Abstract
Several arenaviruses cause hemorrhagic fever (HF) in humans, and evidence indicates that the worldwide-distributed prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) is a neglected human pathogen of clinical significance. Moreover, arenaviruses pose a biodefense threat. No licensed anti-arenavirus vaccines are available, and current anti-arenavirus therapy is limited to the use of ribavirin, which is only partially effective and is associated with anemia and other side effects. Therefore, it is important to develop effective vaccines and better antiviral drugs to combat the dual threats of naturally occurring and intentionally introduced arenavirus infections. The development of arenavirus reverse genetic systems is allowing investigators to conduct a detailed molecular characterization of the viral cis-acting signals and trans-acting factors that control each of the steps of the arenavirus life cycle, including RNA synthesis, packaging and budding. Knowledge derived from these studies is uncovering potential novel targets for therapeutic intervention, as well as facilitating the establishment of assays to identify and characterize candidate antiviral drugs capable of interfering with specific steps of the virus life cycle. Likewise, the ability to generate predetermined specific mutations within the arenavirus genome and analyze their phenotypic expression would significantly contribute to the elucidation of arenavirus–host interactions, including the basis of their ability to cause severe HF. This, in turn, could lead to the development of novel, potent and safe arenavirus vaccines.
Keywords: Arenaviruses, Lassa virus, Junin virus, Lymphocytic choriomeningitis virus, Reverse genetics, Antivirals
1. Introduction
Riboviruses, viruses with RNA genomes, contain the largest number of viruses with significant impact on human health. Several of these agents are associated with significant biohazard concerns and can be manipulated only under the highest biosafety level conditions (BSL4), which complicates research aimed at understanding their biology and pathogenesis, as well as the identification and characterization of antiviral drugs to combat them.
The term reverse genetics, as used in the field of molecular virology, refers to the generation or “rescue” of viruses entirely from cloned cDNAs. This approach was first implemented for the generation and genetic manipulation of positive strand RNA viruses, whose deproteinized genomes are able to act as mRNA and initiate the virus life cycle once delivered into appropriate susceptible cells. In contrast, the template of the polymerases of negative strand (NS) RNA viruses is exclusively a nucleocapsid consisting of the genomic RNA tightly encapsidated by the virus nucleoprotein (N), which associated with the virus polymerase proteins forms a ribonucleoprotein (RNP) complex. This RNP is active in transcription and replication and is the minimum unit of infectivity. Consequently, deproteinized genomes of NS RNA viruses cannot function as mRNAs and are not infectious. Thus, generation of biologically active synthetic NS viruses from cDNA will require trans complementation by all of the viral proteins involved in virus replication and transcription.
These considerations initially posed unique hurdles to the development of reverse genetics systems for NS RNA viruses (Neumann et al., 2002, Conzelmann, 2004). However, during the last 15 years, following the pioneering work of Palese's group (Luytjes et al., 1989), significant progress has been made in this area and reverse genetics systems have been now developed for all ribovirus families. These developments have revolutionized the analysis of cis-acting sequences and trans-acting proteins required for virus replication, transcription, assembly and cell egress. In addition, these developments have paved the way for engineering these viruses for vaccine and gene therapy purposes. Moreover, using reverse genetics approaches it has been possible to develop cell-based minireplicon or minigenome (MG) assays to conduct studies under BSL2 conditions to dissect many aspects of the molecular and cell biology associated with the life cycle of BSL4 viruses. MG-based assays have also provided the foundations for the development of novel screening strategies to identify and validate compounds that could potentially be developed into effective antiviral drugs.
2. Impact of arenaviral infections in human health
Arenaviruses cause chronic infections of rodents with a worldwide distribution (Buchmeier et al., 2007). Asymptomatically infected animals move freely in their natural habitat and may invade human dwellings. Humans are infected most likely through mucosal exposure to aerosols, or by direct contact between infectious materials and abraded skin. These infections are common and in some cases severe.
The family Arenaviridae consists of one unique genus (Arenavirus) with 22 recognized species that are classified into two distinct groups: Old World (OW) and New World (NW) (Buchmeier et al., 2007). This classification was originally established based on serological cross-reactivity, but it is well supported by recent sequence-based phylogenetic studies. Genetically, OW arenaviruses constitute a single lineage, while NW arenaviruses segregate into clades A, B, and C. The OW arenavirus Lassa virus (LASV) and several NW arenaviruses including Junin virus (JUNV), cause hemorrhagic fever (HF) in humans, which represents a serious public health problem in endemic areas (McCormick and Fisher-Hoch, 2002, Peters, 2002, Geisbert and Jahrling, 2004, Buchmeier et al., 2007, Enria et al., 2008, Khan et al., 2008) (Table 1 ).
Table 1.
Geographic distribution and natural reservoirs of arenaviruses known to cause disease in humans
| Arenavirus | Geographic distribution | Natural reservoir | Human disease |
|---|---|---|---|
| LCMV | Europe and Americas | Mus domesticus | LCM |
| Possibly also other regions | Mus musculus | Congenital disorders | |
| LASV | West Africa | Mastomys species | Lassa fever (LF) |
| JUNV | Argentine pampas | Calomys musculinus | Argentine HF (AHF) |
| MACV | Bolivia (Beni region) | Calomys callosus | Bolivian HF (BHF) |
| GTOV | Venezuela | Sigmodon alstoni | Venezuelan HF (VHF) |
| Zygodontomys brevicauda | |||
| SABV | Brazil | Unknown | Unassigned |
| WWAV | USA (NM, CA) | Neotoma albigula | Unassigned |
Acronyms: LCMV, lymphocytic choriomeningitis virus; LASV, Lassa virus; JUNV, Junin virus; MACV, Machupo virus; GTOV, Guanarito virus; SABV, Sabia virus; WWAV, white water arroyo virus; LF, Lassa fever; HF, hemorrhagic fever; AHF, Argentine HF; BFH, Bolivian HF; VHF, Venezuelan HF.
LASV is estimated to infect several hundred thousand individuals yearly in its endemic regions of West Africa, with the highest incidence affecting the “Mano River Union (MRU) countries”, resulting in a high number of Lassa fever (LF) cases associated with significant mortality and high morbidity (Khan et al., 2008). In recent years, increased air travel between Africa and other areas has led to the importation of cases into the United States, Europe, Japan, and Canada (Holmes et al., 1990, Freedman and Woodall, 1999, Isaacson, 2001). JUNV, which is endemic to the humid pampas of Argentine, causes Argentine HF (AHF) that if left untreated has a high (15–30%) case-fatality rate (Enria et al., 2008). Moreover, it is worth noting that compelling evidence indicates that the worldwide-distributed prototypic arenavirus LCMV is a neglected human pathogen of clinical significance, especially in cases of congenital infection leading to hydrocephalus, mental retardation and chorioretinitis in infants (Jahrling and Peters, 1992, Barton and Mets, 1999, Barton and Mets, 2001, Mets et al., 2000, Barton et al., 2002). In addition, LCMV poses a special threat to immunocompromised individuals, as tragically illustrated by recent cases of transplant-associated infections by LCMV with a fatal outcome in the USA (Fischer et al., 2006, Peters, 2006).
The arenavirus potential for person-to-person transmission together with the likelihood that aerosolized forms of these viruses would be highly infectious to humans, have led to the inclusion of HF arenaviruses and LCMV within Category A biological agents that pose a significant biodefense concern (Borio et al., 2002). These concerns are aggravated by the lack of licensed vaccines or effective treatment. Therefore, it is important to develop novel effective anti-arenavirus therapies and vaccines.
Evidence indicates that morbidity and mortality associated with LASV infection, and likely other HF arenavirus infections, are associated with, at least partly, the failure of the host's innate immune response to restrict virus replication and to facilitate the initiation of an effective adaptive immune response (McCormick and Fisher-Hoch, 2002). Accordingly, the extent of viremia is a highly predictive factor for the outcome of LF patients. This scenario would suggest that therapeutic interventions resulting in reduced virus load might suffice, despite the lack of complete virus clearance, to promote the recovery of appropriate host defense responses to control virus multiplication and associated disease.
Besides their significant in human health, arenaviruses are also important model systems to study virus–host interactions. Studies using the prototypic arenavirus LCMV have led to major advances in virology and immunology that apply universally to other microbial infections and viral infections of humans (Oldstone, 2002, Zinkernagel, 2002). The outcome of LCMV infection of its natural host the mouse varies dramatically depending on the species, age, immune status and genetic background of the host, as well as the route of infection, and the strain and dose of infecting virus (Oldstone, 2002, Zinkernagel, 2002, Buchmeier et al., 2007). This provides investigators with a rather unique model system where to investigate parameters that critically affect the phenotypic heterogeneity often associated with infection by the same virus.
3. Current strategies to combat arenaviruses
3.1. Arenavirus vaccines
Efforts to develop arenavirus vaccines have been focused on JUNV and LASV, the causative agents of AHF and LF. The live attenuated Candid 1 strain of JUNV was shown to induce a strong neutralizing antibody response in several animal models, as well as in humans, that correlated with protection against highly virulent strains of JUNV (Enria and Barrera Oro, 2002, Enria et al., 2008). In a double blind study involving agricultural workers in the endemic area of Argentine Candid 1 strain proved to be an effective vaccine, without serious adverse effects (Maiztegui et al., 1998). However, the Candid 1 vaccine is licensed only as an investigational new drug in the U.S. and studies addressing the stability of its attenuation, long-term immunity and safety have not been conducted. Sequence comparison between the Candid 1 and virulent strains of JUNV have shown multiple mutations within NP, GP and L are associated with the vaccine strain, but their relative contributions to attenuation have not been determined.
LASV is the arenavirus with the highest impact on public health, due to its high incidence within the vast endemic area of West Africa where estimates of virus exposure, based on antibody prevalence, range from 4% to 6% in Guinea to 15% to 20% in Nigeria (Khan et al., 2008). Therefore, the development of an effective and safe vaccine would represent a major accomplishment, not only in West Africa but also in other geographic regions due to the risk of imported cases of LF posed by increased international travel. Contrary to the situation found with JUNV and AHF, the neutralizing antibody response is not predictive of survival of LF (McCormick and Fisher-Hoch, 2002). Therefore, it is likely that the mechanisms responsible for the success of the JUNV Candid 1 strain vaccine for AHF may not serve the same purpose in the case of LF.
Different approaches have been pursued aiming at the development of a safe and effective vaccine against LASV, including DNA immunization protocols (Rodriguez-Carreno et al., 2005), and the use of recombinant salmonella (Djavani et al., 2001). Recombinant vaccinia viruses expressing LASV NP or GP have been shown to provide both guinea pigs and non-human primates with protective cell-mediated immunity against lethal challenge (Fisher-Hoch et al., 2000). However, the high prevalence of HIV in West Africa and the safety profile of vaccinia virus raise serious doubts about the potential implementation of this approach. Alphavirus-based vectors have been used to induce protective immunity in guinea pigs (Pushko et al., 2001), and more recently a recombinant vesicular stomatitis virus (VSV), in which the LASV GP replaced the VSV G, was shown to provide protection against lethal challenge (Geisbert et al., 2005). In both of these studies the LASV challenge was done 28 days after immunization, so it remains to be determined whether these recombinant vaccines provide long-term protection – a highly relevant issue, due to a cumulative lifetime risk of exposure to LF vaccine within the West Africa human population.
