Abstract
Adamantane scaffolds for affinity maturation of prostate cancer specific ligands of low molecular mass are described. These scaffolds are modular and can be used for conjugation of up to three ligands and an additional effector molecule by standard peptide coupling techniques. The potential of the scaffolds is demonstrated with the multimerization of GPI 1, a prostate cancer specific small molecule. A detailed study of multimerized GPI conjugates with NIR-fluorophores and their binding properties to different prostate cancer cell lines shows the specific binding of these conjugates to cell types positive for prostate specific membrane antigen (PSMA). We demonstrate that these conjugates allow the sensitive imaging of prostate cancer cells with NIR methodology and suggest that our adamantane scaffolds might be generally useful for affinity maturation of small molecules targeting cell surface epitopes.
Keywords: medicinal chemistry, membrane proteins, multivalency, tumor targeting, hydrocarbons
Introduction
Finding a cure for most cancer types has been difficult, and thus far, in most cases unsuccessful. One of the major challenges in current cancer therapy is the differentiation of tumor and “normal” cells. Cancer cells are different from other cells in a number of ways and one might target these differences for diagnostic and therapeutic purposes. In this context, cancer specific receptors, which are present ideally on the surface of cancer cells, have attracted considerable attention as tumor markers.1,2 If conjugated to (generally unspecific) effector molecules such as contrast agents or cytotoxic moieties, ligands for cancer specific receptors can guide effectors to a tumor site and permit the sensitive detection of cancer for diagnostic purposes or the specific elimination of cancer cells for therapeutic applications.3-6
In contrast to monoclonal antibodies which have been the gold standard for tumor targeting in the past,7 small cancer-specific molecules have very favourable pharmacokinetic properties particularly for diagnostic applications where their fast blood clearance improves the signal to noise ratio.8 In addition, they have better tumor penetration, are relatively easy to synthesize and they may be useful in multimodality imaging.9 On the other hand ligands of low molecular weight do often have inadequate binding affinity and/or specificity for a given tumor marker.
Prostate cancer is the most widely diagnosed cancer in men in the United States, and an estimated 28,000 previously diagnosed will die of the disease in 2008.10 In this paper, we report the application of the multimerized small molecule concept to prostate cancer imaging with targeted NIR-dyes. Near-infrared (700 nm to 900 nm) fluorescent light for in vivo imaging has recently received considerable interest.11 It is invisible to the human eye but has a high transmission through living tissue. It is safe at the fluence rates used, and most tissues have low NIR autofluorescence. Targeted NIR fluorescence imaging systems are thus valuable tools for prostate cancer diagnosis and are expected to be particularly valuable for image-guided prostate cancer surgery.11,12
Results and Discussion
Our group13 and others14-16, have chosen prostate-specific membrane antigen (PSMA), a membrane bound glycoprotein,17 as a target of choice for prostate cancer because of its transmembrane location and the fact that it is overexpressed on malignant prostate cells and their metastases18. Although also expressed on normal prostate epithelial cells, such normal cells are not needed after childbearing, making PSMA an ideal target.
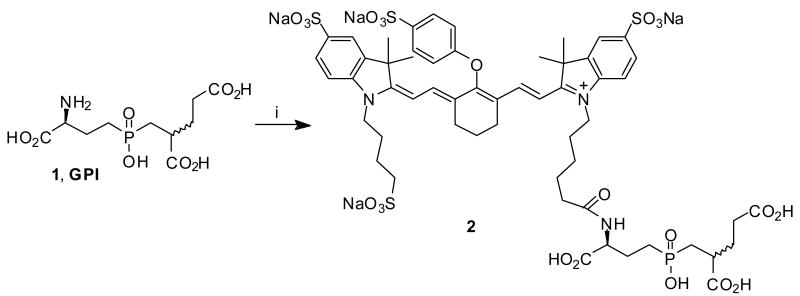
We have previously described the development of a high-affinity (9 nM), small molecule specific for the active site of the tumor marker PSMA. This ligand, termed GPI 119 (Scheme 1), was engineered to contain a primary amine for conjugation to effector molecules, without losing its binding affinity for the target receptor.13 Conjugates of GPI 1 (and other PSMA-specific small molecules) with contrast agents, are therefore valuable tools for prostate tumor imaging.16,20-23
Scheme 1.a.
a Reagents and conditions: i) Et3N, 10a-NHS, DMSO, 12h, 80%.
However, it turned out that GPI cannot effectively compete with high concentrations of endogenous phosphate, limiting its use in vivo. Although GPI 1 itself as well as it's conjugates (like 2) have good affinities to LNCap cells (a PSMA positive prostate cancer cell line) in phosphate free media like TBS, no binding was detected in phosphate buffer or serum (see Table 1). We have addressed this issue for conjugates of GPI 1 with 99mTc-chelates for SPECT-imaging with a multimerization approach20 and report here on the application to prostate cancer imaging with NIR-technology.
Table 1.
Live cell binding affinity of PSMA ligands for the prostate cancer cell lines LNCaP and PC3 (PSMA- and PSMA+) in different media
| Molecule | Cell line | Kd (nM) TBS[a] | Kd (nM) PBS[a] | Kd (nM) Serum[a] |
|---|---|---|---|---|
| 1 | LNCaP | 10.5 ± 1.4[Ref. 13] | NBD | NBD |
| 2 | LNCaP | 3.6 ± 0.6 | NBD | NBD |
| 4 | LNCaP | 0.6 ± 0.1 | 0.8 ± 0.1 | 0.9 ± 0.1 |
| 4 | PC3(PSMA-) | NBD | NBD | NBD |
| 4 | PC3(PSMA+) | 0.2 ± 0.1 | 0.3 ± 0.1 | 0.6 ± 0.1 |
| 5 | LNCaP | 0.2 ± 0.1 | 0.3 ± 0.1 | 0.3 ± 0.1 |
| 5 | PC3(PSMA-) | NBD | NBD | NBD |
| 6 | LNCaP | NBD | NBD | - |
| 6 | PC3(PSMA+) | NBD | NBD | - |
| 8a | LNCaP | 16.4 ± 3.1 | NBD | NBD |
| 8b | LNCaP | 2.4 ± 0.7 | 4.8 ± 1.3 | 4.8 ± 1.1 |
| 8c | LNCaP | 0.4 ± 0.1 | 0.5 ± 0.1 | 0.7 ± 0.1 |
| 8c | PC3(PSMA-) | NBD | NBD | NBD |
| 9a | LNCaP | 5.4 ± 1.1 | NBD | NBD |
| 9b | LNCaP | 1.3 ± 0.2 | 1.9 ± 0.5 | 1.7 ± 0.2 |
| 9c | LNCaP | 0.5 ± 0.1 | 0.5 ± 0.1 | 0.4 ± 0.1 |
| 9c | PC3(PSMA-) | NBD | NBD | NBD |
| 10a | LNCaP | NBD | - | - |
| 10a | PC3(PSMA+) | NBD | - | - |
| 10b | LNCaP | NBD | - | - |
| 10b | PC3(PSMA+) | NBD | - | - |
| 11 | LNCaP | 0.4 ± 0.1 | - | - |
| 11 | PC3(PSMA+) | 0.5 ± 0.1 | - | - |
| 12 | LNCaP | NBD | - | - |
| 12 | PC3(PSMA+) | NBD | - | - |
| 13 | LNCaP | NBD | - | - |
| 13 | PC3(PSMA+) | NBD | - | - |
Mean ± S.D.; NBD: No binding detected, serum: 100 % calf serum.
