Abstract
Background
Genomes store information for building and maintaining organisms. Complete sequencing of many genomes provides the opportunity to study and compare global information properties of those genomes.
Results
We have analyzed aspects of the information content of Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Saccharomyces cerevisiae, and Escherichia coli (K-12) genomes. Virtually all possible (> 98%) 12 bp oligomers appear in vertebrate genomes while < 2% of 19 bp oligomers are present. Other species showed different ranges of > 98% to < 2% of possible oligomers in D. melanogaster (12–17 bp), C. elegans (11–17 bp), A. thaliana (11–17 bp), S. cerevisiae (10–16 bp) and E. coli (9–15 bp). Frequencies of unique oligomers in the genomes follow similar patterns. We identified a set of 2.6 M 15-mers that are more than 1 nucleotide different from all 15-mers in the human genome and so could be used as probes to detect microbes in human samples. In a human sample, these probes would detect 100% of the 433 currently fully sequenced prokaryotes and 75% of the 3065 fully sequenced viruses. The human genome is significantly more compact in sequence space than a random genome. We identified the most frequent 5- to 20-mers in the human genome, which may prove useful as PCR primers. We also identified a bacterium, Anaeromyxobacter dehalogenans, which has an exceptionally low diversity of oligomers given the size of its genome and its GC content. The entropy of coding regions in the human genome is significantly higher than non-coding regions and chromosomes. However chromosomes 1, 2, 9, 12 and 14 have a relatively high proportion of coding DNA without high entropy, and chromosome 20 is the opposite with a low frequency of coding regions but relatively high entropy.
Conclusion
Measures of the frequency of oligomers are useful for designing PCR assays and for identifying chromosomes and organisms with hidden structure that had not been previously recognized. This information may be used to detect novel microbes in human tissues.
Background
The discovery of the structure of DNA [1] was a fundamental event in biology because it illuminated the mechanism by which information can be encoded, passed on to future generations and even constructed by natural selection. With the sequencing of the human and other genomes [2,3], we are now able to compare the information encoded in the genomes of a wide variety of organisms and study the mechanisms that change genomes over time, including sequence mutations, recombination, duplication and deletion [4,5]. A great deal of research has focused on the coding and regulatory regions of genomes and regularly uses informational content measurements to analyze the structure of loci in the genome [6-8]. In addition, recent interest has turned to analyzing regions that do not encode proteins [9].
Relatively little is known about the global informational properties of most genomes. The conditional entropy was measured for human chromosome 22 [10] along with the frequency distributions of 3- through 7-mers in chromosomes 21 and 22 [11]. Stanley et al. analyzed the distribution of 1- to 4-mers within a wide variety of organisms and found that some tend to cluster within genomes (usually in non-coding regions) and others tend to "repel" each other [12,13]. Entropy measures have been applied to yeast and C. elegans whole chromosomes [14]. Recently, McHardy et al. showed that the distribution of 5-mer and 6-mer oligonucleotides in a > 1 kb fragment of DNA is characteristic of an organism and can be used in metagenomic studies to classify and construct the potentially millions of genomes in an environmental sample [15]. We are only beginning to compare informational properties between human chromosomes and across species.
The set of all oligonucleotides (oligos) of length n defines an n-dimensional discrete space, Sn where each point is a possible n-mer and each dimension has only four states (A, C, G or T). Here we have measured the proportion of sequence space covered by oligos of length 1 – 20 in H. sapien, M. musculus, D. melanogaster, C. elegans, A. thaliana, S. cerevisiae, and E. coli k12. We have also measured the frequency of unique n-mers in those species and the compactness of the human genome in n-mer space. We also have identified all 5- to 20-mers that appear in the human genome more than 30,000 times, as well as the set of 15-mers that do not appear in the human genome and are more than 1 nucleotide different from all 15-mers in the human genome. In addition, we have measured the information content of the human genome for different oligo lengths and compared the information content to the proportion of coding regions in each human chromosome. The results match empirical observations and give a global view of the informational properties across a wide variety of genomes. Finally, we profiled the 10-mer space coverage for a wide range of 433 microbial genomes and found that the extent of sequence space coverage is largely determined by genome size and GC content.
Results
Sequence space coverage
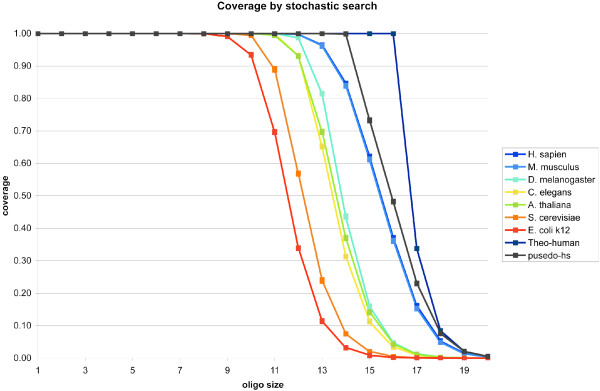
We randomly generated 5 sets of 100,000 probes for each oligo length n, and then determined the proportion of those n-mers present in the genomes of Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Saccharomyces cerevisiae, and Escherichia coli k12. The n-mer space coverage for each genome is plotted against oligo length n in Figure 1 (See additional file 1 for the data). The E. coli genome includes all 8-mers, and less than 0.21% of all 16-mers. In contrast, the human genome includes all 11-mers and less than 0.38% of all 20-mers. Of course, n-mer space coverage reflects genome size to some extent. For example, the human genome has a much higher coverage than the yeast genome for every oligo length. The 7 genomes that we investigated in this study differ the most in their coverage for 13-mer space, ranging from 11.4% coverage for E. coli to 96% coverage for human and mouse. For comparison, we also generated a random "pseudo" human genome with the same length and dinucleotide frequencies (see Appendix for a formal analysis of the expected number of n-mers in this pseudo-human genome). The fact that the true human genome has less coverage of n-mer space than the pseudo-human genome (Figure 1) shows that there are more repeated n-mers in the human genome than one would expect by chance.
Figure 1.
The percentage of all possible n-mers (coverage) that appear in H. sapien, M. musculus, D. melanogaster, C. elegans, A. thaliana, S. cerevisiae, E. coli k12, theoretical and pseudo-human genomes. Theo-human is the maximum coverage a human-length genome could achieve if every n-mer in its genome was unique. The pseudo-human (pseudo-hs) genome is a random genome generated with the same length and dinucleotide frequencies of the human genome. The space coverage of each genome listed above is plotted against the length of the oligomer analyzed, ranging from 1 to 20.
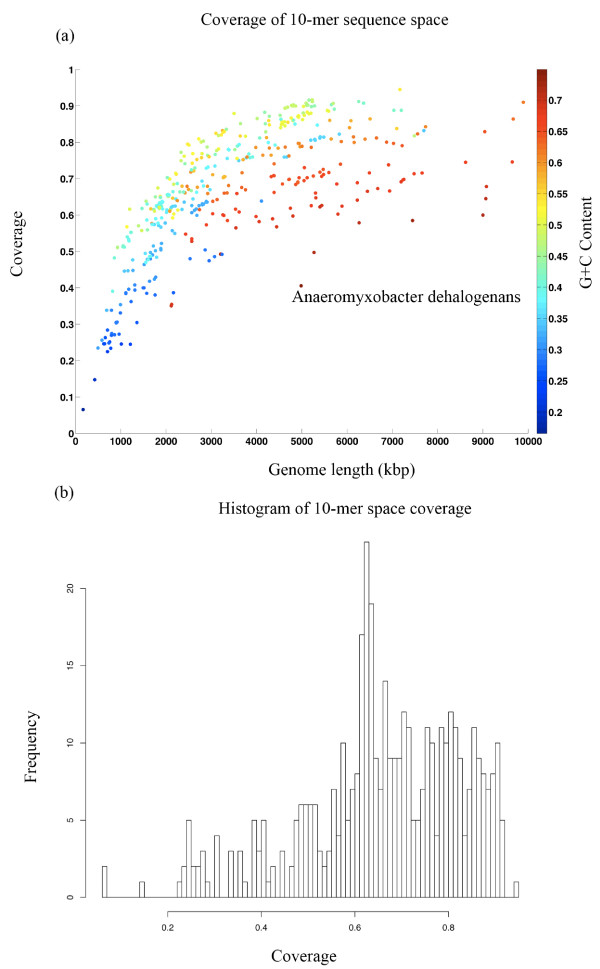
The coverage analysis of genomes can be used to analyze the complexity of any fully sequenced genome. We applied this analysis to 433 fully sequenced microbial genomes (Figure 2, see additional file 2 for the species, coverage, GC content and genome sizes). A multivariate regression of log genome size and deviation of GC content from 0.5 frequency, on 10-mer sequence space coverage, shows that variation in coverage can almost entirely be explained by genome size and GC content (adjusted R2 = 0.92, p < 0.01). However, Anaeromyxobacter dehalogenans is an outlier with lower coverage than would be predicted by the model (actual coverage = 0.406, predicted coverage = 0.563, 99.9% predicted interval: 0.412–0.713). In addition, we confirmed the significant association between log genome size and GC content such that organisms with smaller genomes have lower GC content (Figure 2, linear regression p < 0.001) [16].