Another approach pursued in the development of a vaccine against LF is induction of heterologous immunity by using a closely related but less pathogenic virus. Thus, LCMV has been shown to be able to protect non-human primates against LF (Kiley et al., 1979). More recently, a reassortant virus between LASV and the genetically closely related NW arenavirus Mopeia (MOPV), called ML29, was found to protect guinea pigs against LASV induced disease (Lukashevich et al., 2005). However, safety profiles for LCMV and MOPV in humans have not been determined.
The development of reverse genetics systems for JUNV and LASV, similar to the one described for LCMV, would facilitate the elucidation of the genetic determinants of JUNV and LASV virulence. This, in turn, should help with the design of safer live attenuated vaccines by minimizing concerns related to possible reversion to virulence, establishment of persistent infection and in general conditions associated with high risk for replicating viruses including immunocompromised individuals. The power of reverse genetics as a tool for arenavirus vaccine development is illustrated by recent findings obtained with a recombinant LCMV expressing the surface GP of VSV serotype Indiana instead of its own GP (rLCMV/INDG) (Pinschewer et al., 2003b). In mice, rLCMV/INDG exhibited a highly attenuated phenotype that remained stable upon long-term propagation in immunocompromised mice (Bergthaler et al., 2006), but it was able to elicit very strong, long-term T cell-mediated immunity against lethal challenge with wild-type LCMV (Bergthaler et al., 2006).
3.2. Antiviral drugs
Current therapeutic measures against AHF are centered around the transfusion of immune plasma in defined doses of neutralizing antibodies during the prodromal phase of illness (Enria et al., 2008), which is complicated by the difficulties of maintaining appropriate stocks of immune plasma, and the difficulty of detecting AHF in its early stages, when immune plasma therapy exerts its maximal efficacy.
In vitro and in vivo studies have documented the prophylactic and therapeutic value of the nucleoside analogue ribavirin (Rib) (1-b-d-ribofuranosyl-1,2,4-triazole-3-carboxamide) against several arenaviruses (Andrei and De Clercq, 1993, Damonte and Coto, 2002). Moreover, Rib has been shown to reduce significantly both morbidity and mortality in humans associated with LASV (McCormick et al., 1986, Andrei and De Clercq, 1993, Damonte and Coto, 2002), and experimentally in Machupo (Kilgore et al., 1997) and JUNV infections (McKee et al., 1988), if given early in the course of clinical disease. The mechanisms by which Rib exerts its anti-arenaviral action remain poorly understood, but likely involve targeting different steps of the virus life cycle (Parker, 2005, Leyssen et al., 2008). Recent evidence indicates that Rib can be used as substrate by the RdRp of some riboviruses leading to C to U and G to A transitions (Crotty et al., 2000, Cameron and Castro, 2001, Parker, 2005). This mutagenic activity of Rib has been linked to its antiviral activity via lethal mutagenesis (Cameron and Castro, 2001). However, Rib inhibited very strongly LCMV replication without exerting any noticeable mutagenic effect on the virus genome RNA (Ruiz-Jarabo et al., 2003). In humans, treatment causes a reversible anemia, and the drug has also caused birth defects in laboratory animals, so that it may not be prescribed to pregnant women. In addition, oral Rib is significantly less effective than the one administered intravenously, which poses logistic complications in regions with limited clinical infrastructure.
Several Rib-related inhibitors of IMP dehydrogenase, including ribamidine (1-beta-d-ribofuranosyl-1,2,4-tiazole-3-carboxamide) (Andrei and De Clercq, 1993), as well as acyclic and carbocyclic adenosine analogue inhibitors of S-adenosylhomocysteine (SAH) hydrolase (Andrei and De Clercq, 1990) have been show to have anti-arenavirus activity. Moreover, a variety of sulfated polysaccharides exhibited significant and specific anti-arenavirus activity (Andrei and De Clercq, 1990). Likewise, phenotiazine compounds (Candurra et al., 1996) and myristic acid analogues (Cordo et al., 1999), as well as brassinosteroids (Wachsman et al., 2000), have been reported to have activity against several arenaviruses. In general these compounds displayed only modest and rather non-specific effects, often associated with significant toxicity.
Recently a high-throughput screening (HTS) based on a virus-induced cytopathic effect (CPE) assay successfully identified a potent small-molecule inhibitor of HF New World arenaviruses that acts by targeting the viral GP (Bolken et al., 2005). Likewise, cell-based HTS based on the use of pseudotyped virion particles bearing the GP of highly pathogenic arenaviruses identified several small-molecule inhibitors of virus cell entry mediated by LASV GP (Lee et al., 2008). These findings illustrate how complex chemical libraries used in the context of appropriate screening assays can be harnessed into a powerful tool to identify candidate antiviral drugs with highly specific activities. To this goal, recent progress in the molecular and cell biology of arenaviruses, including the development of reverse genetics systems, have opened new avenues for the identification of steps in the arenavirus life cycle that are amenable to drug targeting and the development of appropriate screening strategies to identify such drugs.
4. Molecular and cell biology of arenaviruses
Arenaviruses are enveloped viruses with a bi-segmented, negative-sense, single-stranded RNA genome and a life cycle restricted to the cell cytoplasm. Each genomic RNA segment, L (ca 7.3 kb) and S (ca 3.5 kb), uses an ambisense coding strategy to direct the synthesis of two polypeptides in opposite orientation, separated by a non-coding intergenic region (termed IGR) with a predicted folding of a stable hairpin structure (Buchmeier et al., 2007) (Fig. 1 ). The S RNA encodes the viral nucleoprotein, NP (ca 63 kDa) and the virion surface glycoprotein precursor, GPC (ca 75 kDa). The L RNA encodes the viral RNA-dependent RNA polymerase (RdRp, or L polymerase) (ca 200 kDa), and a small RING finger protein Z (ca 11 kDa).
Fig. 1.
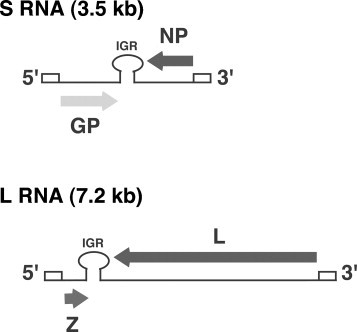
LCMV genome organization.
Viral mRNAs have extra non-templated nucleotides (nt) and appear to have a cap structure at their 5′-ends, but the origin of both the cap and 5′-non-templated nt extensions remain to be determined. NP and L polypeptides are translated from mRNAs with antigenomic sense polarity, whereas GPC and Z ORFs are not translated directly from genomic RNA species, but rather from genome-sense mRNAs that are transcribed using as templates the S and L antigenome RNA species, respectively. Transcription termination of subgenomic non-polyadenylated viral mRNAs was mapped to multiple sites within the distal side of the intergenic region (IGR), suggesting that the IGR acts as a bona fide transcription termination for the viral polymerase, which has been confirmed by more recent studies using reverse genetics approaches (Pinschewer et al., 2005, Lopez and Franze-Fernandez, 2007).
Post-translational cleavage of GPC generates the three components that form the GP complex (GPc): the stable signal peptide (SSP, 58aa), GP1 (Mr 40–46 kDa), and GP2 (Mr 35 kDa) (Buchmeier et al., 2007). The arenavirus SSP is unique in that it remains stably associated with the GP complex following cleavage by signal peptidase and plays crucial roles in the trafficking of GP through the secretory pathway (Eichler et al., 2003, Eichler et al., 2004, York et al., 2004). Generation of GP1 and GP2 of the OW arenaviruses LCMV and LASV was found to be mediated by the SKI-1/S1P cellular protease (Beyer et al., 2003, Kunz et al., 2003, Pinschewer et al., 2003b). This finding has been extended to the NW arenaviruses Guanarito, Machupo and Junin (Rojek et al., 2008). Notably, the protease recognition site present in Guanarito GPC deviates from the originally reported S1P consensus sequence (Rojek et al., 2008), indicating that S1P specificity is broader than previously thought.
The surface of the virus envelope is decorated by evenly spaced projections, or spikes, that consist of complexes of GP1 and GP2. GP1 is located at the top of the spike and is held in place by ionic interactions with the N-terminus of the transmembrane GP2, that forms the stalk of the spike (Neuman et al., 2005, Buchmeier et al., 2007). GP1 is the virion attachment protein that mediates virus interaction with host cell surface receptors and subsequent virus cell entry via receptor mediated endocytosis (Kunz et al., 2002).
Once the viral ribonucleoprotein has been delivered into the cytoplasm of the infected cell, the associated polymerase directs the biosynthetic processes involved in RNA replication and gene transcription (Fig. 2 ). Primary transcription initiated at the genome promoter located at the genome 3′-end results in synthesis of NP and L mRNA from the S and L segments, respectively. Subsequently the virus polymerase can adopt a replicase mode and moves across of the IGR to generate a copy of the full-length antigenome RNA (agRNA). This agRNA will serve as template for the synthesis of the GP (agS) and Z (agL) mRNAs. The agRNA species serve also as templates for the amplification of the corresponding genome RNA species. Assembly and cell release of infectious progeny involves the association of the viral ribonucleoprotein core with the surface GP complex, a process that requires the participation of Z (Capul et al., 2007). Correct processing of GPC is required for the production of infectious virions (Beyer et al., 2003, Kunz et al., 2003, Pinschewer et al., 2003b), which bud from the plasma membrane.
Fig. 2.
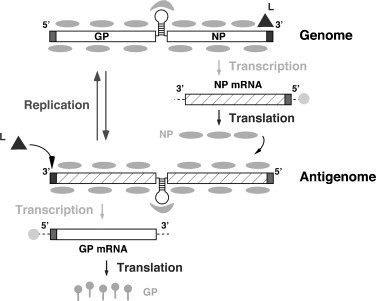
Basic aspects of arenavirus RNA replication and gene transcription illustrated for the S segment. The L polymerase associated with the virus RNP initiates transcription from the genome promoter located at the genome 3′-end. Primary transcription results in synthesis of NP mRNA (or L mRNA in the case of the L segment). Subsequently the virus polymerase can adopt a replicase mode and moves across of the IGR to generate a copy of the full-length antigenome (ag) S RNA (or ag L RNA). This ag S RNA will serve as template for the synthesis of the GP mRNA (the ag L RNA will serve as template for the synthesis of the Z mRNA). The agRNA species serve also as templates for the amplification of the corresponding genome RNA species.
5. Arenavirus genetics
5.1. Reassortment
The segmented nature of the arenavirus genome permits the generation of reassortants between two parental viruses. For this, cells coinfected with both viruses serve as a mixing vessel to facilitate the generation of infectious progeny carrying all possible combinations of L/S genomes. Genetic or biochemical characterization of clonal populations derived from the infectious progeny makes it possible to identify the origins of the RNA segments, and thereby map specific phenotypes of the parental viruses to either the L or S segment. For example, the ability of LCMV to cause growth hormone deficiency in C3H mice (Riviere et al., 1985a), or immunosuppression and persistence in adult immunocompetent mice (Matloubian et al., 1990, Salvato et al., 1991) have been mapped to the S segment. Similarly, the lethality of the WE strain of LCMV in guinea pigs (Riviere et al., 1985b), and differences in virulence between LASV strains (Lukashevich et al., 2005) have been mapped to the L segment.