Nature often takes advantage of multimerization to decrease ligand off-rate and improve affinity of cell surface binders.24,25 We designed suitable scaffolds for the assembly of multiple targeting ligands and contrast agents in the hope that multimerization would improve the performance of our cancer specific ligands.
Several different multivalent scaffolds have been used successfully by other groups in the past particularly for applications in carbohydrate/lectin interactions26,27 but also for peptide/protein interactions28 and in the context of tumor targeting29,30. Among these scaffolds are small molecules with few conjugation sites (∼2-10) and larger systems like dendrimers31 and polymers32.
We wanted to study the effect of GPI-multimerization with a well defined system and focused therefore on relatively small scaffolds with few conjugation sites. For our purposes a suitable scaffold had to match certain design criteria. 1. It had to expose our ligands in the right geometry to cell surface binding epitopes. 2. It had to provide a conjugation site for contrast agents, preferably a primary amine that is compatible with well-established conjugation protocols. 3. The scaffold had to permit a rigid spacing of several highly charged ligands (GPI 1 has four negative charges) and contrast agents to avoid charge crowding that would result in problems for the conjugation of ligands or contrast agents.
Thus, we focused our attention on bridgehead functionalized adamantane as a core structure for the assembly of our multivalent imaging agents. With their rigid tetrahedral structure, these adamantane derivatives meet the above-mentioned criteria.33 Most importantly, they provide a tripodal arrangement of tumor specific ligands using three bridgehead functionalities and an additional fourth bridgehead position for attachment of effector molecules pointing in the opposite direction. This tripodal arrangement of cell specific ligands is an ideal recognition motif for cell surfaces, as a number of receptors share a common binding motif with threefold geometry.25,34-36 In addition, a number of adamantane derivatives (particularly amino adamantanes) are well-known pharmaceuticals of low toxicity.37,38 We have reported the use of adamantane as a multivalent scaffold in a pharmaceutical context20 and additional applications as a scaffold for dendritic structures and in material science are well known.39-42
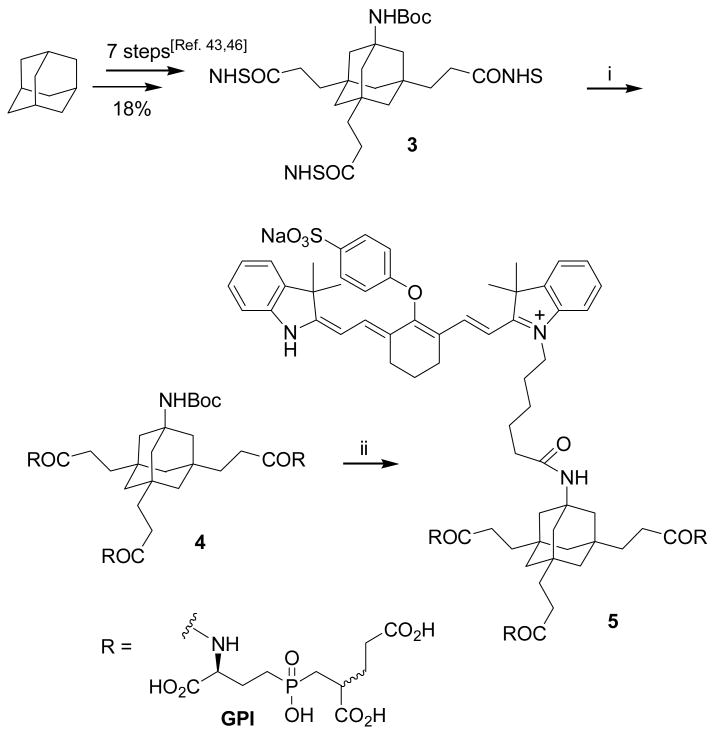
In preliminary work we have shown that tetrasubstituted adamantane derivatives such as 3 (Scheme 2) can be synthesized in gram quantities and good yields from adamantane as an inexpensive starting material.43 These tetrafunctional adamantanes have three carboxylic acids for conjugation of cancer specific ligands and a primary amine for conjugation of contrast agents. In our first experiments (Scheme 2), we conjugated GPI 1 to the Boc-protected adamantane derivative 3 and obtained the trimeric GPI-conjugate 4. The Boc-group was subsequently cleaved under standard conditions with TFA and the commercially available NIR-dye 10b was attached to the resulting free amine via NHS-ester methodology. To synthesize 5, a tenfold excess of 10b-NHS was necessary to obtain a decent yield. During the conjugation of NIR-dye 10b, we noticed a consistent mass loss of 136 which did not affect the fluorescence properties of the dye. We attribute this mass loss to the cleavage of a (CH2)4-SO3- group from 10b.
Scheme 2.a.
a Reagents and conditions: i) Et3N, 1, DMSO, rt, 18h, 50%; ii) 1. TFA, rt, 5h; 2. Et3N, 10b-NHS, DMSO, rt, 18h, two steps 86%.
