Abstract
The COP9 signalosome (CSN) is an evolutionarily conserved protein complex formed by 8 subunits (CSN1 through CSN8). Deneddylating cullin family proteins is considered the bona fide function of the CSN. It has been proposed that the CSN regulates the assembly and disassembly of the cullin-based ubiquitin ligases via its deneddylation activity. Here we report that down-regulation of CSN8 by RNA interference destabilized differentially other CSN subunits and reduced the amount of CSN holo-complexes, leading to increases in neddylated cullin proteins and reduction of F-box protein Skp2 in HEK293 cells. Moreover, suppression of CSN8 enhanced the degradation of a proteasome surrogate substrate and cyclin kinase inhibitor p21cip. Reduced transcript levels of cyclin kinase inhibitor p21cip and p27kip were also observed upon down-regulation of CSN8. These data suggest that the homeostatic level of CSN8/CSN suppresses proteasome proteolytic function and regulates transcription.
Keywords: COP9 signalosome, cullin, nedd8, proteasome, transcription
1. Introduction
The COP9 signalosome (CSN) is a highly conserved multiprotein complex composed of eight unique subunits (CSN1 through CSN8) in higher eukaryotes (Wei & Deng 2003). The CSN was first identified in Arabidopsis thaliana as a negative regulator of photomorphogenesis (Wei et al 1994). The subsequent characterization of the CSN in other eukaryotes has revealed its pleiotropic functions in regulating invertebrate development (Freilich et al 1999), cell cycle (Denti et al 2006, Doronkin et al 2003, Rosel & Kimmel 2006), mating in budding yeast (Maytal-Kivity et al 2002), kinase signaling (Tomoda et al 2005), nuclear transport (Liu et al 2003), and initiation of T cell proliferation (Menon et al 2007, Panattoni et al 2008). At the biochemical level, several activities have been associated with the CSN complex: deubiquitination and deneddylation activities, and the associated kinase activity. First, protein kinases co-purified with human CSN can phosphorylate c-Jun, IκB, NF-κB precursor, and p53 (Bech-Otschir et al 2001, Seeger et al 1998) and CSN can interact with several protein kinases such as inositol 1,3,4-triphosphate 5/6-kinase, protein kinase D, and casein kinases (Uhle et al 2003, Wilson et al 2001). Second, the CSN recruits deubiquitination enzymes and thereby either deconjugates the ubiquitin from mono-ubiquitinated substrates or removes the polyubiquitin chain from the substrates (Groisman et al 2003, Zhou et al 2003). Third, the CSN has an intrinsic isopeptidase activity that resides in the JAMM motif of CSN5. This activity removes Nedd8 moiety from the substrates, mainly the cullin family proteins, via a process known as deneddylation (Cope et al 2002, Lyapina et al 2001). The attachment of Nedd8 to the cullin family proteins (neddylation) is required for the assembly of functional cullin-based E3 ubiquitin ligases while deneddylation mediated by the CSN disassembles and perhaps recycles the E3 ligases (Wolf et al 2003). Therefore, the dynamics of these two processes is thought to be essential to cullin-based E3 ligases. Indeed, loss of function of the CSN resulted in reduction of SCF (Skp1-Cullin 1-F-box) E3 ligase components such as cullins, Rbx1 and particularly F-box proteins, and ultimately led to an abnormal accumulation of the substrates (He et al 2005, Hetfeld et al 2005, Wang et al 2002, Wee et al 2005, Wu et al 2003).
The CSN holo-complex is composed of 8 subunits with a 1:1 stoichiometry and has a molecular weight of approximately 500 kDa (Wei & Deng 2003). Each subunit has either a PCI or MPN domain and is paralogous to one of the eight subunits constituting the lid of the 19S proteasome. Moreover, the CSN and the lid of the 19S proteasome share a similar pattern of interaction among analogous subunits, suggesting that these two complexes may have similar architecture and quaternary structure (Fu et al 2001). Several studies showed that the CSN may directly interact with the 26S proteasome (Kwok et al 1999, Peng et al 2003, Seeger et al 1998). The interaction may even influence proteasome activities, likely through competing with the lid of the 19S proteasome (Huang et al 2005). Therefore, the CSN is postulated as either an alternative lid for the 26S proteasome or a scaffold for the coordination between cullin-based E3 ligases and the proteasome to allow more efficient degradation of specific substrates.
In addition to the holo-complex, small complexes (mini-complex) containing some of the 8 CSN subunits were also identified in Arabidopsis (Wang et al 2002), S. pombe (Mundt et al 2002), Drosophila (Oron et al 2002), and mammalian cells (Tomoda et al 2005). The mini-complexes seem to be cell-type specific, as shown by varied constituents of the subunits with accordingly various molecular masses among species. Although the functional relevance of these mini-complexes remain elusive, Tomoda et al (2005) have revealed that a CSN5-containing mini-complex mediated p27kip down-regulation in leukemia cells independently of the deneddylation activity of the CSN, suggesting that the mini-complexes may have unique cellular functions.
In light of its ability to regulate the activity of a large family of ubiquitin E3 ligases as well as its potential to be an alternative lid of the 19S proteasome, the CSN has been proposed as an important regulator of the ubiquitin-proteasome system (UPS). Indeed, it was reported that the CSN regulated the proteasome-mediated degradation of a number of endogenous proteins, such as retinoblastoma protein (Rb) (Ullah et al 2007), p27kip (Denti et al 2006), IkBα (Schweitzer et al 2007), estrogen receptor α(ERα) (Callige et al 2005), microtubule end-binding protein 1(Peth et al 2007), and Rad1-Rad9-Hus1checkpoint protein complex (Huang et al 2007). However, whether the CSN has an impact on the global UPS-mediated proteolytic function and whether down-regulation of the CSN has an impact on transcriptional regulation remain unclear. Here we report that down-regulation of CSN8 via specific small interfering RNA (siRNA) led to reduced function of the CSN holo-complex and unexpectedly resulted in significant reduction of the protein levels of a surrogate misfolded protein as well as p21cip and p27kip. Both increased proteasomal proteolytic function and depressed transcription appear to be responsible.
2. Materials and methods
2.1. Cell culture, siRNA, and transient transfection
Human embryonic kidney (HEK) 293 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and penicillin-streptomycin (100 U/ml; Invitorgen). The siRNA specific for human CSN8 mRNA (siCSN8: 5′-GGCUGUGAAAGGCAUAUUA-3′) and the scramble siRNA (siSCR: 5′-ACUACCGUUGUUAUAGGUG-3′) were purchased from Ambion (Austin, TX). The siCSN8 used for fluorescent microscopy were labeled by Cy3 at 5′ end of the sense strand. For a transient knockdown of CSN8, cells were seeded 24 hours before siRNA transfection on 60-mm dishes and grown to 30-50% confluency. Lipofetamine™ 2000 transfection reagents (Invitrogen, Carlsbad, CA) were used for siRNAs (30 nM or otherwise specified) transfection by following the manufacturer’s protocol. Six hours after the transfection, the siRNA-containing medium was replaced with regular medium and unless otherwise indicated, the cells were cultured for another 54 hours before being harvested for assessments reported here.
