Abstract
We synthesized tiny stable silver nanoparticles (2.6 ± 1.1 nm) and used its small surface area and functional groups to control single molecule detection (SMD) volumes on single nanoparticles. These new approaches allowed us to develop intrinsic single molecule nanoparticle optical biosensors (SMNOBS) for sensing and imaging of single human cytokine molecules, recombinant human tumor necrosis factor-α (TNFα), and probing its binding reaction with single monoclonal antibody (MAB) molecules in real-time. We found that SMNOBS retained their biological activity over months and showed exceptionally high photostability. Our study illustrated that smaller nanoparticles exhibited higher dependence of optical properties on surface functional groups, making it a much more sensitive biosensor. Localized surface plasmon resonance spectra (LSPRS) of SMNOBS showed a large red shift of peak wavelength of 29 ± 11 nm, as single TNFα molecules bound with single MAB molecules on single nanoparticles. Utilizing its LSPRS, we quantitatively measured its binding reaction in real time at SM level, showing stochastic binding kinetics of SM reactions with binding equilibrium times ranging from 30 to 120 min. SMNOBS exhibited extraordinarily high sensitivity and selectivity, and a notably wide dynamic range of 0-200 ng/mL (0-11.4 nM). Thus, SMNOBS is well suited for the fundamental study of biological functions of single protein molecules and SM interactions of chemical functional groups with the surface of nanoparticles, as well as development of effective disease diagnosis and therapy.
Keywords: Silver nanoparticles, biosensors, single molecule detection, single molecule reactions, tumor necrosis factor-α (TNFα), cytokines, tumor markers
Introduction
Cytokines are secreted regulatory proteins and play vital roles in controlling cell survival, growth, differentiation, and function by binding with specific receptors and initiating immune regulation pathways.1 At the cellular level, it takes only a few cytokine molecules to induce a significant cellular response,2 underscoring the importance of developing new tools to detect and image individual cytokine molecules and to characterize their functions in real time. Tumor necrosis factor-α (TNFα), a 17.5 kDa protein, is a pro-inflammatory cytokine that can mediate a variety of biological effects, such as immune regulation, antitumor activity, viral replication, and infection resistance.3, 4 Studies have shown that a variety of pathological conditions, including cancer, heart disease, diabetes and autoimmune diseases, lead to overproduction of TNFα, and adequate doses of TNFα are vital to effectively treat diseases (e.g., cancer) without severe side effects.3, 4 Unfortunately, despite extensive research over decades, the underlying mechanisms about how TNFα mediates these crucial biological functions still remain incompletely understood and it is thus important to develop ultrasensitive assays for accurate analysis of TNFα.
Several commercially available methods have been used for detecting TNFα: enzyme-linked immunosorbent assays (ELISA),5, 6 radioimmunoassay (RIA),7 cytotoxicity assay8, flow cytometry,9 and RT-PCR10. The ELISA method offers high sensitivity with a detection limit of 5 pg/mL.5, 6 However, these conventional methods typically involve multiple staining and washing steps, which are time consuming and cannot be used in quantitative analysis of TNFα in real time. Fluorescence quantum dots (QDs), protein microarray11 and electrochemical immunoassay12 have been developed to detect TNFα. Nanoparticle-based detection schemes and sensors have also been reported for detecting other proteins, such as prostate-specific antigen (PSA) and streptavidin in solution.13-16 However, these new techniques still cannot achieve real-time measurements and single molecule detection sensitivity.
Unlike bulk measurements, single molecule detection (SMD) offers the unique opportunity to investigate distinctive functions of individual molecules in real-time. Fluorescence microscopy and spectroscopy have been used as popular tools for SMD in solution and in living cells.17-23 Unfortunately, fluorescence probes (e.g., fluorophors, GFP, QDs) suffer photodecomposition and blinking, offering limited lifetime for probing dynamic events of interest and for quantitative analysis.17-23
Noble metal nanoparticles (Ag, Au) have unique optical properties, which depend on their size, shape, surrounding environment, and dielectric constant of the embedding medium.24-26 Localized surface plasmon resonance spectra (LSPRS) of single nanoparticle highly depends upon their surrounding environments and surface properties,24-26 which forms the basis of using single nanoparticles to detect and sense molecules that are approaching or attached on the surface of nanoparticles. Unlike fluorescent probes and QDs, these noble metal nanoparticles do not suffer photodecomposition and do not blink under dark-field optical illumination, and Ag nanoparticles possess exceptionally high quantum yield of Raleigh scattering that are orders of magnitude higher than fluorophors (e.g., R6G).27, 28 Recently, we have demonstrated the possibility of using these intrinsic optical properties of Ag nanoparticles for imaging single living cells in real-time with sub-100 nm spatial resolution and millisecond time resolution,27-32 and for imaging single receptor molecules on single living cells.29
Nonetheless, in our previous study, we used larger Ag nanoparticles (diameter = 12 nm), which was designed to detect low distribution of proteins on living cell surface with a dynamic range of 0-50 molecules per cell.29 Each nanoparticle has a cross-sectional area (πr2) of 113 nm2 and surface area (4πr2) of 452 nm2. Thus, if more than a single protein molecule occupies an area of the cell surface that is smaller than cross-section of single nanoparticle sensors (113 nm2), only a single protein will be detected. We utilized low distribution of proteins (0.21-0.37 molecule/μm2) on single living cells to control SMD volume.29 Such nanoparticle biosensors limited the possibility of real-time imaging and sensing individual protein molecules on single living cells where the expression level of proteins were high and might vary among cells and over time.
Therefore, it is essential to reduce the size of nanoparticles to prepare smaller single nanoparticle biosensors in order to detect and sense clusters of multiple individual molecules within the area smaller than 113 nm2. Furthermore, smaller nanoparticles offer smaller surface area, which limits the number of functional molecules to be attached on the surface of individual nanoparticles and offers a higher probability of attaching single functional molecules per nanoparticle. Note that, the primary criteria of SMD are to design individual detection volumes that can confine single molecules or distinguish individual molecules, and to develop a detection scheme that provides sufficient high signal-to-noise-ratio.18, 22 SMD represents not only the detection of a single molecule at a time, but also detection of multiple molecules at single molecule resolution. The latter is essential to study single molecule interactions and reactions. The smaller detection volume allows one to detect individual molecules at higher concentrations, which offers the possibility of analysis single molecules in bulk solution with a large dynamic range and probing interactions of single molecules in bulk solution for better understanding of an array of chemical and biochemical reactions at the single molecule level. Notably, the smaller detection volume also reduces background noise and hence enhances signal-to-noise-ratio of SMD. Thus, a wide variety of approaches for SMD have always aimed to reduce the detection volume of individual molecules. 17-23
In this study, we synthesized smaller Ag nanoparticles (diameter = 2.6 ± 1.1 nm; cross-sectional area = 5.3 nm2; surface area = 21.2 nm2; volume = 9.2 nm3) and controlled mole ratios of functional groups on the surface of nanoparticles, which allowed single monoclonal antibody (MAB) molecules to be attached onto individual nanoparticles and hence created intrinsic single molecule nanoparticle optical biosensors (SMNOBS). Such tiny nanosensors offer detection of single molecules presented in a volume of individual nanoparticles (9.2 nm3) and in a cross-sectional area of individual nanoparticles (5.3 nm2), which is smaller than the cross-sectional area of antibody molecules. We found that the SMNOBS resisted photodecomposition and could be used for imaging and quantitative analysis of single protein molecules (TNFα) and its binding reactions for hours.
Due to its high surface-area-to-volume ratio, it remains extremely challenging to prepare small Ag nanoparticles (< 5 nm) that are stable (non aggregating) in solution over time,33 even though a variety of synthetic methods have been reported to prepare size- and shape-controlled Ag nanoparticles beyond 10 nm in diameter,28, 34, 35 Thus, this study represents a major advance in preparing photostable noble metal dots for imaging and sensing of low and high concentrations of proteins of interest and probing their interactions at the single molecule resolution, which has not yet been reported previously.
Experimental Section
Reagents and Supplies
Silver nitrate (≥ 99.9%), sodium borohydride (≥ 98%), sodium citrate dihydrate (≥ 99%), hydrogen peroxide (30%), polyvinylpyrrolidone (PVP), 11-mercaptoundecanoic acid (MUA ≥ 95%), 6-mercapto-1-hexanol (MCH) (≥ 97%) and 30 kDa poly(ethylene glycol) (PEG) were purchased from Sigma-Aldrich. 1-Ethyl-3-[3-dimethylaminopropyl]-carbodiimide hydrochloride (EDC ≥ 99%) and N-hydroxysulfosuccinimide (Sulfo-NHS ≥ 98.5%) were purchased from Pierce. Recombinant human TNFα (TNFSF1A; MW = 17.5 KDa) and monoclonal anti-human TNFα antibody (TNFSF1A antibody) were purchased from R&D systems. All solutions, including 10 mM phosphate buffer saline (PBS) (pH = 7.4, 10 mM of phosphate buffer and NaCl), were prepared using nanopure 18 MΩ deionized water (Barnstead).
Synthesis and Characterization of SMNOBS
2.6 nm Ag Nanoparticles
AgNO3 (0.11 mM), sodium citrate (1.91 mM), PVP (0.052 mM), and H2O2 (25.0 mM) in nanopure water (42.3 mL) were prepared freshly, mixed and stirred constantly. As NaBH4 (150 μL, 100 mM) was added into the mixture, the solution color turned to light yellow. After stirring for another 3 hr, the solution was filtered using 0.2 μm membrane filters. The Ag nanoparticle solution was immediately characterized using UV-vis spectroscopy (Hitachi U3310), our dark-field single nanoparticle optical microscopy and spectroscopy (SNOMS), and dynamic light scattering (DLS) (Nicomp 380ZLS particle sizing system). TEM samples were immediately prepared and further characterized using high-resolution transmission electron microscopy (HRTEM) (FEI Tecnai G2 F30 FEG), showing diameter of Ag nanoparticles at 2.6 ± 1.1 nm. Using the same approaches as we reported previously for computing concentrations of nanoparticles,29, 31, 36 we found that the concentration of 2.6 nm Ag nanoparticles was 154 nM.