Figure 2.
(a) Coverage of 10-mer sequence space as a function of genome size in 433 fully sequenced microbial genomes. The legend for the color-coding of GC content appears on the right. Smaller genomes have lower GC content. Anaeromyxobacter dehalogenans is an outlier with unusually low coverage for its genome size and GC content (outside of the 99.9% predicted interval). (b) A histogram for the proportion of the 10-mer sequence space covered by each of the 433 fully sequenced microbial genomes.
Frequency of unique n-mers
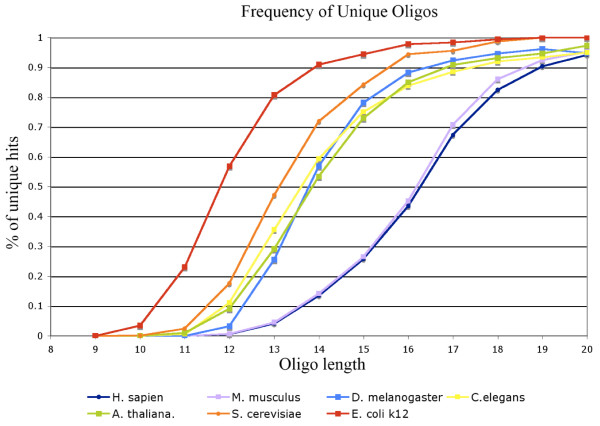
To calculate the percentage of n-mers that appear only once in a genome, we implemented a program to estimate the unique hits among our stochastic search results (Figure 3). The human and mouse genome has only 5% unique hits among all the 13-mers they contain, while Drosophila, Arabidopsis, and yeast have only a single copy of more than 50% of their 13-mers. Figure 3 implies that, in order to have at least an 80% chance of hybridizing to a unique locus, a PCR primer for human and mouse genomes should be greater than 17 bp, while for Drosophila, C. elegans, and Arabidopsis, it should be greater than 15 bp.
Figure 3.
The percentage of n-mers that appeared exactly once (unique hits), out of all the n-mers detected in each genome. Slightly less than 50% of 16-mers detected in humans are unique. Whereas, for E. coli, a little more than 50% of 12-mers were unique.
Accuracy of stochastic estimates
To assess the accuracy of our stochastic estimates for coverage in longer n-mers, we compared the stochastic results with exhaustive search results of all possible n-mers (Table 1). Due to the limitations of computational power, we could only carry out exhaustive searches up to 15-mer space. The 95% confidence interval of our stochastic sampling is small and it contains the true coverage. The confidence intervals also show that accuracy decreases as the dimension of our sampling space increases. However, it is clear from Table 1 that our 100,000 probes are statistically powerful enough to estimate the percentage of coverage for oligos up to length 20.
Table 1.
Estimates and exhaustive calculations of the human genome coverage of n-mer Space
| n-mer | Mean Coverage from Stochastic Sampling1 | 95% Confidence Interval | Coverage from Exhaustive search | |
| 12 | 99.726% | 99.718% | 99.733% | 99.730% |
| 13 | 96.416% | 96.319% | 96.514% | 96.458% |
| 14 | 84.470% | 84.308% | 84.633% | 84.444% |
| 15 | 62.041% | 61.865% | 62.217% | 62.124% |
| 16 | 37.065% | 36.934% | 37.197% | NA2 |
| 17 | 16.156% | 16.058% | 16.254% | NA |
| 18 | 5.332% | 5.278% | 5.386% | NA |
| 19 | 1.529% | 1.508% | 1.551% | NA |
| 20 | 0.382% | 0.369% | 0.396% | NA |
1100,000 n-mers were used to estimate coverage of the genome, repeated 5 times to estimate the 95% confidence interval.
2The set of all possible n-mers greater than 15 bp is too large to be exhaustively searched given our current computational resources.
Detecting non-human organisms with non-human 15-mers
One potential use of genome coverage data is to use the n-mers that do not appear in the human genome as probes to detect microbes or genetic alterations in human samples. In order to do this, we must choose an oligo length long enough that there are some n-mers that do not appear in the human genome (i.e., > 13-mers), and long enough that some non-human n-mers are likely to appear in non-human organisms, yet short enough that that a large fraction of the non-human n-mers could be probed in a human sample (i.e., < 16-mers). We focused on 15-mers because 38% (406.7 M) of all possible 15-mers do not appear in the human genome. However, 404.1 M of these differ from the reference human genome by a single nucleotide and so may appear as single nucleotide polymorphisms (SNPs) in human samples. An oligonucleotide array of the remaining panel of 2.6 M 15-mers, that are at least 2 SNPs different from human sequences, could be constructed using current technology (e.g. Roche NimbleGen arrays). We simulated an oligonucleotide array hybridization experiment using these 2.6 M non-human 15-mers to determine the likelihood of detecting any of the fully sequenced microbes. This hypothetical array would detect 75% (2314 of 3065) viruses with a median of 3 positive non-human 15-mers (range: 0 to 1,705) and 100% (of 433) bacterial species with a median of 3,873 positive non-human 15-mers (range: 1 in the obligate endosymbiont Candidatus Carsonella ruddii PV, up to 92,127 in Burkholderia 383). Of course, some of the fully sequenced microbes are closely related to each other and so these estimates are not based on completely independent samples.
Ultra-frequent n-mers
Some n-mers appear at extremely high frequency in the human genome. Among this set are some recognizable functional motifs. For example, the TATA box (5'-TATAAA-3') is ranked in the top 2% of all the possible 6-mers and the E-box (5'-CACCTG-3') is ranked in the top 11% of all possible 6-mers. We identified all the 5- to 20-mers that appear more than 30,000 times in the human genome. The top 20 ultra-frequent n-mers are shown in Table 2. A full list of high frequency n-mers is available in additional file 3.
Table 2.