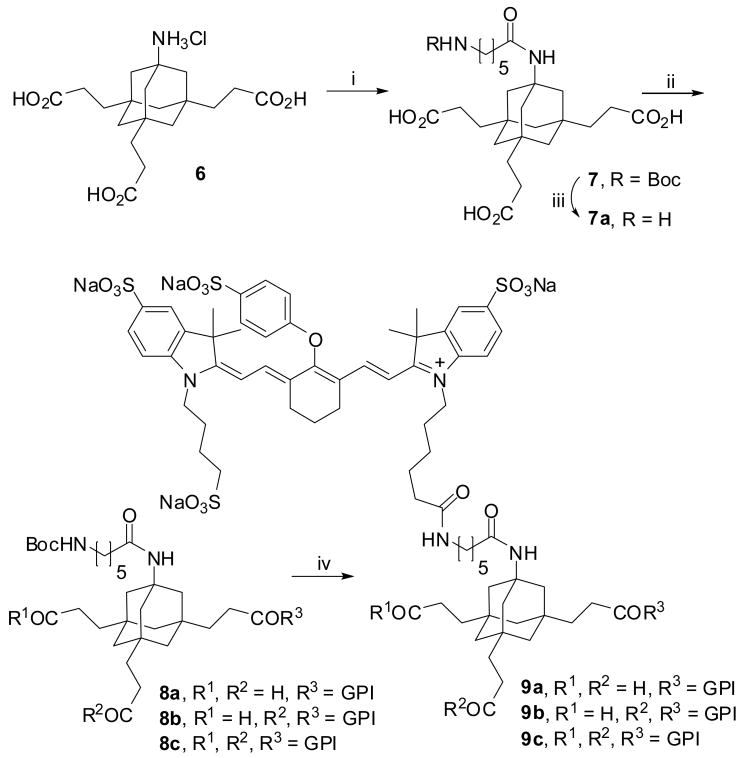
We then tried to conjugate the commercially available tetrasulfonated NIR-dye 10a to deprotected 4; this dye is less hydrophobic than 10b and would be a better choice from a pharmacokinetics point of view for further in vivo investigations. The coupling yield was very low in these attempts, so we introduced a spacing moiety to avoid the steric hindrance at the amino adamantane position during NHS-coupling. This was achieved according to Scheme 3 by transformation of triacid 6 to the corresponding trimethyl ester, which was acylated with Boc-protected γ-aminohexanoic acid and subsequently hydrolyzed with aqueous LiOH to give 7. Conjugation to GPI 1 was done via NHS-ester coupling of 7. The coupling efficiency for this reaction is not high and, next to trimer 8c, a significant portion of the monomeric and dimeric coupling products 8a and 8b were observed. This turned out to be an advantage because all three derivatives were separable by HPLC and allowed us to study the multimeric binding process systematically. In a final deprotection step the Boc-group was removed with TFA to give the free primary amines, ready for conjugation to contrast agents. The NIR labeling was accomplished in one step with a fourfold excess of 10a-NHS ester producing compounds 9a, 9b and 9c.
Scheme 3.a.
a Reagents and conditions: i) 1. TMSCH2N2, rt, 16h, Et3N, 74%; 2. BocNH(CH2)5CO2H, HBTU, DIPEA, 16h, THF, 95%; 3. LiOH, H2O, dioxane, rt, 20h, 71%; ii) 1. EDC, NHS, rt, 12h, 84%; 2. 1, DMSO, rt, 18h, 8a: 17%, 8b: 25%, 8c: 34%; iii) TFA, rt, 5h; iv) 1. TFA, rt, 5h; 2. Et3N, 10a-NHS, DMSO, rt, 18h, two steps each: 9a: 19%, 9b: 23%, 9c: 14%.
The absolute affinity of each compound for the surface of living prostate cancer cells was measured using homologous competition (Table 1). In this previously described assay, 99mTc radiolabeled derivatives of the PSMA specific small molecules were used as radiotracers.20 To show the comparability of the assay with other techniques, we used the well known PSMA-ligand PMPA 1144 as a reference compound and observed similar affinities (400 ± 100 pM) to those described in the literature (275 ± 80 pM).14
The data from the assay using monomeric versions of GPI (1, 2, 8a and 9a) confirmed our finding that high concentrations of phosphate in PBS buffer and serum compete efficiently for the active site of PSMA. In contrast, dimeric GPI-conjugates 8b and 9b and their trimeric homologues 4, 5, 8c and 9c show almost the same binding affinities to LNCAP cells (PSMA+) in two different buffer systems and even in serum. It should be noted that affinities of PSMA ligands are not dependent on the nature of the dye or it's linker moiety to the adamantane core (compare 5 and 9c, for example). The binding affinity of the GPI conjugate 2 is increased by an order of magnitude upon trimerization with the adamantyl scaffold leading to subnanomolar cell surface binders 4, 5, 8c and 9c in TBS, PBS and serum.
We performed several control experiments to confirm the specificity of our trimeric conjugates for cells expressing PMSA on their surface. As a negative control, all compounds were tested against PC3(PSMA-) cells which do not express PSMA on their surface; no binding was detected. As a further crosscheck for specificity, the trimeric conjugate 4 was also tested against PC3(PSMA+) cells and showed again subnanomolar binding in all three media. The binding data for compounds 6, 10a, 10b, 12 and 13 shows clearly that the adamantane scaffold alone (6), the dyes alone (10a and 10b) as well as the conjugates of both (12 and 13) do not contribute significantly to the binding of our trimeric conjugates 4, 5, 8c and 9c.
Encouraged by these positive results, we studied the binding of NIR-labeled compounds 2 and 5 to endogenous PSMA on the cell surface by in vitro NIR fluorescence imaging (Figure 2). Living cells were incubated with the molecules for 20 minutes at room temperature in TBS, PBS or serum. As depicted in Figure 2 for 2 and 5, only the multimeric versions of GPI bound to 100% of the PSMA-positive LNCaP in all conditions. This corresponds nicely with the binding data from Table 1 and allows the sensitive detection of prostate cancer cells with NIR-technology. To prove the specificity of binding, we also checked all compounds using PSMA-negative PC3 cells (shown in Figure 2 for 9c, bottom row). No binding was detected.
Figure 2.

NIR Fluorescence imaging of endogenous PSMA on living cells. Fluorescence images have identical exposure time and normalization. Shown are phase contrast (left) and NIR fluorescence (right) images. NIR fluorescence imaging of endogenous PSMA on living prostate cancer cells was first performed using PSMA-positive LNCaP cells. Cells were incubated with 2 μM of compounds 2 (top row), 5 (2nd row), and 10b (4th row) in 100 % calf serum for 20 min at room temperature prior to washing and image acquisition. NIR fluorescence imaging of living, PSMA-negative PC3 cells is also shown for compounds 5 (3rd row) and 10b (bottom row).