Functional Ag Nanoparticles (AgMMUA)
MUA (10 mM) and MCH (90 mM) in 0.5 mL ethanol were added to 50 mL of freshly prepared Ag nanoparticle aqueous solution to have final concentrations of MUA, MCH and Ag nanoparticles at 0.1 mM, 0.9 mM, and 152 nM, respectively. The mole ratio of MUA to MCH is 1: 9 and the mole ratio of MUA : MCH : Ag nanoparticle is 649: 5844: 1. The solutions were then stirred for 24 hr to attach MUA and MCH onto the surface of nanoparticles via their interaction of –SH groups with nanoparticles (Scheme 1A). The AgMMUA nanoparticles were washed twice using nanopure water to remove excess MUA and MCH using centrifugation (Beckman Optima L90k, 50 Ti rotor, 30,000 rpm at 4°C for 60 min). The AgMMUA nanoparticle solutions were immediately characterized using UV-vis spectroscopy, SNOMS, and DLS. NMR samples were immediately prepared by washing AgMMUA nanoparticles with nanopure water three times using centrifugation, drying the samples using lyophilizer (VirTis) and dissolving 50 mg of AgMMUA nanoparticles in 1 mL D2O, and further characterized by NMR (400 MHz, Bruker).
Scheme 1.
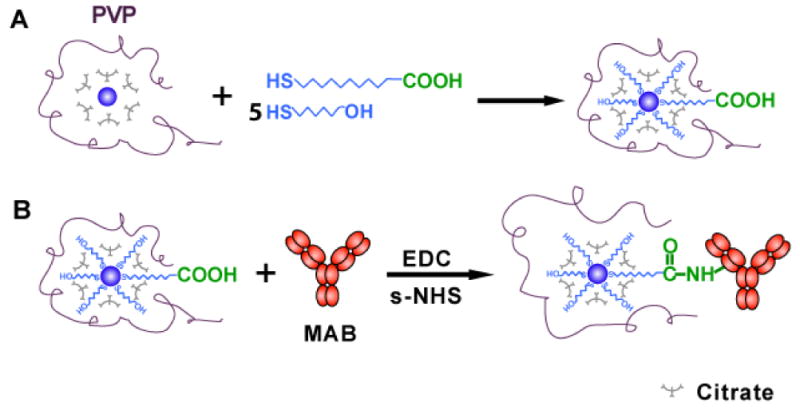
Schematic illustration of synthesis of photostable SMNOBS (AgMMUA-MAB) for sensing single TNFα molecules:
(A) Functionalizing Ag nanoparticles with monolayer of mixed MUA and MCH using the interaction of their thiol groups (-SH) with the surface of Ag nanoparticles to prepare AgMMUA;
(B) Linking the amine group of a MAB with the carboxyl group of the MUA attached on the nanoparticles via a peptide bond using EDC and sulfo-NHS as mediators to prepare AgMMUA-MAB nanoparticles.
Two other mole ratios of MUA : MCH : Ag nanoparticles at 3247 : 3274 : 1, and 390 : 6104 : 1 were attempted to prepare AgMMUA nanoparticles. In these two cases, mole ratios of MUA : MCH were 1 : 1 and 1 : 15, respectively, while Ag nanoparticle concentration and total concentration of MUA and MCH remained the same.
AgMMUA-MAB nanoparticles (SMNOBS)
A two-step method was used to conjugate the carboxyl group of AgMMUA with amine group of MAB via a peptide bond using EDC and sulfo-NHS as mediators (Scheme 1B). EDC (77 μmol) and sulfo-NHS (7.7 μmol) were added to AgMMUA aqueous solution (50 mL, 154 nM), forming AgMMUA-s-NHS esters. After stirring at room temperature for 40 min, AgMMUA-s-NHS was desalted using a Centriprep YM-30 (Millipore) by centrifugation at 1500 rcf (relative-centrifuge-force) for 5 min to remove excess EDC and sulfo-NHS, and then re-dissolved in 10 mM phosphate buffer (PB). PEG (0.05% w/v) was added to prevent nonspecific adsorption of MAB onto the surface of nanoparticles. In the second step, MAB was added to 50 mL AgMMUA-s-NHS in 10 mM PB solution at a mole ratio of MAB to AgMMUA of 0.97. The solution was mixed using a rotary shaker at room temperature for 2 hr and then at 4 °C for 12 hr. The final product (AgMMUA-MAB nanoparticles) was washed using 10 mM PBS buffer to remove excess MAB using centrifugation with 30,000 rpm at 4 °C for 60 min. The pellet was resuspended in 10 mM PBS with 0.05% w/v PEG, and stored at 4 °C for the future use. The size and optical properties of AgMMUA-MAB nanoparticles were characterized using UV-vis spectroscopy, SNOMS and DLS.
Characterization of Photostability of Single Ag Nanoparticles and SMNOBS
We characterized the photostability of single Ag and AgMMUA-MAB nanoparticles (SMNOBS) by acquiring sequential optical images of single Ag and AgMMUA-MAB nanoparticles using EMCCD and Micromax CCD camera with exposure time at 100 ms and readout time of 40.6 ms while these nanoparticles were constantly irradiated under a dark-field microscope illuminator (30 W halogen) for 12 hr.27-29 The illumination power at the sample stage (focal plane of dark field) was (0.070 ± 0.001) Watt during the experiment. We measured scattering intensity of individual nanoparticles and background over time.
Bioactivity, Sensitivity, Specificity, and Dynamic Range of SMNOBS
Bulk Analysis of TNFα in Solution
Both TNFα and AgMMUA-MAB solutions were prepared in 10 mM PBS buffer. The UV-vis spectra of the mixture (AgMMUA-MAB nanoparticles incubated with TNFα) were measured over 48 hr to monitor the binding kinetics and affinity of AgMMUA-MAB with TNFα. The spectra were measured at room temperature every 5 min for the first 2 hr and at every 2.5 hr after that until 48 hr. The mixture was stored at 4 °C to prevent denaturation of the proteins between spectroscopy measurements. The AgMMUA-MAB nanoparticles (50 nM) were used to detect several concentrations of TNFα solution (1, 10, 100, 200 ng/mL or 5.7×10-2, 0.57, 5.7, 11.4 nM) to construct a calibration curve and to determine the dynamic range of SMNOBS. In this study, we used concentration unit of ng/mL, instead of nM, in order to compare with literature data of TNFα, in which weight/volume percent is much more common than molar concentration.4, 37, 38 Blank control experiments were carried out by replacing BSA with TNFα, incubating BSA with AgMMUA-MAB, and measuring UV-vis spectra of the mixture over time.
Real-time SMD of TNFα
Single AgMMUA-MAB nanoparticles were placed on a clean slide by incubating 2 μL of 50 nM AgMMUA-MAB on the slide for 5-10 min, removing the solution by a pipette, and thoroughly rinsing the surface of slide using PBS buffer. The self-made microchamber was created on the slide, allowing AgMMUA-MAB nanoparticles to be incubated with 10 μL PBS buffer. Dark-field optical images and spectra of single AgMMUA-MAB nanoparticles were acquired using SNOMS equipped with Nuance multispectral imaging system. TNFα solution (10 μL, 10 ng/mL) was carefully injected into the microchamber and timer was simultaneously started. Dark-field optical images and spectra of single AgMMUA-MAB nanoparticles were acquired using SNOMS in real time at every 10 min for 8 hr as the single MAB molecules attached onto the nanoparticles bound with single TNFα molecules in solution. Acquisition time of each image and spectra of single nanoparticles at a given time is 90 s.
We carried out blank control experiments to characterize specificity and selectivity of SMNOBS by replacing TNFα with BSA, incubating BSA with single AgMMUA-MAB nanoparticles, and acquiring dark-field optical images and spectra of single AgMMUA-MAB nanoparticles for 8 hr using SNOMS.
Dark-field single nanoparticle optical microscopy and spectroscopy (SNOMS) has been used in our previous studies for real-time imaging and spectroscopic characterization of single nanoparticles in/on single living cells and in zebrafish embryos, and for SMD.21, 27-32 In this study, Nuance Multispectral Imaging System (Cambridge Research & Instrumentation) and other detectors (EMCCD, 5MHz Micromax CCD camera and color digital camera) coupled with a SpectraPro-150 (Roper Scientific) were also used for imaging and acquiring localized surface plasmon resonance spectra (LSPRS) of single nanoparticles (Ag, AgMMUA, AgMMUA-MAB, AgMMUA-MAB-TNFα).
Statistical Analysis
We investigated over 100 nanoparticles for each measurement of single nanoparticles. At least three measurements were performed in each solution. Thus, at least 300 nanoparticles in each solution were studied to gain sufficient statistics to determine their size distribution and color distribution that represented the bulk nanoparticle solution at the single nanoparticle level. All other measurements, including characterization of photostability and LSPR spectra of single nanoparticles and control experiments, were repeated at least three times.
Results and Discussion
Synthesis and Characterization of SMNOBS
We synthesized small, stable Ag nanoparticles by reducing AgNO3 with sodium citrate and hydrogen peroxide in the presence of a stabilizer (PVP), and characterized these nanoparticles using HRTEM, showing that they were nearly spherical with a diameter of 2.6 ± 1.0 nm (86% of 1-4 nm and 14% of 5-7 nm) (Figure 1A & B). The nanoparticles were stable in solution (non-aggregating) for months, suggesting that PVP and citrate played a role in preventing nanoparticles from aggregation.
Figure 1.
