RNA was isolated from cells using TRIzol (Invitrogen) and the RNA was subsequently treated with RNase-free DNase I, quantified by absorbance at 260 nm, and cDNA was synthesized from 1 μg of total RNA using oligo(dT). Quantitative reverse transcription–polymerase chain reaction (RT-PCR) was carried out in an ABI 7900 HT Sequence Detection System using TaqMan master mix and the protocol of the manufacturer (Applied Biosystems, Foster City, CA). All data were normalized using the endogenous GAPDH control. The sequences for the primers and probes are available upon request.
miR-15a isolation and detection
miR-15a was isolated from cells using mirVana miRNA Isolation Kit from Ambion (Austin, TX). Subsequently, the mirVana qRT-PCR Primer Set for miR-15a was used for detection of miR-15a expression according manufacturer's instruction. Primer set for U6 snRNA was used to normalize for RNA content among different experimental samples.
Flow cytometry and cell-cycle analysis
K562 cells were transfected with miRNA mimics. Forty-eight hours later, 106 cells were collected and washed with phosphate-buffered saline (PBS). Cells were then suspended in NP-40 solution (0.1% sodium citrate, 10 mM NaCl, 0.03% NP-40, pH 6.0) with 200 μg/mL RNase and stained with 20 μg/mL propidium iodide (PI). Cells were analyzed for DNA content. The cell population in each cell cycle was calculated with FlowJo software version 4.6.1 (TreeStar, Ashland, OR).
Western blotting
Protein samples were prepared from 2 × 106 K562 cells or CD34+ cells undergoing differerentiation lysed in 40 μL RIPA buffer (1× PBS, 1% NP40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]) with the addition of the protein inhibitors. The same amount proteins were loaded onto SDS-polyacrylamide electrophoresis gel and followed by Western blot with anti–c-Myb antibody (Upstate-Millipore, Charlottesville, VA). The same blots were stripped by Re-blot Solution (Chemicon, Temecula, CA) and relabeled with antibodies against β-actin as loading controls.
Chromatin immunoprecipitation assay (ChIP)
K562 cells (5.0 × 106 cells) were cross-linked with 0.4% formaldehyde. Then the cells were lysed, DNA-protein complexes were immunoprecipitated, and the formaldehyde–cross-linked DNA was reverse cross-linked with a ChIP assay kit (Upstate-Millipore) according to the manufacturer's protocol. DNA-Myb complexes were immunoprecipitated with anti-c-Myb. ChIP assays were also completed with anti-acetyl histone H4 as a positive control, and normal mouse immunoglobulin G (IgG) as a negative control. The purified DNA was then amplified by real-time PCR by standard protocols with SYBR green and miR-15a promoter-specific primers that flanked c-Myb binding site 1, −2248 to −1998 relative to the transcription start site (forward, 5′-CCATCCTTTGCTGG CACTAA-3′; reverse, 5′-GGTCCCCCTCATCACATACA-3′), or c-Myb binding 2, −653 to −464 bp relative to the transcription start site (forward, 5′-GGGAGAATTAGTTTGTTTAGAG CTTTT-3′; reverse, 5′-CACATCAAGGGAAAAACATGAA-3′).
Results
Three microRNAs potentially interact with the 3′-UTR of the Human c-myb mRNA
Several prediction programs have been developed to identify potential miRNA targets.36 An online search of the TargetScanS Target Database demonstrated that 14 miRNAs potentially bind to the 3′-UTR of the human c-myb mRNA. The binding sites of these miRNAs are conserved across species. In this study, we have focused on 3 miRNAs, miR-15a, −107 and −150. There are 2 potential target sites for miR-15a (Figure 1A) and miR-150 (Figure 1B), and one for miR-107 (Figure 1C). To experimentally validate the computational data, 1191 bp of the human c-myb 3′-UTR was subcloned downstream of the f-luc open reading frame (Figure 2A). This reporter construct (pBub1/Myb3U) was cotransfected in the HEK293 T-cell line with pRL-CMV (to normalize for transfection differences) and a control nontargeting RNA oligonucleotide (miR-Control), or miR-15a, −107, or −150. Interestingly, the relative luciferase activity was markedly diminished in cells cotransfected with the pBub1/Myb3U construct and miR-15a (74.1 ± 1.5%) or miR-107 (68.2 ± 5.3%), and was modestly decreased (22.1 ± 4.2%) in cells cotransfected with the pBub1/Myb3U construct and miR-150 mimic (Figure 2B).
Figure 1.
The human c-myb 3′-UTR harbors putative binding sites complementary to miR-15a, miR-107 and miR-150. (A) Complementarity between miR-15a and the 2 putative c-myb 3′-UTR sites targeted (634-661 bp and 661-687 bp downstream from the c-myb stop codon). (B) Complementarity between miR-107 and the single putative c-myb 3′-UTR site targeted (660-686 bp downstream from the c-myb stop codon). (C) Complementarity between miR-150 and the 2 putative c-myb 3′-UTR sites targeted (801-835 bp and 876-910 bp downstream from the c-myb stop codon). Sequence conservation (italics) across human, mouse, rat, and dog is also indicated.