5.2. Reverse genetics
Despite tremendous success in mapping a variety of arenavirus phenotypes to either the L or S segment, the use of reassortant viruses does not make it possible to assess the individual contribution of each of the potential multiple mutations that often are found between the L and S segments of two parental viruses. Likewise, the reassortment approach cannot be used to generate viruses with predetermined mutations in their genomes. This inability to genetically manipulate the arenavirus genome has hampered studies aimed at understanding its molecular and cell biology, as well as the role played by each viral gene product in virus–host interactions during both acute and persistent infections and associated diseases. Reverse genetics systems that support RNA replication and expression of arenavirus minigenomes have been developed for LCMV, as well as for LASV (Hass et al., 2004) and the NW arenavirus Tacaribe (TCV) (Lopez et al., 2001), and have provided investigators with a novel and powerful approach for the investigation of the trans-acting factors and cis-acting sequences that control arenavirus replication and gene expression, as well as assembly and budding (Fig. 3 ). The LCMV MG system is based on intracellular synthesis, via either plasmid-supplied T7 RNA polymerase or RNA polymerase I (pol-I), of RNA analogues (MG) that contains the 5′- and 3′-UTR and IGR cis-acting sequences required for the control of RNA synthesis mediated by the LCMV polymerase, and the ORF of a reporter gene in lieu of the NP ORF. Encapsidation of the pol-I or T7-derived LCMV MG RNA by plasmid-supplied NP generates a template that is recognized by the intracellularly reconstituted LCMV polymerase to direct synthesis of full-length anti-minigenome (aMG) RNA (replicate) and a subgenomic mRNA (transcript) that codes for the reporter gene of choice (Lee et al., 2000). Incorporation into the transfection mix of GPC and Z-expressing plasmids allows for assembly and budding of infectious VLPs (Lee et al., 2002, Perez et al., 2003).
Fig. 3.
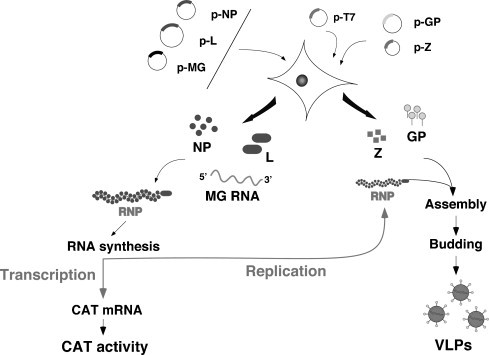
Schema of the LCMV MG rescue system. LCMV RNA analogues, or MG, with correct termini are synthesized intracellularly using either a T7 RP, or RNA pol-I, expression system. Cotransfection with plasmids expressing the viral trans-acting factors L and NP allow for the generation of viral RNP (MG RNA encapsidated by NP) that is substrate for the plasmid-supplied virus L polymerase. The virus polymerase directs the synthesis of two RNA species, the aMG RNA (replicative species) and a subgenomic mRNA that directs expression of the reporter gene of choice, CAT in this case. Incorporation of plasmids expressing Z and GP results in assembly and budding of infectious LCMV virus like particles (VLP).
Results from the LCMV MG system identified NP and L as the minimal viral trans-acting factors required for efficient RNA synthesis, both transcription and replication, mediated by the virus polymerase (Lee et al., 2000). Similar findings have been now documented for LASV (Hass et al., 2004) and TCV (Lopez et al., 2001). Notably, both genetic and biochemical evidence indicated that oligomerization of L is required for polymerase function (Sanchez and de la Torre, 2005). The activity of the LCMV genomic promoter recognized by the virus polymerase requires both sequence specificity within the highly conserved 3′-terminal 19 nt of arenavirus genomes, and the integrity of the predicted panhandle structure formed via sequence complementarity between the 5′-and 3′-termini of viral genome RNAs (Perez and de la Torre, 2002). Detailed structure-function studies using the LASV MG system (Hass et al., 2006) revealed that the LASV promoter, and likely that of all arenaviruses, regulates transcription and replication in a coordinated manner. It is composed of two functional elements: a sequence-specific region from residue 1–12 and a variable complementary region from residue 13–19. The first region appears to interact with the replication complex mainly via base-specific interactions, while in the second region, base pairing between 3′- and 5′-termini of the promoter determines its activity.
The use of the LCMV MG rescue system also provided direct experimental evidence that the IGR is a bona fide transcription termination signal (Pinschewer et al., 2005). This finding has also been documented for TCV. Contrary to a widely accepted model for the control of arenavirus RNA replication and gene transcription, intracellular levels of NP do not play a key role in the control of the balance between RNA replication and transcription by the arenavirus polymerase (Pinschewer et al., 2003a). The mechanisms determining whether the arenavirus polymerase complex behaves in a replicase or transcriptase mode remain to be elucidated.
Z was not required for intracellular transcription and replication of a LCMV MG, but rather Z exhibited a dose-dependent inhibitory effect on both transcription and replication of LCMV MG (Lee et al., 2000, Cornu and de la Torre, 2001, Cornu and de la Torre, 2002, Cornu et al., 2004). This inhibitory effect of Z has been also reported for TAV (Lopez et al., 2001) and LASV (Hass et al., 2004). Z has been postulated to participate in virion morphogenesis (Salvato et al., 1992, Salvato, 1993). For most enveloped viruses, a matrix (M) protein is involved in organizing the virion components prior to assembly, but arenaviruses do not have an obvious counterpart of M. However, we (Perez et al., 2003) and others (Strecker et al., 2003, Urata et al., 2006) have shown that Z is the main driving force of arenavirus budding and that this process is mediated by the Z proline-rich late domain motifs (LDM) PTAP and PPPY, similar to those known to control budding of several other viruses including HIV and Ebola, via interaction with specific host cell proteins (Freed, 2002). Consistent with this observation, Z exhibits features characteristic of bona fide budding proteins: the ability to bud from cells by itself and to efficiently substitute for other LDM (Perez et al., 2003). Targeting of Z to the plasma membrane, the location of arenavirus budding, strictly required its myristoylation (Perez et al., 2004). These data support a role of Z as the arenavirus counterpart of the M proteins found in many other NS RNA viruses, a notion that is consistent with recent ultrastructural data on arenavirus virions determined by cryo-electron microscopy (Neuman et al., 2005).
Infectious LCMV has been rescued entirely from cloned cDNAs using approaches based on both the T7 RNA polymerase (Sanchez and de la Torre, 2006) and RNA pol-I (Flatz et al., 2006) to launch the intracellular synthesis of S and L antigenome RNA species, which are subsequently replicated and transcribed by the virus polymerase, which is intracellularly reconstituted via plasmid-supplied viral L and NP (Fig. 4 ). The ability to generate rLCMV with predetermined specific mutations and analyze their phenotypic expression in its natural host, the mouse, would allow us to tackle a number of long-standing questions of the virus molecular biology, its relationship with the host cell and the specific role of the four known viral genes in arenavirus pathogenesis.
Fig. 4.
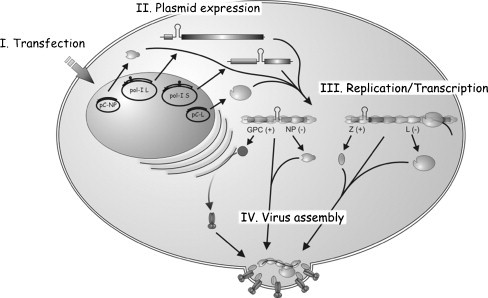
Rescue of infectious rLCMV entirely from cloned cDNAs. Cells are transfected with plasmids that directed T7RP-or pol-I mediated intracellular synthesis of L and S antigenomic (Lag and Sag) RNA species, together with pol-II expression plasmids for the viral trans-acting factors L and NP (and also T7RP if required). At different times after transfection, cell culture supernatants are tested for production of infectious rlCMV. Production of rLCMV is readily and consistently detected at 48 h after transfection, which is followed by a rapid increase in virus production reaching titers of 107 PFU/ml between 60 and 72 h post-transfection. Notably, we observed similar rescue efficiencies using either genomic or antigenomic polarities of the L and S RNAs. This finding indicates that annealing between viral mRNAs and genome, or antigenome, RNA species do not pose a significant problem for the rescue of LCMV, and likely arenaviruses in general. Likewise, both the T7RP and pol-I based rescue systems exhibited similar efficiencies.
6. Arenavirus MG systems as platforms for screening anti-arenaviral drugs
MG rescue systems similar to those already established for LCMV, LASV and TCV could now be developed for other arenaviruses of interest. These MG systems provide investigators with excellent platforms to identify inhibitors of arenavirus replication and propagation and to dissect their mechanisms of action. Thus, the anti-arenavirus activity of Rib has been recreated in the LCMV (Ruiz-Jarabo et al., 2003) and LASV (Hass et al., 2004) MG rescue systems. In contrast, the myristoylation inhibitor 2-hydroxymyristic acid (2-OHM) that was shown to inhibit Z-mediated arenavirus budding (Perez et al., 2004) did not affect, as predicted, MG RNA replication and expression, which further supports the feasibility of using cell-based MG rescue systems to identify drugs that interfere with specific steps of the arenavirus life cycle, and subsequently delineate the underlying mechanisms of action. Likewise, the use of an LCMV MG system was instrumental in assessing the specificity, efficacy and effects of siRNAs targeting different viral mRNAs (Sanchez et al., 2005). The following sections discuss some examples of the use of MG systems to uncover novel anti-arenaviral strategies.
6.1. Targeting the arenavirus promoter
The sequence and structural constraints of the arenavirus genome promoter (Perez and de la Torre, 2002) identified a new potential target for drugs capable of disrupting the interaction between the promoter and the virus polymerase complex, which is expected to have a strong deleterious impact on virus viability by affecting the essential biosynthetic processes of viral transcription and replication. The potential of RNA as a prime target for therapeutic intervention is gaining great interest (Hermann, 2000, Hermann and Westhof, 2000, Sucheck et al., 2000, Sucheck and Wong, 2000, Wilson and Li, 2000, Sucheck and Shue, 2001). The function of many RNA molecules depends on defined three-dimensional structure. The large variety of structures associated with RNA folding create unique binding pockets that provide potential targets for small molecules. Small molecules may interfere with RNA functions by a number of mechanisms including inhibition of the formation of competent RNA–protein complexes.