2.2. GFPu stable cell lines
An enhanced green fluorescence protein (GFP) modified by carboxyl fusion of a ubiquitination signal sequence (degron CL1, ACKNWFSSLSHFVIHL), referred to as GFPu or GFPu, has been used as a surrogate substrate for the UPS (Bence et al 2001). HEK293 cells stably transfected with a GFPu expressing plasmid were previously described (Liu et al 2006).
2.3. Mouse CSN8 cDNA plasmid and the CSN8 rescue experiment
A His-tagged mouse CSN8 construct was newly generated for the CSN8 rescue experiment. Briefly, CSN8 cDNA was cloned by reverse transcriptase (RT-) PCR using total RNA extracted from mouse tissues and inserted into the pShuttle-CMV vector (Stratagene, La Jolla, CA). A His tag was added to the amino terminus of CSN8 to differentiate the transgenic CSN8 from the endogenous CSN8. To overexpress mouse CSN8 in HEK cells to be treated by human siCSN8, the His-tagged mouse CSN8 expressing vector was introduced into the GFPu HEK-293 cells using the Fugene 6 transfection reagent (Roche, Indianapolis, IA). Twenty-four hours after the mouse CSN8 cDNA transfection, cells were resuspended and seeded for siRNA transfection.
2.4. Immunolabeling and fluorescent microscopy
After removal of culture medium, cells cultured in chamber slides were fixed with 4% of paraformaldehyde for 20 minutes. After washed with PBS 3 times (5 minutes each), the cells were permeabilized with 1% of Triton X-100 in PBS for 1 hour, quenched with 0.1M glycine in PBS for one hour, and blocked with 0.5% BSA for 1 hour. The rabbit anti-CSN8 primary antibody (BIOMOL, Plymouth Meeting, PA), and the Alexa-Flour 488 (Figure 1E) or Alexa-Flour 568 (Figure 5D) conjugated donkey anti-rabbit Ig secondary antibody (Molecular Probes, Carlsbad, CA) were used to label CSN8. The mouse anti-GFP primary antibody (Santa Cruz, Santa Cruz, CA) and the Alexa-Flour 488 donkey anti-mouse Ig secondary antibody (Molecular Probes, Carlsbad, CA) were used to label GFPu. DAPI (Sigma, St. Louis, MO) was used to stain nuclei. The immunolabeling was visualized and imaged using fluorescence confocal microscopy.
Fig. 1.
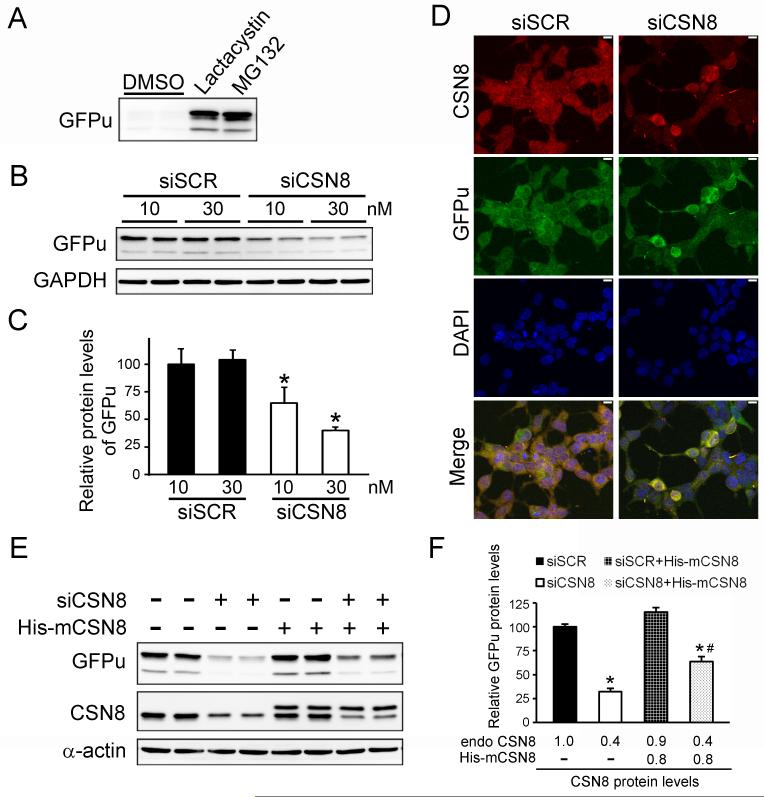
Suppression of CSN8 in HEK293 cells by siRNA. (A) CSN8 mRNA levels in cells at 60 hours after transfection with no siRNA (Non), a scramble siRNA (siSCR, 30 nM), or a siRNA specific for human Csn8 (siCSN8, 30 nM) were assessed by Northern blot analysis. GAPDH was probed as loading controls. (B) A summary of the densitometric data of (A). (C) CSN8 protein levels in total cell lysates from cells with indicated treatments were analyzed by Western blots. GAPDH protein levels were probed as loading controls. (D) A summary of the densitometric data of (C). (E) Representative confocal micrographs of cells transfected with indicated siRNA. Cells that were successfully transfected by Cy3-siCSN8 display red fluorescence dots in the juctanuclear area. CSN8 was immunostained green. The nuclei were labeled with DAPI (blue). Note that CSN8 is significantly reduced in Cy3-positive cells. Scale bar = 10 μm.
Fig. 5.
Specific down-regulation of CSN8 decreases the protein levels of a UPS surrogate substrate (GFPu). (A) A Western blot image of GFPu in HEK293 cells stably expressing GFPu at 2 hours after administration of proteasome inhibitor Lactacystin (6 μM) or MG132 (2 μM). (B, C) Knockdown of CSN8 decreased GFPu protein levels in a dose-dependent manner. Representative western blot images (B) and a summary of densitometry data (C) are presented. *: p<0.05 vs siSCR group (n=4 for each group). (D) Confocal micrographs of cells transfected with indicated siRNA. Cells were immunostained for CSN8 (red) and GFP (green). The nuclei were stained blue with DAPI. Scale bar = 10 μm. (E, F) Overexpression of mouse CSN8 rescued human siCSN8 induced decreases in GFPu protein levels in HEK293 cells. Representative Western blot images (E) and the quantitative data of GFPu protein levels (F) are shown. The relative endogenous (endo) and transgenic CSN8 (His-mCSN8) protein levels were shown in the bottom of the bar graphs (F). *: p<0.05 vs siSCR, #: p<0.05 vs siCSN8. n=4 for each group.
2.5. Non-denaturing polyacrylamide gel electrophoresis (native-PAGE)
This was performed as previously described with minor modifications (Tomoda et al 2005). Briefly, cells were lysed in the lysis buffer (50 mM Tris, pH8.0, 120 mM NaCl, 1 mM EDTA, 10% glycerol) containing 0.1 mM PMSF and 1X protease inhibitor cocktail (Roche, Indianapolis, IN). Equal amounts (∼100 μg) of proteins were separated on the pre-casted native 4-20% gradient gels (BioRad, Hercules, CA) at 4°C in the SDS-free running buffer and followed by conventional Western blot analysis using antibodies specific for indicated CSN subunits.