Ultra-frequent n-mers in the human genome
| 5-mer | Freq | 6-mer | Freq | 7-mer | Freq | 8-mer | Freq |
| AAAAA | 38,658,471 | AAAAAA | 19,638,479 | AAAAAAA | 12,559,969 | AAAAAAAA | 9,155,123 |
| ATTTT | 23,349,997 | AAAAAT | 9,299,025 | AAATAAA | 3,521,836 | ATATATAT | 1,863,212 |
| TATTT | 19,344,297 | TATTTT | 8,283,181 | AAAAAAT | 3,335,157 | TGTGTGTG | 1,701,426 |
| AGAAA | 18,271,461 | AAATAA | 7,271,302 | AAAGAAA | 3,255,464 | TTTTAAAA | 1,574,448 |
| AAATT | 16,119,174 | AGAAAA | 7,027,241 | TTTTAAA | 3,218,950 | AAAATAAA | 1,562,386 |
| TTATT | 15,608,707 | TTTATT | 7,015,408 | TTATTTT | 3,132,222 | AAAGAAAA | 1,467,003 |
| TTTTC | 15,579,017 | TTTTAA | 6,910,624 | ATATATA | 3,034,879 | AAAAGAAA | 1,444,255 |
| CAAAA | 15,364,161 | TTTCTT | 6,890,187 | TTAAAAA | 2,961,520 | AAAAATTA | 1,392,585 |
| TTCTT | 15,093,160 | TTTAAA | 6,820,966 | TTTTCTT | 2,913,461 | TAAAAATA | 1,363,753 |
| TCTTT | 15,053,533 | AAAATT | 6,559,595 | TTTTATT | 2,814,631 | CCAGCCTG | 1,308,899 |
| CTTTT | 14,643,705 | TTCTTT | 6,476,513 | TAAAAAT | 2,811,575 | CAGCCTGG | 1,266,975 |
| CATTT | 13,897,384 | ATAAAA | 6,284,494 | AAAAATT | 2,773,210 | CTTTTTTT | 1,238,232 |
| AAACA | 13,686,083 | TTTTTG | 5,979,892 | TTCTTTT | 2,693,599 | AAAAAGAA | 1,228,463 |
| TTTGT | 13,634,981 | ATTTTA | 5,829,423 | AGAAAAA | 2,614,232 | CAAAAAAA | 1,223,343 |
| TAAAT | 13,334,177 | AAATAT | 5,784,598 | AAAATTA | 2,572,747 | AAAACAAA | 1,216,247 |
| ATATA | 13,333,472 | AAAAGA | 5,708,574 | ACAAAAA | 2,554,220 | TTTTTCTT | 1,175,794 |
| TGAAA | 13,099,712 | TTTTGT | 5,622,008 | TTTGTTT | 2,514,354 | ATTTATTT | 1,171,982 |
| ATATT | 13,067,844 | TATATA | 5,551,066 | TGTGTGT | 2,500,827 | TTTTGTTT | 1,155,387 |
| AAAAC | 12,161,846 | TGTTTT | 5,550,449 | TTTTTTA | 2,460,695 | TTAAAAAA | 1,145,762 |
| AGAGA | 12,078,839 | CTTTTT | 5,515,339 | TTTTTTG | 2,436,880 | AAAAAATA | 1,143,587 |
| 9-mer | Freq | 10-mer | Freq | 11-mer | Freq | ||
| AAAAAAAAA | 7,276,886 | AAAAAAAAAA | 5,952,617 | AAAAAAAAAAA | 4,945,619 | ||
| TGTGTGTGT | 1,424,846 | TGTGTGTGTG | 1,168,929 | TGTGTGTGTGT | 1,067,659 | ||
| ATATATATA | 1,253,711 | ATATATATAT | 967,302 | CTGTAATCCCA | 804,201 | ||
| CCAGGCTGG | 1,043,029 | TGTAATCCCA | 857,278 | ATATATATATA | 785,762 | ||
| CTGGGATTA | 921,482 | GCTGGGATTA | 840,776 | TGTAATCCCAG | 782,505 | ||
| CCTGTAATC | 917,990 | GGATTACAGG | 835,847 | CCTGTAATCCC | 762,833 | ||
| GGATTACAG | 916,302 | CTGTAATCCC | 826,256 | GTAATCCCAGC | 744,961 | ||
| GAGGCTGAG | 911,055 | GGAGGCTGAG | 810,203 | CCTCAGCCTCC | 729,303 | ||
| GCTGGGATT | 899,822 | CTGGGATTAC | 802,982 | GAGGCTGAGGC | 602,716 | ||
| TGTAATCCC | 897,744 | CCTCAGCCTC | 795,109 | GCCTGTAATCC | 579,838 | ||
| GGAGGCTGA | 891,406 | AGGCTGAGGC | 644,447 | AAAATACAAAA | 564,984 | ||
| GTAATCCCA | 887,639 | CCAGCCTGGG | 644,416 | CCTCCCAAAGT | 561,106 | ||
| AGGCTGAGG | 881,036 | GCCTGTAATC | 629,425 | GGGAGGCTGAG | 556,004 | ||
| TTGGGAGGC | 847,581 | AAAAAAAAAG | 626,192 | AGGCTGAGGCA | 555,758 | ||
| CTTTTTTTT | 794,742 | TTTGTATTTT | 619,817 | CACTTTGGGAG | 554,429 | ||
| TTTTATTTT | 780,751 | TTTGGGAGGC | 619,461 | CTTTGGGAGGC | 553,151 | ||
| CAAAAAAAA | 758,731 | AAATACAAAA | 611,794 | AGTAGCTGGGA | 546,426 | ||
| AAAAAAAAT | 741,719 | CAAAAAAAAA | 602,719 | AAAAATACAAA | 542,436 | ||
| AAATACAAA | 740,229 | ACTTTGGGAG | 600,711 | CAGGCTGGAGT | 536,630 | ||
| CCCAGGCTG | 740,090 | TCAGCCTCCC | 600,206 | TCCCAAAGTGC | 534,617 | ||
| 12-mer | Freq | 13-mer | Freq | 14-mer | Freq | ||
| AAAAAAAAAAAA | 4,144,156 | AAAAAAAAAAAAA | 3,468,084 | AAAAAAAAAAAAAA | 2,889,704 | ||
| TGTGTGTGTGTG | 928,266 | TGTGTGTGTGTGT | 867,556 | GTGTGTGTGTGTGT | 775,928 | ||
| TGGGATTACAGG | 744,980 | CTGTAATCCCAGC | 687,709 | CCTGTAATCCCAGC | 639,010 | ||
| CTGGGATTACAG | 737,944 | CTGGGATTACAGG | 684,576 | ATATATATATATAT | 503,574 | ||
| GCTGGGATTACA | 727,608 | ATATATATATATA | 571,291 | CTGGGATTACAGGC | 478,945 | ||
| ATATATATATAT | 664,692 | GCCTGTAATCCCA | 520,611 | AGCACTTTGGGAGG | 459,948 | ||
| GGAGGCTGAGGC | 562,149 | GCCTCCCAAAGTG | 493,408 | GCACTTTGGGAGGC | 448,768 | ||
| GCCTGTAATCCC | 532,230 | GGAGGCTGAGGCA | 489,998 | AAGTGCTGGGATTA | 448,388 | ||
| GCCTCCCAAAGT | 527,552 | CTCCCAAAGTGCT | 486,025 | AAAGTGCTGGGATT | 445,915 | ||
| TGCCTCAGCCTC | 523,666 | GCACTTTGGGAGG | 474,289 | CTCCCAAAGTGCTG | 443,681 | ||
| CCTCCCAAAGTG | 522,504 | CAGCACTTTGGGA | 471,479 | GGAGGCTGAGGCAG | 437,319 | ||
| TCCCAAAGTGCT | 517,353 | AGTGCTGGGATTA | 468,425 | CAAAGTGCTGGGAT | 436,625 | ||
| AAAAATACAAAA | 513,200 | TGCACTCCAGCCT | 466,972 | CAGGCTGGAGTGCA | 436,504 | ||
| GGGAGGCTGAGG | 507,919 | GAGGCTGAGGCAG | 465,893 | CCAGCACTTTGGGA | 434,713 | ||
| GCACTTTGGGAG | 501,469 | AAGTGCTGGGATT | 464,827 | TGCTGGGATTACAG | 428,229 | ||
| CCAGGCTGGAGT | 499,800 | ATCCCAGCACTTT | 460,277 | CCAGGCTGGAGTGC | 425,904 | ||
| AGGCTGAGGCAG | 492,943 | GCACTCCAGCCTG | 454,751 | CCTGCCTCAGCCTC | 423,801 | ||
| AGGCTGGAGTGC | 486,794 | TGTAATCCCAGCA | 453,696 | TCCCAGCACTTTGG | 423,684 | ||
| AGTGCTGGGATT | 486,236 | CCAGGCTGGAGTG | 453,312 | TGTAATCCCAGCAC | 417,426 | ||
| CAGGCTGGAGTG | 484,927 | TCCCAGCACTTTG | 448,301 | AGTGCTGGGATTAC | 416,557 | ||
| 15-mer | Freq | 16-mer | Freq | 17-mer | Freq | ||
| AAAAAAAAAAAAAAA | 2,397,399 | AAAAAAAAAAAAAAAA | 1,981,757 | AAAAAAAAAAAAAAAAA | 1,634,441 | ||
| TGTGTGTGTGTGTGT | 729,915 | TGTGTGTGTGTGTGTG | 662,157 | TGTGTGTGTGTGTGTGT | 624,344 | ||
| GCCTGTAATCCCAGC | 448,640 | TAATCCCAGCACTTTG | 408,698 | TAATCCCAGCACTTTGG | 386,886 | ||