Conclusion
In conclusion, we have developed adamantane scaffolds for affinity maturation of PSMA-specific ligands of low molecular mass. Trimerization of our ligands allowed the sensitive and specific imaging of prostate cancer cells in serum with NIR-methodology, which is particularly useful for intraoperative imaging. It should be noted that our adamantane based multimerization platform has a modular character and can be conjugated to any contrast agent or cytotoxic agent via standard peptide coupling techniques. We have demonstrated this aspect with the synthesis of 99mTc-conjugated trimeric PSMA-ligands for SPECT imaging of prostate cancer20 and are currently transferring our imaging techniques to animal models.
Our adamantane scaffolds orient up to three ligands in a tripodal recognition motif and permits the conjugation of effector molecules without disturbing the binding process. Due to the fact that molecular recognition via complexes of threefold geometry is a common feature of several important receptors, we suggest that the adamantane scaffold might be useful as a multimerization platform for addressing several other cell surface binding epitopes besides PSMA. Because it is not clear at the moment if the multimerization effect is due to multireceptor binding or an enhancement of the local concentration of the ligand we are currently evaluating the optimal spacer length and rigidity between the adamantane core and cell surface specific ligands.45
Experimental section
Compounds 1,19 3,43 643,46 and 1144 were synthesized according to previously described procedures. The NHS esters of IRDye®800RS (10b) and IRDye®800CW (10a) were purchased from LI-COR (Lincoln, NE). All fluorophores were stored as dry powder and at -80° until use. HPLC grade triethylammonium acetate (TEAA), pH 7 was from Glen Research (Sterling, VA), HPLC grade water was from American Bioanalytic (Natick, MA). Solvents and chemicals obtained from commercial sources were analytical grade or better and used without further purification. NMR spectra were recorded on a Bruker-Karlsruhe AMX 400 spectrometer (400 MHz/100.6 MHz). Chemical shifts, δ, are represented in part per million (ppm) and coupling constants, J, in Hertz (Hz) from tetramethylsilane (TMS, 0 ppm) as the internal standard for CDCl3 and residual solvent peaks for DMSO-d6. Mass spectra were obtained with a VG/70-250F (VG analytical) instrument in FAB mode in a p-nitrobenzylalkohol matrix or a Micromass LCT TOF-ES spectrometer (Waters).
[800CW]GPI (2)
5 μL of a 30 mM solution of 1 in DMSO and 5 μL of a 200 mM solution of Et3N in DMSO were mixed together. After 10 minutes, 5 μL of a 51 mM solution of IRDye®800CW-NHS ester (10a-NHS) in DMSO were added. The mixture was stirred for 12h, in the dark and at room temperature. The compound was purified by preparative HPLC (Symmetry column, method C, flow rate = 15 mL/min, retention time = 9.2 min). After freeze drying, 2 was obtained as a green powder, with an 80% yield. The purity of the compound was assessed by LC-MS (Symmetry column, method C, retention time = 6.1 min). ES-TOF(-): calculated m/z 1294.3 [M + 2H]- found 1294.3 [M + 2H]-, 646.6 [M+H]-2/2.
AdamGPI-Boc trimer (4)
50 mg (0.16 mmoles) of 1 dissolved in 0.4 mL of dry DMSO were added to 0.2 mL of pure Et3N. After 10 minutes, a solution of 3 (15 mg, 0.02 mmoles) in 0.4 mL of dry DMSO was added. The reaction mixture was stirred at room temperature overnight. The compound was purified by preparative HPLC (Symmetry column, method A, flow rate = 15 mL/min, retention time = 13.4 min). After freeze drying, 13.3 mg (0.01 mmoles, 50%) of 4 were obtained. The purity of the compound was assessed by LC-MS (Symmetry column, method A, retention time = 10.9 min). HRMS ES-TOF(-): calculated C54H84N4O29P3: m/z 1345.4434 [M-H]-; found 1345.4442 [M-H]-, 672.2807 [M-2H]-2/2.
[800RS]AdamGPI Trimer (5)
After addition of 2 mL of TFA to 13.3 mg (0.01 mmoles) of 4, the solution was stirred at room temperature for 5 hours. The progress of the reaction was followed by LC-MS (SunFire column, method A, retention time = 9.2 min). After freeze drying, 11.8 mg (0.009 mmoles, 95%) of the amine as a white powder were obtained. ES-TOF(-): calculated C49H76N4O27P3: m/z 1245.4 [M-H]-; found 1245.4 [M-H]-, 622.2 [M-2H]-2.
5 μL of a 5 mM solution of the amine in DMSO were added to 5 μL of a 200 mM solution of Et3N in DMSO. After 10 minutes, 50 μL of a 5 mM solution of IRDye®800RS NHS ester in DMSO were added. The mixture was stirred overnight, in the dark and at room temperature. The compound was purified by preparative HPLC (Symmetry column, method C, flow rate = 15 mL/min, retention time = 27.9 min). After freeze drying, 5 was obtained as a green powder, with a 90% yield. The purity of the compound was assessed by LC-MS (Symmetry column, method C, retention time = 21.4 min). ES-TOF(-): calculated m/z 966.4 [M-2H]2-; found 966.4 [M-2H]2-.
Boc-Adamantane triacid (7)
16 mL (32.1 mmoles) of a 2 M solution of (trimethylsilyl)-diazomethane in diethyl ether was added dropwise, at room temperature, to a solution of 2.59 g (6.41 mmoles) of 6 in 50 mL of toluene/methanol (4:1). The resulting suspension was stirred for 16 h at room temperature and 0.9 mL acetic acid was added. The reaction mixture was evaporated and the residue was dissolved in 100 mL of 1M aqueous NaOH. The resulting suspension was extracted three times with 100 mL of dichloromethane. The combined extracts were dried over Na2SO4 and the solvent was removed in vacuo to give 1.95 g (4.76 mmoles, 74%) of the pure trimethylester. 1H-NMR (CDCl3): δ 1.01 (d, 3H, J = 11.9 Hz), 1.07 (d, 3H, J = 11.9 Hz), 1.19 (s, 6H), 1.52 (dd, 6H, J = 8.3 Hz, J = 10.3 Hz), 2.25 (dd, 6H, J = 8.3 Hz, J = 10.3 Hz), 3.65 (s, 9H). 13C-NMR (CDCl3): δ 28.2, 35.0, 35.3, 37.4, 37.5, 44.9, 45.2, 51.8, 53.8, 174.5. MS (FAB): m/z (%) = 410.2 [M + H]+ (90%). Anal. (C22H35NO6) C, H, N.