2.6. Gel filtration chromatography
Cells were lysed in the extraction buffer (EB) containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 2 mM ATP, 2.5 mM EDTA, 1 mM DTT, 10% glycerol, 0.1% NP-40, and freshly added inhibitors respectively for proteases, phosphatases, and metalloproteases. The lysate was centrifuged at 13,000×g for 15 min twice at 4°C. The supernatant was filtered through 0.2-μm filters (Gelman Sciences, East Hills, NY). 800 μg of total proteins were loaded onto a pre-equilibrated Superose 6 (HR10/30) gel filtration column (GE Healthcare, Piscataway, NJ). The column was then eluted with EB and fractions of 0.25 ml were collected and analyzed by Western blots.
2.7. Cell lysate and Western blot analysis
Cultured cells were lysed in 1X SDS sampling buffer (50mM Tris-Cl at pH 6.8, 2% SDS and 10% glycerol). The extracts were sonicated on ice and boiled for 5 minutes. The supernatant was obtained following a 14000×g centrifugation for 5 minutes at 4°C. The protein concentration was measured by the Bicinchoninic Acid (BCA) method. Equal amount of samples were resolved by SDS-PAGE, transferred to PVDF membrane, probed with appropriate antibodies, and followed by detection with enhanced chemiluminence (ECL-Plus) reagents (GE Healthcare, Piscataway, NJ) and a VersaDoc3000 imaging system (BioRad, Hercules, CA). The signal was quantified with the Quantity One software (BioRad, Hercules, CA) as previously described (Chen et al 2005).
2.8. RNA analyses
Total RNA was extracted with the Tri-Reagent (Molecular Research Center, Cincinnati, OH) following the manufacturer’s protocol. The steady state transcript level of CSN8 was estimated by Northern blot analysis, using p32-labeled human csn8-specific cDNA probes generated by using the nick-translation kit (Roche, Indianapolis, IA). The steady state transcript level of GFPu was quantified by RNA dot blot analysis with probes derived from GFPu cDNA. Radioactively labeled blots were exposed to a phosphor screen and detected with a phosphoimager. The transcript levels of p21cip and p27kip were semi-quantitated by RT-PCR at the minimum cycles that can detect PCR products, using specific primers towards p21cip and p27kip. Relative transcript levels were obtained with normalization to GAPDH transcript levels.
2.9. Statistical analysis
All quantitative data are presented as mean ± SD. Differences between experimental groups were evaluated for significance using Student’s t-test for unpaired two group comparison or one-way ANOVA when appropriate. The P value <0.05 is considered statistically significant.
3. Results
3.1. Down-regulation of CSN8 differentially decreased other subunits and altered CSN complex formation
CSN8 is the smallest subunit of the CSN and appears to be the only CSN subunit not found in S. pombe (Rosel & Kimmel 2006, Wei & Deng 2003). To study the role of CSN8 in CSN complex formation and CSN activity in mammalian cells, we used the small interfering RNA (siRNA) technology to transiently silence csn8 gene expression. HEK293 cells were transfected with the siRNA specific for human CSN8 (siCSN8). Very efficient knockdown was achieved with siCSN8 at a dose of 10 nM or higher. A reduction of 60-70% of CSN8 mRNA and protein was detected at 60 hours after siCSN8 (30 nM) administration (Fig. 1A to 1D). As expected, the knockdown of CSN8 showed a dose-dependent manner while transfection of a scramble siRNA (siSCR) did not produce a significant effect (Fig. 1C and 1D). Immunostaining the cells transfected with Cy3-labeled siCSN8 confirmed that siCSN8 was successfully delivered into more than 80% of the cells and the decrease in CSN8 protein occurred exclusively in siCSN8 transfected cells (i.e., showing Cy3 red fluorescence; Fig. 1E), further demonstrating the high efficiency and specificity of CSN8 knockdown.
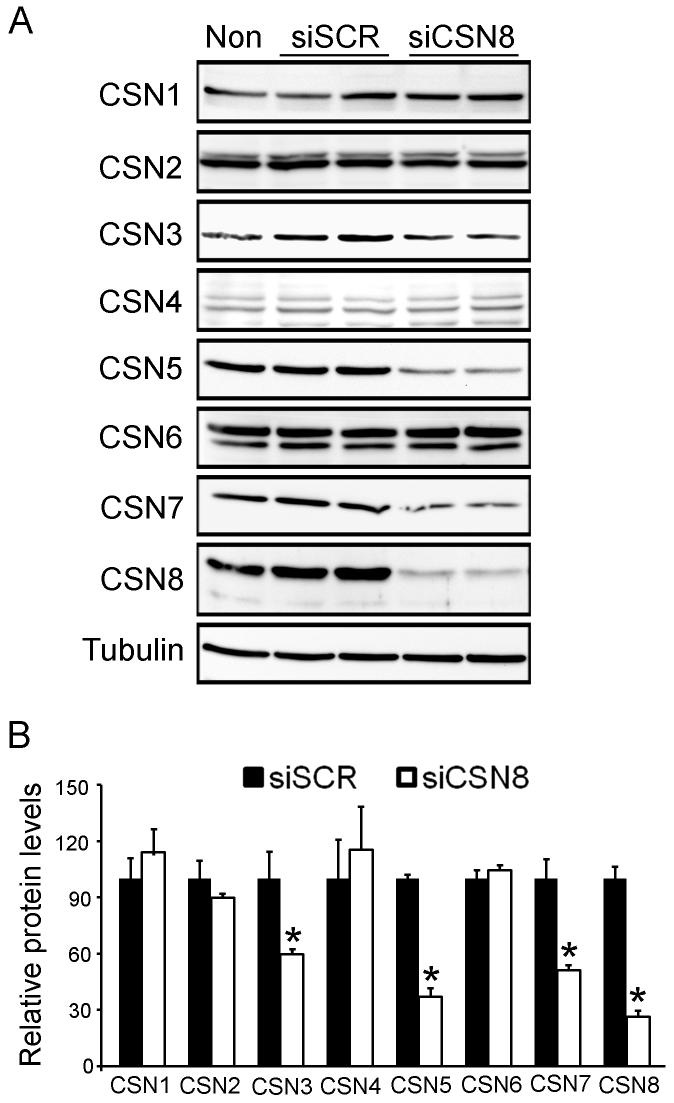
To examine the role of CSN8 in stabilizing other CSN subunits, we examined their protein levels in CSN8 knockdown cells. The decrease in CSN8 suppressed protein levels of CSN3, CSN5 and CSN7 but had little effect on CSN1, CSN2, CSN4, and CSN6 protein levels (Fig. 2).
Fig. 2.