| ATATATATATATATA | 446,009 | ATATATATATATATAT | 402,936 | ATCCCAGCACTTTGGGA | 385,642 | ||
| AGCACTTTGGGAGGC | 435,271 | CCAAAGTGCTGGGATT | 400,385 | AATCCCAGCACTTTGGG | 381,085 | ||
| TAATCCCAGCACTTT | 430,431 | GCCTCCCAAAGTGCTG | 398,210 | AAAGTGCTGGGATTACA | 375,216 | ||
| CAAAGTGCTGGGATT | 423,190 | TCCCAGCACTTTGGGA | 395,536 | CTCCCAAAGTGCTGGGA | 373,151 | ||
| CCTCCCAAAGTGCTG | 420,378 | CCCAAAGTGCTGGGAT | 392,938 | AAGTGCTGGGATTACAG | 369,056 | ||
| ATCCCAGCACTTTGG | 412,897 | TGTAATCCCAGCACTT | 390,400 | GCCTCCCAAAGTGCTGG | 368,350 | ||
| CCAGCACTTTGGGAG | 409,630 | CCTCCCAAAGTGCTGG | 388,489 | CAAAGTGCTGGGATTAC | 364,621 | ||
| CCAGGCTGGAGTGCA | 409,063 | CTGTAATCCCAGCACT | 385,038 | CCCAGCACTTTGGGAGG | 363,569 | ||
| TGTAATCCCAGCACT | 407,346 | AAAGTGCTGGGATTAC | 383,514 | ATATATATATATATATA | 363,206 | ||
| CCCAGCACTTTGGGA | 406,238 | CCCAGCACTTTGGGAG | 383,068 | AGTGCTGGGATTACAGG | 358,629 | ||
| TCCCAGCACTTTGGG | 403,080 | GTGCTGGGATTACAGG | 367,247 | TCTACTAAAAATACAAA | 340,685 | ||
| AAGTGCTGGGATTAC | 399,124 | TGCACTCCAGCCTGGG | 361,996 | CTACTAAAAATACAAAA | 340,602 | ||
| CCTGCCTCAGCCTCC | 398,260 | TACTAAAAATACAAAA | 360,995 | CTCCTGCCTCAGCCTCC | 328,239 | ||
| TGCTGGGATTACAGG | 398,070 | GAGGCTGAGGCAGGAG | 349,118 | TTGTATTTTTAGTAGAG | 325,500 | ||
| GTGCTGGGATTACAG | 394,430 | CTACTAAAAATACAAA | 348,511 | TTCTCCTGCCTCAGCCT | 324,096 | ||
| GCACTCCAGCCTGGG | 376,640 | TCCTGCCTCAGCCTCC | 346,669 | TCTCTACTAAAAATACA | 319,930 | ||
| TTTTGTATTTTTAGT | 374,747 | TTGTATTTTTAGTAGA | 346,193 | GGCTGAGGCAGGAGAAT | 318,655 | ||
| 18-mer | Freq | 19-mer | Freq | 20-mer | Freq | ||
| AAAAAAAAAAAAAAAAAA | 1,345,821 | AAAAAAAAAAAAAAAAAAA | 1,104,496 | AAAAAAAAAAAAAAAAAAAA | 901,140 | ||
| GTGTGTGTGTGTGTGTGT | 569,504 | TGTGTGTGTGTGTGTGTGT | 536,547 | TGTGTGTGTGTGTGTGTGTG | 489,597 | ||
| TCCCAAAGTGCTGGGATT | 374,035 | TAATCCCAGCACTTTGGGA | 361,679 | CTCCCAAAGTGCTGGGATTA | 341,542 | ||
| TAATCCCAGCACTTTGGG | 368,428 | AATCCCAGCACTTTGGGAG | 353,065 | AATCCCAGCACTTTGGGAGG | 335,334 | ||
| ATCCCAGCACTTTGGGAG | 363,933 | ATCCCAGCACTTTGGGAGG | 345,608 | GCCTCCCAAAGTGCTGGGAT | 328,159 | ||
| TGTAATCCCAGCACTTTG | 356,809 | TGTAATCCCAGCACTTTGG | 338,285 | GTAATCCCAGCACTTTGGGA | 323,542 | ||
| CTGTAATCCCAGCACTTT | 354,805 | CAAAGTGCTGGGATTACAG | 337,559 | CCCAAAGTGCTGGGATTACA | 322,532 | ||
| TCCCAGCACTTTGGGAGG | 354,247 | GCCTCCCAAAGTGCTGGGA | 336,291 | CTGTAATCCCAGCACTTTGG | 320,216 | ||
| GTAATCCCAGCACTTTGG | 345,648 | CCTGTAATCCCAGCACTTT | 330,885 | CAAAGTGCTGGGATTACAGG | 315,005 | ||
| CCCAGCACTTTGGGAGGC | 345,007 | CCCAAAGTGCTGGGATTAC | 329,491 | TTTTGTATTTTTAGTAGAGA | 300,256 | ||
| CCTGTAATCCCAGCACTT | 343,923 | CTCTACTAAAAATACAAAA | 313,148 | ATTCTCCTGCCTCAGCCTCC | 280,115 | ||
| TTTTGTATTTTTAGTAGA | 333,004 | TCTCTACTAAAAATACAAA | 307,152 | ATATATATATATATATATAT | 279,576 | ||
| ATATATATATATATATAT | 332,710 | ATATATATATATATATATA | 302,986 | TTTTTGTATTTTTAGTAGAG | 272,798 | ||
| TTTGTATTTTTAGTAGAG | 320,328 | GAGGCTGAGGCAGGAGAAT | 296,297 | CCACTGCACTCCAGCCTGGG | 239,735 | ||
| TCTCTACTAAAAATACAA | 312,108 | GGAGGCTGAGGCAGGAGAA | 291,488 | GCCTGTAATCCCAGCACTTT | 238,538 | ||
| AGGCTGAGGCAGGAGAAT | 311,358 | TTTTTGTATTTTTAGTAGA | 290,073 | GGCCTCCCAAAGTGCTGGGA | 224,951 | ||
| GAGGCTGAGGCAGGAGAA | 308,372 | CCAGGCTGGAGTGCAGTGG | 273,189 | GTCTCTACTAAAAATACAAA | 195,075 | ||
| TCTCCTGCCTCAGCCTCC | 298,682 | CACTGCACTCCAGCCTGGG | 256,220 | TTCTCCTGCCTCAGCCTCCC | 191,155 | ||
| TTTTTGTATTTTTAGTAG | 296,585 | GCCTGTAATCCCAGCACTT | 247,562 | GCCACTGCACTCCAGCCTGG | 183,392 | ||
| CCAGGCTGGAGTGCAGTG | 292,150 | GGCCTCCCAAAGTGCTGGG | 230,427 | TTTTTTTTTTTTTTTTGAGA | 167,522 | ||
Density in sequence space
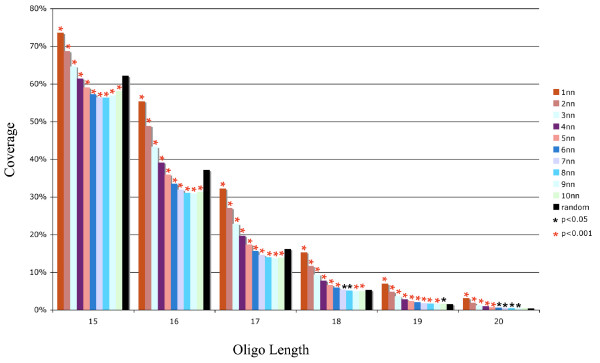
Based on our simulation, the pseudo-human genome has much higher space coverage than the true human genome for every oligo length (Figure 1). This leads to the hypothesis that human genomes are much more compact in sequence space than would be expected by chance. To explore this hypothesis, we examined how many 1 bp variants of human n-mers are also in the human genome. For each of the n-mers found in the human genome, we generated its 3n different 1 bp variants and then scanned the human genome for the presence of these variants. We found that significantly more 1 bp variants of human oligos were also in the human genome compared to random n-mers (Figure 4). The same is true for 2–4 bp variants of human oligos (Figure 4). Thus, the human genome is more compact, or dense, in sequence space than a random genome. When we generated variants that were more distant from the original human n-mer (up to 10 bp variants), we found in some cases that distant oligos were less likely to be in the human genome than a random oligo (Figure 4).
Figure 4.
The density of the human genome in sequence space. For every randomly generated n-mer that was detected in the human genome, we generated all single basepair variants (3n variants for each n-mer) and tested them to see if they were also represented in the human genome (1nn). We also generated 3n of the 2 bp variants (2nn), 3n of the 3 bp variants, and so on up to variants that differed in 10 bp from the original human n-mer. The sequences that are only a few SNPs away from the original human n-mer are significantly more likely to be in the human genome compared to a random n-mer (black bars, "random"). This shows that the human genome is relatively compact in sequence space. The standard error for all points is < 0.003.