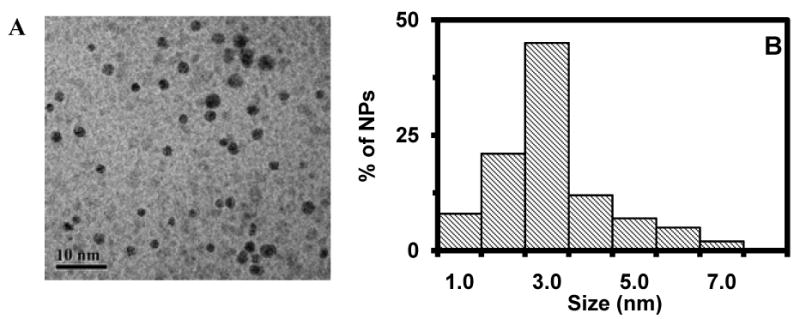
Characterization of size of single Ag nanoparticles:
(A) HRTEM image and (B) histogram of size distribution of Ag nanoparticles, showing the average diameter of Ag nanoparticles at 2.6 ± 1.0 nm. The scale bar is 10 nm.
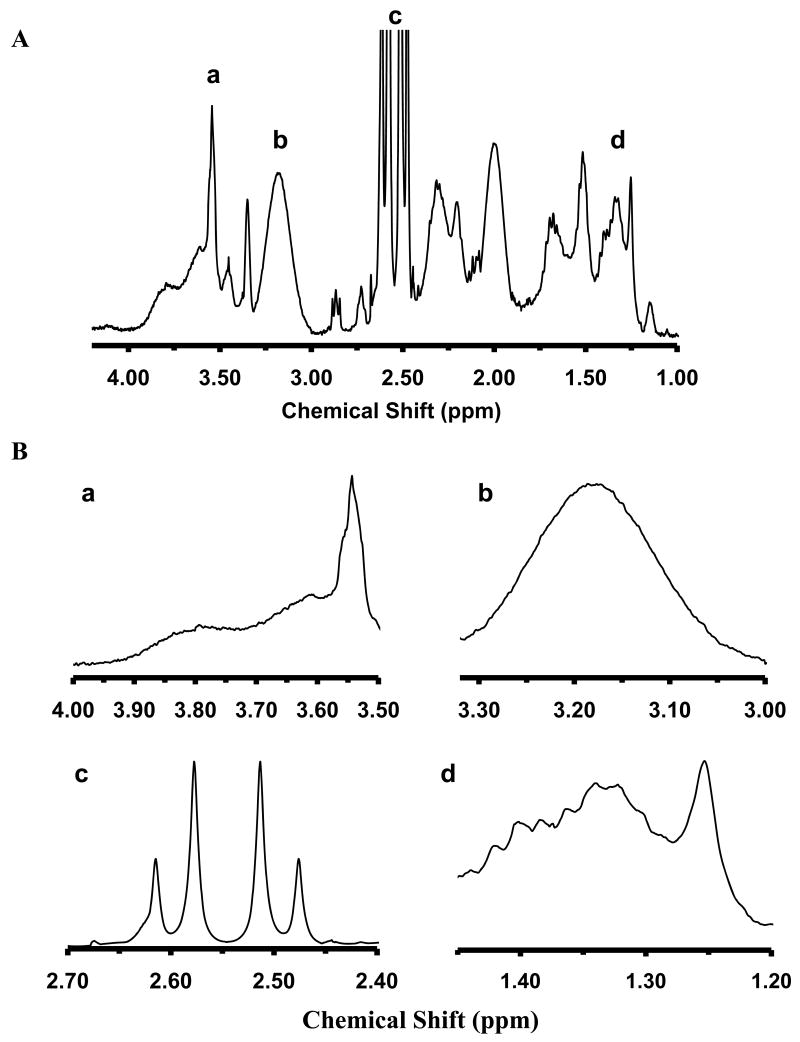
To prepare AgMMUA, we incubated the Ag nanoparticles (152 nM) with high concentrations of MCH (0.9 mM) and MUA (0.1 mM) and attached a monolayer of mixed MUA and MCH onto Ag nanoparticles by replacing electrostatistically adsorbed citrates on the nanoparticles with the interacting thiol groups (-SH) of MUA and MCH with the nanoparticles via replacement reactions (Scheme 1A). We used NMR to characterize the functional groups on the surface of AgMMUA nanoparticles and found that the molar ratio of attached functional groups of citrate : MCH : MUA : PVP was 28 : 5 : 1 : 0.04 (Figure 2), showing that MCH and MUA successfully replaced some of the citrate groups on the surface of nanoparticles and part of PVP molecule was also attached onto the nanoparticles.
Figure 2.
NMR characterization of functional groups of AgMMUA nanoparticles:
(A) Representative NMR spectra of AgMMUA nanoparticles;
(B) Zoom-in of individual peaks of NMR spectra in (A): (a) δ = 3.55 ppm, integration = 2.00, (2H, -(CH2)-OH of MCH); (b) δ = 3.18 ppm, integration = 8.65, (2H, from PVP ring); (c) δ = 2.55 ppm, integration = 25.23, (–CH2- next to thiol from both MUA and MCH and 4H from citrate. (d) δ = 1.20-1.45 ppm, integration = 6.05, (12H from -(CH2)6-CH2COOH of MUA and 4H from –(CH2)2-CH2OH of MCH). Molar ratio of citrate: MCH: MUA: PVP = 28: 5: 1: 0.04
With such a molar ratio of attached functional groups, we calculated the number of functional molecules that could be attached on the surface of a single nanoparticle (2.6 nm in diameter; surface area = 21.2 nm2) using a close-packing model. We found that approximately only a single MUA molecule was physically possible to be attached on a single Ag nanoparticle because the cross-sectional (footprint) areas of a citrate, MUA and MCH molecule on the surface of nanoparticle are 0.514, 0.0458 and 0.0458 nm2, respectively (Figures 1S-4S in supporting information (SI)). Note that carboxyl groups of a citrate molecule sat on the surface of nanoparticles (length of molecule that laid flat on the surface = 0.717 nm),39, 40 which resulted in a footprint area of 0.514 nm2 calculated using a close-packing model (footprint area = square of length). In contrast, thiol groups of MUA and MCH were attached onto the surface of nanoparticles,41, 42 allowing MUA and MCH molecules to stand straight and self-assemble on the surface of nanoparticles with a footprint area of 0.0458 nm2 calculated using the close-packing model (Diameter (D) of MUA or MCH = 0.214 nm; footprint area = D2) (Figures 1S-3S in SI). Thus, the citrate molecule occupied a larger surface area of nanoparticles than the MUA and MCH. Note that a single carboxyl group of a citrate molecule attached on the surface of nanoparticle offers the lowest energy level of citrate molecular conformation compared with those of two or three carboxyl groups attached on the surface of nanoparticles. The detailed calculations and modeling are illustrated in Figures 4S-6S in SI.
Notably, we found that mole ratios of MUA and MCH used to incubate with Ag nanoparticles were crucial to preparation of stable AgMMUA nanoparticles and controlling the ratio of functional groups on the surface of nanoparticles. As described in the experimental section, we used three mole ratios of MUA to MCH at 1:1, 1:9 and 1:15 to incubate with the Ag nanoparticles and to attach the mixed monolayer of MUA and MCH onto the surface of nanoparticles. We kept total moles of MUA and MCH and moles of Ag nanoparticles the same for each ratio of MUA to MCH. When we attempted a mole ratio of MUA to MCH of 1:1, we found that nanoparticles were aggregated, which resulted in unsuccessful preparation of AgMMUA. In contrast, the two other mole ratios led to successful preparations of AgMMUA. However, the ratio of MUA to MCH of 1:15 led to low levels of MUA attached on the surface of nanoparticles.
The plausible explanations of these interesting observations are given in the following. Note that the surface-adsorbed charged citrate layer of the nanoparticles provides the charge repulsion to prevent nanoparticles from aggregating. The presence of larger amounts of MUA, which is a charged steric hindrance molecule, favors a unimolecular nucleophilic substitution (SN1) mechanism of replacement reaction of citrate with MUA and MCH. Such a SN1 replacement mechanism might create less surface charges as citrate molecules departed from the surface of nanoparticles while MUA molecules were not yet attached on the surface, leading to the rapid aggregation of nanoparticles. The presence of a suitable amount of non-charged short chain MCH favors a SN2 replacement reaction, allowing Ag nanoparticles to remain charged during the replacement reaction and preventing Ag nanoparticles from aggregating.29, 33 The hydroxyl group and shorter chain of MCH also aid the solubility of AgMMUA in aqueous solution and help MUA to stand straight on the surface of nanoparticles, making the carboxyl group of MUA more accessible to MAB and favoring conjugation of MUA with MAB.29
We then conjugated the amine group of MAB with the carboxyl group of MUA attached on Ag nanoparticles via a peptide bond using EDC and sulfo-NHS as mediators to prepare AgMMUA-MAB nanoparticles (SMNOBS) (Scheme 1B). Since each nanoparticle was attached to a single MUA molecule (a single carboxyl group per nanoparticle was available to conjugate with a MAB), it ensured that a single nanoparticle was conjugated with one MAB molecule. Even if more than one MUA molecule were attached on the surface of a single nanoparticle, the small size of the nanoparticles would allow only a single MAB molecule to be conjugated with the attached MUA molecule because of steric effects (insufficient space to accommodate more than a single bulky MAB molecule on the surface of a 2.6 nm Ag nanoparticle). Thus, by using the small surface area of single nanoparticles, we effectively created SMD volumes that allowed individual molecules of MAB to be conjugated with individual nanoparticles and successfully prepared SMNOBS for sensing single TNFα molecules.