The study of small-molecule RNA effectors has primarily focused on the aminoglycosides (Hermann, 2000, Hermann and Westhof, 2000, Wilson and Li, 2000, Sucheck and Shue, 2001). The antibiotic activity associated with aminoglycoside targeting of bacterial 16S ribosomal RNA is a well-known success case. Despite their benefits, the efficacy of naturally occurring aminoglycosides has been limited due to the emergence of resistant microbes, oral inactivity, toxicity, and weak binding common to carbohydrates. During recent years there has been great progress in developing aminoglycoside-based analogues (mimetics) to circumvent these issues (Sears and Wong, 1999, Sears and Wong, 2001, Ye and Zhang, 2002). These advances have permitted the generation of complex combinatorial libraries of aminoglycosides that can be subjected to robust novel screening procedures (Hermann, 2000, Hermann and Westhof, 2000, Sucheck et al., 2000, Sucheck and Wong, 2000, Wilson and Li, 2000, Sears and Wong, 2001, Sucheck and Shue, 2001, Ye and Zhang, 2002). These developments are facilitating the identification of drugs that bind specifically to a variety of RNA folds, thus opening new ways to expand the existing repertoire of protein-targeted therapeutics. The potential of aminoglycosides as antiviral molecules by acting on RNA has been illustrated by their ability to disrupt selectively the HIV-1 Rev-RRE (Zapp et al., 1993) and Tat-TAR (Wang et al., 1998) interactions. Similar approaches could be used to select aminoglycosides that interact with the arenavirus promoter and disrupt its activity. The sequence conservation of the arenavirus promoter predicts that aminoglycoside-based small molecules selected based on their activity against the promoter of the prototypic arenavirus LCMV would be also active against LASV and other HF arenaviruses.
The high sequence and structural conservation of the core of the arenavirus genome promoter raises also the possibility of using a method for in vitro selection of combinatorial oligonucleotide libraries referred to as SELEX (“Systematic Evolution of Ligands by Exponential enrichment”) (Ellington and Szostak, 1990, Robertson and Joyce, 1990, Tuerk and Gold, 1990) to select nucleic acid-based molecules (aka aptamers) for target proteins. Aptamers usually exhibit tight binding to their protein targets, high specificity and minimal to no immunogenicity. They are therefore considered to be novel promising therapeutic drugs. SELEX could be used to select for aptamer molecules to target the arenavirus L polymerase and disrupt either the L-virus promoter or L–L interactions, both of which are strictly required for initiation of RNA synthesis.
6.2. Targeting the IGR
Early studies documented that the 3′ ends of the non-polyadenylated LCMV mRNAs were heterogeneous and mapped within the IGR (Meyer et al., 2002). All arenavirus IGR sequences are predicted to fold into single or double stem-loop structures (Meyer et al., 2002, Buchmeier et al., 2007), suggesting a structure-dependent transcription termination mechanism reminiscent of rho-independent termination in prokaryotes (Yarnell and Roberts, 1999, Tortorici et al., 2001). This prediction has received direct experimental support from results obtained using arenavirus MG systems (Pinschewer et al., 2005, Lopez and Franze-Fernandez, 2007). Unexpectedly, LCMV MGs without IGR were dramatically impaired in their ability to passage reporter gene activity via infectious VLP (Pinschewer et al., 2005), suggesting that in addition to its role in the control of RNA synthesis, the IGR plays a role in virus assembly or budding, or both, required for the efficient propagation of LCMV infectivity. In this regard, it is worth noting that LCMV genome and antigenome RNA species present in arenavirus RNPs were found to be sensitive to cleavage by very low concentrations of Rnases, suggesting that the arenavirus genome and antigenome are not tightly encapsidated, and are thereby potentially vulnerable to targeting. There is significant sequence heterogeneity between the IGR of the L and S segments of the each arenavirus species and between the L and S segments of different species. However, as with the virus genome promoter, IGRs are predicted to have a conserved folding that could provide a suitable target.
6.3. Targeting the L–L interaction required for arenavirus polymerase activity
The arenavirus L protein has the characteristic sequence motifs that are conserved among the RdRp (L proteins) of NS RNA viruses. Sequence alignment shows six conserved domains (Poch et al., 1989, Tordo et al., 1992). The proposed polymerase module of L is located within domain III, which contains highly conserved amino acids within motifs designated A and C. The SDD sequence characteristic of motif C of segmented NS RNA viruses, and the presence of the highly conserved D residue within motif A, are strictly required for the function of the LCMV L polymerase (Sanchez and de la Torre, 2005). These studies also showed that many of the mutant L proteins exhibited a strong dominant-negative (DN) effect under assay conditions in which the wild-type and mutant L proteins did not compete for template RNA or other viral or cellular trans-acting proteins required for polymerase activity (Sanchez and de la Torre, 2005). These results were highly suggestive of L–L interaction being required for LCMV polymerase activity.
Consistent with these genetic data, results from co-immunoprecipitation studies using L proteins containing two different tags, HA and Flag, provided biochemical evidence for L–L interaction (Sanchez and de la Torre, 2005). Intragenic complementation has been documented for the L genes of several NS RNA viruses (Repik et al., 1976, Smallwood et al., 2002). Consistent with this, direct L–L physical interaction has been demonstrated for the paramyxoviruses Sendai (Smallwood et al., 2002) and parainfluneza virus 3 (PIV3) (Smallwood and Moyer, 2004), and this L oligomerization was required for polymerase activity. These results suggest that disruption of this L–L interaction would have a potent inhibitory effect on polymerase activity.
Cell-based MG rescue assays are particularly appealing for identifying small molecules capable of disrupting L–L interaction, thus resulting in inhibition of arenavirus polymerase activity. Protein–protein interactions have been generally regarded as difficult or refractory drug targets for small molecules, because of the widely shared assumption that they are unlikely to disturb large protein interfaces. However, recent evidence suggests that the affinity between interacting proteins may be governed only by a minor part of the interface region (Wells and de Vos, 1996). Targeting these highly “localized” surfaces may be sufficient for inhibiting protein–protein interactions. Notably, combinatorial libraries of small molecules generated via solution-phase methodology have been uniquely successful in the identification of small molecules that inhibit protein–protein interactions for a series of significant biological targets (Boger et al., 1998, Boger et al., 2003, Boger, 2003). Similar approaches should be applicable to screens to identify small-molecule inhibitors of arenavirus polymerases.
6.4. Targeting the S1P-mediated proteolytic processing of arenavirus GPC
S1P protease is encoded by the membrane bound transcription-factor protease site 1 gene and is an endoplasmic reticulum (ER)/early Golgi membrane-anchored serine protease (Sakai et al., 1998, Seidah et al., 1999). S1P cleaves within the motif R-X-(hydrophobic)-XI. Despite its broad consensus sequence, S1P exhibits exquisite substrate specificity and is involved in proteolytic processing of a defined set of cellular proteins including brain derived neurotrophic factor (BDNF) precursor protein (Seidah et al., 1999) and the sterol regulatory element-binding proteins (SREBP-1 and SREBP-2), which are membrane-associated transcription factors that regulate genes involved in lipid metabolism and cholesterol synthesis (Brown and Goldstein, 1997, Goldstein et al., 2002, Rawson, 2003a, Rawson, 2003b). S1P contributes also to the regulation of ATF6 and other transcription factors controlling the unfolded protein response (UPR) of the cell to bring the folding capacity of the ER in line with the folding demand (Ye et al., 2000, Schroder and Kaufman, 2005, Zhang and Kaufman, 2006, Zhang et al., 2006).
S1P-mediated processing of the GPs of OW arenaviruses LCMV and LASV has been shown to be crucial for the production of infectious progeny and for cell-to-cell propagation (Lenz et al., 2001, Beyer et al., 2003, Kunz et al., 2003, Pinschewer et al., 2003b). Notably, studies on LCMV and JUNV infection in cells deficient in S1P have indicated that the emergence of variants capable of growing independent of S1P-mediated processing of GPC appears to be highly unlikely (Kunz and de la Torre, unpublished). These findings strongly support the view that inhibitors of S1P protease represent promising drug candidates. It is worth noting that S1P also mediates processing of the major GP precursor of Crimean-Congo hemorrhagic fever virus (CCHFV), a member of the Nairovirus genus with the family Bunyaviridae (Sanchez et al., 2002, Vincent et al., 2003). Therefore, compounds exerting anti-arenaviral activity by inhibiting the processing of the virus GPC precursor would be likely to be effective also against CCHFV, a tick-borne pathogen that causes HF in humans throughout regions of Africa, Asia and Europe.
The key role of S1P in the regulation of lipid metabolism and cholesterol biosynthesis has raised considerable interest in developing specific inhibitors of S1P activity. The non-peptide general serine protease inhibitor aminoethyl benzenesulfonyl fluoride (AEBSF) was reported to inhibit S1P (Basak et al., 2004), but other studies questioned this finding (Elagoz et al., 2002, Bodvard et al., 2007). S1P was not blocked by specific inhibitors of other proprotein convertases, but 3,4-dichloroisocoumarin (DCI) at low nanomolar concentrations efficiently inhibited S1P (Bodvard et al., 2007). Likewise, mutants of the preprosegment of S1P and variants of the serpin α1-antitrypsin that were designed to react with S1P by insertion of reactive loop sequences have been also evaluated as candidate protein-based inhibitors (Pullikotil et al., 2004). These studies identified several molecules that significantly blocked S1P-mediated processing of SREBP-1 and ATF6 and the GP of CCHF virus when expressed in cells providing proof-of-principle.
More recent efforts that have focused on the development of peptide-based irreversible S1P inhibitors have identified decanoylated chloromethylketone (CMK)-derivatized peptides containing the RRLL recognition sequence as potent suicide inhibitors of S1P (Pasquato et al., 2006). These inhibitors also blocked the activity of the furin in vitro, but showed specificity towards S1P when used in cells at <100 μM. Because they cause irreversible inhibition of the catalytic activity of S1P against all targets, whether host cell or pathogen-derived, their use would pose difficulties for a therapeutic approach that aims at specific inhibition of S1P-mediated processing of viral GPs without affecting S1P's activity towards its cellular substrates. Consequently, strategies aimed at targeting S1P-mediated processing of the arenavirus GPC should pursue the identification of compounds that specifically block processing of arenavirus GPCs.
Many viruses, including Japanese encephalitis (Su et al., 2002), murine retroviruses (Dimcheff et al., 2003, Dimcheff et al., 2004), hepatitis C (Pavio et al., 2003), paramyxoviruses (Sun et al., 2004), human cytomegalovirus (Isler et al., 2005), SARS corona virus (Versteeg et al., 2007), and West Nile virus (Medigeshi et al., 2007) induce the UPR. Expression of the viral envelope GP in the ER is often a major trigger for the UPR, due to an overwhelming of the folding capacities of the ER resulting in the accumulation of unfolded viral GP. Some aspects of the UPR, such as an increase in the ER's folding capacity and metabolic adaptations, are likely favorable for viral infection, while others, such as the suppression of protein translation and increased ER protein degradation, appear to be disadvantageous. Non-cytolyic viruses, such as arenaviruses, that readily persist may have evolved mechanisms to differentially modulate the different branches of the UPR to favor viral replication and gene expression maintaining aspects of the UPR that are beneficial for the host cell. Arenaviruses may have evolved to use S1P instead of other proteases of the proprotein convertase family commonly used for processing of viral GPs, e.g. furin, because the interaction of GP with S1P allows the virus to manipulate the host cell's UPR. This feature could pose additional barriers to the emergence of viral variants capable of using an alternative protease for the processing of GPC, which would enhance the antiviral power of inhibitors of S1P.