Entropy rate
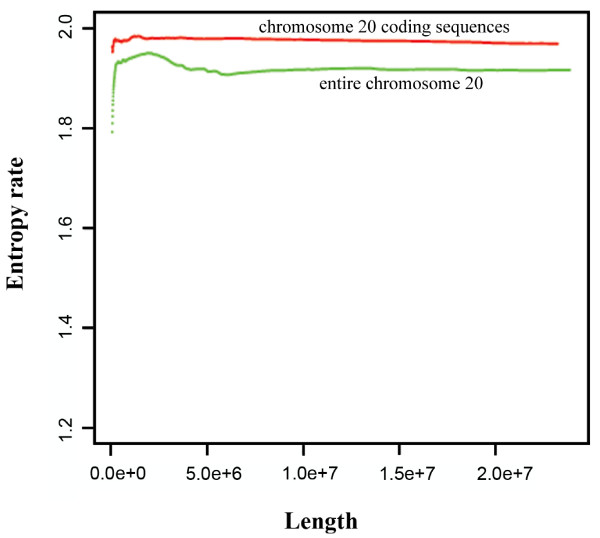
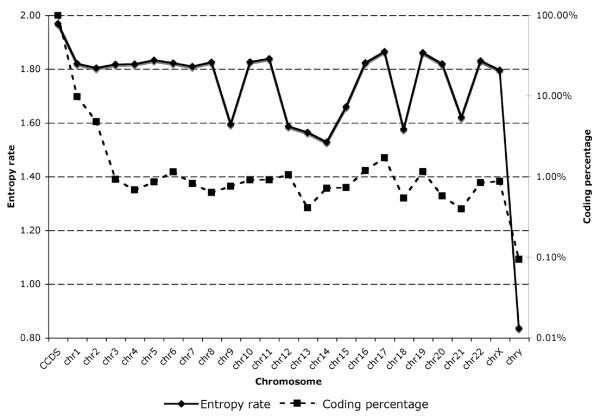
Previous studies have examined the entropy, or information content, within regions of a genome [6-8]. We calculated the information content of each human chromosome. This facilitates comparisons between chromosomes. We used the Lempel-Ziv 77 algorithm to estimate the entropy rate of both coding sequences and whole genomic sequences for the human genome [17]. Coding sequences have a higher entropy rate (information content) than genomic sequences (Figure 5). Note that the highly repetitive regions of telomeres and centromeres are generally coded as non-specific bases ("N") in the human genome and so are excluded from our calculations. Entropy was calculated from either the entire chromosome, for chromosomes < 130 Mb, or the first 130 Mb of longer chromosomes. This is sufficient to generate stable estimates of the entropy (Figure 5). Figure 6 shows both the entropy and the percent of the chromosome filled by coding regions for each human chromosome.
Figure 5.
Entropy rate, using the Lempel-Ziv 77 algorithm, for the coding sequence (red) and the genomic sequence for chromosome 20 (green), as a function of the length of the sequence analyzed. The entropy calculation converges after 10 million bases.
Figure 6.
The entropy, or information content (solid line, left Y axis) and percent of the sequence coding for proteins (dashed line, right Y axis, log scale) for each human chromosome as well as the full set of coding regions (CCDS). Given the higher entropy rate of coding regions to non-coding regions, we expect a correlation between the two measurements. However, chromosomes 1, 2, 9, 12, and 14 have a lower information content than might be expected for the percent of those chromosomes occupied by protein coding regions. Chromosome 20 appears to have a higher entropy than would be expected given its gene poor content. This may be a signal of extensive non-protein coding, yet functional RNA on chromosome 20.
Discussion
We have provided an overview and comparison of some of the informational properties of fully sequenced genomes. We found that virtually all oligomers of length less than 13 are represented in the human genome but only a vanishingly small proportion (< 1.53%) of oligos of length greater than 19. The mouse genome is the same in these respects. Similarly, very few oligos less than 13 bp are unique in the human genome, but the vast majority of oligos of length greater than 19, except repeat elements, are unique in the human genome. This is consistent with practical experience in the design of primers for PCR. Some of the most frequent n-mers in the human genome are microsatellites and ALU elements, as would be expected (Table 2). These ultra-frequent n-mers should be useful as high density markers in the genome and as primers for assays such as random amplified polymorphic DNA (RAPD) [18] in which a large number of regions of the genome may be amplified in a single PCR reaction. In fact, both microsatellites and ALU elements have been exploited for DNA fingerprinting [19-21]. The ultra-frequent n-mers we found are akin to the pyknons identified by Rigoutsos et al., except that pyknons need only appear 40+ times in the human genome, must be at least 16 bp long, and have the additional constraint that they appear in both protein coding and non-coding regions [22].
Whiteford et al. have analyzed a measure of the frequency of unique oligos in a variety of genomes [23]. However, their measure is subtly different from ours. The Whiteford measure of uniqueness essentially shatters a genome into n-mers and asks what proportion of those n-mers only occur once in the genome. This is appropriate for analyzing high throughput, short sequencing reads, since high copy-number n-mers will represent a large portion of the reads. Our measure asks what proportion of distinct n-mers only occur once in a genome? Thus, increasing the copy number of an n-mer already present in the genome would not change our statistic but would decrease the Whiteford measure of the frequency of unique n-mers. Our analysis suggests that the 25+bp reads of current high throughput sequencers are unlikely to produce sequences that would appear by chance in a genome other than the genome being sequenced (Figure 1). These longer n-mers should only be shared between species due to descent from a common ancestor.
The human genome is not spread evenly across sequence space but is rather compacted in closely related sequences (Figure 4). Compaction in sequence space may be the result of molecular evolution. Duplication events followed by divergence between the duplicated regions is thought to be a common mechanism for genome evolution [5] and would lead to such compaction. Similarly, transposons and other repeat elements lead to structure in the non-coding region of the genome that can be detected in the difference between the entropy of the coding regions versus the non-coding regions (Figure 5). Previous work has used Renyi entropy to address the problem of failure of convergence for entropy measures on short DNA sequences [24]. We have used the simpler and more traditional definition of entropy because convergence is not a problem for the analysis of whole human chromosomes (Figure 5).
The difference between the entropy of coding versus non-coding regions of the genome has long been known [14] and may help to explain why chromosomes 13, 18, 21 and the Y chromosome appear to have relatively low entropy compared to the rest of the genome (Figure 6). The correlation between nucleotides at varying distances ("mutual information") is also known to be higher in coding regions than non-coding regions [25]. However, there are a number of chromosomes for which the entropy does not track with the proportion of coding regions in chromosome. Chromosomes 1, 2, 9, 12, and 14 have relatively high proportions of coding regions without relatively high entropy while chromosome 20 has a relatively low proportion of coding regions without relatively low entropy (Figure 6). This may be a signal of functional, non-coding RNA on chromosome 20, for chromosome 20 does not have an unusually low frequency of repeats, an unusual G+C content, or an unusual density of CpG islands [2,3,26]. All of chromosome 20 is conserved as a single segment in the mouse chromosome 2 [2], suggesting it contains little junk DNA. However, the anomaly of non-protein coding information content on chromosome 20 cannot yet be explained by an over-abundance of miRNAs. Out of the 475 currently confirmed miRNAs in the human genome, 11 are located on chromosome 20 [27,28]. This is no more than would be expected by chance (Binomial probability of 11 or more miRNAs, p = 0.46).
Gaps remain in most of the sequenced genomes, but these are unlikely to significantly affect most of our analyses. In the sequence files, missing nucleotides are coded as N's and are skipped over by our algorithms. Build 35 of the human genome was missing 225 Mbp of the human sequence (7% of the genome), 200 Mbp of which is made up of heterochromatin [29]. Heterochromatin is highly repetitive sequence, including telomeres and centromeres. Since the sequenced part of the genome often extends past the borders of heterochromatin [29], it is likely that most of the n-mers in the heterochromatin (for n < 20) would have been counted in our analyses of coverage and uniqueness. The remaining 25 Mbp of euchromatic gaps are often associated with segmental duplications and copy number variations between subjects used for the reference sequencing [30]. Again, many of the n-mers in those gaps are probably represented elsewhere in the sequenced genome. However, the absence from our analysis of the 7% of the human genome with highly repetitive heterochromatin means that our estimates of entropy in the human genome (Figure 6) are probably slightly higher than the true values.
Coverage of sequence space is probably not subject to selection in and of itself, except in specialized cases of diversifying selection, such as occurs in the evolution of major histone compatibility complex (MHC) [31] and some testis genes [32]. However, coverage of sequence space may be a metric of evolvability because it represents the library of genetic sequences that may be duplicated, recombined and modified to generate new genes and functions. All things being equal (including genome size and mutation rates), we would predict that a population of organisms with greater coverage of sequence space should evolve more quickly to new environmental pressures than a population of organisms with fewer sub-sequences. This could be tested in evolvability experiments on bacteria with different degrees of sequence space coverage but similar mutation rates and genome sizes.