It is worth noting that the surface charges of nanoparticles hinder a single protein molecule from being conjugated with multiple nanoparticles, which overcomes one of primary problems of uneven conjugation of fluorescence dyes with protein molecules. Even though multiple amine groups per protein (MAB) molecule are available to conjugate with single carboxyl groups on multiple nanoparticles, the charge repulsion of nanoparticles impede multiple nanoparticles from being conjugated with the same protein molecule. If such conjugation reactions indeed occurred, it would cause the aggregation of nanoparticles, leading to their precipitation from the solution. If single protein molecules were conjugated with multiple nanoparticles, it would also be easily identified because its scattering intensity would be doubled.
Unlike previous studies, the smaller nanoparticles provide intrinsic SMD volumes per nanoparticle and our dark-field single nanoparticle microscopy and spectroscopy enables the sensing and detection of single protein molecules on single nanoparticles. Such ultrasmall nanoparticles avoid the need of using a lower reaction ratio of MAB to AgMMUA to ensure single MAB molecules to be conjugated with single nanoparticles. Thus, the higher reaction ratios of MAB to AgMMUA could be used to allow more AgMMUA nanoparticles to be conjugated with single MAB molecules. The smaller nanoparticles also avoid the need of using lower concentrations of analytes (e.g., proteins) in solution or lower levels of proteins on the cell surface to ensure SMD. These intrinsically superior features allow the SMNOBS to detect and sense proteins of interest in low and high concentrations at the single molecule resolution, offering a large dynamic range. Furthermore, the size of single nanoparticles (2.6 nm) is much smaller than that of an antibody or green fluorescence protein, allowing nanoparticles to more effectively label protein molecules of interest with less steric effects. The tiny sizes of SMNOBS also offer the possibility of them being delivered into living organisms for sensing biomolecules of interest and probing their function in small organelles (e.g., nuclei, mitochondrion) in real time.
Photostability of Single Ag Nanoparticles and SMNOBS
We characterized the photostability of single Ag nanoparticles (Figure 3A&B) and SMNOBS (AgMMUA-MAB) (Figure 3C&D) by acquiring sequential optical images of single Ag and AgMMUA-MAB nanoparticles while these nanoparticles were constantly irradiated under a dark-field microscope illuminator (30 W halogen) over 12 hr. Note that the nanoparticles were exposed to white-light illumination power of (0.070 ± 0.001) Watt at the sample stage (focal plane of dark field) during the experiment. Unlike nanoparticles that were directly radiated by well-focused laser beams,43, 44 we did not observe any significant generation of heat on nanoparticle surfaces that might lead to the denaturation of protein molecules.
Figure 3.
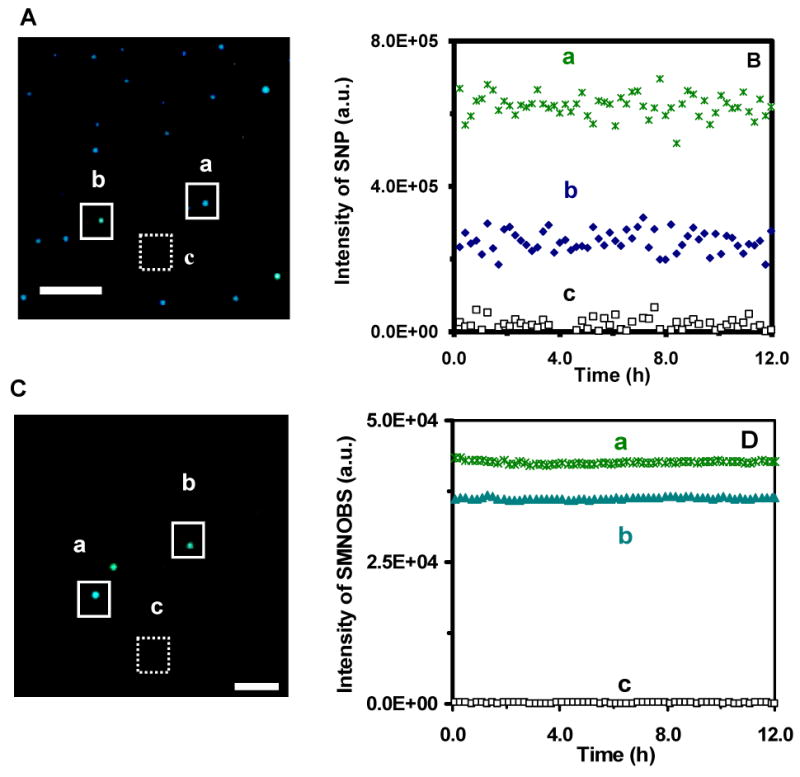
Characterization of photostability of single Ag nanoparticles and AgMMUA-MAB nanoparticles (SMNOBS):
(A) Dark-field optical image of single Ag nanoparticles, showing two representative colors, (a) blue and (b) green of Ag nanoparticles, and (c) background, as squared, respectively.
(B) Plots of scattering intensity of (a) single green nanoparticle, (b) single blue nanoparticle, and (c) background in (A) versus illumination time, exhibiting that single Ag nanoparticles resist photodecomposition and blinking.
(C) Dark-field optical image of single AgMMUA-MAB nanoparticles, showing two representative colors, (a) blue green and (b) green of Ag nanoparticles, and (c) background, as squared, respectively.
(D) Plots of scattering intensity of (a) single blue green nanoparticle, (b) single green nanoparticle, and (c) background in (C) versus illumination time, exhibiting that single AgMMUA-MAB nanoparticles resist photodecomposition and blinking. Scale bar is 5 μm in (A) and 2 μm in (C), showing the distances among nanoaprticles, but not the sizes of single nanoparticles because they are under optical diffraction limit.
We measured the scattering intensity of individual nanoparticles within a 20×20 pixel area (squared in Figure 3A&C: a & b) and average background intensity of several detection areas with the same size of detection volume (20×20 pixel) in the absence of nanoparticles (squared in Figure 3A&C: c). We subtracted the average background intensity from the integrated intensity of single nanoparticles and individual background areas and plotted the subtracted integrated intensity of individual nanoparticles and background as a function of time (Figure 3B&D). The plots show that the scattering intensity of single nanoparticles remains constant over time and slight fluctuations of scattering intensity of single nanoparticles are similar to those observed from background, suggesting that these fluctuations might be attributable to intensity fluctuation of the microscope illuminator or to dark noise of the CCD camera. Thus, these results demonstrate that single Ag and AgMMUA-MAB nanoparticles are photostable and do not suffer photodecomposition and blinking.
Effects of Functional Groups on Size and Optical Properties of Single Nanoparticles
UV-vis spectra of Ag, AgMMUA and AgMMUA-MAB nanoparticle solutions in Figure 4A show that the spectra became broader with the absorbance decreased and the peak wavelength shifted to longer wavelengths (red-shift), as Ag nanoparticles were functionalized with monolayer of mixed MUA and MCH, and as AgMMUA was conjugated with MAB. The changes of absorption spectra (e.g., peak wavelength, absorbance and FWHM) of 2.6 nm Ag nanoparticles as they were functionalized were much greater (at least twice) than those we reported previously for 12 nm Ag nanoparticles.29 This result suggests that the smaller Ag nanoparticles exhibit higher dependence of their optical properties on surface properties (surface functional groups), which may be attributable to the higher surface-area-to-volume ratio of smaller nanoparticles. The surface-area-to-volume ratio of 2.6 nm Ag nanoparticles is 2.3; in contrast, it is 0.5 for 12 nm Ag nanoparticles. Thus, the surface properties of smaller nanoparticles contribute even more significantly to their optical properties than is the case for larger nanoparticles. In other words, the optical properties of smaller nanoparticles are more highly dependent on their surface properties (e.g., surface functional groups and their surrounding environment) than that of the larger nanoparticles, because of their larger surface-area-to-volume ratio.24, 25 Therefore, the optical properties of smaller nanoparticles are more sensitive to changes of surface functional groups, making it a much more sensitive biosensor.
Figure 4.
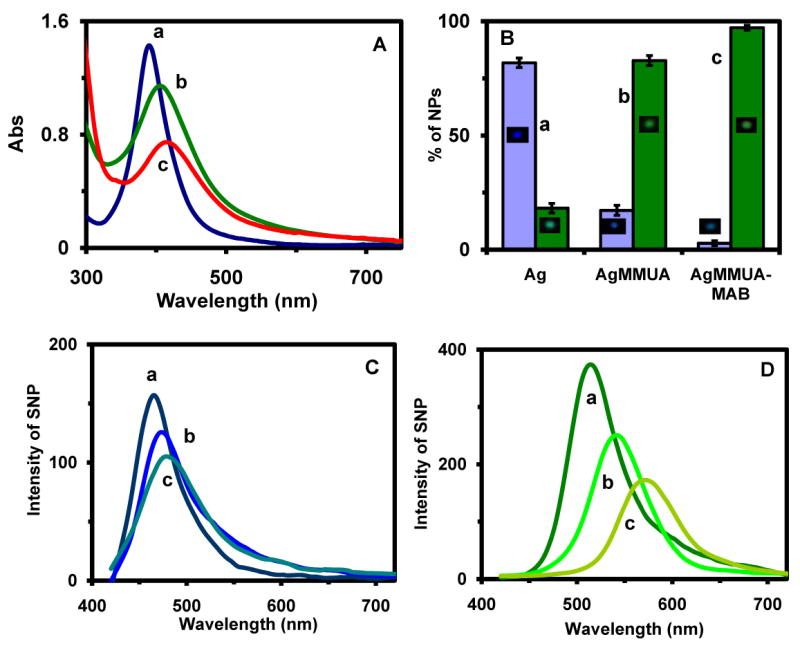
Characterization of optical properties of single Ag, AgMMUA and AgMMUA-MAB nanoparticles:
(A) UV-Vis spectra of 300 nM of (a) 2.6 nm Ag nanoparticles in nanopure water; (b) AgMMUA in nanopure water; (c) AgMMUA-MAB in 10 mM PBS (pH 7.0), showing that the peak wavelengths at 388 nm (FWHM = 66 nm; A = 1.42), 406 nm (FWHM = 116 nm; A = 1.14), and 410 nm (FWHM = 138 nm; A = 0.74), respectively.