Figure 2.
miR-15a can inhibit reporter activity. (A) Schematic representation of the f-luc reporter constructs used. The pBub1/Myb3U plasmid contains the full-length 3′-UTR of the human c-myb fused to f-luc. The promoter of the firefly luciferase (900-bp fragment from Bub1 promoter), stop codon as well as the target sites of the 3 putative microRNAs is also shown. (B) HEK293 T cells were cotransfected with 300 ng pBub1/Myb3U, 15 ng pRL-CMV, and a 50-nM concentration of a given RNA oligonucleotide. Forty-eight hours after transfection, luciferase activities were measured. f-luc activity was normalized to r-luc expression and the mean activities plus or minus SE from 3 independent experiments are shown.
miR-15a decreases human c-Myb expression in K562 human myeloid leukemia cells
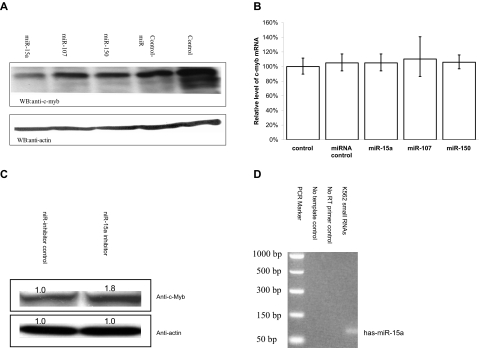
K562 cells endogenously express relatively high c-myb mRNA levels and are widely used to study hematopoietic cells growth and differentiation. To determine whether the candidate miRNAs might effect endogenous c-Myb expression, K562 cells were transfected with the miRNAs of interest. c-Myb levels, before and after transfection, were then quantitated by Western blotting. Of the candidates examined, only miR-15a was able to effect a significant reduction in K562 c-Myb protein content compared with controls (Figure 3A).
Figure 3.
miR-15a can decrease endogenous c-Myb protein levels in K562 cells. (A) K562 cells were transfected with 50-nM concentration of a given RNA oligonucleotide. Forty-eight hours after transfection, c-Myb protein Western blot analysis was performed with cell lysates obtained from the transfected K562 cells. (B) Quantitative real-time PCR (QRT-PCR) assay performed on RNA isolated from K562 cells transfected with the indicated RNA oligonucleotide molecules. Data are presented as a function of c-myb mRNA copies relative to GAPDH mRNA. Mock (control) cells were subjected to nucleoporation, but in the absence of RNA. The mean c-myb mRNA level are from 3 independent transfection experiments. (C) K562 cells were transfected with 100 nM has-miR-15a inhibitor (miR-15a-In) or miR inhibitor negative control (miR-In-Control). Forty-eight hours after transfection, c-Myb protein Western blot analysis was carried out using cell lysates from the transfected K562 cells. (D) Detection of the expression of miR-15a in K562 cells using mirVana qRT-PCR primer set (Ambion) according the manufacturer's instructions.
To investigate whether miR-15a targeted c-myb mRNA for degradation, real-time PCR was carried out on RNA isolated from miR-transfected K562 cells. In accord with classical miRNA activity, c-myb mRNA levels in cells transfected with miR-15a were not significantly decreased compared with control cell steady state mRNA levels (Figure 3B). Together, these results demonstrate that miR-15a decreased c-Myb protein expression by inhibiting mRNA translation, not by targeting c-myb mRNA for degradation. To further confirm that miR-15a can negatively regulate c-myb expression, miR-15a oligonucleotide inhibitors were transfected into K562 cells. Myb protein levels were then measured in transfected cells by Western blotting. A modest but statistically significant increase in c-Myb protein levels was observed in cells treated with the miR-15a inhibitors, providing additional evidence that miR-15a might participate in a physiologically relevant manner in the regulation of c-Myb expression (Figure 3C). Finally, we examined K562 cells for the presence of miR-15a and found that it was indeed expressed in wild-type cells (Figure 3D).
miR-15a inhibits reporter luciferase activities in a dose-dependent manner by binding to its seed regions of c-myb 3′-UTR
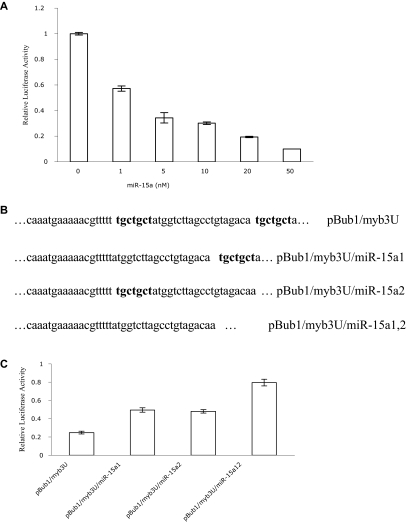
To test the potency of the miR-15a, pBub1/Myb3U was cotransfected into HEK293 T cells with increasing concentrations of miR-15a mimics, or nontargeting control miRNA. Luciferase activities were subsequently determined. Dose-response experiments demonstrated a clear correlation between luciferase activity and amount of miR-15a transfected into cells. Compared with controls, relative luciferase activity was significantly decreased with as little as 1 nM miR-15a. Maximal decrease was obtained with a 50-nM concentration of this miRNA (Figure 4A). In contrast, increasing concentrations of miR control had little effect on luciferase activity even when the concentration of these RNAs was 100 nM (data not shown).