A solution of 232 mg (1.0 mmoles) of 6-(Boc-amino)hexanoic acid and 410 mg (1.0 mmoles) of the above mentioned trimethylester in 20 mL of dry THF was treated with 0.35 mL (2.0 mmoles) of diisopropylethylamine and 391 mg (1.0 mmoles) HBTU. After stirring for 16 hours at room temperature the reaction mixture was heated at 60 °C for 90 minutes. Then dichlormethane and brine was added and the organic phase was washed twice with 20 mL of 1M aqueous HCl, twice with 20 ml of 5% aqueous NaHCO3, twice with 20 mL of brine, and then dried over Na2SO4. The solvent was removed in vacuo to give 592 mg (0.95 mmoles, 95%) of the acylated trimethylester as a yellow oil. 1H-NMR (CDCl3): δ 1.01 (d, 3H, J = 12.2 Hz), 1.13 (d, 3H, J = 12.2 Hz), 1.41 (s, 9H), 1.49-164 (m, 16H), 2.04 (dd, 2H, J = 7.0 Hz, J = 14.5 Hz), 2.24 (t, 6H, J = 8.2 Hz), 3.08 (dd, 2H, J = 6.6 Hz, J = 14.5 Hz), 3.63 (s, 9H), 4.56 (br, 1H), 5.19 (s, 1H). 13C-NMR (CDCl3): δ 25.3, 26.4, 28.2, 28.5, 29.9, 34.9, 37.5, 40.4, 44.9, 45.2, 51.7, 53.7, 79.1, 156.1, 172.3, 174.5. MS (FAB): m/z (%) = 623.5 [M + H]+ (20%). Anal. (C33H54N2O9) C, H, N.
A solution of 71 mg (1.7 mmoles) of LiOH · H2O in 10 ml H2O were added to a suspension of 300 mg (0.48 mmoles) of the acylated trimethylester in 10 mL of dioxane and the resulting solution was stirred 20 h at room temperature. The solvent was removed in vacuo, and the residue was dissolved in 20 mL of 1M aqueous NaOH. The aqueous solution was washed twice with 20 mL of CH2Cl2, acidified to pH 1 with 4M aqueous HCl and extracted three times with 50 mL of ethyl acetate. The combined organic extracts were dried over Na2SO4. Evaporation of the solvent gave 258 mg of crude product that was crystallized from acetonitrile/ethyl acetate (1:2) to give 196 mg (0.34 mmoles, 71%) of pure acid 7. 1H-NMR (DMSO-d6): δ 0.99 (d, 3H, J = 12.2 Hz), 1.04 (d, 3H, J = 12.2 Hz), 1.30-1.43 (m, 21H), 1.50 (s, 6H), 1.97 (t, 2H, J = 7.9 Hz), 2.14 (t, 6H, J = 8.1 Hz), 2.87 (dd, 2H, J = 6.7 Hz), 6.74 (t, 1H, J = 5.4 Hz), 7.27 (s, 1H), 11.97 (s, 3H). 13C-NMR (DMSO-d6): δ 25.2, 27.9, 28.3, 34.2, 36.1, 37.5, 44.4, 44.6, 52.5, 162.1, 171.6, 175.0. MS (FAB): m/z (%) = 581.5 [M + H]+ (50%). Anal. (C30H48N2O9) C, H, N.
Boc-AdamGPI-Trimer (8c)
To a solution of 190 mg (0.33 mmoles) of 7 and 113 mg (0.99 mmoles) of N-hydroxysuccinimide in 10 mL of dry dioxane were added 191 mg (1.00 mmoles) of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride at room temperature. The resulting solution was stirred at room temperature for 12 hours, and then the solvents were evaporated in vacuo. The residual solid was dissolved in ethyl acetate and washed three times with water. Evaporation of the solvent gave 239 mg (0.27 mmoles, 84%) of the tri-NHS-ester as a colorless solid, which was used without further purification. 1H-NMR (CDCl3): δ 1.12 (d, 3H, J = 12.1 Hz), 1.30-1.75 (m, 30H), 2.30 (dd, 2H, J = 6.9 Hz), 2.56-2.61 (m, 6H), 2.83 (br, 12H), 3.09 (t, 2H, J = 6.0 Hz), 4.60 (d, 1H, J = 16.3 Hz), 5.62-5.69 (m, 1H). 13C-NMR (CDCl3): δ 25.4, 25.6, 25.8, 28.5, 34.9, 35.2, 36.8, 36.9, 37.5, 44.2, 44.5, 154.5, 169.4, 169.5, 172.0.
10.36 mg (0.027 mmoles) of 1 dissolved in 0.9 mL of ultra-dry DMSO were added to 0.3 mL of Et3N 200 mM in dry DMSO. After 10 minutes, 2.16 mg (0.0027 mmoles) of tri-NHS-ester dissolved in 0.27 mL of dry DMSO were added. The reaction mixture was stirred at room temperature overnight. The compound was purified by preparative HPLC (Sunfire column, method B, flow rate = 30 mL/min, retention time = 18.1 min). After freeze drying, 1.6 mg (0.0011 mmoles, 40%) of 8c were obtained. The purity of the compound was assessed by LC-MS (Sunfire column, method B, retention time = 10.8 min). ES-TOF(-): calculated C60H95N5O30P3: m/z 1458.5 [M-H]-; found 1458.5 [M-H]-, 728.8 [M-2H]-2/2. Note: during the purification by preparative HPLC, the dimer (8b) and monomer (8a) of the molecule were also separated with retention times of 21.3 and 24.2 min respectively. The yields were 30% and 20% respectively. The purity was assessed by LC-MS using the conditions mentioned above. Dimer (8b): retention time: 11.9 min, ES-TOF(-): calculated C50H79N4O23P2: m/z 1165.4 [M-H]-; found 1165.4 [M-H]-, 582.3 [M-2H]-2/2. Monomer (8a): retention time: 13.8 min, ES-TOF(-): calculated C40H63N3O16P: m/z 872.4 [M-H]-; found 872.4 [M-H]-.
[800CW]Adam GPI-Monomer (9a)
A solution of 8a (0.4 mg, 0.0005 mmoles) in 1.5 mL of TFA was stirred at room temperature for 5 hours. The progress of the reaction was followed by LC-MS (SunFire column, method A, retention time = 11.8 min). After freeze drying, 0.36 mg (95%) of the free amine as a pale yellow powder was obtained. ES-TOF(-): calculated C35H55N3O14P: m/z 772.3 [M-H]-; found 772.3 [M-H]-.