When we performed the analysis of coverage versus genome size in 10-mer sequence space. Anaeromyxobacter dehalogenans stands out as having an exceptionally low sequence coverage for its genome size and GC content (outside of the 99.9% predicted range). A. dehalogenans is an anaerobic bacterium with a GC content of 75%. It is able to reduce a variety of metals including ferric iron and Uranium (VI) and has been studied for its potential role in bioremediation [33,34].
One potential use of our results would be to develop assays to detect non-human organisms and sequences in human tissue samples. We found that with a panel of 2.6 M 15-mers that are at least 2 SNPs different from human 15-mers, we could easily detect all bacterial genomes and 75% of fully sequenced viruses. This approach is inspired by the negative selection algorithm used by the immune system: generate random amino acid sequence (peptide) detectors and then remove those that match self. Patterns of positive probes on an array of non-human 15-mers are likely to be enough to identify known microbes. To identify an unknown microbe, any non-human probe that hybridized to DNA from a human sample could be used as a PCR primer to sequence in both directions from that probe and thereby generate longer sequences of the non-human DNA. This would be important both for identifying pathogens in the etiology of diseases as well as for identifying symbiotic microbes that have received little attention because they either do not cause disease or they only cause disease through their absence. Such an array could also identify non-human sequences generated through insertions, deletions and translocations in cancer where such lesions may be targeted for therapy [35,36]. A number of other approaches have been taken to identifying non-human organisms in human samples. Cellular organisms can be identified by sequencing the 16S rRNA genes in the sample [37-40], though this misses viruses. DeRisi and colleagues have developed an oligonucleotide array with 70-mers of highly conserved sequences within most fully sequenced virus families [41]. This was used to identify the SARS virus as a coronavirus[42,43], though the array may not identify novel viruses that are dissimilar from the known viral families. A brute force metagenomics approach involves sequencing all the DNA or RNA in the sample and removing any sequences that match the human genome [44-47]. Currently, efforts are proceeding to sequence all the microbes found in the human body [48].
We have focused on an elegant and relatively simple metric of genome complexity: sequence space coverage. It can be calculated exhaustively or, more efficiently, through sampling based on a set of randomly generated oligos. While sequence space coverage is clearly influenced by genome size and GC content, we have also shown that the human genome is more compact in sequence space than a random genome. This is probably the signature of molecular evolution. Our measurements of sequence space coverage and entropy allow for the comparison between genomes and between chromosomes within a genome. This has led to the detection of outliers that may help to reveal properties of organisms and chromosomes that are not currently understood. Coverage data can also be used in a negative selection algorithm to develop assays to detect novel microbes in tissue samples.
Methods
We used the Human Genome NCBI build 36 version 1, the reference Mouse sequence for C57BL/6J (NCBI build 36), and Escherichia coli strain K-12 substrain MG1655 (accession number U00096). See additional file 4 for all the build numbers and accession IDs of the genomes for Figure 1. All genomes sequences can be found on ftp://ftp.ncbi.nih.gov/genbank/genomes/. The build number for all the bacteria genomes for Figure 2 can be found in our additional file 5. Genome statistics are based on all chromosomes and reverse complement strands for each organism.
Exhaustive search
To find all the oligonucleotides of length n that are present in a genome is difficult for large n, due to computational memory constraints. We exhaustively searched the genomes for all possible 1- to 15-mers. The algorithm follows:
1. Given oligo length n, create a boolean array of size 4n
2. For each chromosome and its reverse complement:
a. For each nucleotide location i in the chromosome:
i. Convert the n-mer at location i into an array index (coding it as a 2n bit number with each nucleotide represented by 2 bits).
ii. Set the Boolean array [index] = true
3. Divide the number array locations set to true by the size of the array (4n).
Stochastic sampling
The set of all oligonucleotides of length n has cardinality of 4n. The size of sequence space increases exponentially with respect to the oligo length. For example, if n = 18, the set contains 8G different oligos. For large n, it is impossible to save all the entries in the space to the memory of any 64-bit machine. Therefore, we choose to use stochastic sampling to estimate the sequence space coverage of the genomes as follows:
1. Randomly generate 100,000 n-mers and save them into a hash table
2. For each n-mer in the hash table:
a. For each chromosome and its reverse complement:
i. Scan the chromosome and record the number of times the n-mer appears in the genome
3. Calculate the coverage by dividing the total number of n-mers that appeared at least once in the genome by 100,000.
We found the stochastic sampling algorithm accurately estimates the true coverage of the genomes (Table 1).
Identification of Ultra-Frequent Oligomers
In order to identify all oligomers of length 10–20 bp that appear > 30,000 times in the human genome we:
1. Identified all 10-mers that appeared in the human genome > 30,000 times by exhaustive search
2. Let K = the number of n-mers previously identified as appearing > 30,000 times. We generated 4K (n+1)-mers by concatenating one of the four possible nucleotides (A, C, G or T) to the end of the K n-mers that appeared > 30,000 times in the human genome.
3. We counted the number of times the 4K (n+1)-mers appear in the human genome and discarded any that appear less than 30,000 times.
4. If n+1 < 20, we incremented n and looped to step 2 with the new list.
This algorithm is guaranteed to find all n-mers that appear at least 30,000 times in the human genome. If a 20-mer occurs > 30,000 times in the human genome, then all prefixes of that 20-mer must occur at least as many times as the 20-mer. So the 10-mer prefix must also occur > 30,000 times and so would have been identified in step 1. The 11- to 19-mer prefixes, as well as the final 20-mer, would then have been generated in step 2 and not discarded in step 3. Therefore, all 20-mers that appear > 30,000 times are identified by the algorithm. This algorithm can be extended to arbitrary length oligomers for any positive lower bound on the frequency of the oligomers. We chose this approach, rather than the brute force approach of counting the frequency of all n-mers in the human genome because the above algorithm need only maintain a small list of the ultra-frequent n-mers at any step and so is more computationally efficient.
Lempel-Ziv estimators of entropy rate
The Lempel-Ziv algorithm [49] is a computationally efficient scheme for universal data compression. The algorithm requires no a priori knowledge of source statistics (hence is "universal"), is particularly elegant, has a very low computational complexity, and produces very compact descriptions of large alphabets. These virtues have established the algorithm firmly as the standard data compression algorithm for the transmission and storage of large files over the Internet and on computers.
Given a sequence of letters from an alphabet, the algorithm parses the string sequentially to produce a dictionary of new phrases, in order of occurrence in the sequence. Specialized to the genome, the algorithm proceeds as follows.
The four phrases corresponding to the single nucleotides 'A', 'C', 'G' and 'T' form the first entries in the dictionary of phrases. The parsing procedure now proceeds recursively finding each new phrase in the sequence in turn.
The Lempel-Ziv parsing procedure:
1. Search for the longest oligomer subsequence S that has appeared in the dictionary.
2. Add S concatenated with the next nucleotide in the genome as a new, previously unobserved, phrase into the dictionary.
3. If the sequence is exhausted without discovering a new phrase, exit.
4. Else, with the pointer set to the location following the last observed nucleotide, go back to step 1.
Let d(n) denote the number of dictionary items that have been generated after parsing n consecutive symbols in the genome string. Then, the number of bits needed to describe the dictionary up to this point provides a simple compression mechanism to describe the entire string up to this point. The pervasive utility of Lempel-Ziv rests upon the observation that such a description is compact and efficient. The theoretical basis for the Lempel-Ziv algorithm is discussed briefly in the Appendix.
All the Java code for implementing the above algorithms and calculating the statistics for the figures and tables is freely available from the authors.
Statistical methods
All statistical analyses were performed in R. For the analysis of 10-mer sequence space coverage, Figure 2 shows that the association with genome size is non-linear, so we log transformed genome size. In addition, the chance of evenly covering sequence space in a random genome decreases as the frequency of nucleotides is skewed from a uniform distribution. Thus, we transformed GC content by taking the absolute value of (0.5 – GC content). These transformed variables were used as predictors of coverage in a multivariate linear regression that included an interaction term between the predictors. Both variables and their interaction were significant at p < 0.01. We used Cook's distance to identify the top 1% of genomes that have the strongest influence on the regression analysis. We excluded those five genomes and refit the multivariate regression model. We then used that model to predict the coverage values of the five excluded genomes based on their log genome sizes and GC content deviation from 0.5. Only one of the five genomes had a 10-mer coverage value that fell outside the 99.9% prediction range and was therefore identified as an outlier in the model.