(B) Histograms of color distribution of single nanoparticles in (A): (a) 2.6 nm Ag nanoparticle in nanopure water: (82 ± 2)% of blue and (18 ± 2)% green nanoparticles; (b) AgMMUA in nanopure water: (17 ± 2)% of blue and (83 ± 2)% green nanoparticles; and (c) AgMMUA-MAB in 10 mM PBS (pH 7.0): (3 ± 1)% of blue and (97 ± 1)% green nanoparticles.
(C) LSPR spectra of a representative single blue: (a) Ag nanoparticle (λmax = 465 nm; FWHM = 53 nm), (b) AgMMUA nanoparticle (λmax = 474 nm; FWHM = 63 nm), and (c) AgMMUA-MAB nanoparticle (λmax = 477 nm; FWHM = 74 nm).
(D) LSPR spectra of a representative single green: (a) Ag nanoparticle (λmax = 513 nm; FWHM = 64 nm), (b) AgMMUA nanoparticle (λmax = 543 nm; FWHM = 69 nm), and (c) AgMMUA-MAB nanoparticle (λmax = 570 nm; FWHM = 76 nm).
Using dynamic light scattering (DLS), we found that the size distributions of Ag, AgMMUA and AgMMUA-MAB nanoparticles suspended in solution were 2.6 ± 1.1, 3.6 ± 1.5, and 18.6 ± 5.2 in diameters, respectively. These results are in excellent agreement with those computed using modeling systems of a single MUA and MAB on a nanoparticle (Table I), further demonstrating that AgMMUA and AgMMUA-MAB were successfully prepared. The results show that each nanoparticle had one MUA and one MAB molecule on its surface. If more than one molecule of MUA were on the surface of a nanoparticle, two MUA molecules would likely align on opposite sides of the nanoparticle due to charge repulsion, which would lead to a larger diameter of AgMMUA at 5.3 nm. If more than one MAB were on the surface of a nanoparticle, it would have led to the diameter of AgMMUA-MAB at 33.7 nm. As stated previously, it is physically impossible for more than one antibody molecule to attach on the surface of a tiny 2.6 nm nanoparticle.
Table I.
Characterization of Size and Optical Properties of Ag, AgMMUA and AgMMUA-MAB Nanoparticles (NP).
| Nanoparticles |
* Diameter of Bulk NPs (nm) |
LSPRS of Single NP (nm) |
||||
|---|---|---|---|---|---|---|
| Experimental | Theoretical | Blue | Green | |||
| λmax (nm) |
FWHM (nm) |
λmax (nm) |
FWHM (nm) |
|||
| Ag | 2.6 ± 1.1 | N/A | 465 ± 3 | 52 ± 3 | 513 ± 6 | 63 ± 5 |
| AgMMUA | 3.6 ± 1.5 | 4.9 | 474 ± 6 | 64 ± 4 | 543 ± 7 | 69 ± 6 |
| AgMMUA-MAB | 18.6 ± 5.2 | 19.1 | 477 ± 7 | 77 ± 6 | 570 ± 9 | 73 ± 8 |
Experimental measurements and theoretical calculations of average diameters of bulk nanoparticles. Lengths of MUA, MCH and MAB (anti-TNFα) are 1.621, 1.068 and 14.2 nm, calculated using ChemSketch 3D view and protein data bank, respectively. We assume that MUA and MCH molecules titled on the surface of nanoparticles with anchoring angle of 58°, as described previously41, 42 and in Supporting Information. Thus, the vertical distances of MUA and MCH from the surface of nanoparticles are 1.375 and 0.906 nm, respectively.
Histograms of the distribution of colors (LSPRS) of single nanoparticles in solution (Figure 4B: Ag) illustrate a nearly uniform Ag nanoparticle solution with (82 ± 2)% of nanoparticles being blue (λmax = 465 ± 3 nm) and (18 ± 2)% of them being green (λmax = 513 ± 6 nm) (Figure 4C & D: a), respectively. As MUA and MCH functional groups replaced citrate groups (Figure 4B: AgMMUA), colors (LSPRS) of single nanoparticles shifted to longer wavelengths and (83 ± 2) % of nanoparticles became light green (λmax = 543 ± 7 nm; Figure 4D: b) with (17 ± 2) % of them remaining blue (λmax = 474 ± 6 nm; Figure 4C: b), suggesting that majority of Ag nanoparticles were functionalized with MUA and MCH. As AgMMUA nanoparticles were conjugated with MAB (Figure 4B: AgMMUA-MAB), the color of individual nanoparticles shifted further to longer wavelengths and (97 ± 1) % of nanoparticles became yellow green (λmax = 570 ± 9 nm; Figure 4D: c) with only (3 ± 1) % of them remaining blue (λmax = 457 ± 7 nm; Figure 4C: c), indicating a nearly complete conjugation reaction.
Representative LSPR spectra of single Ag, AgMMUA and AgMMUA-MAB nanoparticles are shown in Figure 4C & D and the quantitative changes of peak wavelength and FWHM are summarized in Table I. The scattering intensity of single blue Ag nanoparticles (Figure 4C: a) is lower than that of the green nanoparticle (Figure 4D: a), suggesting that the blue nanoparticle is smaller than the green nanoparticle, which shows size-dependent colors (LSPRS) of single nanoparticles. Notably, the FWHM of single blue Ag nanoparticle (52 ± 3 nm) are 11 nm smaller than that of single green Ag nanoparticles (63 ± 5 nm), suggesting that the smaller size (1-4 nm in diameter) of single blue nanoparticles results in a shorter dephasing time, T2, than that of slightly larger nanoparticles (5-7 nm) of green nanoparticles45.
Bulk Analysis of Bioactivity, Specificity and Dynamic Range of SMNOBS in Solution
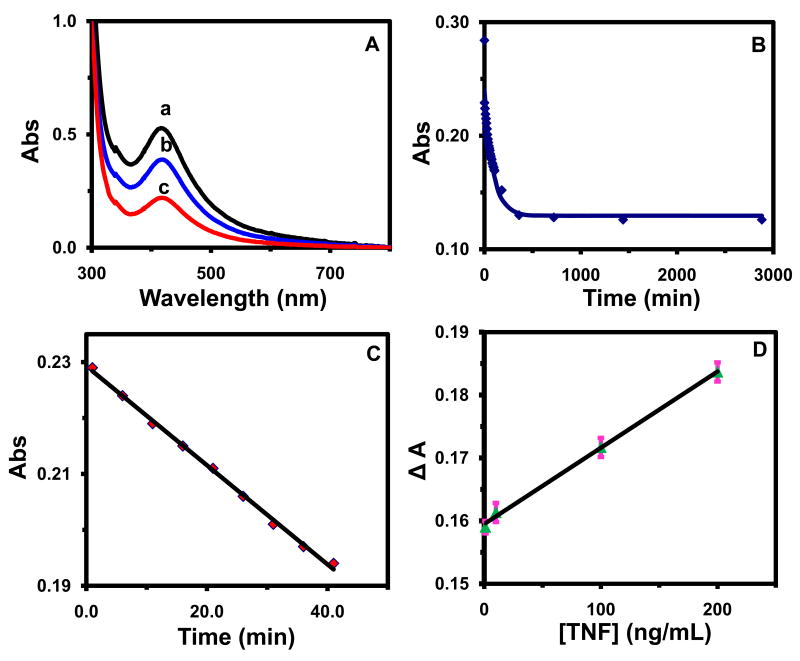
We characterized the bioactivity of AgMMUA-MAB nanoparticles by measuring their binding affinity with TNFα and comparing it with those reported in the literature. UV-vis spectra of AgMMUA-MAB nanopaticles exhibit stable peak wavelengths at 417 ± 3 nm (Figure 5A: a) with the extinction coefficient (molar absorptivity, ε) of 4.6×106 M-1cm-1, which was determined by the Beer-Lambert Law with a dilution series of AgMMUA-MAB nanoparticle solutions. As AgMMUA-MAB nanoparticles were incubated with TNFα, absorbance of the spectra decreased over incubation time with its peak wavelength (417 nm) and FWHM remaining essentially unchanged (Figure 5A), suggesting that AgMMUA-MAB nanoparticles bound with TNFα and the bound AgMMUA-MAB-TNFα nanoparticles did not contribute to the absorption, which might be attributable to precipitation of nanoparticles from the solution due to the cross-linking of nanoparticles29.
Figure 5.
Characterization of bioactivity and dynamic range of AgMMUA-MAB nanoparticles (SMNOBS) with TNFα molecules:
(A) UV-vis spectra of 50 nM AgMMUA-MAB nanoparticles incubated with 500 nM TNFα in the PBS buffer solution at (a) 0 min, (b) 20 min and (c) 24 hr, showing peak wavelength of 417 ± 3 nm and a decease in absorbance over time;
(B) Plot of peak absorbance of the solution corrected with baseline in (A) versus the incubation time, showing the exponential decay: points are experimental measurements and a solid line is generated by fitting the data points with an equation, A = 0.13 + 0.11e-0.011 t and R2 = 0.98;
(C) Plot of peak absorbance in (B) versus the incubation time at the early reaction time (0-40 min), showing a linearity with a slope of -8.9×10-4 min-1, intercept of 0.23, and R2 = 0.99. R2 in (B) and (C) is the multiple correlation coefficient and linear regression coefficient of determination, respectively.
(D) Calibration curve of AgMMUA-MAB nanoparticles for sensing TNFα: Plot of decreases in absorbance of AgMMUA-MAB nanoparticles (ΔA) versus TNFα concentrations, showing the dynamic range of SMNOBS at 0-200 ng/mL with a slope of 1.2×10-4 mL/ng and R2 = 0.99.
