SUMMARY
The fungus Cryptococcus neoformans is a leading cause of mortality and morbidity among HIV-infected individuals. We utilized the completed genome sequence and optimized methods for homologous DNA replacement using high-velocity particle bombardment to engineer 1,201 gene knockout mutants. We screened this resource in vivo for proliferation in murine lung tissue and in vitro for three well-recognized virulence attributes — polysaccharide capsule formation, melanization, and growth at body temperature. We identified dozens of previously uncharacterized genes that affect these known attributes as well as 40 infectivity mutants without obvious defects in these traits. The latter mutants affect predicted regulatory factors, secreted proteins, and immune-related factors, and represent powerful tools for elucidating novel virulence mechanisms. In particular, we describe a GATA family transcription factor that inhibits phagocytosis by murine macrophages independently of the capsule, indicating a previously unknown mechanism of innate immune modulation.
INTRODUCTION
Systematic profiling of gene deletion strain collections has been essential for the comprehensive study of biological processes in the nonpathogenic yeast Saccharomyces cerevisiae (Bader et al., 2003). Likewise, forward genetic screens of large mutant collections of bacterial pathogens have been extremely fruitful in identifying virulence pathways in prokaryotic organisms (Saenz and Dehio, 2005). In contrast, the study of fungal pathogenesis has historically been hindered by a relative lack of genetic tools and techniques in these eukaryotic species. With the release of complete genome sequences for numerous fungal pathogens (Jones et al., 2004; Loftus et al., 2005; Nierman et al., 2005), there are now unprecedented opportunities to create genome-scale resources and to apply systematic methods to the study of fungal virulence.
Cryptococcus neoformans is the most common cause of systemic fungal infections in people infected with HIV (Bicanic and Harrison, 2004). Whereas primary pulmonary infections are typically mildly symptomatic and self-resolving in patients with intact immune systems, these infections commonly disseminate through the bloodstream to infect the meninges in HIV-positive patients. In sub-Saharan Africa and Southeast Asia, where over 80% of the world’s HIV-infected population lives, studies indicate that 10–40% of HIV-related deaths are caused by cryptococcal meningitis (Corbett et al., 2002; French et al., 2002). The high cost and limited availability of antifungal drugs often prohibit effective medical treatment in these areas (Warnock, 2006) and mortality rates in parts of sub-Saharan African approach 100% (Bekondi et al., 2006).
There are three well-established mediators of C. neoformans virulence: a polysaccharide capsule, cell wall-associated melanin, and the ability to grow at human body temperature. The capsule has been shown to have both anti-phagocytic and immunomodulatory properties (Del Poeta, 2004; Monari et al., 2006). Melanin is believed to protect the fungus from a broad range of toxic insults, both in the environment and the host (Nosanchuk and Casadevall, 2003).
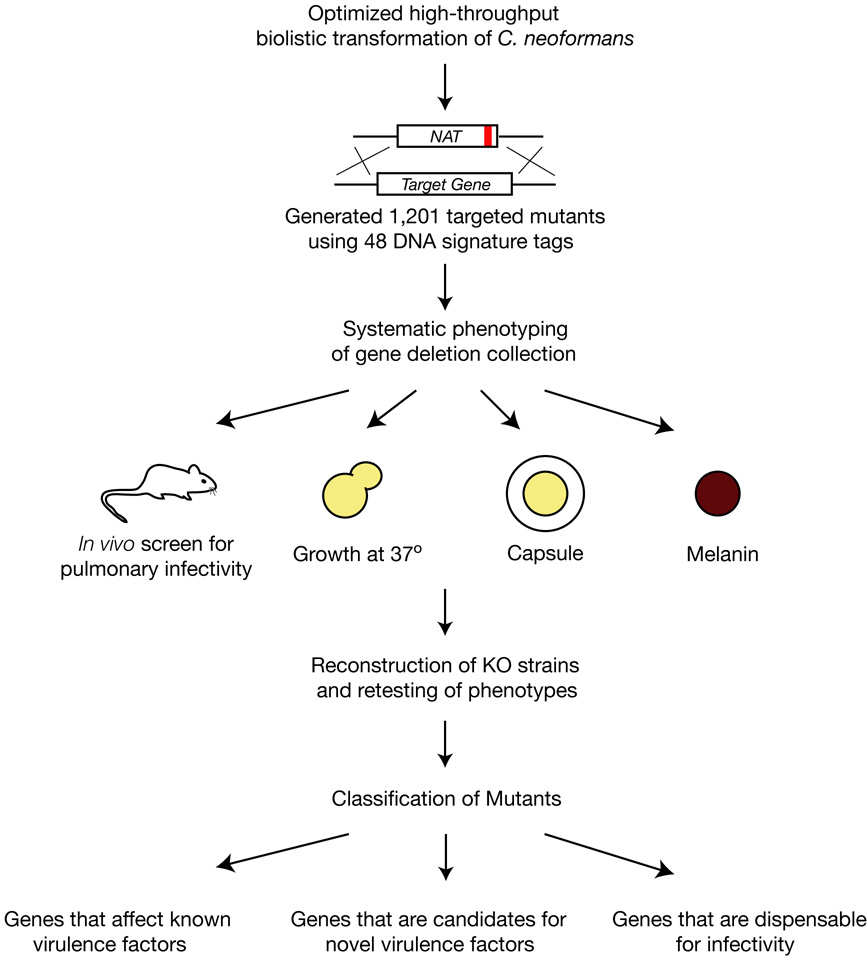
Molecular studies of C. neoformans virulence pathways have focused almost exclusively on the regulation of capsule and melanization (Table S1). Attempts to identify novel virulence mechanisms using random insertional mutagenesis have been hampered by biases in the insertion sites, chromosomal rearrangements, and relatively low ORF hit rates (Idnurm et al., 2004; Nelson et al., 2001). The release of the complete C. neoformans genome sequence now makes it possible to use directed gene knockout mutants to screen for novel virulence mechanisms. To this end, we constructed a large library of 1,201 signature-tagged, targeted gene deletion strains. Below, we describe insights gained from systematic profiling of this library in a murine infection model and in three in vitro assays for virulence-associated attributes (Figure 1). We also describe detailed analysis of a key transcriptional regulator that has pointed to the existence of an unsuspected capsule-independent mechanism for inhibition of phagocytosis.
Figure 1.
Schematic overview of systematic phenotypic profiling strategy utilized in this work.
RESULTS
Generation of signature-tagged C. neoformans gene deletion collection
Approximately 1,500 genes in C. neoformans were targeted for deletion by homologous recombination. The majority (~900) were selected because of the absence of obvious homologs in the nonpathogenic fungus S. cerevisiae. The remainder encode proteins with sequence motifs implicating them in a broad variety of cellular processes. Because these targets were chosen prior to the availability of detailed gene models and were based on BLAST-based annotations (Chow et al., 2007), there was a bias against genes that are C. neoformans-specific. We utilized fusion PCR to create deletion constructs in a high-throughput manner and optimized biolistic transformation for efficient recombination. For reasons described below, each mutant strain contains one of 48 unique signature tag DNA sequences adjacent to the deletion marker. The resulting set of 1,201 verified mutants represents deletions of 1,180 genes (in some cases, different domains of the same gene were targeted separately). For comparison, the genome of C. neoformans is predicted to contain ~6,500 genes (Loftus et al., 2005), and ~190 targeted gene deletions in C. neoformans have previously been reported in the literature (Table S1). The deletion collection is currently available without restriction through the Fungal Genetic Stock Center (www.fgsc.org) or ATCC (www.atcc.org).
Quantitative screen for infectivity mutants in a murine inhalation model of disease
To identify C. neoformans genes involved in infectivity, we adopted signature-tagged mutagenesis (STM), a strategy that enables high-throughput, parallel analysis of pathogen mutants in experimental infections (Saenz and Dehio, 2005). In this method, individual mutants are marked in their genomes with a unique DNA sequence (termed a signature tag or a barcode). This allows a pool of many mutants to be tested in a single animal infection by comparing the relative representation of each mutant in the pool before and after selection in the host. In practice, this is accomplished by isolating DNA from mutant pathogen pools before and after infection and quantifying the abundance of each tag in the sample.