10 μL of a 10 mM solution of amine in DMSO and 10 μL of a 200 mM solution of Et3N in DMSO were mixed together. After 10 minutes, 40 μL of a 11.9 mM solution of IRDye®800CW NHS ester in DMSO were added. The mixture was stirred overnight, in the dark and at room temperature. The compound was purified by preparative HPLC (Sunfire column, method C, flow rate = 30 mL/min, retention time 11.3 min). After freeze drying, 9a was obtained as a green powder, with a 20% yield. The purity of the compound was assessed by LC-MS (Sunfire column, method C, retention time = 7.7 min). ES-TOF(-): calculated m/z 1756.6 [M + 2H]-; found 1756.6 [M + 2H]-, 877.8 [M+H]-2/2.
[800CW]AdamGPI-Dimer (9b)
A solution 8b (1.3 mg, 0.0011 mmoles) in 1.5 mL of TFA was stirred at room temperature for 5 hours. The progress of the reaction was followed by LC-MS (SunFire column, method A, retention time = 10.6 min). After freeze drying, 1.1 mg (95%) of the free amine as a pale yellow powder was obtained. HRMS ES-TOF(-): calculated C45H71N4O21P2: m/z 1065.4086 [M-H]-; found 1065.4120 [M-H]-.
10 μL of a 10 mM solution of the amine in DMSO were added to 10 μL of a 200 mM solution of Et3N in DMSO. After 10 minutes, 40 μL of an 11.9 mM solution of IRDye®800CW NHS ester in DMSO were added. The reaction mixture was stirred at room temperature overnight and in the dark. The compound was purified by preparative HPLC (Sunfire column, method C, flow rate = 30 mL/min, retention time = 6.2 min). After freeze drying, 9b was obtained as a green powder (yield: 24%). The purity of the compound was assessed by LC-MS (Sunfire column, method C, retention time = 6.7 min). ES-TOF(-): calculated m/z 2049.6 [M + 2H]-; found 1024.3 [M+H]-2/2.
[800CW]AdamGPI-Trimer (9c)
A solution of 8c (1.6 mg, 0.0011 mmoles) in 1.5 mL of TFA was stirred at room temperature for 5 hours. The progress of the reaction was followed by LC-MS (SunFire column, method A, retention time = 9.22 min). After freeze drying, 1.4 mg (95%) of the free amine as a pale yellow powder was obtained. HRMS ES-TOF(-): calculated C55H87N5O28P3: m/z 1358.4750 [M-H]-; found 1358.4435 [M-H]-, 678.7416 [M-2H]-2/2.
10 μL of a 10 mM solution of the amine in DMSO were added to 10 μL of a 200 mM solution of Et3N in DMSO. After 10 minutes, 40 μL of an 11.9 mM solution of IRDye®800CW NHS ester in DMSO were added. The reaction mixture was stirred at room temperature overnight and in the dark. The compound was purified by preparative HPLC (Sunfire column, method C, flow rate = 30 mL/min, retention time = 9.4 min). After freeze drying, 9c was obtained as a green powder (yield: 15%). The purity of the compound was assessed by LC-MS (Sunfire column, method C, retention time = 5.8 min). ES-TOF(-): calculated m/z 2343.7 [M + 2H]-; found 1171.4 [M + H]-2/2.
Cell Lines
Human prostate cancer cells (PC-3 and LNCaP) were purchased from the ATCC (Manassas, VA. The cells lines were cultured at 37°C, in a humidified atmosphere containing 5% CO2, in RPMI 1640 medium (Mediatech Cellgro) supplemented with 10% of fetal bovine serum (Gemini Bio-tech products, Woodland, CA) and 5% of penicillin/streptomycin (Cambrex Bio Science, Walkersville, MD).
Affinity assays were performed as previously described.20 A homologous competition assay was employed using the 99mTc-labeled version as radiotracer. Cells were washed 2 times with ice-cold tris-buffered saline (TBS), pH 7.4 and incubated for 20 minutes at 4°C with 0.02 MBq (0.5 μCi) of radiotracer in the presence or absence of the test compound. Cells were then washed 3 times with TBS using a Millipore vacuum manifold (Catalog # MSVMHTS00) and the well contents was transferred directly to 12 × 75 mm plastic tubes placed in gamma counter racks. Transfer was accomplished using a modified (Microvideo Instruments, Avon, MA) 96-well puncher (Millipore MAMP09608) and disposable punch tips (Millipore MADP19650). Well contents were counted on a model 1470 Wallac Wizard (Perkin Elmer, Wellesley, MA) ten-detector gamma counter. To avoid internalization of the radioligand due to constitutive endocytosis13, live cell binding was performed at 4°C., and curves fit using Prism version 4.0a (GraphPad, San Diego, CA) software.
In Vitro Fluorescence Study
For fluorescence study of endogenous PSMA, exponentially growing LNCaP and PC-3 cells at a confluence of 75% on glass coverslips were incubated with 0.2 mL of TBS, PBS or 100 % calf serum containing 2 μM of the compound to be tested for 20 minutes at room temperature. The cells were washed three times with TBS, PBS or serum and fixed with 2% paraformaldehyde in PBS for 10 minutes at room temperature. The cells were then permeabilized with PBS supplemented with 0.1% Tween-20 (PBS-T). The coverslips were mounted using Fluoromount-G and imaged on a previously described four-channel NIR fluorescence microscope.47
Supplementary Material
Additional experimental data for all final compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
Figure 1.
Compounds for control experiments listed in Table 1.
Acknowledgments
We thank Barbara Clough for editing and Grisel Vasquez for Administrative Assistance. This work was funded by NIH grant R01-CA-115296 and grants from the Lewis Family Fund, Ellison Foundation, DAAD PPP 315-sc-ab, and DFG MA 2519/3.
Abreviations
- NIR
near infrared
- PSMA
prostate specific membrane antigen
- PBS
phosphat buffered saline
- TBS
Tris (2-amino-2-(hydroxymethyl)propane-1,3-diol) buffered saline
- SPECT
single photon emission computed tomography
- NHS
N-hydroxysuccinimide
- DMSO
dimethylsulfoxide
- TFA
trifluoroacetic acid
- HBTU
O-Benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluoro-phosphate
- DIPEA
diisopropylethylamine
- EDC
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride
Contributor Information
John V. Frangioni, Email: jfrangio@bidmc.harvard.edu.
Wolfgang Maison, Email: wolfgang.maison@org.chemie.uni-giessen.de.