Down-regulation of CSN8 differentially alters the protein levels of other CSN subunits. (A) Representative Western blot images. Total proteins were extracted from the cells transfected with indicated siRNA, fractionated with SDS-PAGE, and analyzed with Western blots for the indicated proteins. β-Tubulin was probed as loading controls for quantitative analyses. (B) A summary of densitometry data of (A). The mean density of a given protein in siSCR treated cells was respectively set as 100% and used to normalize all the densitometry data of the protein. *: p<0.05 vs siSCR group (n=4 for each group).
To study the effect of the down-regulation of CSN8 on the assembly of CSN complexes, we first performed native-PAGE which had previously proved to be effective in separating the holo-complex (slower gel mobility) and the mini-complex (faster gel mobility) of the CSN (Fukumoto et al 2005, Tomoda et al 2005). Cell lysates extracted from siCSN8 or siSCR transfected cells were subjected to the native-PAGE followed by immunoblotting using antibodies against indicated subunits (Fig. 3A). As expected, knockdown of CSN8 markedly reduced the abundance of CSN8 in both the holo-complex and the mini-complex. Notably, the subunits that were down-regulated by CSN8 knockdown showed different distribution patterns between the holo-complex and the mini-complex. The abundance of CSN5 was reduced in both the holo-complex and the mini-complex in siCSN8-transfected cells. However, there was an increase of CSN7 in the mini-complex although less CSN7 resided in the holo-complex in siCSN8-transfected cells (Fig. 3A). Upon CSN8 knockdown, more CSN7 appeared in the smaller species of the mini-complexes while more CSN5 appeared in the relatively larger species of the mini-complexes (Fig. 3A).
Fig. 3.
Down-regulation of CSN8 alters CSN complex formation. (A) The distribution of CSN holo- and mini-complexes in total cell lysates was analyzed by native-PAGE followed by Western blots for the indicated CSN subunits. (B) The distributions of CSN2, CSN5, and CSN8 were analyzed by gel filtration chromatography. Fractions 17 - 38 were resolved by SDS-PAGE and analyzed by Western blot for indicated proteins.
To confirm the findings from the native-PAGE, we also performed gel filtration chromatography on cell lysates and analyzed the distribution of CSN subunits by immunoblotting. The holo-complex with a higher molecular weight is eluted in earlier fractions while the mini-complex comes out later. Consistently, CSN8 knockdown reduced the abundance of CSN8 and CSN5 in both holo- and mini-complexes. Interestingly, although total CSN2 protein level was not altered upon CSN8 down-regulation (Fig. 2), less CSN2 resided in the holo-complex while more CSN2 appeared in the mini-complex (Fig. 3B).
Taken together, our data suggest that CSN8 is essential to CSN holo-complex formation and reduction of holo-complex formation by reducing CSN8 may alter the equilibrium of the mini-complexes.
3.2. Down-regulation of CSN8 diminished the bona fide activity of the CSN
We next tested whether knockdown of CSN8 is sufficient to interfere with CSN function, by examining the levels of neddylated cullin1 (Cul1) and Cul4A in siRNA transfected cells. Knockdown of CSN8 resulted in a concomitant increase in the neddylated form of both Cul1 and Cul4A in total cell lysates, compared with cells transfected with siSCR (Fig. 4A and 4B). In addition to accumulation of the neddylated form of the cullin family proteins, loss of function of the CSN has previously been found to destabilize SCF components, such as the substrates adaptors F-box proteins (Denti et al 2006, He et al 2005, Wee et al 2005). Hence, we analyzed the protein levels of 2 mammalian F-box proteins, Skp2 and β-TrCP. The protein abundance of Skp2 was significantly reduced by approximately 50% in siCSN8 transfected cells, compared with siSCR transfected cells (Fig. 4C and 4D). This reduction was attenuated by the treatment with the proteasome inhibitor MG132 (10 μM), suggesting that the decrease of Skp2 levels is due to increased proteasome-dependent degradation. However, the protein levels of another F-box protein β-TrCP were not affected by knockdown of CSN8, suggesting that knockdown of CSN8 may only have effects on the stability of a subset of F-box proteins.
Fig. 4.
Suppression of CSN8 alters the bona fide activity of the CSN. (A, B) Representative Western blot images (A) for Cul1 and Cul4A and a summary of densitometry data (B) are presented respectively in the left and right panels. α-Actin was probed as loading controls. *: p<0.05 vs siSCR group (n=4 for each group). (C, D) Representative western blot images (C) for Skp2 and β-TrCP and a summary of densitometry data (D) are presented respectively in the upper and lower panels. β-Tubulin serves as a loading control. *: p<0.05 vs siSCR group (n=4 for each group).
Taken together, our data demonstrate that CSN8 is required for the deneddylation activity of the CSN but down-regulation of CSN8 destabilizes only a subset of F-box proteins.
3.3. Down-regulation of CSN8 reduced a proteasome surrogate substrate GFPu
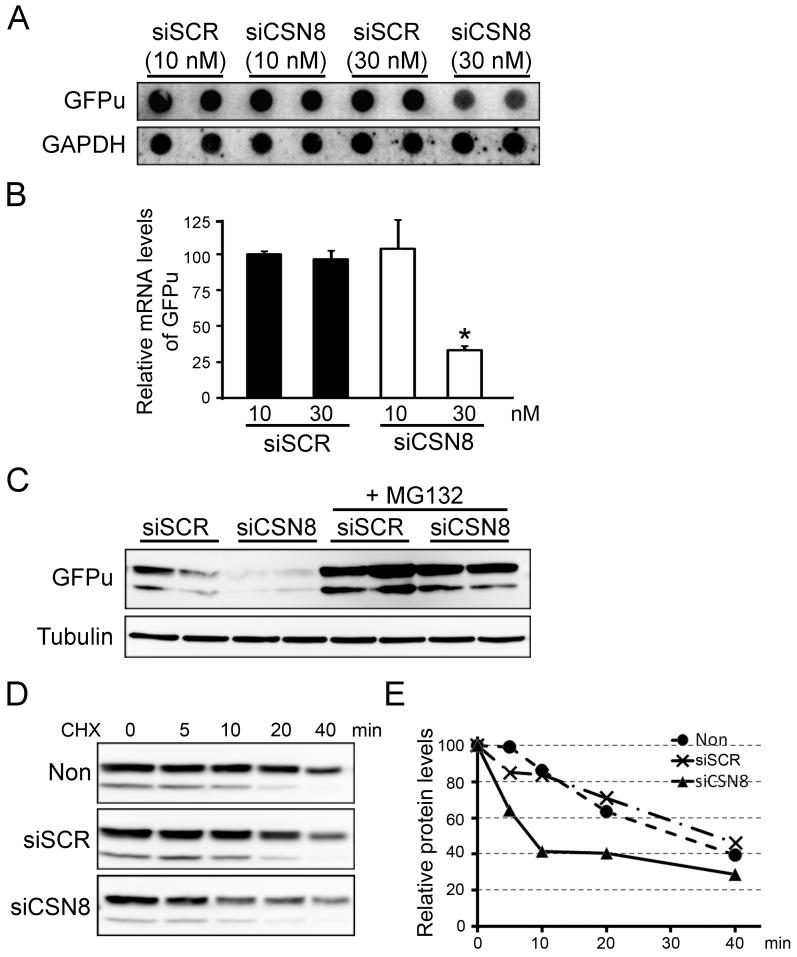