6.5. Targeting arenavirus budding
As with other bona fide viral budding proteins, Z-mediated budding of arenaviruses requires the interaction of Z with specific cellular factors within the endosomal/MVB pathway (Pornillos et al., 2002). The identification and characterization of LFV Z-host protein interactions involved in virus budding may uncover novel antiviral targets and facilitate the development of screening strategies to identify drugs capable of disrupting budding and preventing virus propagation. In this regard, we have already identified TSG101 and CHMP3, members respectively of the ESCRT-I and ESCRT-III complexes, as Z-interacting cellular proteins required for budding. Notably, down-regulation of TSG101 via siRNA (Perez et al., 2003) or the use of dominant-negative forms of CHMP3 (Perez and de la Torre, unpublished) resulted in dramatically reduced levels of Z-mediated budding.
The ability of Z to direct self-budding in the absence of other viral proteins should facilitate the development of assays amenable to both genetics and chemical HTS to identify host cellular proteins required for Z-mediated budding, as well as small-molecule inhibitors of this process. To this end, the emergence of RNA interference as a pathway that allows the modulation of gene expression has enabled functional genetic screens in mammalian cell types (Aza-Blanc et al., 2003, Berns et al., 2004, Paddison et al., 2004). Likewise, combinatorial chemical libraries have emerged as a leading source of compounds, which can be screened to identify small-molecule inhibitors of Z-mediated budding, using cell-based assays.
Methods currently used to assess budding mediated by wild-type or mutant viral proteins rely mainly on the recovery by high-speed ultracentrifugation or immunoprecipitation of VLPs present in culture supernatants of cells previously transfected with a plasmid expressing the protein of interest. The presence of the corresponding protein in the recovered VLPs is detected based on the use of radiolabeled proteins or by Western blot using appropriate antibodies. These methods are not easily amenable to HTS to identify inhibitors of Z-mediated arenavirus budding. An ideal HTS assay would permit assessment of Z budding by examining tissue culture supernatants directly, such as with an enzymatic assay that could identify cellular mediators of Z budding.
Recent work with Ebola virus VP40 described a luciferase-based budding assay that might be suitable for the development of HTS (McCarthy et al., 2006). This assay, however, had some significant limitations: it measured the incorporation of firefly luciferase (FFL) into VP40 VLPs, and thereby did not directly measure the budding capability of VP40, and it required ultracentrifugation of the VLPs for detection of budding. These results, however, raised the possibility of using chimeric proteins consisting of the bona fide viral budding protein fused to an appropriate reporter gene such as FFL as tools to develop assays in which budding could be rapidly and quantitatively assessed by measuring levels of gene reporter activity. In this regard, recent findings have shown that the fusion of the smaller (185 amino acids) luciferase from Gaussia princeps (GLuc) (Tannous et al., 2005) to Z resulted in a chimeric protein (Z-GLuc) that retain wild-type Z budding activity that could be monitored by direct measuring of GLuc activity in TCS of Z-GLuc transfected cells (A.A. Capul and J.C. de la Torre, manuscript submitted). Initial studies have shown that this Z-GLuc based budding assay consistently exhibits a high (average 10-fold) signal-to-noise ratio, as described (Ghosh et al., 2005), suggesting that it should be amenable for the development of HTS to identify small-molecule inhibitors of Z-mediated budding.
7. Generation of recombinant arenaviruses for the development of HTS to identify novel anti-arenaviral drugs
The development of HTS to simultaneously screen a broad class of compounds against the functions of multiple viral targets required for the completion of the arenavirus life cycle would be of great value for the identification of novel anti-arenaviral drugs. To this aim the use of rLCMV expressing appropriate reporter genes should facilitate the development of assays amenable to HTS formats.
A variety of recombinant LCM viruses (rLCMV) have been rescued entirely from cloned cDNAs (Pinschewer et al., 2003b, Flatz et al., 2006, Sanchez and de la Torre, 2006). In contrast, the rescue of rLCMV expressing additional heterologous gene products posed unexpected difficulties. Several strategies successfully employed with other NS RNA viruses to express additional gene products did not work in the case of LCMV (S. Emonet and J.C. de la Torre, unpublished). These included: (1) incorporation into the virus genome of an additional transcription unit containing the ORF of interest; (2) use of a viral or cellular internal ribosome entry site (IRES) to direct cap-independent synthesis of a protein coded by an ORF located downstream within a polycistronic mRNA; and (3) use of the self-cleaving 2A protease of picornaviruses for efficient processing of an initially synthesized polypeptide to generate independently the proteins coded by the ORFs located up- and downstream of the 2 A. Recent findings, however, have supported the feasibility of rescuing a rLCMV containing three genome segments: 1 L and 2 S (S. Emonet and J.C. de la Torre, unpublished). This has opened the possibility of rescuing a three-segment rLCMV, in which each of the two S segments contains a gene of interest (GOI) instead of one of either GPC or NP. The rationale behind this approach is that the physical separation of GP and NP into two different S segments would represent a strong selective pressure to select and maintain a virus capable of packaging 1 L and 2 S segments (Fig. 5 ). This strategy has been used to generate a variety of tri-segmented (2 S + 1 L) rLCMV, where in each of the S segments one of the two virus genes (GPC or NP) was replaced by a gene of interest, including reporter genes such as CAT or luciferase. Notably, the genotype, growth properties and reporter gene expression of these rLCMV were stably maintained during serial passage. This new development provides investigators with a fantastic tool for the development of HTS assays to globally identify inhibitors of arenavirus multiplication.
Fig. 5.
Use of a tri-segments genome strategy to incorporate additional genes within the arenavirus genome. Each of the two S segments was altered to replace one of the viral ORF by the ORF of a gene of interest (GOI). The physical separation of GP and NP into two different S segments would represent a strong selective pressure to select and maintain a virus capable of packaging 1 L and 2 S segments.
8. Perspectives
The findings described in the previous sections have illustrated the potential of using arenavirus MG rescue systems as platforms for drug screening. MG cell-based assays offer a number of benefits in the discovery and analysis of antiviral compounds. They permit the effect of an antiviral compound to be observed in the context of living cells, so that any compounds that show antiviral activity necessarily are able to enter and act within living cells. They also allow the immediate identification of compounds with undesirable cytotoxicity, using well-established assays. Because viral functions related to infectivity are not required for MG RNA replication and expression, these systems are safer and easier to work with than infectious pathogenic arenaviruses. It is worth noting that the implementation of siRNA-based screens in the context of cell-based MG rescue assays can identify host cell proteins that play key roles in arenavirus RNA replication and gene expression and thereby open potential novel targets that could be used in the development of effective anti-arenaviral drugs. The ability to generate rLCMV with predetermined genetic modifications would also facilitate studies aimed at identifying mechanisms of antiviral drug action and the possible generation and selection of drug-resistant viral variants.
The development of reverse genetics systems for several arenaviruses has also opened new research avenues to study the biology of this virus family, which contains several members highly relevant to human health. In addition, the prototypic arenavirus LCMV remains a very robust model system to study virus–host interactions in the context of infection of the natural host, the mouse. The ability to manipulate the LCMV genome and generate rLCMV with predetermined mutations allows investigators to gain a detailed understanding of the roles played by different viral genes in specific phenotypic outcomes, ranging from acute fatal meningitis to immunosuppression and chronic infections associated with neurobehavioral abnormalities.
Acknowledgements
Work from the author's laboratory was supported by NIH grants AI47140 and AI-065359 to JCT.
References
- Andrei G., De Clercq E. Inhibitory effect of selected antiviral compounds on arenavirus replication in vitro. Antiviral Res. 1990;14:287–299. doi: 10.1016/0166-3542(90)90009-v. [DOI] [PubMed] [Google Scholar]
- Andrei G., De Clercq E. Molecular approaches for the treatment of hemorrhagic fever virus infections. Antiviral Res. 1993;22:45–75. doi: 10.1016/0166-3542(93)90085-w. [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P., Cooper C.L., Wagner K., Batalov S., Deveraux Q.L., Cooke M.P. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell. 2003;12:627–637. doi: 10.1016/s1097-2765(03)00348-4. [DOI] [PubMed] [Google Scholar]
- Barton L.L., Mets M.B. Lymphocytic choriomeningitis virus: pediatric pathogen and fetal teratogen. Pediatr. Infect. Dis. J. 1999;18:540–541. doi: 10.1097/00006454-199906000-00013. [DOI] [PubMed] [Google Scholar]
- Barton L.L., Mets M.B. Congenital lymphocytic choriomeningitis virus infection: decade of rediscovery. Clin. Infect. Dis. 2001;33:370–374. doi: 10.1086/321897. [DOI] [PubMed] [Google Scholar]
- Barton L.L., Mets M.B., Beauchamp C.L. Lymphocytic choriomeningitis virus: emerging fetal teratogen. Am. J. Obstet. Gynecol. 2002;187:1715–1716. doi: 10.1067/mob.2002.126297. [DOI] [PubMed] [Google Scholar]
- Basak S., Stewart N.A., Chretien M., Basak A. Aminoethyl benzenesulfonyl fluoride and its hexapeptide (Ac-VFRSLK) conjugate are both in vitro inhibitors of subtilisin kexin isozyme-1. FEBS Lett. 2004;573:186–194. doi: 10.1016/j.febslet.2004.07.069. [DOI] [PubMed] [Google Scholar]