References
- 1.Maison W, Frangioni JV. Improved Chemical Strategies for the Targeted Therapy of Cancer. Angew Chem, Int Ed. 2003;42:4726–4728. doi: 10.1002/anie.200301666. [DOI] [PubMed] [Google Scholar]
- 2.Becker KF, Hofler H. Mutant cell surface receptors as targets for individualized cancer diagnosis and therapy. Current Cancer Drug Targets. 2001;1:121–128. doi: 10.2174/1568009013334205. [DOI] [PubMed] [Google Scholar]
- 3.Dillman RO. Monoclonal antibodies in the treatment of malignancy: basic concepts and recent developments. Cancer Invest. 2001;19:833–841. doi: 10.1081/cnv-100107745. [DOI] [PubMed] [Google Scholar]
- 4.Haubner R, Kuhnast B, Mang C, Weber WA, Kessler H, Wester HJ, Schwaiger M. [18F]Galacto-RGD: Synthesis, Radiolabeling, Metabolic Stability, and Radiation Dose Estimates. Bioconjug Chem. 2004;15:61–69. doi: 10.1021/bc034170n. [DOI] [PubMed] [Google Scholar]
- 5.Houshmand P, Zlotnik A. Targeting tumor cells. Curr Opin Cell Biol. 2003;15:640–644. doi: 10.1016/s0955-0674(03)00106-6. [DOI] [PubMed] [Google Scholar]
- 6.Liu S, Edwards DS. 99mTc-Labeled Small Peptides as Diagnostic Radiopharmaceuticals. Chem Rev. 1999;99:2235–2268. doi: 10.1021/cr980436l. [DOI] [PubMed] [Google Scholar]
- 7.Reichert JM, Valge-Archer VE. Development trends for monoclonal antibody cancer therapeutics. Nat Rev Drug Discov. 2007;6:349–356. doi: 10.1038/nrd2241. [DOI] [PubMed] [Google Scholar]
- 8.Imai K, Takaoka A. Comparing antibody and small-molecule therapies for cancer. Nat Rev Cancer. 2006;6:714–727. doi: 10.1038/nrc1913. [DOI] [PubMed] [Google Scholar]
- 9.Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat Rev Cancer. 2006;6:583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 10.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 11.Frangioni JV. In vivo near-infrared fluorescence imaging. Curr Opin Chem Biol. 2003;7:626–634. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka E, Choi HS, Fujii H, Bawendi MG, Frangioni JV. Image-guided oncologic surgery using invisible light: completed pre-clinical development for sentinel lymph node mapping. Ann Surg Oncol. 2006;13:1671–1681. doi: 10.1245/s10434-006-9194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humblet V, Lapidus R, Williams LR, Tsukamoto T, Rojas C, Majer P, Hin B, Ohnishi S, De Grand AM, Zaheer A, Renze JT, Nakayama A, Slusher BS, Frangioni JV. High-affinity near-infrared fluorescent small-molecule contrast agents for in vivo imaging of prostate-specific membrane antigen. Mol Imaging. 2005;4:448–462. doi: 10.2310/7290.2005.05163. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Neale JH, Pomper MG, Kozikowski AP. NAAG peptidase inhibitors and their potential for diagnosis and therapy. Nat Rev Drug Discov. 2005;4:1015–1026. doi: 10.1038/nrd1903. [DOI] [PubMed] [Google Scholar]
- 15.Smith-Jones PM, Vallabhajosula S, Navarro V, Bastidas D, Goldsmith SJ, Bander NH. Radiolabeled monoclonal antibodies specific to the extracellular domain of prostate-specific membrane antigen: preclinical studies in nude mice bearing LNCaP human prostate tumor. J Nucl Med. 2003;44:610–617. [PubMed] [Google Scholar]
- 16.Banerjee SR, Foss CA, Castanares M, Mease RC, Byun Y, Fox JJ, Hilton J, Lupold SE, Kozikowski AP, Pomper MG. Synthesis and evaluation of technetium-99m- and rhenium-labeled inhibitors of the prostate-specific membrane antigen (PSMA) J Med Chem. 2008;51:4504–4517. doi: 10.1021/jm800111u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mesters JR, Barinka C, Li W, Tsukamoto T, Majer P, Slusher BS, Konvalinka J, Hilgenfeld R. Structure of glutamate carboxypeptidase II, a drug target in neuronal damage and prostate cancer. EMBO J. 2006;25:1375–1384. doi: 10.1038/sj.emboj.7600969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulke N, Varlamova OA, Donovan GP, Ma D, Gardner JP, Morrissey DM, Arrigale RR, Zhan C, Chodera AJ, Surowitz KG, Maddon PJ, Heston WDW, Olson WC. The homodimer of prostate-specific membrane antigen is a functional target for cancer therapy. PNAS. 2003;100:12590–12595. doi: 10.1073/pnas.1735443100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valiaeva N, Bartley D, Konno T, Coward JK. Phosphinic acid pseudopeptides analogous to glutamyl-gamma-glutamate: synthesis and coupling to pteroyl azides leads to potent inhibitors of folylpoly-gamma-glutamate synthetase. J Org Chem. 2001;66:5146–5154. doi: 10.1021/jo010283t. [DOI] [PubMed] [Google Scholar]
- 20.Misra P, Humblet V, Pannier N, Maison W, Frangioni JV. Production of multimeric prostate-specific membrane antigen small-molecule radiotracers using a solid-phase 99mTc preloading strategy. J Nucl Med. 2007;48:1379–1389. doi: 10.2967/jnumed.107.040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mease RC, Dusich CL, Foss CA, Ravert HT, Dannals RF, Seidel J, Prideaux A, Fox JJ, Sgouros G, Kozikowski AP, Pomper MG. N-[N-[(S)-1,3-Dicarboxypropyl]carbamoyl]-4-[18F]fluorobenzyl-L-cysteine, [18F]DCFBC: a new imaging probe for prostate cancer. Clin Cancer Res. 2008;14:3036–3043. doi: 10.1158/1078-0432.CCR-07-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foss CA, Mease RC, Fan H, Wang Y, Ravert HT, Dannals RF, Olszewski RT, Heston WD, Kozikowski AP, Pomper MG. Radiolabeled small-molecule ligands for prostate-specific membrane antigen: in vivo imaging in experimental models of prostate cancer. Clin Cancer Res. 2005;11:4022–4028. doi: 10.1158/1078-0432.CCR-04-2690. [DOI] [PubMed] [Google Scholar]