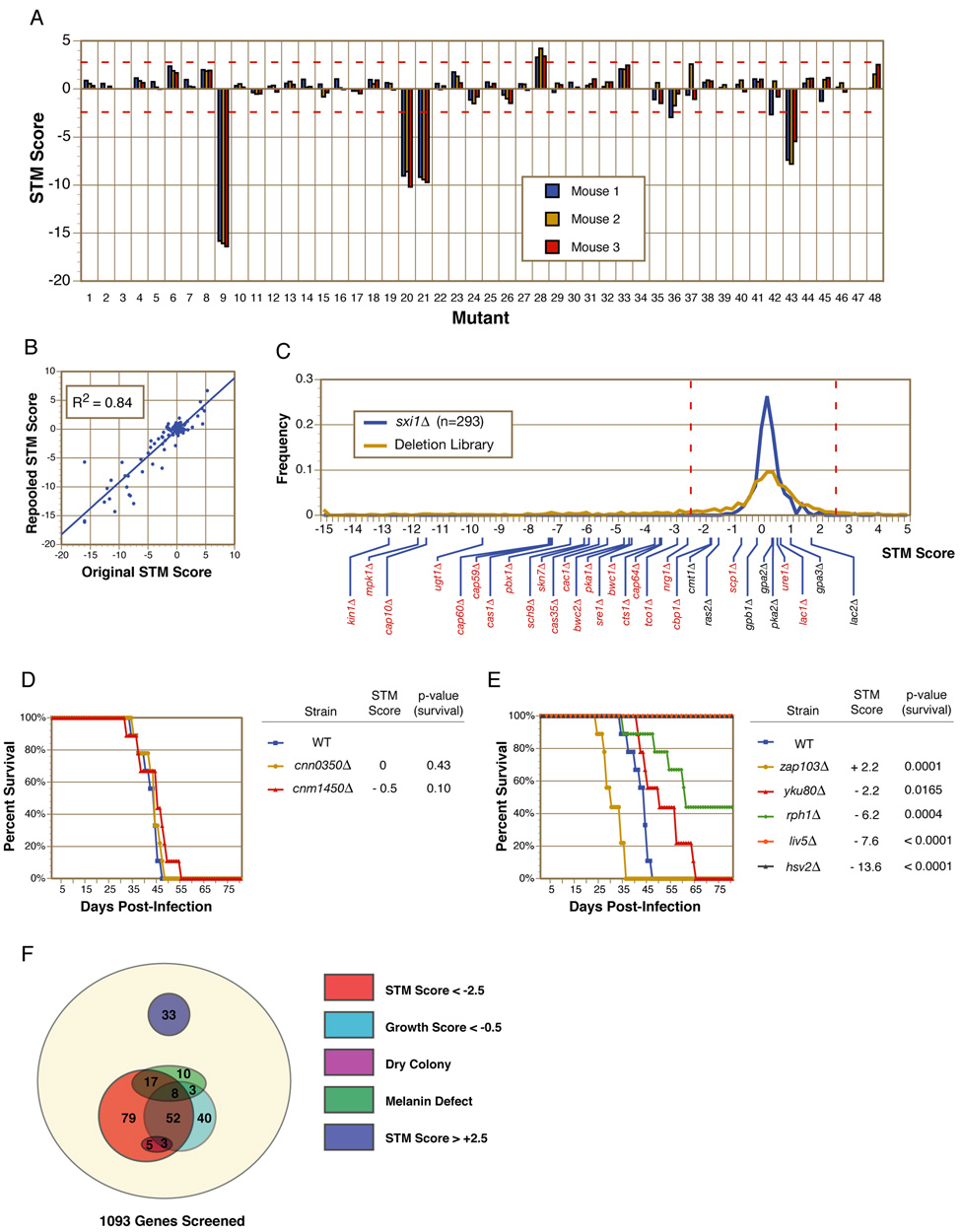
We developed a robust, quantitative STM assay utilizing a previously established mouse model of pulmonary cryptococcal infection (Cox et al., 2000). C. neoformans deletion mutants were grown independently and then pooled into groups of 48 for intranasal infection of mice. Infections were allowed to progress until the mice displayed signs of significant morbidity (~5–6 weeks), at which time the mice were sacrificed and C. neoformans cells were recovered from the lung tissue. Quantitative PCR using tag-specific primers together with a common primer in the gene deletion cassette was used to measure the relative abundance of each mutant in both the input and recovered pools. The ratio of these two values ([recovered] / [input]) was calculated on a log2 scale and termed the STM score (see Experimental Procedures). As shown in Figure 2A, STM scores demonstrated remarkable consistency among the three mice infected with each pool of signature-tagged mutants. In total, 1,100 deletion mutants were screened using the STM assay.
Figure 2. In vivo signature-tagged mutagenesis (STM) analysis of gene deletions.
(A) Mutants were pooled in groups of 48 and used to intranasally infect three mice. Quantitative PCR with signature tag-specific primers was used to calculate an STM score for each mutant. Red-dotted lines indicate cutoffs used to identify STM phenotypes of interest. (B) 104 mutants assayed by STM were sorted into new pools containing different competitors and retested. Original STM scores show a strong correlation with repooled STM scores. (C) The frequencies of STM scores observed in this screen were plotted for both the deletion collection and the sxi1Δ control strains. Red-dotted lines indicate cutoffs used to identify STM phenotypes of interest. Indicated below are the STM scores of gene knockouts in the collection previously reported to have (red) or not to have (black) virulence defects. (D–E) Survival curve analyses of mice infected with individual strains demonstrate the correlation between STM phenotypes and virulence phenotypes. Both hypovirulent and hypervirulent strains were accurately identified by STM. P-values indicate the significance of the virulence phenotype. (F) Shown is the distribution of in vivo and in vitro phenotypes in the deletion collection. Significant overlaps were seen between mutants with defects in the three virulence attributes and mutants with in vivo infectivity defects.
We next determined if the STM score for a given mutant was affected by the composition of mutants in that pool. 104 mutants were removed from their original pools and sorted into new pools containing different mutants. Infections were repeated and STM scores were recalculated for each mutant. The original STM scores were highly correlated with the re-pooled STM scores (R2=0.84), demonstrating that the STM scores were largely independent of the identities of the competing mutants and were highly reproducible from pool to pool (Figure 2B).
As an additional control for reproducibility of the assay, we constructed 43 independent disruptions of the gene SXI1, which is required for mating but dispensable for virulence (Hull et al., 2004); each mutant was marked with a different signature tag. The 43 sxi1Δ mutant strains were tested a total of 293 times throughout the STM screen and, as expected, the distribution of STM scores for the sxi1Δ strains was centered near zero (median=0.1) and showed a tight range (SD=0.5) (Figure 2C). The distribution of STM scores for the deletion library as a whole was also centered on zero (median=0.0); however, in this case, the distribution of scores was much broader (SD=3.3), suggesting a wide range of fitness in the assay.
STM phenotypes correlate with virulence
STM screens are designed to measure the relative competitive fitness of mutants in a mixed infection. Thus, the term “infectivity” in this context refers to this ability to proliferate/survive within host tissue. “Virulence,” on the other hand, is classically defined as the ability of an individual strain not only to proliferate, but to cause disease as well. To assess the ability of our STM screen phenotypes to predict virulence defects, we performed two additional tests.
First, we asked whether previously identified virulence mutants included in our deletion collection were detected by the screen. Among 24 previously-reported virulence mutants, 21 displayed STM scores significantly below zero, ranging from −12.8 to −1.9 (labeled on the x-axis of Figure 2C and Table S2). Two published virulence mutants, lac1Δ and ure1Δ, had normal STM scores. These mutants, however, are known to replicate normally in mouse lungs and have late defects in dissemination (Noverr et al., 2004; Olszewski et al., 2004); such mutants would not be anticipated to have defects in our screen. The final virulence mutant, scp1Δ, was retested in our assay using a different pool of mutants, and a significantly lower STM score was detected (−3.6 vs. −0.8). These results demonstrate a robust sensitivity of the STM screen for virulence genes known to affect propagation in pulmonary tissue (21 mutants identified of 22 mutants with known lung defects). Conversely, the sxi1Δ STM data described above did not yield a single negative outlier and all seven mutants in the collection containing deletions of genes known to be dispensable for virulence had normal STM scores suggesting a screen specificity for infectivity defects approaching 100% (Figure 2C, Table S2).
In a second test, we conducted classical time-to-endpoint survival analyses for seven previously-uncharacterized mutants with a wide range of STM scores. In these experiments, virulence was directly measured by determining the rate at which mice infected with a single strain of C. neoformans reach a defined disease endpoint (see Supplementary Materials). Monotypic intranasal infections of 8–10 mice per mutant and a wild-type control were performed and the time to significant morbidity was assessed for each mouse. Survival curves were plotted for the mutant strains and compared to that of the wild-type control. Two mutants with STM scores close to zero were indistinguishable from wild-type in these survival curves (Figure 2D). Four mutants with STM scores significantly below zero were attenuated for virulence (Figure 2E) with the degree of attenuation closely correlated with the magnitude of the STM defect. Interestingly, a mutant with an STM score significantly above zero (zap103Δ) was hypervirulent (Figure 2E). Together, these studies suggest that the STM assay is a sensitive and specific measure for infectivity and that in this pulmonary infection model of cryptococcosis, altered infectivity correlates well with altered virulence.
Based on these results, we chose STM scores of < −2.5 and > +2.5 as the cutoff points for reduced infectivity and increased infectivity, respectively. Application of these criteria to our data yielded 164 (15%) mutants in the former category and 33 (3%) in the latter.
Systematic in vitro phenotyping of the deletion collection identifies mutants defective in known virulence attributes
To complement the in vivo STM screen, we systematically assayed the deletion collection in vitro for three known virulence-associated phenotypes: 1) growth in minimal medium at 37°C, 2) melanization, and 3) synthesis of the polysaccharide capsule.
A high-throughput, 96-well format assay was developed to measure the growth of each mutant in synthetic minimal medium (YNB) at 37°C. As expected, a large overlap existed between mutants that displayed poor growth at 37°C and mutants with STM defects (P=8.2E-30) (Figure 2F). Using hypergeometric distribution analysis, we determined that mutants that displayed growth scores greater than −0.5 did not show a significant enrichment for mutants with virulence phenotypes (P > 0.01) (Figure S1); thus, we used a growth score of −0.5 as a cutoff for determining which mutants were categorized as having significant growth defects. Some mutants displayed severe growth defects but not STM defects. We attributed at least a portion of this inconsistency to the fact that minimal medium does not perfectly mimic the nutritional environment found within the host. For example, mutants in peroxisome function (pex1Δ and pex6Δ) have been shown previously to have normal virulence despite being unable to grow on YNB with glucose as the sole carbon source (Idnurm et al., 2007).