It has been reported that CSN regulates the stability of several endogenous regulatory proteins such as Rb (Ullah et al 2007), p27kip (Denti et al 2006), IkBα(Schweitzer et al 2007) and ERα (Callige et al 2005), through controlling the activity of their respective ubiquitin E3 ligases. However, whether the CSN has effects on the global proteolytic function of the proteasome per se remains to be determined. To monitor proteasome proteolytic function, a GFP modified by carboxyl fusion of a ubiquitination signal, degron CL1 (referred to as GFPu) has previously been generated and validated as surrogate substrate both in cultured cells and in intact animals (Bence et al 2001, Chen et al 2005, Dong et al 2004, Kumarapeli et al 2005). In absence of changes in synthesis, GFPu protein abundance inversely reflects proteasome proteolytic function. A clonal stable HEK-293 cell line stably expressing GFPu was previously created (Liu et al 2006). In this cell line, GFPu protein levels were markedly increased upon the treatment with different proteasome inhibitors (Lactacystin at 6 μM or MG132 at 2 μM), indicating that the cell line behaves as expected (Fig. 5A). To probe the impact of CSN8 down-regulation on UPS proteolytic function, we transfected this stable cell line with either siCSN8 or siSCR and analyzed the abundance of GFPu protein. Intriguingly, GFPu protein levels were reduced by about 35% at a low dose (10 nM) of siCSN8 and by 60% at a higher dose (30 nM), compared with siSCR administration (Fig. 5B and 5C). Consistently, fluorescent confocal images from immunostained transfected cells also revealed a marked reduction of GFPu levels in CSN8-deficient cells (Fig. 5D). To exclude the potential off-target effect of siCSN8, we co-transfected the cells with His-tagged mouse CSN8, which has 3-nucleotide mismatch with the human siCSN8 and therefore can not be silenced by the human siCSN8. As expected, co-transfection of mouse CSN8 did not affect the knockdown effects of siCSN8 on the endogenous CSN8, but largely attenuated the reduction of GFPu protein (Fig. 5E and 5F).
3.4. Reduced GFPu protein levels were caused by both enhanced protein degradation and suppressed transcription in the siCSN8 transfected cells
To determine how CSN8 silence affects the GFPu expression, we first performed RNA dot blot analysis to examine the steady state transcript levels of GFPu. Transfection of siCSN8 at a higher dose (30 nM) suppressed the transcript levels of GFPu, while the low dose (10 nM) of siCSN8 did not change GFPu transcript levels (Fig. 6A and 6B). Consistently, blocking the proteasome function by MG132 (10 μM) largely, but not completely, prevented the siCSN8 (30 nM) induced decrease of GFPu protein levels (Fig. 6C). To further determine whether the reduction of GFPu in siCSN8 transfected cells is due to increased protein degradation, we performed cycloheximide (CHX, 100 μM) chase analyses on GFPu protein levels in siRNA transfected cells. In siCSN8-transfected cells, 60% of GFPu was degraded within 10 min after CHX administration, while 80% of GFPu was still present in mock-treated or siSCR-transfected cells. These indicate that the degradation of GFPu protein was markedly enhanced in siCSN8 treated cells (Fig. 6D and 6E). Therefore, our data suggest that reduced GFPu protein levels are caused by both suppressed synthesis and accelerated degradation.
Fig. 6.
The CSN regulates GFPu at both the transcriptional and post-translational levels. (A) The steady-state transcript levels of GFPu were detected by RNA dot blot analysis using p32-labeled transcript-specific probes derived from GFPu cDNA. GAPDH was similarly probed as loading controls. (B) A summary of densitometry data of (A). *: p<0.05 vs siSCR group. (C) Proteasome inhibition by MG132 attenuated the reduction of GFPu levels in si-CSN8 transfected cells. (D) Cycloheximide (CHX) chase assay for GFPu. Cells transfected with indicated siRNA were treated with CHX (100μM). The levels of GFPu at the indicated time points after CHX treatment were analyzed by Western blots. (E) Within each treatment group, the relative value of the GFPu signals of different time points post-CHX treatment over that of the 0 min time point were plotted.
3.5. Dow-regulation of CSN8 altered the levels of p21cip and p27kip
Since loss of function of the CSN appears to alter the proteasome function and regulate GFPu expression at the transcriptional level, we next tested whether down-regulation of CSN8 has similar effects on the endogenous protein levels.
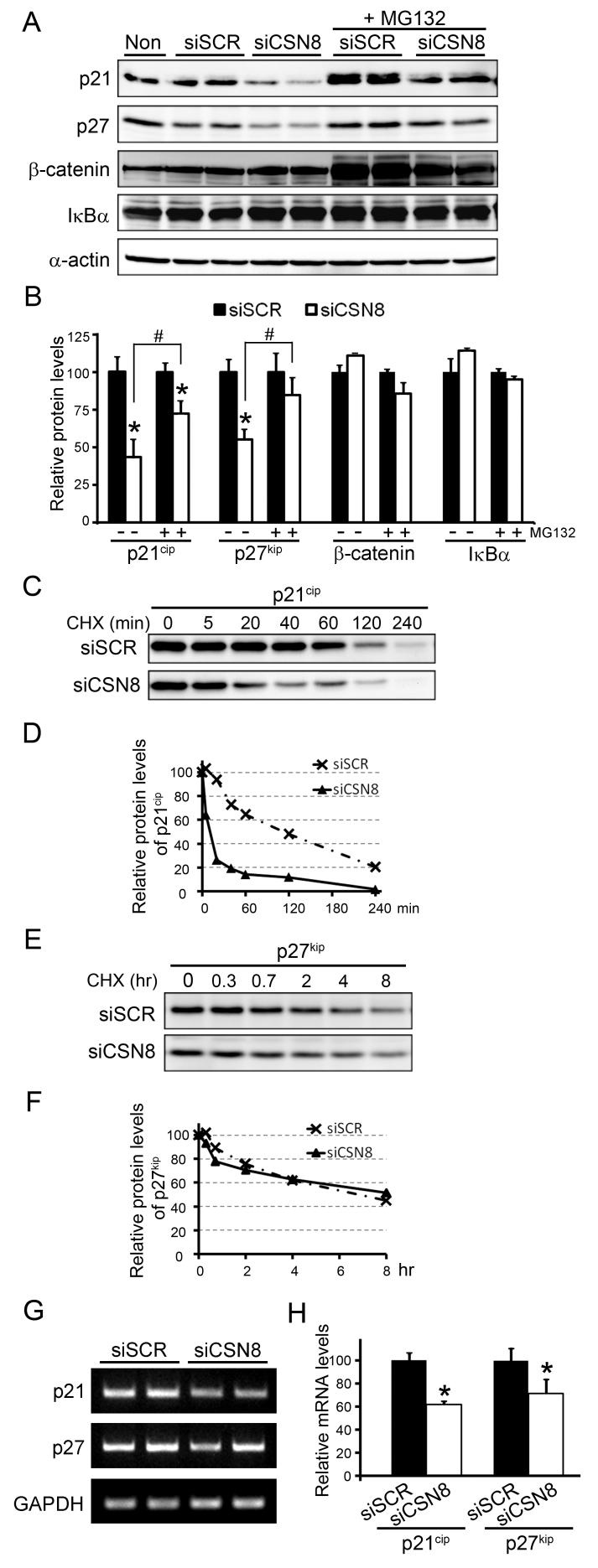
It has been previously shown that p21cip, p27kip, β-catenin, and IκBα are degraded by the proteasome (Bloom et al 2003, Carrano et al 1999, Nakayama et al 2000, Sutterluty et al 1999, Tsvetkov et al 1999). We first examined the abundance of these proteins in siRNA transfected cells. We observed that the protein levels of p21cip and p27kip were significantly reduced in siCSN8 transfected cells (Fig. 7A and 7B). However, the abundance of β-catenin and IκBα was not affected by the silencing of siCSN8 (Fig. 7A and 7B). Furthermore, the protein levels of 21cip and p27kip were either partially or largely restored by the treatment of MG132 (10 μM) (Fig. 7A and 7B), suggesting that the reduction of p21cip and p27kip results from enhanced proteasomal degradation. Indeed, CHX chase experiments showed that the turnover of p21cip was accelerated in siCSN8 transfected cells (Fig. 7C and 7D). However, the CHX chase failed to reveal an increased degradation of p27kip in our experimental conditions (Fig. 7E and 7F). To determine whether CSN8/CSN is involved in regulating gene transcription, we also performed RT-PCR to measure the transcripts of p21cip and p27 kip. The mRNA levels of p21cip and p27 kip were both significantly decreased in siCSN8 transfected cells (Fig. 7G and 7H), indicating that reduced transcription of p21cip and p27kip may attribute to their reduced protein levels. These data suggest that, in addition to regulating proteins degradation, CSN8/CSN may also be involved in regulating transcription.