- Bergthaler A., Gerber N.U., Merkler D., Horvath E., de la Torre J.C., Pinschewer D.D. Envelope exchange for the generation of live-attenuated arenavirus vaccines. PLoS Pathog. 2006;2:e51. doi: 10.1371/journal.ppat.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K., Hijmans E.M., Mullenders J., Brummelkamp T.R., Velds A., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., Agami R., Ge W., Cavet G., Linsley P.S., Beijersbergen R.L., Bernards R. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Beyer W.R., Popplau D., Garten W., von Laer D., Lenz O. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J. Virol. 2003;77:2866–2872. doi: 10.1128/JVI.77.5.2866-2872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodvard K., Mohlin J., Knecht W. Recombinant expression, purification, and kinetic and inhibitor characterisation of human site-1-protease. Protein Exp. Purif. 2007;51:308–319. doi: 10.1016/j.pep.2006.07.015. (Epub 2006 August 2001) [DOI] [PubMed] [Google Scholar]
- Boger D.L. Solution-phase synthesis of combinatorial libraries designed to modulate protein–protein or protein-DNA interactions. Bioorg. Med. Chem. 2003;11:1607–1613. doi: 10.1016/s0968-0896(03)00031-2. [DOI] [PubMed] [Google Scholar]
- Boger D.L., Desharnais J., Capps K. Solution-phase combinatorial libraries: modulating cellular signaling by targeting protein–protein or protein–DNA interactions. Angew. Chem. Int. Ed. Engl. 2003;42:4138–4176. doi: 10.1002/anie.200300574. [DOI] [PubMed] [Google Scholar]
- Boger D.L., Ducray P., Chai W., Jiang W., Goldberg J. Higher order iminodiacetic acid libraries for probing protein–protein interactions. Bioorg. Med. Chem. Lett. 1998;8:2339–2344. doi: 10.1016/s0960-894x(98)00423-5. [DOI] [PubMed] [Google Scholar]
- Bolken T.C., Laquerre S., Zhang Y., Bailey T.R., Pevear D.C., Kickner S.S., Sperzel L.E., Jones K.F., Warren T.K., Amanda Lund S., Kirkwood-Watts D.L., King D.S., Shurtleff A.C., Guttieri M.C., Deng Y., Bleam M., Hruby D.E. Identification and characterization of potent small molecule inhibitor of hemorrhagic fever New World arenaviruses. Antiviral Res. 2005 doi: 10.1016/j.antiviral.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borio L., Inglesby T., Peters C.J., Schmaljohn A.L., Hughes J.M., Jahrling P.B., Ksiazek T., Johnson K.M., Meyerhoff A., O’Toole T., Ascher M.S., Bartlett J., Breman J.G., Eitzen E.M., Jr., Hamburg M., Hauer J., Henderson D.A., Johnson R.T., Kwik G., Layton M., Lillibridge S., Nabel G.J., Osterholm M.T., Perl T.M., Russell P., Tonat K. Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA. 2002;287:2391–2405. doi: 10.1001/jama.287.18.2391. [DOI] [PubMed] [Google Scholar]
- Brown M.S., Goldstein J.L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Buchmeier M.J., Peters C.J., de la Torre J.C. Arenaviridae: the viruses and their replication. In: Knipe D.M., Holey P.M., editors. fifth ed. vol. 2. 2007. pp. 1792–1827. (Fields Virology). [Google Scholar]
- Cameron C.E., Castro C. The mechanism of action of ribavirin: lethal mutagenesis of RNA virus genomes mediated by the viral RNA-dependent RNA polymerase. Curr. Opin. Infect. Dis. 2001;14:757–764. doi: 10.1097/00001432-200112000-00015. [DOI] [PubMed] [Google Scholar]
- Candurra N.A., Maskin L., Damonte E.B. Inhibition of arenavirus multiplication in vitro by phenotiazines. Antiviral Res. 1996;31:149–158. doi: 10.1016/0166-3542(96)06956-2. [DOI] [PubMed] [Google Scholar]
- Capul A.A., Perez M., Burke E., Kunz S., Buchmeier M.J., de la Torre J.C. Arenavirus Z-glycoprotein association requires Z myristoylation but not functional RING or late domains. J. Virol. 2007;81:9451–9460. doi: 10.1128/JVI.00499-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann K.K. Reverse genetics of mononegavirales. Kawaoka Y., editor. Curr. Top. Microbiol. Immunol. 2004;283:1–41. doi: 10.1007/978-3-662-06099-5_1. [DOI] [PubMed] [Google Scholar]
- Cordo S.M., Candurra N.A., Damonte E.B. Myristic acid analogs are inhibitors of Junin virus replication. Microbes Infect. 1999;1:609–614. doi: 10.1016/s1286-4579(99)80060-4. [DOI] [PubMed] [Google Scholar]
- Cornu T.I., de la Torre J.C. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J. Virol. 2001;75:9415–9426. doi: 10.1128/JVI.75.19.9415-9426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu T.I., de la Torre J.C. Characterization of the arenavirus RING finger Z protein regions required for Z-mediated inhibition of viral RNA synthesis. J. Virol. 2002;76:6678–6688. doi: 10.1128/JVI.76.13.6678-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu T.I., Feldmann H., de la Torre J.C. Cells expressing the RING finger Z protein are resistant to arenavirus infection. J. Virol. 2004;78:2979–2983. doi: 10.1128/JVI.78.6.2979-2983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S., Maag D., Arnold J.J., Zhong W., Lau J.Y.N., Hong Z., Andino R., Cameron C.E. The broad-spectrum antiviral ribonucleotide, ribavirin, is an RNA virus mutagen. Nat. Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- Damonte E.B., Coto C.E. Treatment of arenavirus infections: from basic studies to the challenge of antiviral therapy. Adv. Virus Res. 2002;58:125–155. doi: 10.1016/s0065-3527(02)58004-0. [DOI] [PubMed] [Google Scholar]
- Dimcheff D.E., Askovic S., Baker A.H., Johnson-Fowler C., Portis J.L. Endoplasmic reticulum stress is a determinant of retrovirus-induced spongiform neurodegeneration. J. Virol. 2003;77:12617–12629. doi: 10.1128/JVI.77.23.12617-12629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimcheff D.E., Faasse M.A., McAtee F.J., Portis J.L. Endoplasmic reticulum (ER) stress induced by a neurovirulent mouse retrovirus is associated with prolonged BiP binding and retention of a viral protein in the ER. J. Biol. Chem. 2004;279:33782–33790. doi: 10.1074/jbc.M403304200. (Epub 32004 June 33783) [DOI] [PubMed] [Google Scholar]
- Djavani M., Yin C., Lukashevich I.S., Rodas J., Rai S.K., Salvato M.S. Mucosal immunization with Salmonella typhimurium expressing Lassa virus nucleocapsid protein cross-protects mice from lethal challenge with lymphocytic choriomeningitis virus. J. Hum. Virol. 2001;4:103–108. [PMC free article] [PubMed] [Google Scholar]
- Eichler R., Lenz O., Strecker T., Eickmann M., Klenk H.D., Garten W. Identification of Lassa virus glycoprotein signal peptide as a trans-acting maturation factor. EMBO Rep. 2003;4:1084–1088. doi: 10.1038/sj.embor.7400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler R., Lenz O., Strecker T., Eickmann M., Klenk H.D., Garten W. Lassa virus glycoprotein signal peptide displays a novel topology with an extended endoplasmic reticulum luminal region. J. Biol. Chem. 2004;279:12293–12299. doi: 10.1074/jbc.M312975200. [DOI] [PubMed] [Google Scholar]
- Elagoz A., Benjannet S., Mammarbassi A., Wickham L., Seidah N.G. Biosynthesis and cellular trafficking of the convertase SKI-1/S1P: ectodomain shedding requires SKI-1 activity. J. Biol. Chem. 2002;277:11265–11275. doi: 10.1074/jbc.M109011200. (Epub 12001 December 11226) [DOI] [PubMed] [Google Scholar]
- Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Enria D.A., Barrera Oro J.G. Junin virus vaccines. Curr. Top. Microbiol. Immunol. 2002;263:239–261. doi: 10.1007/978-3-642-56055-2_12. [DOI] [PubMed] [Google Scholar]
- Enria D.A., Briggiler A.M., Sanchez Z. Treatment of Argentine hemorrhagic fever. Antiviral Res. 2008;78:132–139. doi: 10.1016/j.antiviral.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S.A., Graham M.B., Kuehnert M.J., Kotton C.N., Srinivasan A., Marty F.M., Comer J.A., Guarner J., Paddock C.D., DeMeo D.L., Shieh W.J., Erickson B.R., Bandy U., DeMaria A., Jr., Davis J.P., Delmonico F.L., Pavlin B., Likos A., Vincent M.J., Sealy T.K., Goldsmith C.S., Jernigan D.B., Rollin P.E., Packard M.M., Patel M., Rowland C., Helfand R.F., Nichol S.T., Fishman J.A., Ksiazek T., Zaki S.R. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N. Engl. J. Med. 2006;354:2235–2249. doi: 10.1056/NEJMoa053240. [DOI] [PubMed] [Google Scholar]
- Fisher-Hoch S.P., Hutwagner L., Brown B., McCormick J.B. Effective vaccine for Lassa fever. J. Virol. 2000;74:6777–6783. doi: 10.1128/jvi.74.15.6777-6783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatz L., Bergthaler A., de la Torre J.C., Pinschewer D.D. Recovery of an arenavirus entirely from RNA polymerase I/II-driven cDNA. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4663–4668. doi: 10.1073/pnas.0600652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed E.O. Viral late domains. J. Virol. 2002;76:4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman D.O., Woodall J. Emerging infectious diseases and risk to the traveler. Med. Clin. North Am. 1999;83:865–883. [PubMed] [Google Scholar]
- Geisbert T.W., Jahrling P.B. Exotic emerging viral diseases: progress and challenges. Nat. Med. 2004;10:S110–121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- Geisbert T.W., Jones S., Fritz E.A., Shurtleff A.C., Geisbert J.B., Liebscher R., Grolla A., Stroher U., Fernando L., Daddario K.M., Guttieri M.C., Mothe B.R., Larsen T., Hensley L.E., Jahrling P.B., Feldmann H. Development of a new vaccine for the prevention of Lassa fever. PLoS Med. 2005;2:e183. doi: 10.1371/journal.pmed.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R.N., DeBiasio R., Hudson C.C., Ramer E.R., Cowan C.L., Oakley R.H. Quantitative cell-based high-content screening for vasopressin receptor agonists using transfluor technology. J. Biomol. Screen. 2005;10:476–484. doi: 10.1177/1087057105274896. [DOI] [PubMed] [Google Scholar]
- Goldstein J.L., Rawson R.B., Brown M.S. Mutant mammalian cells as tools to delineate the sterol regulatory element-binding protein pathway for feedback regulation of lipid synthesis. Arch. Biochem. Biophys. 2002;397:139–148. doi: 10.1006/abbi.2001.2615. [DOI] [PubMed] [Google Scholar]
- Hass M., Golnitz U., Muller S., Becker-Ziaja B., Gunther S. Replicon system for Lassa virus. J. Virol. 2004;78:13793–13803. doi: 10.1128/JVI.78.24.13793-13803.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass M., Westerkofsky M., Muller S., Becker-Ziaja B., Busch C., Gunther S. Mutational analysis of the Lassa virus promoter. J. Virol. 2006;80:12414–12419. doi: 10.1128/JVI.01374-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann T. Strategies for the design of drugs targeting RNA and RNA–protein complexes. Angew. Chem. Int. Ed. Engl. 2000;39:1890–1904. doi: 10.1002/1521-3773(20000602)39:11<1890::aid-anie1890>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Hermann T., Westhof E. Rational drug design and high-throughput techniques for RNA targets. Comb. Chem. High Throughput Screen. 2000;3:219–234. doi: 10.2174/1386207003331652. [DOI] [PubMed] [Google Scholar]
- Holmes G.P., McCormick J.B., Trock S.C., Chase R.A., Lewis S.M., Mason C.A., Hall P.A., Brammer L.S., Perez-Oronoz G.I., McDonnell M.K. Lassa fever in the United States. Investigation of a case and new guidelines for management. N. Engl. J. Med. 1990;323:1120–1123. doi: 10.1056/NEJM199010183231607. [DOI] [PubMed] [Google Scholar]
- Isaacson M. Viral hemorrhagic fever hazards for travelers in Africa. Clin. Infect. Dis. 2001;33:1707–1712. doi: 10.1086/322620. [DOI] [PubMed] [Google Scholar]