To identify mutants defective in melanin synthesis, we grew the deletion collection on agar plates containing L-DOPA (a diphenolic substrate). Wild-type cells turned dark brown over 2–3 days as they synthesized and accumulated melanin. Thirty-eight deletion mutants displayed decreased levels of melanization compared to wild-type, including known melanin mutants such as lac1Δ, cac1Δ, and pka1Δ.(Alspaugh et al., 2002; D'Souza et al., 2001; Noverr et al., 2004). As noted above, the lac1Δ mutant (which lacks the laccase primarily responsible for melanin synthesis) has late defects in dissemination from the lungs but grows normally in lung tissue (Noverr et al., 2004); as such, it was not identified by the STM screen. Likewise, a mutant containing a deletion of the second C. neoformans laccase gene, LAC2, as well as a double knockout mutant, lac1Δ lac2Δ, did not display STM phenotypes (Table S2). This is consistent with melanization playing a role in dissemination, but not lung infectivity. Thus, we were surprised to note that overall there was a significant overlap between mutants with melanization defects and those with STM defects in the lungs (Figure 2F). This association remained significant (P =1.4E-11) even after excluding mutants with growth defects from the analysis. Given that the laccases were dispensable for lung infection, additional defects beyond poor pigment production must be proposed in this subset of melanin-defective mutants to account for their lung infectivity defects. These deficits may affect co-regulated pathways that function independently of and/or redundantly with melanization to promote lung proliferation.
To identify mutants defective in polysaccharide capsule formation, we screened for mutants with a dry colony appearance on rich medium (YPAD) agar plates. A dry colony morphology is characteristic of most known C. neoformans mutants with absent or greatly reduced amounts of capsule (Griffith et al., 2004; Moyrand et al., 2004; Moyrand and Janbon, 2004). Using this simple assay, we identified eleven potentially capsule-defective mutants including six strains (cap59Δ, cap64Δ, cap60Δ, cap10Δ, cas35Δ, and nrg1Δ) that contain deletions in genes previously implicated in capsule synthesis (Cramer et al., 2006; Moyrand et al., 2004; Okabayashi et al., 2007). In addition, PBX1 and PBX2 were first identified in this screen; deletion of PBX1 or PBX2 affects the fidelity of polysaccharide synthesis and their detailed analysis has been described elsewhere (Liu et al., 2007). As expected, all mutants with dry colony morphologies also had STM defects, consistent with the well-established importance of the capsule for causing disease (Figure 2F).
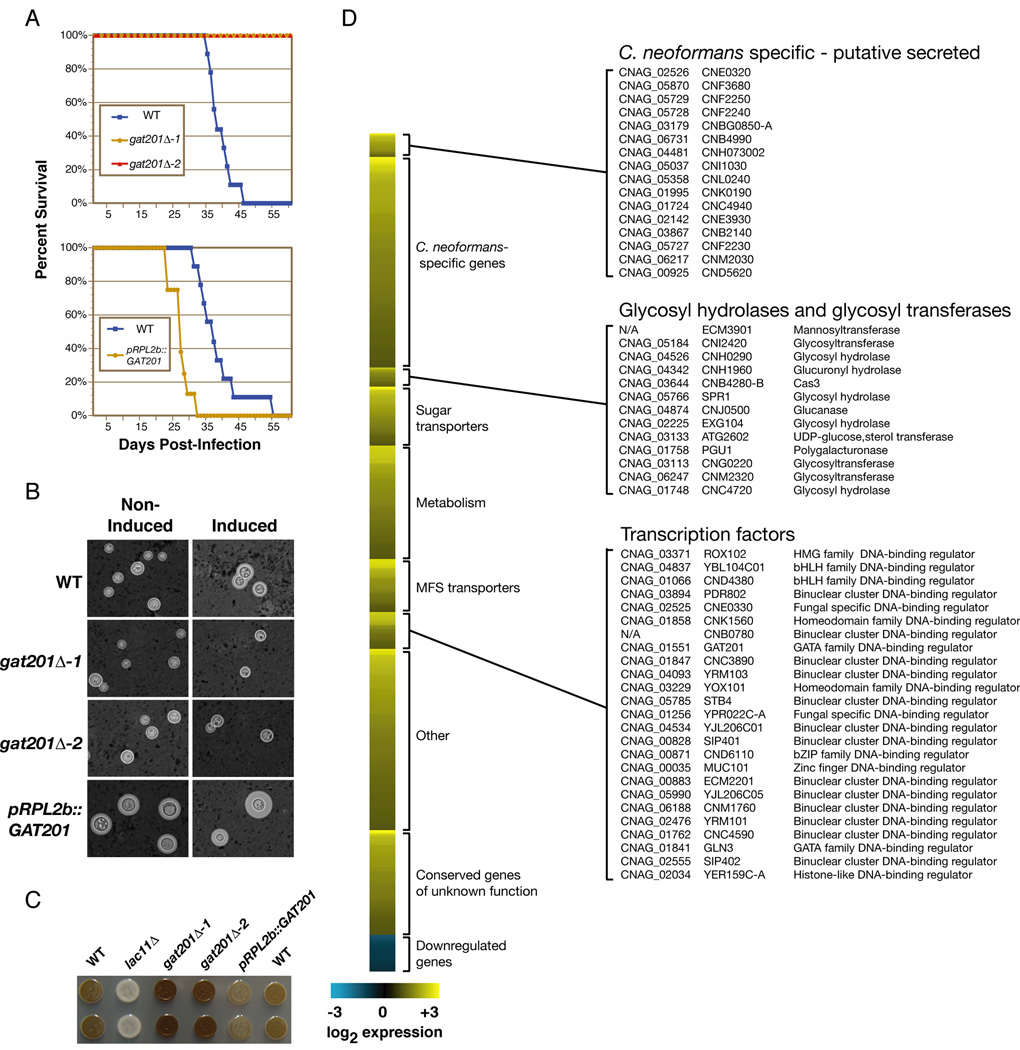
Certain mutants, such as pka1Δ and cac1Δ, are known to produce reduced or structurally-altered capsules (Alspaugh et al., 2002; D'Souza et al., 2001), but do not have dry colony morphologies. To identify this class of mutant, we tested a subset of mutants in the collection for the ability to induce capsule formation using an India ink assay. This stain yields a characteristic halo around cells with enlarged capsules (we note this analysis does not rule out subtle capsule structure defects that would otherwise only be detectable by NMR analysis). We tested 82 mutants that displayed melanin and/or STM defects but normal colony morphology. Only one mutant, containing a deletion of GAT201 (which encodes a GATA-family zinc finger DNA-binding transcriptional regulator, Figure S2), produced no visible capsule by India ink staining under capsule-inducing conditions (Figure 4C). Further investigation of the role of GAT201 in C. neoformans pathogenesis is described below.
Figure 4. Gat201 is a regulator of multiple virulence pathways.
(A) gat201Δ mutant strains (two independent knockout strains) were avirulent during monotypic infection and a GAT201 overexpression strain was hypervirulent. (B) gat201Δ mutant strains did not induce an observable capsule by India ink staining in capsule-inducing conditions, though capsule polysaccharide on the cell surface was detected by antibody staining (Figure 5B). GAT201 overexpression resulted in ectopic capsule formation in normally non-inducing conditions. Quantification of capsule size is shown in Figure S11. (C) gat201Δ mutants strains showed increased melanization while a GAT201 overexpression strain showed decreased melanization. (D) Transcriptional profiles of pRPL2b::GAT201 and wild-type grown at 37°C in minimal medium were compared using whole-genome microarrays. Values represent the average of four microarrays.
In total, among 164 mutants with STM defects, 85 also had defects in growth at 37°C, polysaccharide capsule formation, and/or melanization. The remaining 79 mutants appeared normal in all three in vitro assays (Figure 2F). To verify that the observed phenotypes were linked to the targeted deletions, we reconstructed 107 deletion strains and retested for the original phenotype: 5 of 5 reconstructed strains reproduced the original capsule phenotype; 33 of 35 reproduced the original melanin phenotype; 53 of 67 reproduced an STM score significantly below zero; and 8 of 11 reproduced an STM score significantly above zero (Figure S3). The results overall indicate strong linkage between the targeted mutation and the observed phenotypes. Slightly higher variability in reproduction of STM phenotypes may reflect both higher variability of the assay and the potential for a larger number of unlinked mutations to affect this phenotype.
Novel melanin and capsule genes
Thirty-three novel genes that regulate melanization and five novel genes that regulate capsule formation were identified and verified by knockout reconstruction and retesting (Figure 3, Table 1). As described previously (Chow et al., 2007), gene names were assigned based on homology to known S. cerevisiae genes when available. Four melanin genes and one capsule gene could not be named using this approach and, instead, were designated MLN1–MLN4 and CPL1 respectively. The melanin and capsule genes identified in these screens implicate a number of novel pathways in the regulation of these two virulence attributes. For example, mutants that lack the transcription factor Rim101 and its predicted upstream regulator Rim20 both display melanization defects (Figure S3). Similarly, deletion of components of the Set3 deacetylase complex resulted in significantly larger capsules compared to wild-type as well as defects in melanization (Figure S3–S4).