Fig. 7.
Down-regulation of CSN8 reduces the protein levels of p21cip and p27kip. (A, B) Representative images (A) of Western blot analyses of p27kip, p21cip, β-catenin, and IκBα in cells transfected with indicated siRNA and a summary of densitometry data (B) are presented. *: p<0.05 vs siSCR group (n=4 for each group). (C, D) CHX chase assays for p21cip in siCSN8 and siSCR transfected cells. Cells were treated with CHX (100μM) for the indicated time. The cell lysates were analyzed by Western blots for p21cip (C) and the densitometric readings of p21cip in each sample relative to the values of CHX-untreated cells (0 min) are plotted in (D). (E, F) A CHX chase assay for p27kip was done as described in (C, D). (G, H) Semi-quantitative analyses of the mRNA levels of p21cip and p27kip using semi-quantitative reverse-transcriptional (RT-) PCR. RT-PCR was performed using primers specific the mRNAs indicated. Representative RT-PCR gel images (G) and a summary of the densitometry data (H) are presented. The density of each sample was normalized by that of GAPDH.
4. Discussion
Among the eight CSN subunits, six have a PCI domain and two carry an MPN domain, which are paralogous to 8 subunits of the lid of the 19S proteasome. How the CSN holo-complex is organized remains elusive, but the interactions between subunits via PCI domain are believed to be important to CSN assembly (Wei & Deng 2003). Therefore, every subunit appears to be essential to the formation of the holo-complex. Supporting this notion, we observed that down-regulation of the smallest subunit CSN8 in HEK293 cells reduced the amount of CSN holo-complexes (Fig. 3). Interestingly, down-regulation of CSN8 also altered the abundance of different mini-complexes, suggesting that the disassembled subunits may interact with each other and form new forms of mini-complexes with various gel mobilities. Mini-complexes containing some of the CSN subunits have been reported in Arabidopsis (Karniol et al 1999, Serino et al 1999, Wang et al 2002), Drosophila (Oron et al 2002), and mammalian cells (Tomoda et al 2005, Tomoda et al 2002) but the constituents of the mini-complexes and their functions are not clear. Notably, decreases in CSN8 reduced significantly CSN3, CSN5, and CSN7 but not other 4 CSN subunits (Fig. 2), suggesting that CSN8 may directly interact CSN3, CSN5 and CSN7 during the formation of CSN holo- and mini-complexes and stabilize them. This supports the current model for CSN subunit interactions (Wei & Deng 2003).
It has been proposed that all of the 8 subunits are required for the deneddylation activity, although the isopeptidase activity resides in CSN5 (Wei & Deng 2003). Consistent with this notion, we found here that knockdown of CSN8 significantly diminished CSN deneddylation activity as evidenced by increased levels of neddylated Cul1 and Cul4A (Fig. 4A and 4B). Interestingly, the increase in neddylated Cul4A was accompanied by a decrease in the native form of Cul4A but the increase in neddylated Cul1 was not (Fig. 4A). Loss of CSN subunits in several organisms have also been shown to destabilize F-box proteins such as Skp2 and β-TrCP, compromise SCF E3 ligase activities, and accumulate the respective substrates of the SCF E3’s (Cope & Deshaies 2006, Cope et al 2002, Doronkin et al 2003, Schwechheimer et al 2001, Schweitzer et al 2007). In the present study, we observed that suppression of CSN8 in human HEK293 cells reduced F-box protein Skp2 but not β-TrCP (Fig. 4C and 4D), suggesting that suppression of CSN8 may only destabilize a subset of cullin-based E3 ligases at least in these cells. Despite a decrease in Skp2, p27kip (the substrate for Skp2) did not appear to be stabilized by suppression of CSN8 (Fig. 7A, 7B, 7E and 7F), suggesting that the remaining Skp2 is still sufficient to assemble functional SCFskp2 E3 ligase to target p27kip for degradation in our models. In contrast to our finding, p27kip appeared to be stabilized by CSN4 and CSN5 down-regulation in 293T cells, which may be attributed to both the destruction of SCF ubiquitin ligase and the disruption of CSN5 and p27 interaction and subsequent nuclear export (Denti et al 2006, Tomoda et al 2002). These discrepant observations on p27 degradation between CSN5 knockdown and CSN8 knockdown cells suggest that each CSN subunit may behave differently in regulating the abundance of a specific protein. Additionally, consistent with unaltered levels of β-TrCP, the substrates that are targeted by β-TrCP for proteasomal degradation, such as β-catenin and IκBα, were not accumulated either (Fig. 7A and 7B). In agreement with our data, the degradation patterns of β-TrCP and its substrates, including β-catenin and IκBα, were unaltered in stimulated Csn8-deficient T cells (Menon et al 2007).