A plot of the peak absorbance subtracted from baseline versus time in Figure 5B exhibits high linearity during the first 40 min of the incubation time and then remains constant, suggesting that the binding of AgMMUA-MAB with TNFα is a first-order reaction as illustrated in Eq. 1:
| [1] |
Note that TNFα concentration (500 nM) is 10 times higher than that of AgMMUA-MAB (50 nM). Thus, TNFα concentration remains essentially unchanged over the entire reaction, and a second-order (or multiple-order) reaction can be treated as a pseudo-first-order reaction.
From the decrease of peak absorbance (ΔA = εbC; where b is the light path and C is nanoparticle concentration.) in Figure 5B, we found that 25.6 nM AgMMUA-MAB bound with TNFα as the binding reaction reached equilibrium. Therefore, we determined the equilibrium concentrations of AgMMUA-MAB (24.4 nM), TNFα (474.4 nM) and AgMMUA-MAB-TNFα (25.6 nM) and used them to calculate the equilibrium constant (affinity constant, KB) of AgMMUA-MAB with TNFα as (2.2 ± 0.1) × 106 M-1.
We also calculated the association (binding) (ka) and dissociation (kd) rate constants as 7.7×103 M-1min-1 and 7.1×10-3 min-1 (Figure 5C), respectively, by fitting the experimental data (points) with a simulation of the binding reaction (line) using the equation, A = 0.13 + 0.11 e -0.011t, which was derived using the same approach as we described previously29. Here A and t represent absorbance and reaction time, respectively. From the association (ka) and dissociation (kd) rate constants, we also determined the binding affinity constant, KB = ka/kd = (1.1 ± 0.1) × 106 M-1, which is close to that measured directly using equilibrium concentrations.
The average of KB is (1.7 ± 0.1) × 106 M-1, which agrees well with the affinity constant (1.8×106 M-1) of TNFα with a monoclonal antibody reported previously using other methods46. Even though the binding sites and MAB in the reported study might differ from ours, it offers an effective reference for comparison. Thus, the result suggests that MAB attached on AgMMUA-MAB nanoparticles remain its biological activities and any steric effect of functional nanoparticles does not significantly interfere the binding of attached MAB with TNFα in solution.
We then investigated the calibration curves of SMNOBS (AgMMUA-MAB) by plotting the decrease in peak absorbance of AgMMUA-MAB (ΔA) in response to the presence of various concentrations of TNFα (versus concentrations of TNFα: 0, 1, 10, 100, and 200 ng/mL in 10 mM PBS solution), illustrating a linear calibration curve in Figure 5D. The result shows that ΔA is proportional to the concentration of TNFα. We performed blank control experiments by replacing TNFα with the same concentrations of BSA in PBS buffer and using the same volume of PBS buffer only (in the absence of TNFα). We found that ΔAs were independent of BSA concentration, and remained essentially unchanged with a slight decrease in absorbance, owing to the dilution of AgMMUA-MAB by addition of PBS buffer. The result shows the high specificity and selectivity of SMNOBS, and exhibits a dynamic range of at least 0-200 ng/mL. Since we are much more interested in sensing low concentrations of TNFα using SMNOMS, we did not use higher concentrations of TNFα beyond 200 ng/mL. Previous studies reported that TNFα concentrations in normal arterial plasma of healthy human beings ranged from 1.2 to 130 pg/mL and many of these measurements were highly dependent upon the sensitivity of assays.6, 7, 37 A variety of diseases, such as cancer, diabetes, coronary artery disease, and HIV infection, could increase TNFα concentration 100-1000 fold higher than its normal level,4, 37, 38 which is within the dynamic range of our nanoparticle biosensors. Thus, our SMNOBS are well suited for monitoring disease diagnosis and therapy.
Real-time Sensing of Single Molecules and their Binding Reactions Using SMNOBS
We then immobilized SMNOBS on the surface of microscope slides and used them to capture and detect single TNFα molecules in solution and to study their binding kinetics in real time. Dark-field images of single AgMMUA-MAB nanoparticles incubated with TNFα molecules in PBS buffer solution over 8 hr show the colors of more than 78% of single AgMMUA-MAB nanoparticles changed from green to yellowish green over time (Figure 6A: a-f) and the peak wavelength of their LSPR spectra shifted to the longer wavelength with the average of peak wavelength shifts (Δλmax) at 29 ± 11 nm (Figure 6B & C). The LSPR spectra of the representative nanoparticle circled in Figure 6A show a red shift of peak wavelength from 543 (green) (Figure 6B: a) to 572 nm (Figure 6B: d-f), as the nanoparticle was incubated with TNFα molecules, illustrating that the binding reaction reached its binding equilibrium (bound or On -state) at 30-60 min (Figure 6B: d-e & 6C: a). Plots of peak wavelength (λmax) of LSPRS of representative single nanoparticles versus incubation time in Figure 6C show that SM binding reactions of TNFα with MAB attached on single nanoparticles reach equilibrium (bound-state) at various times, ranging from 30 to 120 min, suggesting stochastic binding kinetics of SM specific interactions.
Figure 6.
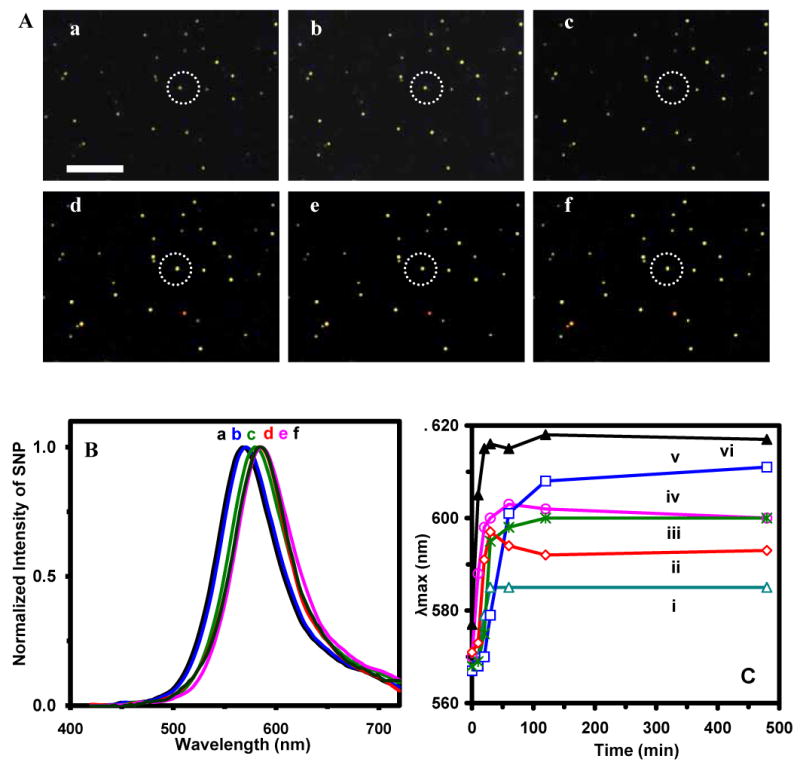
Real-time sensing single TNFα molecules using SMNOBS and probing binding reactions of single protein molecules (TNFα with MAB):
(A) Snap shots of real-time optical images of single AgMMUA-MAB nanoparticles incubated with TNFα at (a) 0, (b) 10, (c) 20, (d) 30, (e) 60, and (f) 240 min; Scale bar is 10 μm, showing the distances among nanoparticles, but not the sizes of single nanoparticles because they are under optical diffraction limit.
(B) LSPR spectra of a representative single AgMMUA-MAB nanoparticle circled in (A) incubated with TNFα at (a)-(f) (0-240 min), showing the red shift of peak wavelength of 29 nm over 30 min of incubation time.
(C) Plots of peak wavelength (λmax) of single nanoparticles in (A) versus incubation time, showing stochastic binding equilibrium times at: (i-ii) 30, (iii) 120, (vi) 30-60, (v)120, and (vi) 30 min, respectively. Acquisition time of each image and spectra of single nanoparticles at a given time in (A) and (B) is 90 s. Points are experimental data and the lines are added to show the trend of data points.
The red shifts of (29 ± 11 nm) peak wavelengths of LSPRS of single AgMMUA-MAB nanoparticles exhibit unprecedentedly high sensitivity of single nanoparticle biosensors for detecting single protein molecules (e.g., TNFα), and demonstrates that LSPRS of single nanoparticles highly depend upon their surrounding environments and surface properties. The red shifts of peak wavelength are likely attributable to the lower LSPR energy, owing to (i) chemical interface damping and the change of dielectric constants and surrounding environments of the nanoparticles, 25, 47 and (ii) an increase in the effective radius and aspect ratio of single nanoparticles25, as single TNFα molecules diffused to the surface of nanoparticles and bound with single MAB molecules on single nanoparticles.
As single TNFα molecules diffused near to the surface of individual AgMMUA-MAB nanoparticles and attempted its binding with AgMMUA-MAB, we observed a gradual red-shift of peak wavelength of LSPRS of single AgMMUA-MAB nanoparticles. Once TNFα bound with AgMMUA-MAB, the peak wavelength of LSPRS remained constant and we did not observe their peak-wavelength shifted back to its initial unbound state (Off-state). The result suggests that either single TNFα molecules did not disassociate from MAB on the surface of nanoparticles or the disassociated single TNFα molecules might be trapped among the surface functional groups (MMUA-PVP) of nanoparticles (Scheme I) and therefore were unable to depart from the surface of nanoparticles. The large binding constant (KB = 1.7×106 M-1) of TNFα-MAB could hinder their disassociation (Off-state), leading to a long association time (On-rate) (> 1.6×106 s = 444 hr), as predicted by recent simulations 48. Similar interesting phenomena were observed in our previous study, in which we directly observed such a complete binding reaction process in our previous study as single nanoparticle biosensors diffused to the surface of cells, searched for receptors on the surface of the cells, attempted its binding with the receptors several times, and finally bound with the receptors.29 We did not observe immediate disassociation of ligand-receptor complex (Off-sate), which could be attributable to the large affinity of the ligand-receptor complex (KB = 4.3×107 M-1) that led to a long association time (on-rate), as predicted by simulations48.