Figure 4.
miR-15a interacts with the 3′-UTR of c-myb directly. (A) Dose-dependent inhibition of luciferase activity by miR-15a. HEK293 T cells were cotransfected with 200 ng pBub1/Myb3U, 10 ng pRL-CMV, and either miR-Control or miR-15a at the concentrations indicated, and relative luciferase activities were calculated as described above. The mean activities plus or minus SE from 3 independent transfection experiments are shown. (B) Constructs for the wild type c-myb UTR reporter and miR-15a seed binding sites (bold) deletion mutants. Only the miR-15a binding regions are shown. (C) Deletion of the seed binding sites of miR-15a in 3′-UTR of c-myb reduces miR-15a activity. HEK293 T cells were cotransfected with 10 ng pRL-CMV, 50 nM miR-15a or miR control and 4 3′-UTR of c-myb containing reporter constructs (300 ng), respectively. miR-15a inhibition activity was calculated as a function of luciferase activity reduction by miR-15a relative to miR control. Data are representative of 3 independent experiments.
To demonstrate that miR-15a interacts with a specific target sequence localized in the human c-myb 3′-UTR, 3 additional reporter constructs were generated in which the 2 7-bp “seed” sequences (ie ACGACGA), which are complementary to the 5′ end of miR-15a (Figure 1A), were deleted individually or simultaneously (Figure 4B). The resulting constructs, pBub1/Myb3U/miR-15a1, pBub1/Myb3U/ miR-15a2 and pBub1/Myb3U/ miR-15a1,2 were cotransfected together with miR-15a into HEK293 T cells. Luciferase activity in respective cells was then measured. Compared with the decrease in luciferase activity observed when the authentic c-myb 3′UTR was cotransfected with miR-15a, deletion of the miR-15a binding sites in the c-myb 3′UTR resulted in a 2- to 3-fold increase in luciferase activity, indicating that miR15a was no longer able to bind the UTR with the same avidity (Figure 4C). All these data are consistent with the hypothesis that miR-15a hybridizes with the predicted sequence in the c-myb 3′-UTR, and that alteration of the sequence to which the miR hybridizes would result in enhanced luciferase activity.
miR-15a blocks cell-cycle transit in G1
c-Myb is required for hematopoietic cell-cycle progression. One might, therefore, assume that if miR-15a bound to c-myb mRNA and repressed its expression, cell-cycle progression would be impeded. To examine this assumption, we determined whether overexpression of miR-15a would perturb K562 cell cycling, and whether this effect could be causally linked to its effect on c-Myb protein expression levels. Flow cytometric analysis of actively growing K562 cells transfected with miR-15a revealed that approximately 48% of cells were in the G0/G1 phase of the cell cycle compared with approximately 35% of control cells. A corresponding decrease in cells comprising the G2/M phase was also noted as approximately 20% of control cells were in this phase of the cell cycle compared with approximately 10% of cells treated with the miR-15a (Figure 5A top panel). Of note, neither miR-107 nor miR-150 mimics had any effect on K562 cell cycling (Figure 5A bottom panel).
Figure 5.
miR-15a blocks K562 cell-cycle progression in G1. (A) K562 cells were transfected with 100 nM of the indicated microRNA mimics. Forty-eight hours after nucleofection, cells were stained with PI and then analyzed for DNA content by FACS. The population of cells in each phase of the cell cycle was calculated with FlowJo software. Since nucleofection resulted in the death of approximately 30% of K562 cells, the cell-cycle analysis excluded the sub-G1 population. A representative result of 3 independent experiments is shown. (B) Exogenous c-Myb expression partially overcomes miR-15a-induced cell-cycle arrest. Representative data from 3 independent experiments are shown.
To further define the mechanism of the miR15a induced cell-cycle block, 2 additional experiments were carried out. First, we investigated the effect of miR-15a on cell-cycle progression of HEK293 and Hela cells. Neither of these cell lines is hematopoietic in origin, and c-Myb expression in both is either very low or undetectable. When these cells were transfected with miR-15a, unlike K562 cells, there was no discernable effect on cell cycling (data not shown). Second, we carried out “rescue” experiments in K562 cells. For these studies, we used pcDNA3-c-Myb, a construct previously reported from our laboratory that expresses full-length c-Myb but does not contain the 3′-UTR.29 As such, pcDNA 3-c-Myb expresses a form of c-Myb that should not be subject to miR-15a's regulatory influence. In K562 cells cotransfected with both miR-15a and pcDNA3-cMyb, the previously observed block in cell-cycle progression was largely but not completely abrogated (Figure 5B). This is most apparent when one notes the population of miR-15a expressing K562 cells in G0/G1 (∼48%; Figure 5A) and compares this to the percentage of G0/G1 cells in miR-15a expressing K562 cells cotransfected with pcDNA3-cMYB (∼35%). The percentage of K562 cells in G0/G1 transfected with a control miRNA was also approximately 35% (Figure 5A). Cotransfecting miR-15a and pcDNA3-c-Myb into K562 cells resulted in an increase of the G0/G1 population by approximately 20% (Figure 5B right panel), compared with miR-15a-transfected K562 cells (Figure 5A top right panel) which showed an approximately 37% increase of cells in G0/G1. In aggregate, these data are consistent with the thesis that miR-15a regulates cell-cycle progression through its effects on c-Myb. They also suggest that its effects are unrelated to potential effects on other Myb family members, since cycling of the nonhematopoietic cells was unaffected.