- Isler J.A., Skalet A.H., Alwine J.C. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 2005;79:6890–6899. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahrling P.B., Peters C.J. Lymphocytic choriomeningitis virus. A neglected pathogen of man. Arch. Pathol. Lab. Med. 1992;116:486–488. [PubMed] [Google Scholar]
- Khan S.H., Goba A., Chu M., Roth C., Healing T., Marx A., Fair J., Guttieri M.C., Ferro P., Imes T., Monagin C., Garry R.F., Bausch D.G. New opportunities for field research on the pathogenesis and treatment of Lassa fever. Antiviral Res. 2008;78:103–115. doi: 10.1016/j.antiviral.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Kiley M.P., Lange J.V., Johnson K.M. Protection of rhesus monkeys from Lassa virus by immunisation with closely related arenavirus. Lancet. 1979;2:738. doi: 10.1016/s0140-6736(79)90659-7. [DOI] [PubMed] [Google Scholar]
- Kilgore P.E., Ksiazek T.G., Rollin P.E., Mills J.N., Villagra M.R., Montenegro M.J., Costales M.A., Paredes L.C., Peters C.J. Treatment of Bolivian hemorrhagic fever with intravenous ribavirin. Clin. Infect. Dis. 1997;24:718–722. doi: 10.1093/clind/24.4.718. [DOI] [PubMed] [Google Scholar]
- Kunz S., Borrow P., Oldstone M.B. Receptor structure, binding, and cell entry of arenaviruses. Curr. Top. Microbiol. Immunol. 2002;262:111–137. doi: 10.1007/978-3-642-56029-3_5. [DOI] [PubMed] [Google Scholar]
- Kunz S., Edelmann K.H., de la Torre J.-C., Gorney R., Oldstone M.B.A. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology. 2003;314:168–178. doi: 10.1016/s0042-6822(03)00421-5. [DOI] [PubMed] [Google Scholar]
- Lee A.M., Rojek J.M., Spiropoulou C.F., Gundersen A.T., Jin W., Shaginian A., York J., Nunberg J.H., Boger D.L., Oldstone M.B., Kunz S. Unique small molecule entry inhibitors of hemorrhagic fever arena viruses. J. Biol. Chem. 2008 doi: 10.1074/jbc.M802089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.J., Novella I.S., Teng M.N., Oldstone M.B., de La Torre J.C. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 2000;74:3470–3477. doi: 10.1128/jvi.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.J., Perez M., Pinschewer D.D., de la Torre J.C. Identification of the lymphocytic choriomeningitis virus (LCMV) proteins required to rescue LCMV RNA analogs into LCMV-like particles. J. Virol. 2002;76:6393–6397. doi: 10.1128/JVI.76.12.6393-6397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz O., ter Meulen J., Klenk H.D., Seidah N.G., Garten W. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12701–12705. doi: 10.1073/pnas.221447598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyssen P., De Clercq E., Neyts J. Molecular strategies to inhibit the replication of RNA viruses. Antiviral Res. 2008;78:9–25. doi: 10.1016/j.antiviral.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez N., Franze-Fernandez M.T. A single stem-loop structure in Tacaribe arenavirus intergenic region is essential for transcription termination but is not required for a correct initiation of transcription and replication. Virus Res. 2007;124:237–244. doi: 10.1016/j.virusres.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Lopez N., Jacamo R., Franze-Fernandez M.T. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: z protein is an inhibitor of these processes. J. Virol. 2001;75:12241–12251. doi: 10.1128/JVI.75.24.12241-12251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashevich I.S., Patterson J., Carrion R., Moshkoff D., Ticer A., Zapata J., Brasky K., Geiger R., Hubbard G.B., Bryant J., Salvato M.S. A live attenuated vaccine for Lassa fever made by reassortment of Lassa and Mopeia viruses. J. Virol. 2005;79:13934–13942. doi: 10.1128/JVI.79.22.13934-13942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W., Krystal M., Enami M., Pavin J.D., Palese P. Amplification, expression, and packaging of foreign gene by influenza virus. Cell. 1989;59:1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- AHF Study Group Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. J. Infect. Dis. 1998;177:277–283. doi: 10.1086/514211. [DOI] [PubMed] [Google Scholar]
- Matloubian M., Somasundaram T., Kolhekar S.R., Selvakumar R., Ahmed R. Genetic basis of viral persistence: single amino acid change in the viral glycoprotein affects ability of lymphocytic choriomeningitis virus to persist in adult mice. J. Exp. Med. 1990;172:1043–1048. doi: 10.1084/jem.172.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S.E., Licata J.M., Harty R.N. A luciferase-based budding assay for Ebola virus. J. Virol. Methods. 2006;137:115–119. doi: 10.1016/j.jviromet.2006.06.007. [DOI] [PubMed] [Google Scholar]
- McCormick J.B., Fisher-Hoch S.P. Lassa fever. In: Oldstone M.B., editor. vol. 262. Springer-Verlag; Berlin, Heidelberg, New York: 2002. pp. 75–110. (Arenaviruses I). [Google Scholar]
- McCormick J.B., King I.J., Webb P.A., Scribner C.L., Craven R.B., Johnson K.M., Elliott L.H., Belmont-Williams R. Lassa fever. Effective therapy with ribavirin. N. Engl. J. Med. 1986;314:20–26. doi: 10.1056/NEJM198601023140104. [DOI] [PubMed] [Google Scholar]
- McKee K.T., Jr., Huggins J.W., Trahan C.J., Mahlandt B.G. Ribavirin prophylaxis and therapy for experimental argentine hemorrhagic fever. Antimicrob. Agents Chemother. 1988;32:1304–1309. doi: 10.1128/aac.32.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medigeshi G.R., Lancaster A.M., Hirsch A.J., Briese T., Lipkin W.I., Defilippis V., Fruh K., Mason P.W., Nikolich-Zugich J., Nelson J.A. West Nile virus infection activates the unfolded protein response leading to CHOP induction and apoptosis. J. Virol. 2007;8:8. doi: 10.1128/JVI.01151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mets M.B., Barton L.L., Khan A.S., Ksiazek T.G. Lymphocytic choriomeningitis virus: an underdiagnosed cause of congenital chorioretinitis. Am. J. Ophthalmol. 2000;130:209–215. doi: 10.1016/s0002-9394(00)00570-5. [DOI] [PubMed] [Google Scholar]
- Meyer B.J., de La Torre J.C., Southern P.J. Arenaviruses: genomic RNAs, transcription, and replication. In: Oldstone M.B., editor. vol. 262. Springer-Verlag; Berlin, Heidelberg: 2002. pp. 139–149. (Arenaviruses I). [DOI] [PubMed] [Google Scholar]
- Neuman B.W., Adair B.D., Burns J.W., Milligan R.A., Buchmeier M.J., Yeager M. Complementarity in the supramolecular design of arenaviruses and retroviruses revealed by electron cryomicroscopy and image analysis. J. Virol. 2005;79:3822–3830. doi: 10.1128/JVI.79.6.3822-3830.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G., Whitt M.A., Kawaoka Y. A decade after the generation of a negative-sense RNA virus from cloned cDNA—what have we learned? J. Gen. Virol. 2002;83:2635–2662. doi: 10.1099/0022-1317-83-11-2635. [DOI] [PubMed] [Google Scholar]
- Oldstone M.B. Biology and pathogenesis of lymphocytic choriomeningitis virus infection. In: Oldstone M.B., editor. vol. 263. 2002. pp. 83–118. (Arenaviruses). [DOI] [PubMed] [Google Scholar]
- Paddison P.J., Silva J.M., Conklin D.S., Schlabach M., Li M., Aruleba S., Balija V., O'Shaughnessy A., Gnoj L., Scobie K., Chang K., Westbrook T., Cleary M., Sachidanandam R., McCombie W.R., Elledge S.J., Hannon G.J. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- Parker W.B. Metabolism and antiviral activity of ribavirin. Virus Res. 2005;107:165–171. doi: 10.1016/j.virusres.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Pasquato A., Pullikotil P., Asselin M.C., Vacatello M., Paolillo L., Ghezzo F., Basso F., Di Bello C., Dettin M., Seidah N.G. The proprotein convertase SKI-1/S1P. In vitro analysis of Lassa virus glycoprotein-derived substrates and ex vivo validation of irreversible peptide inhibitors. J. Biol. Chem. 2006;281:23471–23481. doi: 10.1074/jbc.M513675200. (Epub 22006 June 23421) [DOI] [PubMed] [Google Scholar]
- Pavio N., Romano P.R., Graczyk T.M., Feinstone S.M., Taylor D.R. Protein synthesis and endoplasmic reticulum stress can be modulated by the hepatitis C virus envelope protein E2 through the eukaryotic initiation factor 2alpha kinase PERK. J. Virol. 2003;77:3578–3585. doi: 10.1128/JVI.77.6.3578-3585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M., Craven R.C., de la Torre J.C. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12978–12983. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M., de la Torre J.C. Characterization of the Genomic Promoter of the Prototypic Arenavirus Lymphocytic Choriomeningitis Virus (LCMV) J. Virol. 2002;77:1184–1194. doi: 10.1128/JVI.77.2.1184-1194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M., Greenwald D.L., de la Torre J.C. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J. Virol. 2004;78:11443–11448. doi: 10.1128/JVI.78.20.11443-11448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C.J. Human infection with arenaviruses in the Americas. In: Oldstone M.B., editor. vol. 262. Springer-Verlag; Berlin, Heidelberg: 2002. pp. 65–74. (Arenaviruses I). [DOI] [PubMed] [Google Scholar]
- Peters C.J. Lymphocytic choriomeningitis virus–an old enemy up to new tricks. N. Engl. J. Med. 2006;354:2208–2211. doi: 10.1056/NEJMp068021. [DOI] [PubMed] [Google Scholar]
- Pinschewer D.D., Perez M., de la Torre J.C. Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication. J. Virol. 2003;77:3882–3887. doi: 10.1128/JVI.77.6.3882-3887.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinschewer D.D., Perez M., de la Torre J.C. Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J. Virol. 2005;79:4519–4526. doi: 10.1128/JVI.79.7.4519-4526.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinschewer D.D., Perez M., Sanchez A.B., de la Torre J.C. Recombinant lymphocytic choriomeningitis virus expressing vesicular stomatitis virus glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7895–7900. doi: 10.1073/pnas.1332709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O., Garrus J.E., Sundquist W.I. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 2002;12:569–579. doi: 10.1016/s0962-8924(02)02402-9. [DOI] [PubMed] [Google Scholar]
- Pullikotil P., Vincent M., Nichol S.T., Seidah N.G. Development of protein-based inhibitors of the proprotein of convertase SKI-1/S1P: processing of SREBP-2, ATF6, and a viral glycoprotein. J. Biol. Chem. 2004;279:17338–17347. doi: 10.1074/jbc.M313764200. (Epub 12004 February 17316) [DOI] [PubMed] [Google Scholar]
- Pushko P., Geisbert J., Parker M., Jahrling P., Smith J. Individual and bivalent vaccines based on alphavirus replicons protect guinea pigs against infection with Lassa and Ebola viruses. J. Virol. 2001;75:11677–11685. doi: 10.1128/JVI.75.23.11677-11685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson R.B. Control of lipid metabolism by regulated intramembrane proteolysis of sterol regulatory element binding proteins (SREBPs) Biochem. Soc. Symp. 2003:221–231. doi: 10.1042/bss0700221. [DOI] [PubMed] [Google Scholar]