Even though there are two binding sites on each MAB molecule, we did not observe two well defined binding steps of two TNFα molecules with a single AgMMUA-MAB nanoparticle, showing that each nanoparticle biosensor detected a single TNFα molecule. Charge repulsion of surface functional groups, smaller surface area of nanoparticles, and low concentration of TNFα in solution might be attributable to prohibiting the second TNFα molecule from binding with the same MAB molecule on the surface of single nanoparticles.
We carried out blank control experiments by replacing TNFα with BSA in the experiment and we did not observe any significant shift of LSPRS of single AgMMUA-MAB nanoparticles (Figure 7), showing the high selectivity and specificity of SMNOBS. The results also show that SMNOBS resists photodecomposition and blinking, exhibiting remarkable photostability over hours. This control experiment further demonstrates that the red shift of peak wavelength observed in Figure 6 is not caused by the aggregation of nanoparticles because the nanoparticles were coated on the slide and thoroughly rinsed using PBS buffer before incubating with TNFα solution, and there were no nanoparticles in solution that might aggregate with AgMMUA-MAB nanoparticles on the surface. Taken together, the results show the possibility of using LSPRS of single nanoparticles to monitor single molecule reactions on the surface of single nanoparticles in real time.
Figure 7.
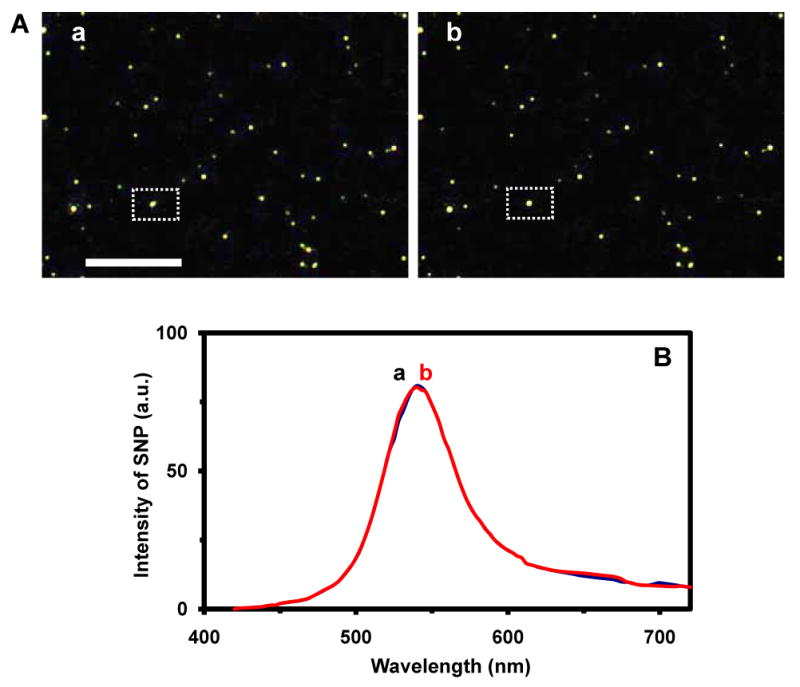
Characterization of selectivity and specificity of SMNOBS (control experiment):
(A) Snap shots of real-time optical images of single AgMMUA-MAB nanoparticles incubated with BSA at (a) 0 and (b) 240 min, showing that the color of single nanoparticles remains unchanged over time. Scale bar is 10 μm, showing the distances among nanoaprticles, but not the sizes of single nanoparticles because they are under optical diffraction limit.
(B) LSPR spectra of a representative single AgMMUA-MAB nanoparticle squared in (A), incubated with BSA at (a) 0 and (b) 240 min, showing LSPRS remains unchanged over 8 hr. Acquisition times of images and spectra are the same as described in Figure 6.
Notably, we did not observe a digital response (bound/unbound or On/Off) of single molecule reaction of TNFα with MAB, that was supposed to be uniquely associated with single molecule reaction, as predicted and reported using fluorescence detection.48, 49 Even though a single molecule reaction is supposed to occur at a single step as described by quantum mechanics, a complete reaction dynamic process takes several steps, because both molecules need to diffuse together, interact (collide several times) to search for the optimum bind sites, and finally bind. Studies of single molecule reactions thus far reported primarily used fluorescence probes which were limited by the photodecomposition and blinking of fluorescence probes, and hence it was impossible to observe such a complete single-molecule reaction process.
Furthermore, current reported studies were primarily conducted in bulk solution, and investigated bulk-molecule reactions at single molecule resolution. In other words, only two reactive molecules existed in a detection volume, but more than two reactive molecules were simultaneously present in the same solution. Thus, single molecule reactions occurred in a diffusion-controlled manner. For instance, when one molecule bound with a target molecule, the other molecule would diffuse toward the volume once occupied by the bound molecule due to its concentration gradient, which will affect the disassociation of a bound molecule. One can imagine, if an ultra-low concentration of analyte is used, it will take a significantly long time for two molecules to meet, interact and bind, and hence one will observe an unreasonably long disassociation time (Off-rate). If a high concentration of analyte is used, one will witness a long association time (On-rate). On and off rates also highly depend on the reaction rate of the molecules, and other factors (e.g., probing volume, temperature) as described above. It is nearly impossible for one to observe on and off rate from a same single-molecule reaction in a reasonable time frame that is limited by lifetime of a fluorescence molecule. Thus, what a data of a diffusion-controlled single molecule reaction in bulk solution should look like still remains an open question. It is worth noting that the study of single molecule reactions in real time, especially probing its complete binding dynamics, is still in its infancy, even though detection of single molecules has become somewhat routine in many research laboratories.
To our knowledge, no other tools have been reported to monitor two individual molecules simultaneously in real time as they diffuse toward binding distances, search for binding sites, interact, and bind. The superior photostability (non-photobleaching and non-blinking), unprecedentedly high quantum yield of Raleigh scattering and unique features of LSPRS of single Ag nanoparticle biosensors, as well as their ultrasmall sizes, offer exciting new opportunities to probe complete binding dynamics and kinetics of single molecule reactions in real time, and to better understand the reactions of bulk molecules at single molecule resolution. Studies are in progress to further investigate the dependence of on and off rate of single molecule reactions on a variety of parameters, such as binding constants, concentrations, detection volumes, temperatures, and temporal resolution of measurements.
Summary
In summary, we have successfully developed AgMMUA-MAB nanoparticles as SMNOBS to image and detect single TNFα molecules, to trace their diffusion and interaction with the surface of functional groups, and to measure binding affinities and kinetics in real time using their unique features of LSPRS. We synthesized ultrasmall stable Ag nanoparticles (2.6 ± 1.1 nm) and used the small surface area of individual nanoparticles and its surface functional groups to control SMD volume. We found that MAB conjugated with single AgMMUA nanoparticles retained its biological activities with KB of (1.7 ± 0.1) × 106 M-1. Using the high dependence of LSPRS of single Ag nanoparticles on their surface functional molecules and high quantum yield of Raleigh scattering of Ag nanoparticles, we detected single TNFα molecules with unprecedentedly high sensitivity and selectivity, and an extraordinarily wide dynamic range (0-200 ng/mL) and with no need of washing steps. We showed that SMNOBS resisted photodecomposition and blinking, and could be used to quantitatively measure binding affinity and stochastic binding kinetics of single protein molecules (e.g., TNFα-MAB) over hours in real time. We demonstrated that SMNOBS were able to sense and detect bulk TNFα molecules in solution, as well as capture and detect single TNFα molecules from the solution as SMNOBS were attached on the surface, suggesting their potential applications as probes for protein microarray and analysis of single protein molecules on cell surfaces and tissue samples. Such biosensors are well suited both for diagnosis of TNFα related diseases and for probing their fundamental roles in a variety of biological functions, as well as probing single molecular interactions of chemical functional groups with the surface of nanoparticles.
Supplementary Material
Six figures (Figures 1S-6S) are included on SI and they are:
Figure 1S: Three-dimension (3D) molecular structure of 11-MUA anion shows the length and width of molecule at 1.621 nm and 0.214 nm, respectively.
Figure 2S: Three-dimension (3D) molecular structure of 6-Mercaptohexanol (MCH) shows the length and width of molecule at 1.068 nm and 0.214 nm, respectively.
Figure 3S: Illustration of mixed monolayer of self-assembly of MUA and MCH molecules on the surface of nanoparticle, indicating the footprint area of individual molecule of 0.214 nm × 0.214 nm = 0.0458 nm2.
Figure 4S: Three-dimension (3D) molecular structure of citrate anion shows the length of molecule at 0.717 nm as one carboxyl group of citrate anion is attached on the surface of Ag nanoparticles.
Figure 5S: Three-dimension (3D) molecular structure of citrate anion shows the length of molecule at 0.442 nm as two carboxyl groups of citrate anion is attached on the surface of Ag nanoparticles.
Figure 6S: Three-dimension (3D) molecular structure of citrate anion shows the length of molecule at 0.558 nm as three carboxyl groups of citrate anion are attached with the surface of Ag nanoparticles.
This material is available free of charge via the Internet at http://pubs.acs.org
Acknowledgments
This work is supported in part by NSF (NIRT: BES 0507036), NIH (R01 GM076440), DoD-MURI (AFOSR #F49620-02-1-0320), and DOE (DE-FG02-03ER63646). Nallathamby is grateful for the support of Dominion Scholar Fellowship. We thank CharFac of U. of Minnesota and U. of New Mexico (NNIN sites funded by NSF) for their assistance to characterize Ag nanoparticles using HRTEM.