In addition to regulating the cullin-based E3 ligases, some evidence suggests that CSN may regulate proteasome proteolytic function by direct interaction with the 26S proteasome (Huang et al 2005). Ignoring any of these features of the CSN may underestimate the impact of the CSN on UPS functions. We then sought to determine the impact of down-regulation of CSN8 on proteasome proteolytic function using a well-established proteasome functional reporter system (Fig. 5A). Interestingly, the degradation of GFPu, a reliable surrogate substrate for the proteasome, was accelerated by silencing CSN8 (Fig. 5B, 6C, 6D and 6E), suggesting enhanced proteasome proteolytic function in CSN8 knockdown cells. The enhanced degradation of GFPu was unlikely the result of an increase in 20S proteasome function, because the proteasome peptidase activity was not altered by knockdown of CSN8 (Supplemental Fig. S1). The overall ubiquitination activity was not altered by suppression of CSN8 either, as determined by in vitro de novo ubiquitin conjugation assay (Supplemental Fig. S2). It remains unclear whether the enhanced proteasome function induced by suppression of CSN8 results from an increase of the association of the 19S with the 20S proteasome. Consistently, the stability of an endogenous protein p21cip was also decreased (Fig. 7A, 7B, 7C and 7D). Although it has been reported that p21cip can be targeted for proteasomal degradation in either ubiquitination-dependent or ubiquitination-independent manner (Bendjennat et al 2003, Bloom et al 2003, Bornstein et al 2003, Chen et al 2007, Li et al 2007), how the CSN controls the turnover of p21cip remains to be further defined. Similar to our finding, the proteasome-mediated degradation of Rbf1 and Rbf2 was increased in cultured cells and embryos with a diminished CSN activity, suggesting that the CSN may protect certain types of regulatory proteins from degradation (Ullah et al 2007).
In addition to regulating UPS-mediated proteolytic function, CSN may also regulate gene expression at the transcription level, as indicated by reduced mRNA levels of both transgene of GFPu and endogenous genes of p21cip and p27kip in CSN8 knockdown cells (Fig. 6A, 6B; Fig. 7G, 7H). Indeed, the CSN was proposed to act as a transcriptional repressor during the development of Drosophila, as supported by altered temporal regulation of gene expression in the csn mutants (Oron et al 2007). Another recent study also revealed that CSN4 can co-occupy the retinoblastoma protein (Rb) target gene promoters with Rbf1 and Rbf2, suggesting an active role for the CSN in transcriptional regulation (Ullah et al 2007). In murine T cells, the CSN appeared to occupy the promoters of certain cell cycle-related genes and CSN8 deficiency led to aberrant gene expression. Interestingly, the expression of p21cip was induced in the stimulated CSN8-deficient T cells, suggesting the transcriptional regulation of the CSN may be cell-type specific (Menon et al 2007). These studies collectively indicate that the CSN may be involved in the transcriptional regulation of gene expression, either directly or associated with other transcription factors.
In summary, the present study demonstrates that CSN8 is essential to the stability of CSN3, CSN5, and CSN7, as well as CSN holo-complex formation and that in absence of CSN8 other CSN subunits may form abnormal mini-complexes. This study also suggests that CSN8/CSN negatively regulates UPS-mediated degradation of a subset of protein substrates and positively regulate the transcription of at least a subset of genes.
Supplementary Material
Acknowledgements
Dr. X. Wang is an Established Investigator of American Heart Association (AHA). This work was in part supported by grants R01HL085629 and R01HL072166 from the National Heart, Lung, and Blood Institute/NIH and the grant 0740025N from AHA (to X. W.) and by the MD/PhD Program of the University of South Dakota. Dr. H. Su is supported by an AHA postdoctoral fellowship (0625738Z). We wish to thank Dr. Ning Wei of Yale University, New Haven, CT, USA for her critical reading and stimulating discussion over this manuscript.
References
- Bech-Otschir D, Kraft R, Huang X, Henklein P, Kapelari B, et al. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. Embo J. 2001;20:1630–9. doi: 10.1093/emboj/20.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–5. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Bendjennat M, Boulaire J, Jascur T, Brickner H, Barbier V, et al. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell. 2003;114:599–610. doi: 10.1016/j.cell.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell. 2003;115:71–82. doi: 10.1016/s0092-8674(03)00755-4. [DOI] [PubMed] [Google Scholar]
- Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752–7. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- Callige M, Kieffer I, Richard-Foy H. CSN5/Jab1 is involved in ligand-dependent degradation of estrogen receptor {alpha} by the proteasome. Mol Cell Biol. 2005;25:4349–58. doi: 10.1128/MCB.25.11.4349-4358.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–9. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, et al. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97:1018–26. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol Cell. 2007;26:843–52. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope GA, Deshaies RJ. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006;7:1. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–11. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- Denti S, Fernandez-Sanchez ME, Rogge L, Bianchi E. The COP9 signalosome regulates Skp2 levels and proliferation of human cells. J Biol Chem. 2006;281:32188–96. doi: 10.1074/jbc.M604746200. [DOI] [PubMed] [Google Scholar]
- Dong X, Liu J, Zheng H, Glasford JW, Huang W, et al. In situ dynamically monitoring the proteolytic function of the ubiquitin-proteasome system in cultured cardiac myocytes. Am J Physiol Heart Circ Physiol. 2004;287:H1417–25. doi: 10.1152/ajpheart.01233.2003. [DOI] [PubMed] [Google Scholar]
- Doronkin S, Djagaeva I, Beckendorf SK. The COP9 signalosome promotes degradation of Cyclin E during early Drosophila oogenesis. Dev Cell. 2003;4:699–710. doi: 10.1016/s1534-5807(03)00121-7. [DOI] [PubMed] [Google Scholar]
- Freilich S, Oron E, Kapp Y, Nevo-Caspi Y, Orgad S, et al. The COP9 signalosome is essential for development of Drosophila melanogaster. Curr Biol. 1999;9:1187–90. doi: 10.1016/S0960-9822(00)80023-8. [DOI] [PubMed] [Google Scholar]
- Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. Embo J. 2001;20:7096–107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto A, Tomoda K, Kubota M, Kato JY, Yoneda-Kato N. Small Jab1-containing subcomplex is regulated in an anchorage- and cell cycle-dependent manner, which is abrogated by ras transformation. FEBS Lett. 2005;579:1047–54. doi: 10.1016/j.febslet.2004.12.076. [DOI] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, et al. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–67. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- He Q, Cheng P, He Q, Liu Y. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 2005;19:1518–31. doi: 10.1101/gad.1322205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetfeld BK, Helfrich A, Kapelari B, Scheel H, Hofmann K, et al. The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr Biol. 2005;15:1217–21. doi: 10.1016/j.cub.2005.05.059. [DOI] [PubMed] [Google Scholar]