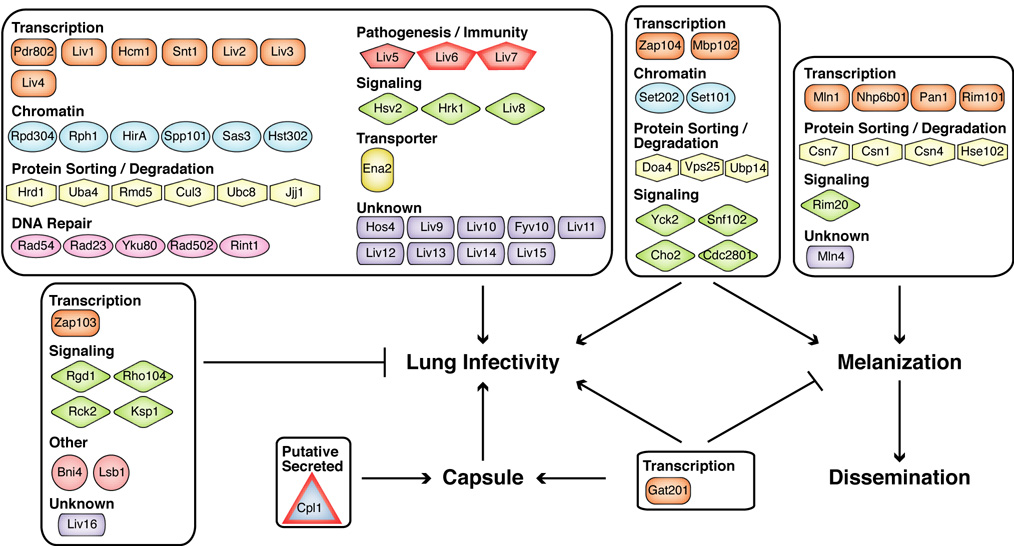
Figure 3. Proteins identified in this work that regulate capsule formation, melanization, and/or infectivity.
Factors previously reported to affect these phenotypes and factors that affect growth in YNB are not included. Proteins predicted to encode signal peptides are outlined in red. The relationships between melanization, dissemination, and lung infectivity are discussed in the text.
TABLE 1. Novel capsule and melanin mutants.
Genes identified in this work that regulate capsule formation, melanization, and/or infectivity. Previously characterized genes can be found in Table S2.
| Gene Deletion Phenotype |
|||||||
|---|---|---|---|---|---|---|---|
| KO# | Gene Name | Broad Annotation | Description | STM Score | Growth Score | Capsule | Melanin |
| Capsule | |||||||
| Transcription | |||||||
| D429 | GAT201 | CNAG_01551 | GATA family DNA-binding regulator | −18.3↓* | −0.18 | − | ++ |
| D226 | SSN801 | CNAG_00440 | Cyclin subunit of Mediator subcomplex | −1.4 | 0.18 | ++ | ++ |
| Chromatin | |||||||
| D110 | HOS2 | CNAG_05563 | Class II HDAC; component of Set3 complex in S. cerevisiae | −16.1↓ | −1.24↓ | ++ | − |
| D1487 | SET302 | CNAG_06591 | SET domain protein; homolog of S. cerevisiae Set3 | −10.9↓ | −0.86↓ | ++ | − |
| Putative secreted proteins | |||||||
| D667 | CPL1 | CNAG_02797† | Potential secreted protein of unknown function | −5.3↓* | 0.04 | − | + |
| Melanin | |||||||
| Transcription | |||||||
| D1248 | ZAP104 | CNAG_05392 | Zinc finger DNA-binding regulator | −7.1↓* | 0.06 | + | − |
| D1512 | MBP102 | CNAG_06721 | APSES family DNA-binding regulator | −3.5↓ | 0.08 | + | − |
| D528 | NHP6B02 | CNAG_02115 | Nonhistone chromosomal protein | −0.9 | −1.86↓ | + | +/− |
| D1218 | MLN1 | CNAG_04837 | bHLH family DNA-binding regulator | −0.3 | −0.04 | + | − |
| D228 | NHP6B01 | CNAG_00445 | Nonhistone chromosomal protein | 1.1 | 0.00 | + | − |
| D356 | PAN1 | CNAG_01173 | LAG1 family DNA-binding regulator | 1.4 | −0.40 | + | − |
| D1258 | RIM101 | CNAG_05431 | Zn finger DNA-binding regulator regulated by proteolysis | 2.8↑ | −0.07 | + | − |
| Chromatin | |||||||
| D1498 | RTF1 | CNAG_06648 | Component of PAF complex required for histone H2B monoubiquitination | −12.7↓ | −1.66↓ | + | − |
| D78 | SET202 | CNAG_03188 | Histone H3 lysine 36 methyltransferase | −8.1↓ | −0.14 | + | − |
| D373 | SET101 | CNAG_01243 | Histone H3 lysine 4 methyltransferase | −4.5↓* | −0.38 | + | +/− |
| Protein sorting / Protein degradation | |||||||
| D282 | DOA4 | CNAG_00757 | Ubiquitin hydrolase | −15.6↓ | −0.06 | + | − |
| D1250 | VAM6 | CNAG_05395 | Vacuolar membrane fusion factor | −14.3↓ | −0.61↓ | + | − |
| D1222 | VPS25 | CNAG_04863 | ESCRT-II Complex subunit | −13.8↓ | −0.06 | + | − |
| D1314 | UBP14 | CNAG_05708 | Ubiquitin carboxyl-terminal hydrolase | −12.3↓ | 0.27 | + | +/− |
| D454 | CSN1201 | CNAG_01697 | COP9 signalosome subunit 12; PINT domain | −8.9↓ | −0.58↓ | + | +/− |
| D936 | CSN7 | CNAG_04177 | COP9 signalosome subunit 7; PINT domain | −1.7 | −0.07 | + | +/− |
| D598 | CSN1 | CNAG_02513 | COP9 signalosome subunit 1; PINT domain | −1.1 | −0.16 | + | +/− |
| D737 | CSN4 | CNAG_03151 | COP9 signalosome subunit 4; PINT domain | −0.2 | 0.00 | + | +/− |
| D1349 | HSE102 | CNAG_05882 | Class E VPS factor involved in endosomal sorting | −0.2 | 0.00 | + | − |
| D671 | RPS3102 | CNAG_02827 | NEDD8 ubiquitin-like protein that regulates Cullins | 4.1↑ | −1.68↓ | + | − |
| Signaling | |||||||
| D245 | YCK2 | CNAG_00556 | Casein kinase homolog involved in endocytosis | −15.4↓ | −0.44 | + | − |
| D1476 | SNF102 | CNAG_06552 | AMP-activated kinase; ortholog of S. cerevisiae Snf1 | −15.0↓* | −0.34 | + | − |
| D731 | CHO2 | CNAG_03139 | Putative protein-S-isoprenylcysteine methyltransferase | −12.3↓ | 0.27 | + | − |
| D221 | CDC2801 | CNAG_00415 | Cyclin-dependent kinase of PITSLRE subfamily | −9.9↓ | −0.11 | + | +/− |
| D825 | RIM20 | CNAG_03582 | ESCRT-III-associated regulator of Rim101 | −0.3 | 0.09 | + | − |
| Nuclear pore | |||||||
| D891 | NUP75 | CNAG_03917 | Nuclear pore protein homologous to human Nup75 | −14.2↓ | −1.83↓ | + | +/− |
| Unknown function | |||||||
| D1408 | MLN2 | CNAG_06224 | CS domain protein of unknown function | −1.4 | −1.24↓ | + | − |
| D1453 | MLN3 | CNAG_06430 | Mus7 domain-containing protein conserved in fungi | −1.6 | −0.78↓ | + | +/− |
| D445 | MLN4 | CNAG_01644 | Protein of unknown function; C. neoformansspecific | −1.4 | −0.18 | + | − |
Down arrows (↓) indicate significant decreases compared to wild-type.
Up arrows (↑) indicate significant increases compared to wild-type.
Asterisks (*) indicate that the STM phenotype was verified in a reconstructed knockout strain.
Crosses (†) indicate gene models that are predicted to encode signal peptides.
Novel infectivity genes
A particularly interesting category of mutants identified in these screens had defects in lung infectivity but were normal for growth, capsule, and melanin. Forty of these mutants were verified by knockout reconstruction and retesting using the STM assay (Figure 3, Table 2). Again, gene names were assigned based on homology to known S. cerevisiae genes when available. Fifteen unnamed genes were designated LIV1–LIV15 (Lung InfectiVity).
TABLE 2. Infectivity mutants without defects in capsule formation, melanization, or growth in YNB.