References
- 1.Nicola NA. Guidebook to cytokines and their receptors. A Sambrook & Tooze Publication at Oxford University Press; Oxford: 1994. and references therein. [Google Scholar]
- 2.Gurevich KG, Agutter PS, Wheatley DN. Cell Signal. 2003;15:447–453. doi: 10.1016/s0898-6568(02)00138-9. [DOI] [PubMed] [Google Scholar]
- 3.Tagawa M. Curr Pharm Des. 2000;6:681–699. doi: 10.2174/1381612003400597. [DOI] [PubMed] [Google Scholar]
- 4.Haworth C, Maini RN, Feldman M. In: The Cytokine Handbook. Thomson A, editor. Academic Press; 1998. pp. 777–801. [Google Scholar]
- 5.Copeland S, Siddiqui J, Remick D. J Immunol Methods. 2004;284:99–106. doi: 10.1016/j.jim.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Klabusay M, Kohutova V, Coupek P, Nenickova M, Tesarova E. Mediators of Inflammation. 2006:1–7. [Google Scholar]
- 7.de Kossodo S, Houba V, Grau GE. J Immunol Methods. 1995;182:107–114. doi: 10.1016/0022-1759(95)00028-9. [DOI] [PubMed] [Google Scholar]
- 8.Watts AD, Onier-Cherix N, Hunt NH, Chaudhri G. J Immunol Methods. 1999;225:179–184. doi: 10.1016/s0022-1759(99)00046-0. [DOI] [PubMed] [Google Scholar]
- 9.Shinn AH, Bravo NC, Maecker HT, Smith JW. J Immunol Methods. 2003;282:169–174. doi: 10.1016/j.jim.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Pico de Coana Y, Barrero C, Cajiao I, Mosquera C, Patarroyo ME, Patarroyo MA. Cytokine. 2004;27:129–133. doi: 10.1016/j.cyto.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Zajac A, Song D, Qian W, Zhukov T. Colloids Surf B Biointerfaces. 2007;58:309–314. doi: 10.1016/j.colsurfb.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Liu G, Engelhard MH, Lin Y. Anal Chem. 2006;78:6974–6979. doi: 10.1021/ac060809f. [DOI] [PubMed] [Google Scholar]
- 13.Haes AJ, Van Duyne RP. J Am Chem Soc. 2002;124:10596–10604. doi: 10.1021/ja020393x. [DOI] [PubMed] [Google Scholar]
- 14.Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 15.Agrawal A, Sathe T, Nie S. In: New Frontiers in Ultrasensitive Bioanalysis: Advanced Analytical Chemistry Applications in Nanobiotechnology, Single Molecule Detection, and Single Cell Analysis. Xu XHN, editor. Wiley; New Jersey: 2007. pp. 71–89. and references therein. [Google Scholar]
- 16.Schultz S, Smith DR, Mock JJ, Schultz DA. Proc Natl Acad Sci U S A. 2000;97:996–1001. doi: 10.1073/pnas.97.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 18.Xu XHN, Song Y, Nallathamby PD. In: New Frontiers in Ultrasensitive Bioanalysis: Advanced Analytical Chemistry Applications in Nanobiotechnology, Single Molecule Detection, and Single Cell Analysis. Xu XHN, editor. Wiley; New Jersey: 2007. pp. 41–65. and references therein. [Google Scholar]
- 19.Yu J, Xiao J, Ren X, Lao K, Xie XS. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 20.Zander C, Enderlein J, Keller RA. Single molecule detection in solution: methods and applications. Wiley-VCH; Berlin: 2002. and references therein. [Google Scholar]
- 21.Xu XHN, Brownlow WJ, Huang S, Chen J. Biochem Biophys Res Commun. 2003;305:79–86. doi: 10.1016/s0006-291x(03)00692-2. [DOI] [PubMed] [Google Scholar]
- 22.Xu XHN, Yeung ES. Science. 1997;275:1106–1109. doi: 10.1126/science.275.5303.1106. [DOI] [PubMed] [Google Scholar]
- 23.Xu XHN, Yeung ES. Science. 1998;281:1650–1653. doi: 10.1126/science.281.5383.1650. [DOI] [PubMed] [Google Scholar]
- 24.Bohren CF, Huffman DR. Absorption and scattering of light by small particles. Wiley; New York: 1983. [Google Scholar]
- 25.Kreibig U, Vollme M. Optical properties of metal clusters. Springer; Berlin: 1995. [Google Scholar]
- 26.Mie G. Ann Phys. 1908;25:377–445. [Google Scholar]
- 27.Lee KJ, Nallathamby PD, Browning LM, Osgood CJ, Xu XHN. ACS Nano. 2007;1:133–143. doi: 10.1021/nn700048y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nallathamby PD, Lee KJ, Xu XHN. ACS Nano. 2008;2:1371–1380. doi: 10.1021/nn800048x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang T, Nallathamby PD, Gillet D, Xu XH. Anal Chem. 2007;79:7708–7718. doi: 10.1021/ac0709706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyriacou SV, Brownlow WJ, Xu XH. Biochemistry. 2004;43:140–147. doi: 10.1021/bi0351110. [DOI] [PubMed] [Google Scholar]
- 31.Xu XHN, Brownlow WJ, Kyriacou SV, Wan Q, Viola JJ. Biochemistry. 2004;43:10400–10413. doi: 10.1021/bi036231a. [DOI] [PubMed] [Google Scholar]
- 32.Xu XHN, Chen J, Jeffers RB, Kyriacou SV. Nano Letters. 2002;2:175–182. [Google Scholar]
- 33.Yamamoto M, Kashiwagi Y, Nakamoto M. Langmuir. 2006;22:8581–8586. doi: 10.1021/la0600245. [DOI] [PubMed] [Google Scholar]
- 34.Métraux GS, Mirkin CA. Adv Mater. 2005;17:412–415. [Google Scholar]
- 35.Wiley B, Sun Y, Xia Y. Acc Chem Res. 2007;40:1067–1076. doi: 10.1021/ar7000974. [DOI] [PubMed] [Google Scholar]
- 36.Xu XHN, Huang S, Brownlow W, Salatia K, Jeffers R. J Phys Chem B. 2004;108:15543–15551. [Google Scholar]
- 37.Heinisch R, Zanetti C, Comin F, Fernandes J, Ramires J, Serrano C., Jr Vascular Health and Risk Management. 2005;1:245–250. [PMC free article] [PubMed] [Google Scholar]
- 38.Skoog T, Dichtl W, Boquist S, Skoglund-Andersson C, Karpe F, Tang R, Bond M, Faire U, Nilsson J, Eriksson P, Hamsten A. European Heart Journal. 2002;23:376–383. doi: 10.1053/euhj.2001.2805. [DOI] [PubMed] [Google Scholar]
- 39.Floate S, Hosseini M, Arshadi MR, Ritson D, Young KL, Nichols RJ. J Electroanal Chem. 2003;542:67–74. [Google Scholar]
- 40.Lin Y, Pan GB, Su GJ, Fang XH, Wan LJ, Bai CL. Langmuir. 2003;19:10000–10003. [Google Scholar]
- 41.Bain CD, Troughton EB, Tao YT, Evall J, Whitesides GM, Nuzzo RG. J Am Chem Soc. 1989;111:321–335. [Google Scholar]
- 42.Strong L, Whitesides GM. Langmuir. 1988;4:546–558. [Google Scholar]
- 43.Hu M, Hartland G. J Phys Chem B. 2002;106:7029–7033. [Google Scholar]
- 44.Richardson H, Hickman Z, Govorov A, Thomas A, Zhang W, Kordesch M. Nano Lett. 2006;6:783–788. doi: 10.1021/nl060105l. [DOI] [PubMed] [Google Scholar]
- 45.Hubenthal F. Progress in Surface Science. 2007;82:378–387. [Google Scholar]
- 46.Liu Y, Zhang W, Yu X, Zhang H, Zhao R, Shangguan D, Li Y, Shen B, Liu G. Sensors and Actuators B. 2004;99:416–424. [Google Scholar]
- 47.Hövel H, Fritz S, Hilger A, Kreibig U, Vollmer M. Phys Rev B. 1993;48:18178–18188. doi: 10.1103/physrevb.48.18178. [DOI] [PubMed] [Google Scholar]
- 48.Beveridge AC, Jett JH, Keller RA. In: New Frontiers in Ultrasensitive Bioanalysis: Advanced Analytical Chemistry Applications in Nanobiotechnology, Single Molecule Detection, and Single Cell Analysis. Xu XHN, editor. Wiley; New Jersey: 2007. pp. 1–27. and references therein. [Google Scholar]
- 49.Xie XS. Single Mol. 2001;2:229–236. and references therein. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Six figures (Figures 1S-6S) are included on SI and they are:
Figure 1S: Three-dimension (3D) molecular structure of 11-MUA anion shows the length and width of molecule at 1.621 nm and 0.214 nm, respectively.
Figure 2S: Three-dimension (3D) molecular structure of 6-Mercaptohexanol (MCH) shows the length and width of molecule at 1.068 nm and 0.214 nm, respectively.
Figure 3S: Illustration of mixed monolayer of self-assembly of MUA and MCH molecules on the surface of nanoparticle, indicating the footprint area of individual molecule of 0.214 nm × 0.214 nm = 0.0458 nm2.
Figure 4S: Three-dimension (3D) molecular structure of citrate anion shows the length of molecule at 0.717 nm as one carboxyl group of citrate anion is attached on the surface of Ag nanoparticles.
Figure 5S: Three-dimension (3D) molecular structure of citrate anion shows the length of molecule at 0.442 nm as two carboxyl groups of citrate anion is attached on the surface of Ag nanoparticles.
Figure 6S: Three-dimension (3D) molecular structure of citrate anion shows the length of molecule at 0.558 nm as three carboxyl groups of citrate anion are attached with the surface of Ag nanoparticles.
This material is available free of charge via the Internet at http://pubs.acs.org