Myb directly regulates miR-15a expression
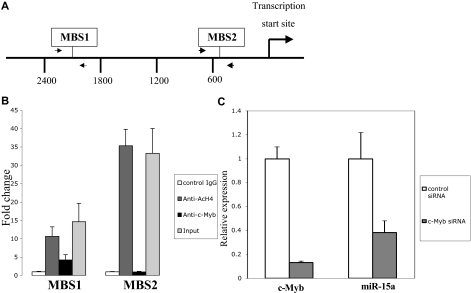
Sequence analysis of the human miR-15a promoter revealed 7 potential Myb binding sites within 3.0 kb of the transcriptional start site. One of the 7 sites is canonical, 5′- pyrimidine AAC (G/T)G-3′. The remaining 6 sites were noncanonical, 5′-AACNG-3′. Two of the possible 7 c-Myb binding sites are conserved among human, mouse and rat genome suggesting that they are the most likely candidates for functional Myb binding. These sites were designated as −2124 MBS1 (c-Myb binding site 1), and −502 MBS2 (Myb binding site 2) respectively, on the basis of their upstream positions with reference to the transcriptional start site (Figure 6A). To determine whether endogenous c-Myb could physically associate with these conserved c-Myb binding sites, chromatin immunoprecipitation (ChIP) analysis, using c-Myb antibody, was performed with extracts from K562 cells. A significant enrichment of the promoter (4-fold elevation) was observed when primers were used to amplify the fragment that spans the region between −2248 and −1998, but not the region between −653 and −464 (Figure 6B). Thus, these data suggest that c-Myb binds to the miR-15a promoter, at least through the site designated as MBS1.
Figure 6.
c-Myb regulates miR-15a expression. (A) Diagram indicates the c-Myb binding sites (MBS) in the miR-15a promoter, as well as the primers (arrows) used for ChIP assays. (B) ChIP assay shows c-Myb binding to miR-15a in K562 cells through Myb binding site 1. Protein-DNA complexes from K562 cells were immunoprecipitated with normal mouse IgG, antiacetylated H4 monoclonal antibody(Ac-H4) or c-Myb monoclonal antibody. Real-time PCR was performed using miR-15a promoter primers flanked c-Myb binding sites. Fold enrichment of a given DNA region was calculated as the ratio between the enrichment obtained with a specific antibody and that obtained with the normal IgG. Results are the average of 3 independent cross-linking ChIP experiments. Error bars indicate standard error. (C) c-myb siRNA down-regulate miR-15a expression in K562 cells. K562 cells were transfected with 100 nM c-myb siRNA or control siRNA. Forty-eight hours after transfection, c-myb and miR-15a expression was examined using a quantitative real-time PCR (QRT-PCR) assay performed on RNA isolated from K562 cells transfected with the indicated RNA oligonucleotide molecules. Results depicted are representative of 3 independents experiments.
To examine the potential physiologic significance of c-Myb protein binding to the miR-15a promoter, miR-15a expression levels were examined in K562 cells after treating cells with a siRNA targeted to the endogenous c-myb mRNA. In these experiments, c-myb mRNA expression declined by approximately 90%, as measured by quantitative real-time PCR, 48 hours after the c-myb targeted siRNA were transfected into K562 cells. The specificity of the knockdown was substantiated by the absence of an effect on β-actin expression (data not shown). Delivery of these same siRNA into K562 cells also resulted in an approximately 70% decrease in miR-15a expression (Figure 6C). To exclude the possibility of off-target effects causing these results, we evaluated additional anti-Myb siRNA and found good correlation between inhibition of c-Myb, and inhibition of miR-15a (data not shown). These data are supportive of our hypothesis that c-Myb protein binding directly regulates miR-15a expression.