Table 2 includes only infectivity mutants that did not display defects in capsule formation, melanization, or growth in YNB. These mutants were tested for six additional phenotypes: decreased urease activity and sensitivity to acidified nitrite (NO), SDS, potassium cyanide (KCN), pH 4, and low-iron.
| Gene Deletion Phenotype |
||||||
|---|---|---|---|---|---|---|
| KO# | Gene Name | Broad Annotation | Description | STM Score | Growth Score | Additional Phenotypes |
| Decreased Infectivity (STM Score < −2.5) | ||||||
| Transcription | ||||||
| D883 | PDR802 | CNAG_03894 | Binuclear cluster DNA-binding regulator | −7.2↓* | −0.08 | None detected |
| D231 | LIV1 | CNAG_00460 | bHLH family DNA-binding regulator | −6.1↓* | −0.35 | None detected |
| D725 | HCM1 | CNAG_03116 | Forkhead DNA-binding regulator | −4.2↓* | −0.46 | None detected |
| D140 | SNT1 | CNAG_00051 | SANT domain protein related to human NCOR corepressor subunit | −4.0↓* | −0.10 | None detected |
| D177 | LIV2 | CNAG_00201 | LAG1 family DNA-binding regulator | −3.9↓* | 0.06 | None detected |
| D1338 | LIV3 | CNAG_05835 | WOR1 family DNA-binding regulator | −3.7↓* | 0.03 | None detected |
| D1420 | LIV4 | CNAG_03116 | Myb family DNA-binding regulator | −2.5↓* | −0.03 | None detected |
| Chromatin | ||||||
| D111 | RPD304 | CNAG_05690 | Class II histone deacetylase | −11.5↓* | −0.3 | None detected |
| D1296 | RPH1 | CNAG_05622 | Histone H3 lysine 36 demethylase | −8.0↓* | −0.25 | None detected |
| D933 | HIRA | CNAG_04158 | Histone chaperone | −6.7↓* | −0.15 | Inc. sensitivity to NO |
| D790 | SPP101 | CNAG_03406 | PHD domain protein of unknown function | −6.1↓* | −0.09 | None detected |
| D1428 | SAS3 | CNAG_06322 | Histone acetyltransferase | −4.4↓* | −0.18 | None detected |
| D62 | HST302 | CNAG_02085 | Homolog of Sir2; Class III histone deacetylase | −2.8↓* | −0.11 | None detected |
| Protein sorting / Protein degradation | ||||||
| D1266 | HRD1 | CNAG_05469 | Transmembrane ubiquitin ligase required for ER-associated degradation | −8.0↓* | −0.38 | Inc. sensitivity to NO |
| D317 | UBA4 | CNAG_00986 | Urm1 activating protein | −6.0↓* | −0.2 | None detected |
| D399 | RMD5 | CNAG_01350 | Ubiquitin ligase of unknown function | −4.3↓* | 0.02 | None detected |
| D729 | CUL3 | CNAG_03138 | Cullin subunit of SCF-like ubiquitin ligase | −3.6↓* | −0.11 | None detected |
| D1166 | UBC8 | CNAG_04611 | Ubiquitin conjugating enzyme | −3.5↓* | 0.16 | None detected |
| D1278 | JJJ1 | CNAG_05538 | DnaJ-like cochaperone; contains U1 snRNA-type Zn finger | −3.4↓* | 0.11 | None detected |
| DNA Repair | ||||||
| D352 | RAD54 | CNAG_01163 | SWI/SNF family ATPase that promotes recombinational DNA repair | −9.3↓* | −0.49 | Inc. sensitivity to NO |
| D288 | RAD23 | CNAG_00772 | Ubiquitin-binding protein required for nucleotide excision repair | −7.6↓* | 0.09 | Inc. sensitivity to NO |
| D832 | YKU80 | CNAG_03637 | Double strand break repair factor and silencing regulator | −4.9↓* | 0.04 | None detected |
| D632 | RAD502 | CNAG_02630 | SWI/SNF family ATPase that promotes recombinational DNA repair | −2.7↓* | −0.04 | None detected |
| D855 | RINT1 | CNAG_03760 | Rad50-interacting protein implicated in DNA repair | −2.5↓* | 0.01 | Inc. sensitivity to KCN |
| Pathogenesis / Immunity | ||||||
| D576 | LIV5 | CNAG_02409 | TB2/DP1 family member homologous to M. grisea pathogenicity protein | −12.6↓* | −0.09 | None detected |
| D166 | LIV6 | CNAG_00170† | Related to human T-cell immunomodulatory protein; FG-GAP repeats | −6.2↓* | 0.14 | None detected |
| D1466 | LIV7 | CNAG_06464† | Protein of unknown function; human homolog is LPS-inducible | −3.2↓* | −0.17 | None detected |
| Signaling | ||||||
| D972 | HSV2 | CNAG_04371 | WD40 protein related to 3-phosphorylated inositide-binding protein | −15.9↓* | 0.18 | None detected |
| D280 | HRK1 | CNAG_00745 | Protein kinase; S. cerevisiae ortholog regulates metabolite transport | −6.8↓* | 0.14 | Inc. sensitivity to SDS |
| D437 | LIV8 | CNAG_01611 | RGS protein homolog - G protein GAP | −2.7↓* | 0.10 | Inc. sensitivity to NO, SDS, KCN |
| Unknown | ||||||
| D70 | HOS4 | CNAG_02727 | Ankyrin repeat protein of unknown function | −7.6↓* | 0.09 | None detected |
| D736 | LIV9 | CNAG_03149 | RING and ubiquitin-binding protein with BRAP2 domain | −7.3↓* | −0.04 | None detected |
| D686 | LIV10 | CNAG_02930 | DUF1765 domain protein of unknown function | −7.1↓* | −0.19 | None detected |
| D171 | FYV10 | CNAG_00181 | LisH, CTLH domain protein of unknown function | −6.5↓* | −0.06 | None detected |
| D1256 | LIV11 | CNAG_05422 | Protein of unknown function | −5.0↓* | −0.49 | None detected |
| D1035 | LIV12 | CNAG_05105 | Protein of unknown function | −3.9↓* | −0.11 | None detected |
| D660 | LIV13 | CNAG_02753 | UPF0121 domain protein of unknown function | −3.2↓* | −0.34 | None detected |
| D1448 | LIV14 | CNAG_06397 | Protein of unknown function | −2.9↓* | 0.06 | None detected |
| D1357 | LIV15 | CNAG_05934 | Translin domain protein of unknown function | −2.8↓* | 0.09 | None detected |
| Transporter | ||||||
| D1091 | ENA2 | CNAG_00531 | Sodium pump | −11.1↓* | 0.11 | None detected |
| Increased Infectivity (STM Score > 2.5) | ||||||
| Transcription | ||||||
| D88 | ZAP103 | CNAG_04352 | Zinc finger DNA-binding regulator | 3.2↑* | −0.14 | |
| Signaling | ||||||
| D1340 | RGD1 | CNAG_05838 | Rho GAP homolog | 5.3↑* | 0.07 | |
| D1490 | RHO104 | CNAG_06606 | Rho family GTPase | 4.4↑* | −0.33 | |
| D1156 | RCK2 | CNAG_00130 | Ca-CaM kinase homolog | 3.7↑* | 0.03 | |
| D488 | KSP1 | CNAG_01905 | Protein kinase | 3.4↑* | −0.04 | |
| Cell wall | ||||||
| D61 | BNI4 | CNAG_02071 | S. cerevisiae ortholog is a scaffold protein involved in cell wall assembly | 4.8↑* | 0.27 | |
| Actin-related | ||||||
| D359 | LSB1 | CNAG_01183 | BAR-SH3 domain; S. cerevisiae homolog binds actin nucleator WASP | 3.6↑* | 0.11 | |
| Unknown | ||||||
| D1179 | LIV16 | CNAG_07193† | Conserved protein of unknown function | 3.8↑* | 0.05 | |
Down arrows (↓) indicate significant decreases compared to wild-type.
Up arrows (↑) indicate significant increases compared to wild-type.
Asterisks (*) indicate that the STM phenotype was verified in a reconstructed knockout strain.
Crosses (†) indicate gene models that are predicted to encode signal peptides.
In addition to capsule, melanin, and growth, a number of other less-studied factors have been implicated in C. neoformans virulence based on genetic studies. These include urease, alternative oxidase, phospholipase, reductive iron uptake, glucosylceramide synthesis, and resistance to nitrogen and oxygen species (Akhter et al., 2003; Cox et al., 2003; Cox et al., 2001; Cox et al., 2000; de Jesus-Berrios et al., 2003; Jung et al., 2008; Shea et al., 2006). In most cases, mutants in these pathways display growth defects on certain media. For example, mutants lacking the phospholipase gene (PLB1) are sensitive to SDS (Siafakas et al., 2007), mutants lacking the flavohemoglobin gene (FHB1) are sensitive to acidified nitrite, and mutants lacking the alternative oxidase gene (AOX1) are sensitive to potassium cyanide (KCN). Thus, in order to identify potential roles for these infectivity genes, we systematically tested the 40 mutants described above for growth on five media conditions as well as for urease activity using an enzymatic assay. This analysis revealed five mutants with sensitivity to acidified nitrite, two with sensitivity to SDS, and two with sensitivity to KCN (Table 2, Figure S5–S7). In contrast, none of the mutants displayed decreased growth in pH 4 media (Figure S5) or in low ferrous iron conditions (Figure S8) and none of the mutants displayed a defect in urease activity (Figures S9).
Two of the mutants that displayed increased sensitivity to acidified nitrite contain deletions of DNA repair genes (rad54Δ and rad23Δ), suggesting that unrepaired DNA damage, perhaps via free radical attack by phagocytes, can be deleterious to infection (Figure S5). Notably, a mutant containing a deletion of LIV8, which encodes an RGS (Regulator of G-protein Signaling) domain, was exquisitely sensitive to acidified nitrite, SDS, and KCN (Figures S5–S7). We note that since these media assays are relatively nonspecific, the sensitivities observed may or may not explain the virulence defects of particular mutants. For example, a mutant containing a deletion of the virulence gene SRE1, which has been shown to be involved in ergosterol biosynthesis (Chang et al., 2007; Chun et al., 2007), displayed sensitivity to both SDS and KCN (Figures S6–S7).