- Rawson R.B. The SREBP pathway—insights from insigs and insects. Nat. Rev. Mol. Cell. Biol. 2003;4:631–640. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- Repik P., Flamand A., Bishop D.H. Synthesis of RNA by mutants of vesicular stomatitis virus (Indiana serotype) and the ability of wild-type VSV New Jersey to complement the VSV Indiana ts G I-114 transcription defect. J. Virol. 1976;20:157–169. doi: 10.1128/jvi.20.1.157-169.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere Y., Ahmed R., Southern P., Oldstone M.B. Perturbation of differentiated functions during viral infection in vivo. II. Viral reassortants map growth hormone defect to the S RNA of the lymphocytic choriomeningitis virus genome. Virology. 1985;142:175–182. doi: 10.1016/0042-6822(85)90431-3. [DOI] [PubMed] [Google Scholar]
- Riviere Y., Ahmed R., Southern P.J., Buchmeier M.J., Oldstone M.B. Genetic mapping of lymphocytic choriomeningitis virus pathogenicity: virulence in guinea pigs is associated with the L RNA segment. J. Virol. 1985;55:704–709. doi: 10.1128/jvi.55.3.704-709.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D.L., Joyce G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Carreno M.P., Nelson M.S., Botten J., Smith-Nixon K., Buchmeier M.J., Whitton J.L. Evaluating the immunogenicity and protective efficacy of a DNA vaccine encoding Lassa virus nucleoprotein. Virology. 2005;335:87–98. doi: 10.1016/j.virol.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Rojek J.M., Lee A.M., Nguyen N., Spiropoulou C.F., Kunz S. Site 1 protease is required for proteolytic processing of the glycoproteins of the South American hemorrhagic fever viruses Junin, Machupo, and Guanarito. J. Virol. 2008;82:6045–6051. doi: 10.1128/JVI.02392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Jarabo C.M., Ly C., Domingo E., de la Torre J.C. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) Virology. 2003;308:37–47. doi: 10.1016/s0042-6822(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Sakai J., Nohturfft A., Goldstein J.L., Brown M.S. Cleavage of sterol regulatory element-binding proteins (SREBPs) at site-1 requires interaction with SREBP cleavage-activating protein. Evidence from in vivo competition studies. J. Biol. Chem. 1998;273:5785–5793. doi: 10.1074/jbc.273.10.5785. [DOI] [PubMed] [Google Scholar]
- Salvato M., Borrow P., Shimomaye E., Oldstone M.B. Molecular basis of viral persistence: a single amino acid change in the glycoprotein of lymphocytic choriomeningitis virus is associated with suppression of the antiviral cytotoxic T-lymphocyte response and establishment of persistence. J. Virol. 1991;65:1863–1869. doi: 10.1128/jvi.65.4.1863-1869.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvato M.S. Molecular biology of the prototype arenavirus, lymphocytic choriomeningitis virus. In: Salvato M.S., editor. vol. 1. Plenum; New York: 1993. pp. 133–156. (The Arenaviridae). [Google Scholar]
- Salvato M.S., Schweighofer K.J., Burns J., Shimomaye E.M. Biochemical and immunological evidence that the 11 kDa zinc-binding protein of lymphocytic choriomeningitis virus is a structural component of the virus. Virus Res. 1992;22:185–198. doi: 10.1016/0168-1702(92)90050-j. [DOI] [PubMed] [Google Scholar]
- Sanchez A.B., de la Torre J.C. Genetic and biochemical evidence for an oligomeric structure of the functional L polymerase of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 2005;79:7262–7268. doi: 10.1128/JVI.79.11.7262-7268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A.B., de la Torre J.C. Rescue of the prototypic Arenavirus LCMV entirely from plasmid. Virology. 2006;350:370–380. doi: 10.1016/j.virol.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Sanchez A.B., Perez M., Cornu T., de la Torre J.C. RNA interference-mediated virus clearance from cells both acutely and chronically infected with the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 2005;79:11071–11081. doi: 10.1128/JVI.79.17.11071-11081.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A.J., Vincent M.J., Nichol S.T. Characterization of the glycoproteins of Crimean-Congo hemorrhagic fever virus. J. Virol. 2002;76:7263–7275. doi: 10.1128/JVI.76.14.7263-7275.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M., Kaufman R.J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Sears P., Wong C.H. Carbohydrate mimetics: a new strategy for tackling the problem of carbohydrate-mediated biological recognition. Angew. Chem. Int. Ed. Engl. 1999;38:2300–2324. (pii) [PubMed] [Google Scholar]
- Sears P., Wong C.H. Toward automated synthesis of oligosaccharides and glycoproteins. Science. 2001;291:2344–2350. doi: 10.1126/science.1058899. [DOI] [PubMed] [Google Scholar]
- Seidah N.G., Mowla S.J., Hamelin J., Mamarbachi A.M., Benjannet S., Toure B.B., Basak A., Munzer J.S., Marcinkiewicz J., Zhong M., Barale J.C., Lazure C., Murphy R.A., Chretien M., Marcinkiewicz M. Mammalian subtilisin/kexin isozyme SKI-1: a widely expressed proprotein convertase with a unique cleavage specificity and cellular localization. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1321–1326. doi: 10.1073/pnas.96.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood S., Cevik B., Moyer S.A. Intragenic complementation and oligomerization of the L subunit of the sendai virus RNA polymerase. Virology. 2002;304:235–245. doi: 10.1006/viro.2002.1720. [DOI] [PubMed] [Google Scholar]
- Smallwood S., Moyer S.A. The L polymerase protein of parainfluenza virus 3 forms an oligomer and can interact with the heterologous Sendai virus L, P and C proteins. Virology. 2004;318:439–450. doi: 10.1016/j.virol.2003.09.045. [DOI] [PubMed] [Google Scholar]
- Strecker T., Eichler R., Meulen J., Weissenhorn W., Dieter Klenk H., Garten W., Lenz O. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles [corrected] J. Virol. 2003;77:10700–10705. doi: 10.1128/JVI.77.19.10700-10705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H.L., Liao C.L., Lin Y.L. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J. Virol. 2002;76:4162–4171. doi: 10.1128/JVI.76.9.4162-4171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucheck S.J., Greenberg W.A., Tolbert T.J., Wong C.H. Design of small molecules that recognize RNA: development of aminoglycosides as potential antitumor agents that target oncogenic RNA sequences this work was supported by the NIH. We thank Professor Peter Voght for his suggestion of the oncogenic RNA sequences as targets. Angew. Chem. Int. Ed. Engl. 2000;39:1080–1084. doi: 10.1002/(sici)1521-3773(20000317)39:6<1080::aid-anie1080>3.0.co;2-b. (pii) [DOI] [PubMed] [Google Scholar]
- Sucheck S.J., Shue Y.K. Combinatorial synthesis of aminoglycoside libraries. Curr. Opin. Drug Discov. Dev. 2001;4:462–470. [PubMed] [Google Scholar]
- Sucheck S.J., Wong C.H. RNA as a target for small molecules. Curr. Opin. Chem. Biol. 2000;4:678–686. doi: 10.1016/s1367-5931(00)00142-3. [DOI] [PubMed] [Google Scholar]
- Sun M., Rothermel T.A., Shuman L., Aligo J.A., Xu S., Lin Y., Lamb R.A., He B. Conserved cysteine-rich domain of paramyxovirus simian virus 5 V protein plays an important role in blocking apoptosis. J. Virol. 2004;78:5068–5078. doi: 10.1128/JVI.78.10.5068-5078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous B.A., Kim D.E., Fernandez J.L., Weissleder R., Breakefield X.O. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Tordo N., De Haan P., Goldbach R., Poch O. Evolution of negative-stranded RNA genomes. Semin. Virol. 1992;3:341–357. [Google Scholar]
- Tortorici M.A., Albarino C.G., Posik D.M., Ghiringhelli P.D., Lozano M.E., Rivera Pomar R., Romanowski V. Arenavirus nucleocapsid protein displays a transcriptional antitermination activity in vivo. Virus Res. 2001;73:41–55. doi: 10.1016/s0168-1702(00)00222-7. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Urata S., Noda T., Kawaoka Y., Yokosawa H., Yasuda J. Cellular factors required for Lassa virus budding. J. Virol. 2006;80:4191–4195. doi: 10.1128/JVI.80.8.4191-4195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg G.A., van de Nes P.S., Bredenbeek P.J., Spaan W.J. The coronavirus spike protein induces ER stress and upregulation of intracellular chemokine mRNA concentrations. J. Virol. 2007;1:1. doi: 10.1128/JVI.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M.J., Sanchez A.J., Erickson B.R., Basak A., Chretien M., Seidah N.G., Nichol S.T. Crimean-Congo hemorrhagic fever virus glycoprotein proteolytic processing by subtilase SKI-1. J. Virol. 2003;77:8640–8649. doi: 10.1128/JVI.77.16.8640-8649.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsman M.B., Lopez E.M., Ramirez J.A., Galagovsky L.R., Coto C.E. Antiviral effect of brassinosteroids against herpes virus and arenaviruses. Antivir. Chem. Chemother. 2000;11:71–77. doi: 10.1177/095632020001100107. [DOI] [PubMed] [Google Scholar]
- Wang S., Huber P.W., Cui M., Czarnik A.W., Mei H.Y. Binding of neomycin to the TAR element of HIV-1 RNA induces dissociation of Tat protein by an allosteric mechanism. Biochemistry. 1998;37:5549–5557. doi: 10.1021/bi972808a. [DOI] [PubMed] [Google Scholar]
- Wells J.A., de Vos A.M. Hematopoietic receptor complexes. Annu. Rev. Biochem. 1996;65:609–634. doi: 10.1146/annurev.bi.65.070196.003141. [DOI] [PubMed] [Google Scholar]
- Wilson W.D., Li K. Targeting RNA with small molecules. Curr. Med. Chem. 2000;7:73–98. doi: 10.2174/0929867003375434. [DOI] [PubMed] [Google Scholar]
- Yarnell W.S., Roberts J.W. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- Ye J., Rawson R.B., Komuro R., Chen X., Dave U.P., Prywes R., Brown M.S., Goldstein J.L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Ye X.-S., Zhang L.H. Aminoglycoside mimetics as small-molecule drugs targeting RNA. Curr. Med. Chem. 2002;9:929–939. doi: 10.2174/0929867024606740. [DOI] [PubMed] [Google Scholar]
- York J., Romanowski V., Lu M., Nunberg J.H. The signal peptide of the Junin arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1-G2 complex. J. Virol. 2004;78:10783–10792. doi: 10.1128/JVI.78.19.10783-10792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapp M.L., Stern S., Green M.R. Small molecules that selectively block RNA binding of HIV-1 Rev protein inhibit Rev function and viral production. Cell. 1993;74:969–978. doi: 10.1016/0092-8674(93)90720-b. [DOI] [PubMed] [Google Scholar]
- Zhang K., Kaufman R.J. The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology. 2006;66:S102–109. doi: 10.1212/01.wnl.0000192306.98198.ec. [DOI] [PubMed] [Google Scholar]
- Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D.T., Back S.H., Kaufman R.J. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- Zinkernagel R.M. Lymphocytic choriomeningitis virus and immunology. Curr. Top. Microbiol. Immunol. 2002;263:1–5. doi: 10.1007/978-3-642-56055-2_1. [DOI] [PubMed] [Google Scholar]