Correlation between miR-15a and c-Myb levels in primary CD34+ cells undergoing erythroid differentiation in vitro
As the above experiments were carried out in cell lines, and we were particularly interested in the potential physiologic relevance of miR-15a, c-myb mRNA interactions in normal cells, we next examined the expression of these 2 mRNA species in normal human CD34+ cells undergoing erythroid maturation in vitro. To this end, normal human bone marrow CD34+ cells were cultured in suspension in the presence of SCF and Epo as detailed in the methods section. We then quantitated the expression of miR-15a, c-myb mRNA, and c-Myb protein on days 1 through 4, the time during which cells would be undergoing erythroid lineage commitment and initial developent; on days 6 and 8, during which time the proerythroblasts expand and undergo maturation; and on day 10 when most cells in culture resemble late-stage erythroblasts (Figure 7). After 24 hours in culture (day 1), miR-15a levels rose rapidly, but then declined precipitously on day 2. They then rose gradually again through day 4, only to begin a gradual decline which reached its perigee at day 10. At this time point, the levels of miR-15a were approximately 50% lower than those measured at day 0 (Figure 7A). c-myb mRNA kinetics were less dramatic than those of miR-15a. Its levels were relatively constant from day 0 through day 3, then declined by approximately 60% on day 4 (compared with day 0). Thereafter, the mRNA levels remained relatively unchanged at approximately 40% to 60% of day 0 value, through day 10 (Figure 7B). Interestingly, c-Myb protein levels rose through day 4, and then began a gradual decline until day 10, at which time Myb protein was approximately 10% of the levels measured at day 0 (Figure 7C,D).
Figure 7.
Expression of miR-15a and c-Myb in human CD34+ cells cultured under conditions favoring erythroid differentiation. Human CD34+ cells were grown in medium supplemented with SCF and Epo. The expression of miR-15a (A) and c-myb (B) was examined on days 1, 2, 3, 4, 6, 8, and 10 using mirVana qRT-PCR miRNA detection method for miR-15a, or general real-time PCR for c-myb. c-Myb protein was measured by western-blotting (C) and quantitation of the c-Myb protein was done by density analysis (D). Representative data from 3 independent experiments are shown.
Inhibition of colony formation of hematopoietic progenitors by miR-15a
Although the inverse relationship between miR-15a and Myb protein was generally observed in the kinetic expression experiments described immediately above, the likelihood of a more complex relationship under physiologic conditions was suggested. Therefore, to substantiate the existence of a functional relationship between miR-15a expression and normal hematopoiesis, we cotransfected a miR-15a mimic or a 10-fold greater amount of a nontargeting miRNA into whole normal human bone marrow mononuclear cells or more purified CD34+ cells. The transfected cells were then assayed for the ability to form CFU derived erythroid (Figure 8A,B), and granulocyte-macrophage derived colonies (Figure 8C). Consistent with the data presented above, CD34+ cell–derived CFU-E (Figure 8A) and CFU-GM colonies (Figure 8C) were inhibited by approximately 3- and 4-fold, respectively, when transfected with the miR-15a mimic, compared with cells transfected with the control miRNA.
Figure 8.
miR-15a inhibits colony formation by normal human bone marrow progenitor cells. Normal human bone marrow mononuclear or CD34+ cells were transfected with control miR or a miR-15a mimic. Effects on CD34+ cell–derived colony-forming unit–erythroid (CFU-E; 3 × 103/35-mm dish; A), marrow mononuclear cell–derived colony-forming unit–erythroid (CFU-E) and burst-forming unit–erythroid (BFU-E; 1.2 × 105/35-mm dish; B); and CD34+ cell–derived colony-forming unit–granulocyte, macrophage (CFU-GM; 3 × 103/35-mm dish; C) are shown.
To determine whether there was a specific stage of development at which miR-15a expression might be particularly important, whole normal marrow mononuclear cells (MNCs) were transfected with either the miR-15a mimic, or control miRNA and then cloned under conditions favoring growth of either CFU-E or BFU-E (Figure 8B). In accord with the results shown in Figure 8A, CFU-E derived colonies from whole MNCs transfected with the miR-15a mimic were inhibited approximately 60% compared with control transfected cells. In contrast, BFU-E derived colonies were inhibited only approximately 20%. In both cases, the differences in colony growth between control and mimic treated cells was statistically significant (P < .01). In aggregate, these results not only demonstrate that miR-15a plays a role in regulating normal human hematopoietic cell differentiation, they further suggest that miR-15a expression may be most important in regulating expression of c-Myb, or other as yet unidentified genes, which govern the transition from the BFU-E to CFU-E stage of development.
Discussion
Noncoding microRNAs are now well recognized as important posttranscriptional regulators of gene expression.16,37–39 Their mechanism of action has been intensively studied, and is known to be exerted primarily through effects on suppressing mRNA translation, or less commonly by promoting mRNA physical destruction, after hybridizing in the 3′UTR of the message.29 Bioinformatic studies have suggested that the major targets of miRNAs are transcription factors.40 This observation, and our laboratory's interest in c-myb and hematopoiesis, prompted us to determine whether c-myb expression was subject to miRNA regulation in developing myeloid cells.