Nonetheless, the 33 mutants that did not display phenotypes in any of these in vitro assays represent the genes that are the strongest candidates for novel C. neoformans virulence factors. Seven genes in this category encode predicted sequence-specific transcription factors. Another five genes encode factors likely involved in chromatin regulation. Of particular note, three genes encode factors with strong homology to proteins previously implicated in pathogenesis or regulation of the human immune response (Figure 3, Table 2). LIV5 encodes a protein homologous to one required for pathogenesis by the plant fungal pathogen Magnaporthe grisea (Fujimoto et al., 2002). LIV7 encodes a putative secreted protein that is homologous to a human protein induced by lipopolysaccharide. Finally, LIV6 encodes a protein highly homologous to the human T-cell immunomodulatory protein (TIP). Secreted TIP has been shown to directly regulate cytokine production by T-cells (Fiscella et al., 2003). Protein structure prediction indicated that Liv6 contains FG-GAP repeats, which are commonly found in alpha integrins, as well as a signal peptide sequence, suggesting that it may be secreted (Figure S2). Deletion of LIV5 and LIV6 were confirmed to result in virulence defects by survival curve analysis (Figure 2E, Figure S10).
We also identified mutants that reproducibly showed significant increases in the STM assay. Eight mutants were confirmed by knockout reconstruction and retesting to display enhanced colonization of lung tissue (Table 2). These include deletions of genes encoding two members of Rho signaling pathways (RGD1 and RHO104), two kinases (RCK2 and KSP1), and a transcription factor (ZAP103). One unnamed gene was designated LIV16. Deletions of ZAP103 and RGD1 were confirmed to result in increased virulence by survival curve analysis (Figure 2E, Figure S10).
Gat201: a key regulator of virulence and capsule-independent anti-phagocytic mechanisms
Among mutants with normal growth at 37°C, the strain containing a deletion of GAT201 exhibited the largest decrease in the STM screen (STM score = −18.3). Indeed, the gat201Δ mutant strain was avirulent in monotypic infection of mice using our standard inoculum (Figure 4A). This mutant was chosen for further investigation.
As described above, capsules of gat201Δ cells could not be visualized by India ink staining under capsule-inducing conditions (Figure 4B). However, a more sensitive assay using a monoclonal antibody to stain the main capsular polysaccharide, glucoronoxylomannan, detected capsule polysaccharide on the cell surface of gat201Δ cells (Figure 5B). These data suggest GAT201 is not required for production of basal levels of capsule polysaccharide but is needed for proper induction of large capsules. gat201Δ cells were also hypermelanized when grown on L-DOPA plates (Figure 4C). Previously-identified hypermelanized mutants have been associated with both increased and decreased virulence and often display additional phenotypes such as altered capsule production (Bahn et al., 2005; Wang et al., 2004). Similarly, our studies indicate GAT201 participates directly or indirectly in the regulation of both capsule induction and melanization.
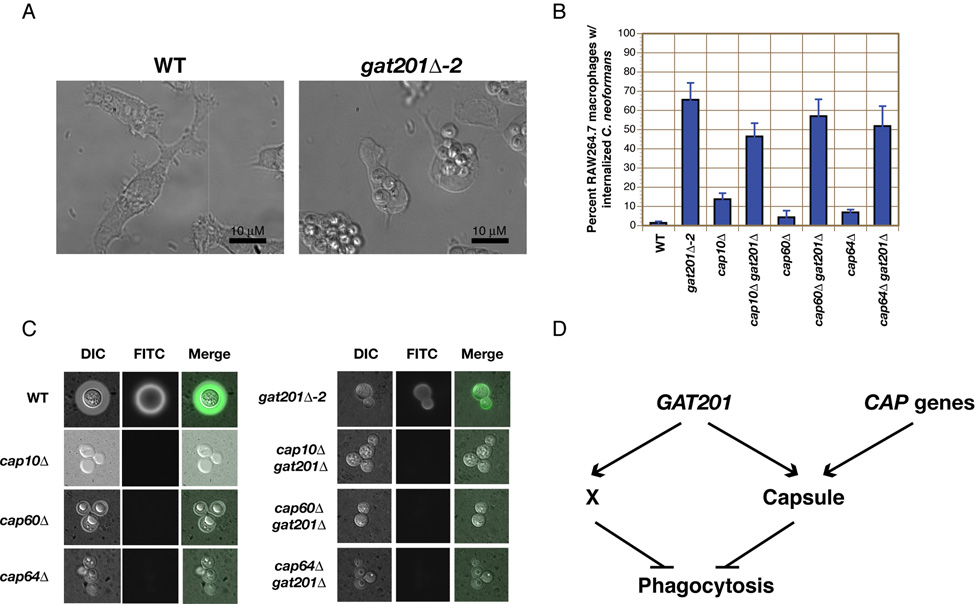
Figure 5. Gat201 inhibits phagocytosis independent of capsule.
(A–B) Phagocytosis of untreated wild-type (WT) and gat201Δ cells by RAW264.7 macrophages was measured in both WT and capsule-deficient backgrounds. The percentage of macrophages containing internalized C. neoformans cells was determined after 24 hours of co-incubation. Data are represented as mean +/− SD from triplicate well s. (C) GXM can be detected on the cell surface of gat201Δ cells, but not in single and double mutants containing deletions of CAP genes. (D) Model of GAT201 regulation of phagocytosis. “X” represents a novel capsule-independent mechanism for inhibiting phagocytosis by macrophages.
Consistent with this notion, overexpression of GAT201 in wild-type cells using the promoter of a constitutive ribosomal protein gene (pRPL2b) was sufficient to induce large capsule formation, even in normally non-inducing growth conditions (Figure 4B, Figure S11). As shown in Figure 4C, the pRPL2b::GAT201 strain also displayed a slight melanization defect. The pRPL2b::GAT201 strain was hypervirulent in mice after monotypic intranasal infection (Figure 4A), demonstrating that overexpression of GAT201 is sufficient to activate key virulence pathways. A reconstructed gat201Δ strain recapitulated the phenotypes of the original gat201Δ strain in these assays (Figure 4A–4C).
Whole-genome, microarray-based expression profiling of the pRPL2b::GAT201 strain identified 543 genes that were significantly upregulated and 24 genes that were significantly downregulated compared to a wild-type strain (Figure 4D). Consistent with the enhanced nutritional and metabolic requirements necessary for ectopic polysaccharide capsule formation, the induced set contained a substantial number of genes encoding proteins involved in carbohydrate regulation, including sugar transporters (40 genes), glycosyl hydrolases (8 genes), and glycosyl transferases (5 genes). In addition, twenty-five genes that encode DNA-binding transcription factors were upregulated in the pRPL2b::GAT201 strain, suggesting that Gat201 exerts at least a portion of its effects by activating the expression of other transcription factors. Using a previously described annotation database (Chow et al., 2007), we identified C. neoformans genes that do not have homologs in nine sequenced ascomycetes and two sequenced basidiomycetes. Over 29% of Gat201-upregulated genes (159 genes) fall into this C. neoformans-specific gene class, which represents a significant enrichment over their general representation on the microarray (P =1.2E-35). Sixteen of these genes are predicted to produce proteins with signal sequences but no transmembrane domain; these potential secreted effectors include all members of a unique four-gene family (CNF2230, CNF2240, CNF2250, and CND5620).
To further probe the role of Gat201 in virulence, we examined the interaction of gat201Δ cells with RAW264.7 mouse macrophages. Phagocytic cells play a central role in the host immune response against C. neoformans, and the ability of C. neoformans to avoid phagocytosis and killing by these cells is crucial to pathogenesis (Del Poeta, 2004). In contrast to most fungi, such as C. albicans and S. cerevisiae, wild-type C. neoformans undergoes negligible phagocytosis by cultured macrophages in the absence of opsonizing antibodies or complement (Kozel and Mastroianni, 1976). The anti-phagocytic properties of C. neoformans have largely been attributed to the polysaccharide capsule (Del Poeta, 2004). We recapitulated these results with wild-type C. neoformans, observing both robust induction of capsule and a very low rate of phagocytosis after 24 hours of co-culturing with RAW264.7 macrophages (<2% of macrophages contained internalized wild-type C. neoformans, Figures 5A–B, Figure S12). Deletion of the capsule-associated genes CAP10, CAP60, or CAP64 resulted in approximately a three- to seven-fold increase in the number of macrophages containing C. neoformans (4–14%). As previously reported (Moyrand and Janbon, 2004), cap10Δ, cap60Δ, and cap64Δ mutants had no detectable capsular material, either by antibody or India ink staining (Figure 5C). Unexpectedly, a dramatic increase in the amount of phagocytosis was seen when RAW264.7 macrophages were co-cultured with gat201Δ cells; 66% of macrophages contained internalized C. neoformans after 24 hours (Figure 5A–B). Since gat201Δ mutants were phagocytosed more readily than CAP gene mutants, yet displayed a weaker capsule defect, the phagocytosis phenotype of gat201Δ cells could not easily be explained by its hypocapsular phenotype. To examine the role of GAT201 in the absence of capsule, we asked whether double mutants lacking both GAT201 and one of the CAP genes were phagocytosed to a greater extent than the CAP gene single mutants. Strikingly, we found this to be the case: high levels of phagocytosis were seen in gat201Δ cap10Δ (47%), gat201Δ cap60Δ (57%), and gat201Δ cap64Δ (52%) double mutant strains (Figure 5B). Unlike gat201Δ single mutants, these double mutant strains had no detectable capsules by antibody or India ink staining and displayed clumpy phenotypes characteristic of cap10Δ, cap60Δ, and cap64Δ single mutants (Figure 5C), indicating that the capsule is not the basis for the difference in phagocytosis between gat201Δ and the CAP mutants.