- 23.Pomper MG, Musachio JL, Zhang J, Scheffel U, Zhou Y, Hilton J, Maini A, Dannals RF, Wong DF, Kozikowski AP. 11C-MCG: synthesis, uptake selectivity, and primate PET of a probe for glutamate carboxypeptidase II (NAALADase) Mol Imaging. 2002;1:96–101. doi: 10.1162/15353500200202109. [DOI] [PubMed] [Google Scholar]
- 24.Kitov PI, Bundle DR. On the nature of the multivalency effect: a thermodynamic model. J Am Chem Soc. 2003;125:16271–16284. doi: 10.1021/ja038223n. [DOI] [PubMed] [Google Scholar]
- 25.Mammen M, Chio SK, Whitesides GM. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem, Int Ed. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Lundquist JJ, Toone EJ. The cluster glycoside effect. Chem Rev. 2002;102:555–578. doi: 10.1021/cr000418f. [DOI] [PubMed] [Google Scholar]
- 27.Lindhorst TK. Artificial multivalent sugar ligands to understand and manipulate carbohydrate-protein interactions. Top Curr Chem. 2002;218:201–235. [Google Scholar]
- 28.Wright D, Usher L. Multivalent binding in the design of bioactive compounds. Curr Org Chem. 2001;5:1107–1131. [Google Scholar]
- 29.Carlson C, Mowery P, Owen R, Dykhuizen E, Kiessling L. Selective Tumor Cell Targeting Using Low-Affinity, Multivalent Interactions. ACS Chem Biol. 2007;2:119–127. doi: 10.1021/cb6003788. [DOI] [PubMed] [Google Scholar]
- 30.Thumshirn G, Hersel U, Goodman SL, Kessler H. Multimeric cyclic RGD peptides as potential tools for tumor targeting: solid-phase peptide synthesis and chemoselective oxime ligation. Chem Eur J. 2003;9:2717–2725. doi: 10.1002/chem.200204304. [DOI] [PubMed] [Google Scholar]
- 31.Voegtle F, Richardt G, Werner N. Dendritische Moleküle. B G Teubner Verlag; Wiesbaden: 2007. [Google Scholar]
- 32.Haag R, Kratz F. Polymer therapeutics: concepts and applications. Angew Chem, Int Ed Engl. 2006;45:1198–1215. doi: 10.1002/anie.200502113. [DOI] [PubMed] [Google Scholar]
- 33.Newkome GR, Kotta KK, Moorefield CN. Design, synthesis, and characterization of conifer-shaped dendritic architectures. Chem Eur J. 2006;12:3726–3734. doi: 10.1002/chem.200501482. [DOI] [PubMed] [Google Scholar]
- 34.Fournel S, Wieckowski S, Sun W, Trouche N, Dumortier H, Bianco A, Chaloin O, Habib M, Peter JC, Schneider P, Vray B, Toes RE, Offringa R, Melief CJ, Hoebeke J, Guichard G. C3-symmetric peptide scaffolds are functional mimetics of trimeric CD40L. Nat Chem Biol. 2005;1:377–382. doi: 10.1038/nchembio746. [DOI] [PubMed] [Google Scholar]
- 35.Gibson SE, Castaldi MP. C3 symmetry: molecular design inspired by nature. Angew Chem, Int Ed Engl. 2006;45:4718–4720. doi: 10.1002/anie.200600776. [DOI] [PubMed] [Google Scholar]
- 36.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 37.Geldenhuys Werner J, M SF, Bloomquist Jeffrey R, Marchand Alan P, Van der Schyf Cornelis J. Pharmacology and structure-activity relationships of bioactive polycyclic cage compounds: A focus on pentacycloundecane derivatives. Med Res Rev. 2005;25:21–48. doi: 10.1002/med.20013. [DOI] [PubMed] [Google Scholar]
- 38.Zah J, Terre'Blanche G, Erasmus E, Malan SF. Physicochemical prediction of a brain-blood distribution profile in polycyclic amines. Bioorg Med Chem. 2003;11:3569–3578. doi: 10.1016/s0968-0896(03)00365-1. [DOI] [PubMed] [Google Scholar]
- 39.Newkome GR, Nayak A, Behera RK, Moorefield CN, Baker GR. Chemistry of micelles series. 22. Cascade polymers: synthesis and characterization of four-directional spherical dendritic macromolecules based on adamantane. J Org Chem. 1992;57:358–362. [Google Scholar]
- 40.Li Q, Jin C, Petukhov PA, Rukavishnikov AV, Zaikova TO, Phadke A, LaMunyon DH, Lee MD, Keana JF. Synthesis of well-defined tower-shaped 1,3,5-trisubstituted adamantanes incorporating a macrocyclic trilactam ring system. J Org Chem. 2004;69:1010–1019. doi: 10.1021/jo035640+. [DOI] [PubMed] [Google Scholar]
- 41.Menger FM, Migulin VA. Synthesis and Properties of Multiarmed Geminis. J Org Chem. 1999;64:8916–8921. doi: 10.1021/jo9912350. [DOI] [PubMed] [Google Scholar]
- 42.Archibald TG, Malik AA, Baum K, Unroe MR. Thermally stable acetylenic adamantane polymers. Macromolecules. 1991;24:5261–5265. [Google Scholar]
- 43.Maison W, Frangioni JV, Pannier N. Synthesis of Rigid Multivalent Scaffolds Based on Adamantane. Org Lett. 2004;6:4567–4569. doi: 10.1021/ol048055j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson PF, Cole DC, Slusher BS, Stetz SL, Ross LE, Donzanti BA, Trainor DA. Design, synthesis, and biological activity of a potent inhibitor of the neuropeptidase N-acetylated alpha-linked acidic dipeptidase. J Med Chem. 1996;39:619–622. doi: 10.1021/jm950801q. [DOI] [PubMed] [Google Scholar]
- 45.Nasr K, Pannier N, Frangioni JV, Maison W. Rigid Multivalent Scaffolds Based on Adamantane. J Org Chem. 2008;73:1056–1060. doi: 10.1021/jo702310g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pannier Nadine, M W. Rigid C3-Symmetric Scaffolds Based on Adamantane. Eur J Org Chem. 2008:1278–1284. doi: 10.1021/jo702310g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakayama A, Bianco AC, Zhang CY, Lowell BB, Frangioni JV. Quantitation of brown adipose tissue perfusion in transgenic mice using near-infrared fluorescence imaging. Mol Imaging. 2003;2:37–49. doi: 10.1162/15353500200303103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional experimental data for all final compounds. This material is available free of charge via the Internet at http://pubs.acs.org.