An online search of the TargetScanS miRNA Target database (http://genes.mit.edu/tscan/targetscanS2005.html(Ben lewis); accessed May 15, 2006) suggested that the 3′-UTR of the c-myb mRNA could potentially be targeted by up to 14 miRNAs. Of these, we selected miR-15a, miR-107, and miR-150, for further study for the following reasons. First, since miR-15a and miR-107 are both putative tumor suppressors,41,42 and might therefore naturally counteract any oncogenic potential of c-Myb. Second, miR-150 is highly expressed in developing T and B lymphocytes43 and, at least in developing B cells, has been reported to regulate c-myb expression in a stage-specific, dose-dependent manner.44 We were therefore interested in identifying potential interactions between of miR-150 and c-Myb during myelopoiesis.
To explore a possible relationship between the expression of these miRNAs and the expression of c-myb, we first examined whether these 3 miRNAs could mediate the repression of a luciferase/c-Myb-3′-UTR reporter construct in HEK293 cells, and endogenous c-myb expression in K562 cells. Interestingly, and in contrast to recent reports, miR-150 only slightly decreased the activity of c-Myb driven luciferase activity. Although miR-15a and miR-107 both reduced luciferase activity in the reporter cell line, only miR-15a did so in conjunction with a down-regulation of endogenous c-Myb protein level, suggesting that the effect of miR-107 on luciferase expression, at least in this cell system, was not due to a miRNA mediated inhibition of c-myb 3′-UTR driving reporter activity.
To further confirm that miR-15a directly regulates c-Myb expression, deletions of the miR-15a seed region were introduced into the c-myb 3′-UTR immediately downstream of the luciferase reporter construct used in these experiments (Figure 2A). These deletions resulted in a dose-dependent loss of the inhibitory effect of miR-15a on luciferase expression, indicating that these sites are recognized by miR-15a and are used for suppression. Furthermore, enforced expression of a miR-15a inhibitor in K562 cells increased c-Myb expression, suggesting that endogenous miR-15a can modulate c-Myb expression in a relevant cell type as well. Consistent with such an effect, our experimental results demonstrated that overexpression of miR-15a in K562 cells caused an expected cell-cycle block in G1, and that this blockade could be partially rescued by overexpression of a c-myb mutant mRNA lacking the miR-15a binding sites. It was perhaps curious that miR-15a had no effect on the cell-cycle progression of HEK293 and Hela cells in spite of the fact that these 2 cell lines could both be efficiently transfected with the miR-15a molecules used in our study. One possible explanation for this finding is that since endogenous c-Myb expression is essentially undetectable in HEK293 cells, and is quite low in HeLa cells, c-Myb expression in these cell types is of little significance and thus unlikely to be the subject of regulatory influences. These results might also be explained by the previously suggested possibility that regulation of c-Myb by miRNAs is lineage specific.
With regard to the potential physiologic significance of miR-15a, we found that its expression increased during PMA-induced megakarycytic differentiation of K562 cells (data not shown). A recent study also showed that miR-15a was elevated during ATRA-induced NB4 myeloid differentiation.45,46 It is well documented that c-myb expression is down-regulated during these processes,47 suggesting the possibility that miR-15a may play a biologically relevant role in the inhibition of c-myb expression during hematopoietic cell differentiation. We also found that miR-15a inhibited both erythroid and myeloid colony formation from CD34+ bone marrow cells in vitro. These results are in accord with previously reported effects of c-myb targeted antisense oligodeoxynucleotides,11 and c-myb knockout studies in murine ES cells,10 suggesting the intriguing possibility that miR-15a might function a natural c-Myb inhibitor in hematopoietic cells. Nonetheless, the kinetic data presented in Figure 7 demonstrate that under physiologic conditions the relationship between miR-15a and c-Myb protein is likely complex, and may well be dependent on the stage of development. In support of this hypothesis, we note that on days 0 to 4, miR-15a levels are, except for the anomalous uptick on day 1, low (Figure 7A), while c-Myb protein levels are high (Figure 7C). Thereafter, miR-15a levels increase while c-myb mRNA declines. Finally, on days 8 and 10, when the cells in culture have essentially completed their maturation routine, the inverse relationship between miR15a and Myb protein is no longer apparent. The erythroid colony data presented in Figure 8 provide more direct evidence to support the hypothesis that miR-15a effects are linked to stage of development. CFU-E were much more susceptible to inhibition by the miR-15a mimic than BFU-E suggesting that miR-15a expression may be most important in regulating expression of c-Myb, or other as yet unidentified genes, as cells transition from the BFU-E to CFU-E stage of development.
In addition to its role as regulator of c-Myb and normal human hematopoiesis, our data also suggest a possible role for miR-15a as a tumor suppressor. In this regard, miR-15a is located at chromosome 13q14 and deletions involving chromosome 13q14 are well described in lymphoid and myeloid malignancies. Other reports have shown that deregulated expression of miR-15a induces apoptosis by targeting the antiapoptotic protein BCL2.33 Our study suggests that miR-15a may contribute to cell-cycle control, and function as an antioncogene, by suppressing c-myb mRNA translation. As BCL2 has also been found to be activated by c-Myb,48 the interaction of miR-15a with c-Myb may constitute another regulatory component of the BCL2 expression pathway. In aggregate, then, these findings lead us to hypothesize that mutations in miR-15a precursors, or miR-15a binding sites in the c-myb 3′-UTR, might play a role in the pathogenesis of human malignancies, those of hematopoietic origin in particular.