DISCUSSION
The incidence of invasive fungal infections in humans has risen dramatically in the last two decades (Warnock, 2006), yet our understanding of the molecular mechanisms relevant to fungal pathogenesis remains limited. The analysis of the C. neoformans gene deletion collection described here demonstrates the potential for harnessing genome sequencing efforts to implement systematic approaches for identifying and dissecting fungal virulence mechanisms. Since biolistic gene knockout methods for C. neoformans were introduced in 1993, ~190 targeted deletions have been reported in the literature (Table S1 summarizes the phenotypes of previously-described gene deletions). We report the construction and analysis of 1,201 targeted gene deletion strains, significantly extending the genetic resources for this pathogen. Systematic profiling of the deletion collection yielded a number of insights into C. neoformans virulence:
Melanization: By identifying and analyzing numerous novel factors required for melanization, we obtained strong evidence that virulence and melanization are co-regulated, but that the lung proliferation defects observed in a large subset of melanin mutants result from defects beyond poor melanization.
Polysaccharide capsule: We confirmed the role of the polysaccharide capsule in virulence. Our screen identified four novel hypocapsular mutants, all of which displayed defects in infection. Gat201, following Nrg1 (Cramer et al., 2006), is the second DNA-binding transcription factor found to regulate capsule formation. Gat201 is normally limiting for synthesis of this virulence factor since overexpression of GAT201 is sufficient to induce capsule synthesis under classically non-inducing conditions. As Gat201 also controls melanization, inhibition of phagocytosis, and virulence, it appears to be a central point of regulation for cryptococcal pathogenesis.
Capsule-independent anti-phagocytic pathways: The anti-phagocytic properties of C. neoformans have largely been attributed to its polysaccharide capsule (Del Poeta, 2004). Our analysis of the Gat201 transcriptional regulator has identified a previously unsuspected capsule-independent mechanism of anti-phagocytosis since deletion of GAT201, both in wild-type and capsule-deficient genetic backgrounds, resulted in a dramatic increase in phagocytosis of untreated C. neoformans by macrophages. Some CAP mutants do produce the minor capsule polysaccharide, galactoxylomannan (GalXM), which is detectable in culture supernatant (Vaishnav et al., 1998). Whether GAT201 functions by controlling the synthesis of GalXM, protein factors, and/or lipids, and whether the effect seen in gat201Δ mutants is due to the lack of an anti-phagocytic factor or the presence of a pro-phagocytic factor remain unclear. Transcriptional profiling of a GAT201 overexpression strain detected a significant enrichment (P =1.2E-35) for transcripts of C. neoformans genes that are not found in other sequenced fungi including a number of predicted secreted proteins—some of these could contribute to C. neoformans-specific virulence pathways, including ones involved in inhibiting phagocytosis by macrophages. Finally, 25 genes encoding DNA-binding transcription factors were upregulated in the overexpression strain, suggesting that Gat201 may control a large, virulence-associated transcriptional regulatory circuit.
Potential novel virulence pathways: Molecular studies of C. neoformans virulence have focused primarily on the regulation of the capsule and melanization. The discovery of 48 previously undescribed genes (40 mutants with decreased infectivity, 8 mutants with increased infectivity), that affect proliferation in lung tissue, but not capsule, growth or melanin, strongly suggests the existence of additional unknown virulence mechanisms and provides tools for their identification. In principle, these genes could code for 1) regulators of novel virulence pathways or 2) direct effectors of virulence. Potential regulators include seven predicted transcription factors, whose targets presumably include pathogenesis factors. Possible direct effectors of virulence include three factors with homology to proteins previously linked to pathogenesis and/or immune modulation (Liv5, Liv6 and Liv7). A substantial number of proteins required for infectivity were identified whose function cannot be inferred from sequence relationships, indicating that novel molecular functions yet to be assigned to specific proteins in any organism are likely to play a role in fungal virulence. Taken together, the identification and categorization of infectivity mutants in a large deletion library highlight the advantages of using a multipronged, systematic genetic analysis to dissect pathogenic mechanisms.
EXPERIMENTAL PROCEDURES
Generation of gene deletion strains
Cryptococcus neoformans var. grubii serotype A clinical strain H99 (a gift from J. Lodge) was used as wild-type. A previously described strategy for gene deletion (Chun et al., 2007) utilizing overlap fusion PCR and biolistic techniques was used to generate knockout strains (see Supplementary Materials). Details on the deletion collection including the targeted regions in each mutant can be found in Table S4.
Signature-tagged mutagenesis
Groups of 48 C. neoformans deletion strains were individually grown in liquid YPAD and combined into a single pool. These pooled inocula were used to intranasally infect three mice each. Three aliquots of each inoculum (~5 × 105 cells) were also plated on Sabouraud agar plates containing 40 µg/mL gentamicin and 50 µg/mL carbenicillin. Genomic DNA (Input DNA) was purified independently from each plate using hexadecyltrimethyl ammonium bromide (CTAB, Sigma). Infected mice were monitored and sacrificed at the disease endpoint. Lungs were removed and homogenized in 5 mL sterile PBS. Serial dilutions of each organ sample were plated on Sabouraud agar plates containing 40 µg/mL gentamicin and 50 µg/mL carbenicillin. Genomic DNA (Output DNA) from each set of lungs was purified using CTAB.
The amounts of each signature tag in a genomic DNA prep were measured using 48 simultaneous quantitative PCR reactions. Each reaction was identical except for a different tag-specific primer (Table S7). The threshold cycle (Ct) was determined for each reaction and an STM score was calculated for each mutant using a variation of the 2−ΔΔCt method for quantitation analysis (Livak and Schmittgen, 2001). STM scores from three mice were averaged to determine a final STM score for each mutant. A detailed description of this STM method can be found in the Supplementary Materials.
In vitro phenotyping
Polysaccharide capsule formation, melanin synthesis, urease activity, and growth on acidified nitrite, SDS, potassium cyanide, pH 4, and low iron medium were assayed using standard conditions (see Supplementary Materials). Growth in minimal medium was quantified by measuring the change in optical density at 600 nm (OD600) after 24 hours growth using an optical plate reader (Molecular Devices). Growth scores were normalized to the median growth in groups of 48 (see Supplementary Materials).
Statistical analysis
Significances of overlaps between STM, capsule, and melanin phenotypes were measured using a hypergeometric test. Correlation of STM and growth defects was analyzed by measuring enrichment for STM defects in a 25-mutant moving average across growth scores using a hypergeometric test.
Significances of mutant STM scores were calculated by measuring the divergence of a mutant STM score from the distribution of sxiΔ STM scores using a z-score calculation. For retesting of STM phenotypes in reconstructed knockouts, an STM score of −1.5 was used as a threshold for significance (P < .001).
Survival curves were analyzed using the Kaplan-Meier method and log rank test.
Macrophage assays
To test for phagocytosis of untreated C. neoformans, RAW264.7 macrophages (4×104 cells/well) were seeded in 96-well tissue culture-treated glass bottom plates (Nunc) in DMEM media and allowed to adhere overnight. C. neoformans cells from overnight cultures grown in liquid YPAD were washed three times with PBS and then lightly sonicated using a Sonifer 450 equipped with a microtip (Branson, two one-second pulses at a power setting of 2) to break up any clumps of cells prior to incubation with macrophages. Sonication did not affect the viability of cells and similar results were obtained in phagocytosis assays that utilized wild-type and gat201Δ cells that were not sonicated (Figure S13). The yeast cells were resuspended in PBS to a concentration of 107 cells/mL, and 10 µL (105 cells) were co-incubated with RAW264.7 cells (in 200 µL fresh DMEM/well) for an MOI of ~ 2:1. Following 24 hours of co-incubation, unphagocytosed C. neoformans cells were removed with three washes of PBS, and yeast and macrophages were fixed with 1% formaldehyde/PBS. Remaining extracellular yeast were labeled with a mixture of an anti-glucoronxylomannan antibody, mAb 339 (Belay et al., 1997), and an anti-beta-glucan mouse monoclonal antibody (Biosupplies Australia) in PBS, a combination which labels both encapsulated and acapsular strains. Following three washes with PBS, the cells were then incubated with FITC-conjugated donkey anti-mouse secondary antibody (Jackson ImmunoResearch) in PBS. Phagocytosis of C. neoformans was quantified on an Axiovert 200M (Zeiss) microscope running Axiovision software. Percentage of macrophages with phagocytosed yeast was determined by counting the number of macrophages with internalized yeast (as defined by the absence of FITC-labeling). At least 200 macrophages were assayed per well, and each strain was tested in triplicate.