Authors' contributions
CCM conceived the study. ZL performed all the analyses. SSV carried out the theoretical entropy analysis. CCM, ZL and SSV wrote the paper. All authors approved the final manuscript.
Appendix
Theoretical basis for the Lempel-Ziv algorithm
The Lempel-Ziv procedure forms a very competitive and computationally appealing entropy estimator for long, complex sequences like the human genome. The technical justification for the Lempel-Ziv algorithm in such contexts is provided by a fundamental convergence theorem: the superior limit of the ratio of the number of bits needed to describe the Lempel-Ziv dictionary of phrases to the length of the sequence almost surely bounds the entropy rate of any ergodic, stationary random sequence from below [50]. More formally, if the entropy rate of the source X is denoted by H(X), then
Theorems of this nature form at once the motivation and the ultimate justification of the procedure. The estimates obtained converge very quickly as seen in Figure 5.
Theoretical analysis of the random pseudo-human genome
Clustering and repetitions of n-mers in the human genome over and beyond the chance fluctuations that govern a truly random sequence will manifest themselves in a lower coverage of sequences in n-mer space than can be accounted for by chance. It is informative to consider the n-mer coverage of a random pseudo-human genome of the same length as the human genome and with the nucleotides A, C, G, and T appearing with equal frequency in the genome. This model is equivalent to a sequence of dinucleotides chosen by independent sampling from the set of sixteen dinucleotides {AA, AC, ..., TG, TT}, each dinucleotide selected with equal probability. This permits a more refined comparison with the human genome in view of features such as the marked depletion of CG dinucleotides in mammalian genomes [51], which are not captured by single nucleotide frequencies alone. As we shall see, however, from an analytical perspective, a consideration of dinucleotide frequencies makes very little difference to the conclusions. The n-mer space coverage of the pseudo-human genome will bound from above the corresponding n-mer space coverage of the human genome (see Figure 1), the discrepancy between the two providing evidence of statistical clustering or bunching of n-mers in the human genome. Such discrepancies in coverage are manifested in the entropy rate: the pseudo-human genome has entropy rate equal to the maximal value log2 4 or 2 bits per symbol and bounds from above the entropy rate of the human genome (Figure 5).
Accordingly, consider the simplest model of a random pseudo-human genome as a sequence of nucleotides drawn by independent sampling from the alphabet {A, C, G, T}, with each nucleotide having equal probability of selection. The 4n possible n-mers each appear with equal probability in the nucleotide sequence. If one considers a random sequence of t n-mers (corresponding to a ''genome'' length of N = nt base pairs), the number of n-mers that are absent in the sequence follows the classical coupon collector's paradigm [52]. The probability that exactly m of the possible n-mers is absent in the random sequence is given by
When t is large, say of the order of one billion as in the human genome, the probabilities Pm are approximated by a Poisson distribution. More precisely, if
where logarithms are to the Napier base e, c is any fixed real constant, and the order term represents a quantity that grows asymptotically no faster than the vanishing term log log(t)2/log(t)2 as t becomes large, then the number of excluded n-mers approaches a Poisson distribution with mean exp(-c) for large values of t. More specifically, the probability that exactly m n-mers are excluded in the random sequence is asymptotically given by
for large values of t. In particular, the probability that all the n-mers are present in the sequence is approximately given by
In view of the very rapid extinction of the double exponential for very small values of c, say, between -3 and +3 for typical genome sizes, a small positive c will result in a probability close to one that all n-mers are present while a small negative c will result in gaps in coverage with overwhelming probability. As the term involving c is sub-dominant in the expression for n in terms of t, a phase transition in n-mer coverage in the random pseudo-human genome occurs around the critical value of
For instance, when t is one billion, of the order of the size of the human genome, the critical value for n is 13: with overwhelming probability all n-mers of length 13 or fewer will be found in the random pseudo-human genome with uniform nucleotide frequencies, while there is a breakdown in coverage for n-mers of length in excess of 14.
The results are not materially affected if the uniform nucleotide frequencies in the random pseudo-human genome are replaced by the actual observed nucleotide frequencies of 0.295, 0.204, 0.205, and 0.296 for the nucleotides A, C, G, and T in the human genome, or even, in a slightly more refined calculation, the pseudo-human genome is constructed by independent sampling from the set of sixteen dinucleotides with each dinucleotide appearing not with equal probability but in the same frequency as in the human genome (the CG dinucleotide, in particular, being markedly depleted [51]). The n-mer coverage for the actual distribution of nucleotide (or dinucleotide) frequencies is bounded below by the coverage of n-mer space by a uniform random sequence over an alphabet of size 5 as the probability 1/5 = 0.2 of selecting a given nucleotide (or probability 1/25 = 0.04 of selecting a given dinucleotide) from an alphabet of size 5 lies below the observed nucleotide (respectively, dinucleotide) frequencies in the human genome. We can get increasingly conservative bounds in this fashion by increasing the alphabet size as, under uniform selection, the nucleotide frequency decreases inversely proportional to alphabet size, while the dinucleotide frequency decreases inversely with the square of the alphabet size. The size of alphabet, however, makes little difference to the point where a phase transition in n-mer coverage is observed as the expression for the phase transition point for n is relatively insensitive to alphabet size (the expression for n depends only on the logarithm of the alphabet size). Consequently, random pseudo-human genomes with nucleotide and dinucleotide frequencies matching that of the human genome exhibit an essentially complete coverage of n-mers with n up to 13, with gaps in coverage occurring abruptly for n-mers of size 14 and beyond. For the analysis in Figure 1, we generated a random pseudo-human genome with the same dinucleotide frequencies as the human genome by using a first-order Markov process with transition probabilities that match the probabilities for each nucleotide following the previous nucleotide in the human genome. The substantial agreement of the stochastic simulation results for the pseudo-human genome in Figure 1 with the theoretical predictions serves to provide a validation of the sampling methodology.
Supplementary Material
Supplementary-1. Space coverage of n-mer space (1–20) for Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Saccharomyces cerevisiae, and Escherichia coli (K-12) genomes by the stochastic searching algorithm.
Supplementary-2. The 10-mer space coverage for 433 fully sequenced microbial with genomes GC content and genome sizes.
A full list of high frequency n-mers for Homo sapiens genome.
Supplementary-4. The NCBI genome sequence accession ID for Drosophila melanogaster, Arabidopsis thaliana, Caenorhabditis elegans, Saccharomyces cerevisiae.
Supplementary-5. The NCBI genome sequence accession ID for 433 microbial.
Acknowledgments
Acknowledgements
We would like to thank Tom Paulson, Dennis Chao, Ramana Davuluri, John Rux and Shane Jensen for helpful advice and discussion. This work was supported by NIH grant P30 CA010815, the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health, the Pew Charitable Trust, and the McLean Contributionship. The sponsors had no role in the study design; collection, analysis, and interpretation of data; writing of the paper; or decision to submit it for publication.
Contributor Information
Zhandong Liu, Email: zhandong@mail.med.upenn.edu.
Santosh S Venkatesh, Email: venkatesh@ee.upenn.edu.
Carlo C Maley, Email: cmaley@alum.mit.edu.
References
- Watson JD. The Double Helix: A Personal Account of the Discovery of the Structure of DNA. Penguin. 1970.