BecausMyb transcription factor directly binds the upstream promoter region of the miR-15a/miR-16-1 cluster, and because this binding leads to expression of miR-15a, we suggest that it is likely that c-Myb regulates miR-16-1 expression as well. Since, as discussed above, miR-15a and miR-16-1 can repress c-Myb expression, we propose the existence of a negative autoregulatory feedback loop between c-Myb, and the miR-15a/miR-16-1 cluster. Furthermore, since c-Myb can up-regulate its own expression by binding in the 5′ flanking region of the c-myb gene,45 we suggest that miR-15a/miR-16-1 might be a natural counterbalance, or negative feedback loop, to fine tune c-myb expression. Disruption of this feedback loop might lead to aberrant c-Myb activity and contribute to malignant transformation.
Finally, although we believe that we have provided compelling data to support our contention that miR-15a regulates c-Myb expression, our data do not exclude the possibility that miR-150 and miR-107 may also interact with c-myb mRNA. Rather, we propose that the 3 miRNAs examined in this study might, independently or simultaneously, regulate c-Myb expression, depending on the cellular context in which they were expressed. For example, c-Myb has recently been reported to be a critical target of mir-150 during B-cell differentiation.43 Recent reports also suggest that miR-16, another member of miR-16/miR-15 family, can induce cell-cycle arrest.49,50 Interestingly, c-myb mRNA also harbors miR-16 target sites. Therefore, it is reasonable to speculate that c-Myb may be important for the phenotype caused by overexpression of miR-16 and be regulated by miR-16 as well. As was noted above, 11 additional miRNAs were predicted to target c-Myb and further studies will be necessary to determine their role in c-Myb regulation.
Acknowledgments
We thank Dr Cezary Swider, Stem Cell and Xenograft Core Facility, University of Pennsylvania School of Medicine, for isolation of CD34+ cells from normal human bone marrow.
This work was supported in part by grants from the National Institutes of Health (CA101859), a Tobacco Settlement Grant from the Pennsylvania Department of Health (the Department specifically disclaims responsibility for any analysis, interpretations or conclusions), and a grant from the United States-Israel Binational Science Foundation (BSF no. 2003028-01).
Footnotes
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.Z. conducted most of the experiments and wrote the initial version of the manuscript; A.K. performed cultures of normal human CD34+ cells; S.J. provided help in data analysis and discussion; and A.M.G. was involved in experiment design, data analysis, and manuscript writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alan M. Gewirtz, MD, Rm 713, BRB II/III, University of Pennsylvania School of Medicine, 421 Curie Blvd, Philadelphia, PA 19104; e-mail: gewirtz@mail.med.upenn.edu.
References
- 1.Trauth KM, Mutschler B, Jenkins N, Gilbert D, Copeland N, Klempnauer K. Mouse A-myb encodes a trans-activator and is expressed in mitotically active cells of the developing central nervous system, adult testis and B lymphocytes. EMBO J. 1994;13:5994–6005. doi: 10.1002/j.1460-2075.1994.tb06945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weston K. Myb proteins in life, death and differentiation. Curr Opin Genet Dev. 1998;8:76–81. doi: 10.1016/s0959-437x(98)80065-8. [DOI] [PubMed] [Google Scholar]
- 3.Carpinelli MR, Hilton DJ, Metcalf D, et al. Suppressor screen in Mpl−/− mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc Natl Acad Sci U S A. 2004;101:6553–6558. doi: 10.1073/pnas.0401496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gewirtz AM, Anfossi G, Venturelli D, Valpreda S, Sims R, Calabretta B. G1/S transition in normal human T-lymphocytes requires the nuclear protein encoded by c-myb. Science. 1989;245:180–183. doi: 10.1126/science.2665077. [DOI] [PubMed] [Google Scholar]
- 5.Jiang J, Best B, Menzel S, et al. cMYB is involved in the regulation of fetal hemoglobin production in adults. Blood. 2006;108:1077–1083. doi: 10.1182/blood-2006-01-008912. [DOI] [PubMed] [Google Scholar]
- 6.Lieu YK, Kumar A, Pajerowski AG, Rogers TJ, Reddy EP. Requirement of c-myb in T cell development and in mature T cell function. Proc Natl Acad Sci U S A. 2004;101:14853–14858. doi: 10.1073/pnas.0405338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukai HY, Motohashi H, Ohneda O, Suzuki N, Nagano M, Yamamoto M. Transgene insertion in proximity to the c-myb gene disrupts erythroid-megakaryocytic lineage bifurcation. Mol Cell Biol. 2006;26:7953–7965. doi: 10.1128/MCB.00718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandberg ML, Sutton SE, Pletcher MT, et al. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell. 2005;8:153–166. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 9.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 2005;23:275–286. doi: 10.1016/j.immuni.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Mucenski ML, McLain K, Kier AB, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 11.Gewirtz AM, Calabretta B. A c-myb antisense oligodeoxynucleotide inhibits normal human hematopoiesis in vitro. Science. 1988;242:1303–1306. doi: 10.1126/science.2461588. [DOI] [PubMed] [Google Scholar]