- Huang J, Yuan H, Lu C, Liu X, Cao X, Wan M. Jab1 Mediates Protein Degradation of the Rad9-Rad1-Hus1 Checkpoint Complex. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Hetfeld BK, Seifert U, Kahne T, Kloetzel PM, et al. Consequences of COP9 signalosome and 26S proteasome interaction. Febs J. 2005;272:3909–17. doi: 10.1111/j.1742-4658.2005.04807.x. [DOI] [PubMed] [Google Scholar]
- Karniol B, Malec P, Chamovitz DA. Arabidopsis FUSCA5 encodes a novel phosphoprotein that is a component of the COP9 complex. Plant Cell. 1999;11:839–48. doi: 10.1105/tpc.11.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarapeli AR, Horak KM, Glasford JW, Li J, Chen Q, et al. A novel transgenic mouse model reveals deregulation of the ubiquitin-proteasome system in the heart by doxorubicin. Faseb J. 2005;19:2051–3. doi: 10.1096/fj.05-3973fje. [DOI] [PubMed] [Google Scholar]
- Kwok SF, Staub JM, Deng XW. Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J Mol Biol. 1999;285:85–95. doi: 10.1006/jmbi.1998.2315. [DOI] [PubMed] [Google Scholar]
- Li X, Amazit L, Long W, Lonard DM, Monaco JJ, O’Malley BW. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol Cell. 2007;26:831–42. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 2003;17:1130–40. doi: 10.1101/gad.1090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Chen Q, Huang W, Horak KM, Zheng H, et al. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. Faseb J. 2006;20:362–4. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, et al. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–5. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V, Piran R, Pick E, Hofmann K, Glickman MH. COP9 signalosome components play a role in the mating pheromone response of S. cerevisiae. EMBO Rep. 2002;3:1215–21. doi: 10.1093/embo-reports/kvf235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Chi H, Zhang H, Deng XW, Flavell RA, Wei N. COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat Immunol. 2007;8:1236–45. doi: 10.1038/ni1514. [DOI] [PubMed] [Google Scholar]
- Mundt KE, Liu C, Carr AM. Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol Biol Cell. 2002;13:493–502. doi: 10.1091/mbc.01-10-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. Embo J. 2000;19:2069–81. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oron E, Mannervik M, Rencus S, Harari-Steinberg O, Neuman-Silberberg S, et al. COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development. 2002;129:4399–409. doi: 10.1242/dev.129.19.4399. [DOI] [PubMed] [Google Scholar]
- Oron E, Tuller T, Li L, Rozovsky N, Yekutieli D, et al. Genomic analysis of COP9 signalosome function in Drosophila melanogaster reveals a role in temporal regulation of gene expression. Mol Syst Biol. 2007;3:108. doi: 10.1038/msb4100150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panattoni M, Sanvito F, Basso V, Doglioni C, Casorati G, et al. Targeted inactivation of the COP9 signalosome impairs multiple stages of T cell development. J Exp Med. 2008;205:465–77. doi: 10.1084/jem.20070725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Shen Y, Feng S, Wang X, Chitteti BN, et al. Evidence for a physical association of the COP9 signalosome, the proteasome, and specific SCF E3 ligases in vivo. Curr Biol. 2003;13:R504–5. doi: 10.1016/s0960-9822(03)00439-1. [DOI] [PubMed] [Google Scholar]
- Peth A, Boettcher JP, Dubiel W. Ubiquitin-dependent proteolysis of the microtubule end-binding protein 1, EB1, is controlled by the COP9 signalosome: possible consequences for microtubule filament stability. J Mol Biol. 2007;368:550–63. doi: 10.1016/j.jmb.2007.02.052. [DOI] [PubMed] [Google Scholar]
- Rosel D, Kimmel AR. The COP9 signalosome regulates cell proliferation of Dictyostelium discoideum. Eur J Cell Biol. 2006;85:1023–34. doi: 10.1016/j.ejcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, et al. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 2001;292:1379–82. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- Schweitzer K, Bozko PM, Dubiel W, Naumann M. CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. Embo J. 2007;26:1532–41. doi: 10.1038/sj.emboj.7601600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Kraft R, Ferrell K, Bech-Otschir D, Dumdey R, et al. A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. Faseb J. 1998;12:469–78. [PubMed] [Google Scholar]
- Serino G, Tsuge T, Kwok S, Matsui M, Wei N, Deng XW. Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell. 1999;11:1967–80. doi: 10.1105/tpc.11.10.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterluty H, Chatelain E, Marti A, Wirbelauer C, Senften M, et al. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat Cell Biol. 1999;1:207–14. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- Tomoda K, Kato JY, Tatsumi E, Takahashi T, Matsuo Y, Yoneda-Kato N. The Jab1/COP9 signalosome subcomplex is a downstream mediator of Bcr-Abl kinase activity and facilitates cell-cycle progression. Blood. 2005;105:775–83. doi: 10.1182/blood-2004-04-1242. [DOI] [PubMed] [Google Scholar]
- Tomoda K, Kubota Y, Arata Y, Mori S, Maeda M, et al. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J Biol Chem. 2002;277:2302–10. doi: 10.1074/jbc.M104431200. [DOI] [PubMed] [Google Scholar]
- Tsvetkov LM, Yeh KH, Lee SJ, Sun H, Zhang H. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr Biol. 1999;9:661–4. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- Uhle S, Medalia O, Waldron R, Dumdey R, Henklein P, et al. Protein kinase CK2 and protein kinase D are associated with the COP9 signalosome. Embo J. 2003;22:1302–12. doi: 10.1093/emboj/cdg127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah Z, Buckley MS, Arnosti DN, Henry RW. Retinoblastoma protein regulation by the COP9 signalosome. Mol Biol Cell. 2007;18:1179–86. doi: 10.1091/mbc.E06-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kang D, Feng S, Serino G, Schwechheimer C, Wei N. CSN1 N-terminal-dependent activity is required for Arabidopsis development but not for Rub1/Nedd8 deconjugation of cullins: a structure-function study of CSN1 subunit of COP9 signalosome. Mol Biol Cell. 2002;13:646–55. doi: 10.1091/mbc.01-08-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee S, Geyer RK, Toda T, Wolf DA. CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat Cell Biol. 2005;7:387–91. doi: 10.1038/ncb1241. [DOI] [PubMed] [Google Scholar]
- Wei N, Chamovitz DA, Deng XW. Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell. 1994;78:117–24. doi: 10.1016/0092-8674(94)90578-9. [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW. The COP9 signalosome. Annu Rev Cell Dev Biol. 2003;19:261–86. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- Wilson MP, Sun Y, Cao L, Majerus PW. Inositol 1,3,4-trisphosphate 5/6-kinase is a protein kinase that phosphorylates the transcription factors c-Jun and ATF-2. J Biol Chem. 2001;276:40998–1004. doi: 10.1074/jbc.M106605200. [DOI] [PubMed] [Google Scholar]
- Wolf DA, Zhou C, Wee S. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol. 2003;5:1029–33. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- Wu K, Yamoah K, Dolios G, Gan-Erdene T, Tan P, et al. DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J Biol Chem. 2003;278:28882–91. doi: 10.1074/jbc.M302888200. [DOI] [PubMed] [Google Scholar]
- Zhou C, Wee S, Rhee E, Naumann M, Dubiel W, Wolf DA. Fission yeast COP9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Mol Cell. 2003;11:927–38. doi: 10.1016/s1097-2765(03)00136-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.