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Li WH, Gu Z, Wang H, Nekrutenko A. Evolutionary analyses of the human genome. Nature. 2001;409:847–849. doi: 10.1038/35057039. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Larkin DM, Everts-van der Wind A, Bourque G, Tesler G, Auvil L, Beever JE, Chowdhary BP, Galibert F, Gatzke L, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Nalla VK, Rogan PK. Automated splicing mutation analysis by information theory. Hum Mutat. 2005;25:334–342. doi: 10.1002/humu.20151. [DOI] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Bernaola-Galvan P, Grosse I, Carpena P, Oliver JL, Roman-Roldan R, Stanley HE. Finding borders between coding and noncoding DNA regions by an entropic segmentation method. Phys Rev Lett. 2000;85:1342–1345. doi: 10.1103/PhysRevLett.85.1342. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holste D, Grosse I, Herzel H. Statistical analysis of the DNA sequence of human chromosome 22. Phys Rev E Stat Nonlin Soft Matter Phys. 2001;64:041917. doi: 10.1103/PhysRevE.64.041917. [DOI] [PubMed] [Google Scholar]
- Zhang L, Sun T. Statistical Properties of Nucleotides in Human Chromosomes 21 and 22. Chaos, Solitons and Fractals. 2005;23:1077–1085. [Google Scholar]
- Stanley RH, Dokholyan NV, Buldyrev SV, Havlin S, Stanley HE. Clustering of identical oligomers in coding and noncoding DNA sequences. J Biomol Struct Dyn. 1999;17:79–87. doi: 10.1080/07391102.1999.10508342. [DOI] [PubMed] [Google Scholar]
- Buldyrev SV, Dokholyan NV, Havlin S, Stanley HE, Stanley RHR. Expansion of Tandem Repeats and Oligomer Clustering in Coding and Noncoding DNA sequences. Physica A. 1999;273:19–32. doi: 10.1016/S0378-4371(99)00338-6. [DOI] [Google Scholar]
- Mantegna RN, Buldyrev SV, Goldberger AL, Havlin S, Peng CK, Simons M, Stanley HE. Systematic analysis of coding and noncoding DNA sequences using methods of statistical linguistics. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1995;52:2939–2950. doi: 10.1103/physreve.52.2939. [DOI] [PubMed] [Google Scholar]
- McHardy AC, Martin HG, Tsirigos A, Hugenholtz P, Rigoutsos I. Accurate phylogenetic classification of variable-length DNA fragments. Nat Methods. 2007;4:63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- Zavala A, Naya H, Romero H, Sabbia V, Piovani R, Musto H. Genomic GC content prediction in prokaryotes from a sample of genes. Gene. 2005;357:137–143. doi: 10.1016/j.gene.2005.06.030. [DOI] [PubMed] [Google Scholar]
- Welch TA. A Technique for High-Performance Data Compression. Computer. 1984;17:8–19. doi: 10.1109/MC.1984.1659158. [DOI] [Google Scholar]
- Atienzar FA, Jha AN. The random amplified polymorphic DNA (RAPD) assay and related techniques applied to genotoxicity and carcinogenesis studies: a critical review. Mutat Res. 2006;613:76–102. doi: 10.1016/j.mrrev.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Stoler DL, Chen N, Basik M, Kahlenberg MS, Rodriguez-Bigas MA, Petrelli NJ, Anderson GR. The onset and extent of genomic instability in sporadic colorectal tumor progression. Proc Natl Acad Sci USA. 1999;96:15121–15126. doi: 10.1073/pnas.96.26.15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass DH, Batzer MA. Inter-Alu polymerase chain reaction: advancements and applications. Anal Biochem. 1995;228:185–193. doi: 10.1006/abio.1995.1338. [DOI] [PubMed] [Google Scholar]
- Bonafe M, Cardelli M, Marchegiani F, Cavallone L, Giovagnetti S, Olivieri F, Lisa R, Pieri C, Franceschi C. Increase of homozygosity in centenarians revealed by a new inter-Alu PCR technique. Exp Gerontol. 2001;36:1063–1073. doi: 10.1016/S0531-5565(01)00112-7. [DOI] [PubMed] [Google Scholar]
- Rigoutsos I, Huynh T, Miranda K, Tsirigos A, McHardy A, Platt D. Short blocks from the noncoding parts of the human genome have instances within nearly all known genes and relate to biological processes. Proc Natl Acad Sci USA. 2006;103:6605–6610. doi: 10.1073/pnas.0601688103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford N, Haslam N, Weber G, Prugel-Bennett A, Essex JW, Roach PL, Bradley M, Neylon C. An analysis of the feasibility of short read sequencing. Nucleic Acids Res. 2005;33:e171. doi: 10.1093/nar/gni170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinga S, Almeida JS. Renyi continuous entropy of DNA sequences. J Theor Biol. 2004;231:377–388. doi: 10.1016/j.jtbi.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Grosse I, Herzel H, Buldyrev SV, Stanley HE. Species independence of mutual information in coding and noncoding DNA. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 2000;61:5624–5629. doi: 10.1103/physreve.61.5624. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Matthews LH, Ashurst J, Burton J, Gilbert JG, Jones M, Stavrides G, Almeida JP, Babbage AK, Bagguley CL, et al. The DNA sequence and comparative analysis of human chromosome 20. Nature. 2001;414:865–871. doi: 10.1038/414865a. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004:D109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Bovee D, Zhou Y, Haugen E, Wu Z, Hayden HS, Gillett W, Tuzun E, Cooper GM, Sampas N, Phelps K, et al. Closing gaps in the human genome with fosmid resources generated from multiple individuals. Nat Genet. 2008;40:96–101. doi: 10.1038/ng.2007.34. [DOI] [PubMed] [Google Scholar]
- Apanius V, Penn D, Slev PR, Ruff LR, Potts WK. The nature of selection on the major histocompatibility complex. Crit Rev Immunol. 1997;17:179–224. doi: 10.1615/critrevimmunol.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- Stevenson BJ, Iseli C, Panji S, Zahn-Zabal M, Hide W, Old LJ, Simpson AJ, Jongeneel CV. Rapid evolution of cancer/testis genes on the X chromosome. BMC Genomics. 2007;8:129. doi: 10.1186/1471-2164-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford RA, Cole JR, Tiedje JM. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl Environ Microbiol. 2002;68:893–900. doi: 10.1128/AEM.68.2.893-900.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Sanford RA, Loffler FE. Uranium(VI) reduction by Anaeromyxobacter dehalogenans strain 2CP-C. Appl Environ Microbiol. 2006;72:3608–3614. doi: 10.1128/AEM.72.5.3608-3614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, Cervantes F, Cornelissen JJ, Fischer T, Hochhaus A, Hughes T, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348:994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Bent SJ, Schneider MG, Davis CC, Islam MR, Forney LJ. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology. 2004;150:2565–2573. doi: 10.1099/mic.0.26905-0. [DOI] [PubMed] [Google Scholar]
- Sghir A, Gramet G, Suau A, Rochet V, Pochart P, Dore J. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl Environ Microbiol. 2000;66:2263–2266. doi: 10.1128/AEM.66.5.2263-2266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GB, Hart CA, Mason JR, Hughes M, Walshaw MJ, Bruce KD. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol. 2003;41:3548–3558. doi: 10.1128/JCM.41.8.3548-3558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroes I, Lepp PW, Relman DA. Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci USA. 1999;96:14547–14552. doi: 10.1073/pnas.96.25.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci USA. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Wang D, Urisman A, Liu YT, Springer M, Ksiazek TG, Erdman DD, Mardis ER, Hickenbotham M, Magrini V, Eldred J, et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:257–260. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacConaill L, Meyerson M. Adding pathogens by genomic subtraction. Nat Genet. 2008;40:380–382. doi: 10.1038/ng0408-380. [DOI] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G, Druce J, Du L, Tran T, Birch C, Briese T, Conlan S, Quan PL, Hui J, Marshall J, et al. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- Xu Y, Stange-Thomann N, Weber G, Bo R, Dodge S, David RG, Foley K, Beheshti J, Harris NL, Birren B, et al. Pathogen discovery from human tissue by sequence-based computational subtraction. Genomics. 2003;81:329–335. doi: 10.1016/S0888-7543(02)00043-5. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv J, Lempel A. Compression of individual sequences by variable rate coding. IEEE Transactions on Information Theory. 1978;IT-24:530–536. doi: 10.1109/TIT.1978.1055934. [DOI] [Google Scholar]
- Cover TM, Thomas JA. Elements of Information Theory. New York: Wiley; 1991. [Google Scholar]
- Karlin S, Brocchieri L, Trent J, Blaisdell BE, Mrazek J. Heterogeneity of genome and proteome content in bacteria, archaea, and eukaryotes. Theor Popul Biol. 2002;61:367–390. doi: 10.1006/tpbi.2002.1606. [DOI] [PubMed] [Google Scholar]
- Feller W. An Introduction to Probability Theory and its Applications. Vol. 1. New York: Wiley; 1965. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary-1. Space coverage of n-mer space (1–20) for Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Saccharomyces cerevisiae, and Escherichia coli (K-12) genomes by the stochastic searching algorithm.
Supplementary-2. The 10-mer space coverage for 433 fully sequenced microbial with genomes GC content and genome sizes.
A full list of high frequency n-mers for Homo sapiens genome.
Supplementary-4. The NCBI genome sequence accession ID for Drosophila melanogaster, Arabidopsis thaliana, Caenorhabditis elegans, Saccharomyces cerevisiae.
Supplementary-5. The NCBI genome sequence accession ID for 433 microbial.