- 12.Metcalf D, Carpinelli MR, Hyland C, et al. Anomalous megakaryocytopoiesis in mice with mutations in the c-Myb gene. Blood. 2005;105:3480–3487. doi: 10.1182/blood-2004-12-4806. [DOI] [PubMed] [Google Scholar]
- 13.Taylor D, Badiani P, Weston K. A dominant interfering Myb mutant causes apoptosis in T cells. Genes Dev. 1996;10:2732–2744. doi: 10.1101/gad.10.21.2732. [DOI] [PubMed] [Google Scholar]
- 14.Nakata Y, Shetzline S, Sakashita C, et al. c-Myb contributes to G2/M cell cycle transition in human hematopoietic cells by direct regulation of cyclin B1 expression. Mol Cell Biol. 2007;27:2048–2058. doi: 10.1128/MCB.01100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosson D, O'Brien T. Constitutive c-myb expression in K562 cells inhibits induced erythroid differentiation but not tetradecanoyl phorbol acetate-induced megakaryocytic differentiation. Mol Cell Biol. 1995;15:772–779. doi: 10.1128/mcb.15.2.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- 17.Baer MR, Augustinos P, Kinniburgh AJ. Defective c-myc and c-myb RNA turnover in acute myeloid leukemia cells. Blood. 1992;79:1319–1326. [PubMed] [Google Scholar]
- 18.Bender TP, Kremer C, Kraus M, Buch T, Rajewsky K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol. 2004;5:721–729. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 19.Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 2003;22:4478–4488. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakamoto H, Dai G, Tsujino K, et al. Proper levels of c-Myb are discretely defined at distinct steps of hematopoietic cell development. Blood. 2006;108:96–903. doi: 10.1182/blood-2005-09-3846. [DOI] [PubMed] [Google Scholar]
- 21.Guerra J, Withers D, Boxer LM. Myb binding sites mediate negative regulation of c-myb expression in T-cell lines. Blood. 1995;86:1873–1880. [PubMed] [Google Scholar]
- 22.Lauder A, Castellanos A, Weston K. c-Myb transcription is activated by protein kinase B (PKB) following interleukin 2 stimulation of Tcells and is required for PKB-mediated protection from apoptosis. Mol Cell Biol. 2001;21:5797–5805. doi: 10.1128/MCB.21.17.5797-5805.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mountz JD, Subler MA. Regulation of c-myb oncogene expression in immature and mature murine T cells. Mol Immunol. 1993;30:787–795. doi: 10.1016/0161-5890(93)90001-r. [DOI] [PubMed] [Google Scholar]
- 24.Phan SC, Feeley B, Withers D, Boxer LM. Identification of an inducible regulator of c-myb expression during T-cell activation. Mol Cell Biol. 1996;16:2387–2393. doi: 10.1128/mcb.16.5.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohwer F, Todd S, McGuire KL. The effect of IL-2 treatment on transcriptional attenuation in proto-oncogenes pim-1 and c-myb in human thymic blast cells. J Immunol. 1996;157:643–649. [PubMed] [Google Scholar]
- 26.Bender TP, Thompson CB, Kuehl WM. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987;237:1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- 27.Thompson MA, Flegg R, Westin EH, Ramsay RG. Microsatellite deletions in the c-myb transcriptional attenuator region associated with over-expression in colon tumour cell lines. Oncogene. 1997;14:1715–1723. doi: 10.1038/sj.onc.1201007. [DOI] [PubMed] [Google Scholar]
- 28.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 29.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 30.Chendrimada TP, Finn KJ, Ji X, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 31.Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m(7)G Cap Binding-like Motif within Human Ago2 Represses Translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 32.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clappier E, Cuccuini W, Kalota A, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL) - the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 35.Lahortiga I, Keersmaecker KD, Vlierberghe PV, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet. 2007;39:593–595. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 36.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 37.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 38.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 39.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 40.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 41.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 45.Garzon R, Pichiorri F, Palumbo T, et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–4157. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 46.Malene BP, Niels B, Jack BC. Derangement of transcription factor profiles during in vitro differentiation of HL60 and NB4 cells. Leuk Res. 2007;31:827–837. doi: 10.1016/j.leukres.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 47.Manfioletti G, Gattei V, Buratti E, et al. Differential expression of a novel proline-rich homeobox gene (Prh) in human hematolymphopoietic cells. Blood. 1995;85:1237–1245. [PubMed] [Google Scholar]
- 48.Salomoni P, Perrotti D, Martinez R, Franceschi C, Calabretta B. Resistance to apoptosis in CTLL-2 cells constitutively expressing c-Myb is associated with induction of BCL-2 expression and Myb-dependent regulation of bcl-2 promoter activity. Proc Natl Acad Sci U S A. 1997;94:3296–3301. doi: 10.1073/pnas.94.7.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linsley PS, Schelter J, Burchard J, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raveche ES, Salerno E, Scaglione BJ, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]