Microarrays
Four replicate cultures each of H99 and pRPL2b::GAT201 were grown overnight to saturation in YNB at 37°C. Cultures were diluted to an OD600 of 0.1 in YNB (pre-warmed to 37°C), grown to an OD600 of 1.0, and then harvested by centrifugation and snap freezing. RNA isolation, array hybridizations, and data analysis using SAM analysis were performed as previously described (Chun et al., 2007, see Supplementary Materials). Microarray data can be found at the Gene Expression Omnibus (GSE10750)
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to J. Lodge, C. Hull, J. Heitman, T. Kozel, and G. Cox for strains, reagents, training, and assistance with protocols, to S. O’Brien for assistance with bioinformatics, and to J. Cox and J. Brown for helpful discussion and comments on the manuscript. This work was supported by an Opportunity Grant from the Herb and Marion Sandler Foundation to HDM and a grant from the NIAID (R01AI065519) to HDM. OWL was supported by a Howard Hughes Medical Institute (HHMI) Predoctoral Fellowship and by the NIAID-sponsored UCSF Microbial Pathogenesis and Host Defense Training Program (T32AI060537). CDC was supported by a National Science Foundation Predoctoral Fellowship. CC was supported by a Department of Biochemistry and Biophysics Boyer Postdoctoral Fellowship and a grant from the Stewart Family Trust. SMN was supported by a HHMI Postdoctoral Fellowship for Physicians, a Burroughs-Wellcome Fund Career Development Award, and an NIH K08 award. OWL and EDC generated and profiled the deletion library. OWL and CC reconstructed and retested deletion strains. CC constructed the overexpression strain and performed microarray experiments. CDC and OWL constructed double mutant strains and performed macrophage phagocytosis experiments. SMN developed protocols for the STM assay, and provided reagents, training, and guidance. Strain requests should be addressed to HDM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akhter S, McDade HC, Gorlach JM, Heinrich G, Cox GM, Perfect JR. Role of alternative oxidase gene in pathogenesis of Cryptococcus neoformans. Infect Immun. 2003;71:5794–5802. doi: 10.1128/IAI.71.10.5794-5802.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, Cox GM, Perfect JR, Kronstad JW, Heitman J. Adenylyl cyclase functions downstream of the Galpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot Cell. 2002;1:75–84. doi: 10.1128/EC.1.1.75-84.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Heilbut A, Andrews B, Tyers M, Hughes T, Boone C. Functional genomics and proteomics: charting a multidimensional map of the yeast cell. Trends Cell Biol. 2003;13:344–356. doi: 10.1016/s0962-8924(03)00127-2. [DOI] [PubMed] [Google Scholar]
- Bahn YS, Kojima K, Cox GM, Heitman J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell. 2005;16:2285–2300. doi: 10.1091/mbc.E04-11-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekondi C, Bernede C, Passone N, Minssart P, Kamalo C, Mbolidi D, Germani Y. Primary and opportunistic pathogens associated with meningitis in adults in Bangui, Central African Republic, in relation to human immunodeficiency virus serostatus. Int J Infect Dis. 2006;10:387–395. doi: 10.1016/j.ijid.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Belay T, Cherniak R, Kozel TR, Casadevall A. Reactivity patterns and epitope specificities of anti-Cryptococcus neoformans monoclonal antibodies by enzyme-linked immunosorbent assay and dot enzyme assay. Infect Immun. 1997;65:718–728. doi: 10.1128/iai.65.2.718-728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicanic T, Harrison TS. Cryptococcal meningitis. Br Med Bull. 2004;72:99–118. doi: 10.1093/bmb/ldh043. [DOI] [PubMed] [Google Scholar]
- Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol Microbiol. 2007;64:614–629. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- Chow ED, Liu OW, O'Brien S, Madhani HD. Exploration of whole-genome responses of the human AIDS-associated yeast pathogen Cryptococcus neoformans var grubii: nitric oxide stress and body temperature. Curr Genet. 2007;52:137–148. doi: 10.1007/s00294-007-0147-9. [DOI] [PubMed] [Google Scholar]
- Chun CD, Liu OW, Madhani HD. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 2007;3:e22. doi: 10.1371/journal.ppat.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett EL, Churchyard GJ, Charalambos S, Samb B, Moloi V, Clayton TC, Grant AD, Murray J, Hayes RJ, De Cock KM. Morbidity and mortality in South African gold miners: impact of untreated disease due to human immunodeficiency virus. Clin Infect Dis. 2002;34:1251–1258. doi: 10.1086/339540. [DOI] [PubMed] [Google Scholar]
- Cox GM, Harrison TS, McDade HC, Taborda CP, Heinrich G, Casadevall A, Perfect JR. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect Immun. 2003;71:173–180. doi: 10.1128/IAI.71.1.173-180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GM, McDade HC, Chen SC, Tucker SC, Gottfredsson M, Wright LC, Sorrell TC, Leidich SD, Casadevall A, Ghannoum MA, Perfect JR. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol Microbiol. 2001;39:166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. Urease as a virulence factor in experimental cryptococcosis. Infect Immun. 2000;68:443–448. doi: 10.1128/iai.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer KL, Gerrald QD, Nichols CB, Price MS, Alspaugh JA. Transcription factor Nrg1 mediates capsule formation, stress response, and pathogenesis in Cryptococcus neoformans. Eukaryot Cell. 2006;5:1147–1156. doi: 10.1128/EC.00145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza CA, Alspaugh JA, Yue C, Harashima T, Cox GM, Perfect JR, Heitman J. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol. 2001;21:3179–3191. doi: 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus-Berrios M, Liu L, Nussbaum JC, Cox GM, Stamler JS, Heitman J. Enzymes that counteract nitrosative stress promote fungal virulence. Curr Biol. 2003;13:1963–1968. doi: 10.1016/j.cub.2003.10.029. [DOI] [PubMed] [Google Scholar]
- Del Poeta M. Role of phagocytosis in the virulence of Cryptococcus neoformans. Eukaryot Cell. 2004;3:1067–1075. doi: 10.1128/EC.3.5.1067-1075.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiscella M, Perry JW, Teng B, Bloom M, Zhang C, Leung K, Pukac L, Florence K, Concepcion A, Liu B, et al. TIP, a T-cell factor identified using high-throughput screening increases survival in a graft-versus-host disease model. Nat Biotechnol. 2003;21:302–307. doi: 10.1038/nbt797. [DOI] [PubMed] [Google Scholar]
- French N, Gray K, Watera C, Nakiyingi J, Lugada E, Moore M, Lalloo D, Whitworth JA, Gilks CF. Cryptococcal infection in a cohort of HIV-1-infected Ugandan adults. AIDS. 2002;16:1031–1038. doi: 10.1097/00002030-200205030-00009. [DOI] [PubMed] [Google Scholar]
- Fujimoto D, Shi T, Christian D, Mantanguihan JB, Leung H. Tagging quantitative loci controlling pathogenicity in Magnaporthe grisea by insertional mutagenesis. Physiol Mol Plant Pathol. 2002;61:77–88. [Google Scholar]
- Griffith CL, Klutts JS, Zhang L, Levery SB, Doering TL. UDP-glucose dehydrogenase plays multiple roles in the biology of the pathogenic fungus Cryptococcus neoformans. J Biol Chem. 2004;279:51669–51676. doi: 10.1074/jbc.M408889200. [DOI] [PubMed] [Google Scholar]
- Hull CM, Cox GM, Heitman J. The alpha-specific cell identity factor Sxi1alpha is not required for virulence of Cryptococcus neoformans. Infect Immun. 2004;72:3643–3645. doi: 10.1128/IAI.72.6.3643-3645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Giles SS, Perfect JR, Heitman J. Peroxisome function regulates growth on glucose in the basidiomycete fungus Cryptococcus neoformans. Eukaryot Cell. 2007;6:60–72. doi: 10.1128/EC.00214-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Reedy JL, Nussbaum JC, Heitman J. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot Cell. 2004;3:420–429. doi: 10.1128/EC.3.2.420-429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, Lian T, Singh A, Kosman DJ, Kronstad JW. Iron Source Preference and Regulation of Iron Uptake in Cryptococcus neoformans. PLoS Pathog. 2008;4:e45. doi: 10.1371/journal.ppat.0040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozel TR, Mastroianni RP. Inhibition of phagocytosis by cryptococcal polysaccharide: dissociation of the attachment and ingestion phases of phagocytosis. Infect Immun. 1976;14:62–67. doi: 10.1128/iai.14.1.62-67.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu OW, Kelly MJ, Chow ED, Madhani HD. Parallel beta-helix proteins required for accurate capsule polysaccharide synthesis and virulence in the yeast Cryptococcus neoformans. Eukaryot Cell. 2007;6:630–640. doi: 10.1128/EC.00398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, Vamathevan J, Miranda M, Anderson IJ, Fraser JA, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monari C, Bistoni F, Vecchiarelli A. Glucuronoxylomannan exhibits potent immunosuppressive properties. FEMS Yeast Res. 2006;6:537–542. doi: 10.1111/j.1567-1364.2006.00072.x. [DOI] [PubMed] [Google Scholar]
- Moyrand F, Chang YC, Himmelreich U, Kwon-Chung KJ, Janbon G. Cas3p belongs to a seven-member family of capsule structure designer proteins. Eukaryot Cell. 2004;3:1513–1524. doi: 10.1128/EC.3.6.1513-1524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyrand F, Janbon G. UGD1, encoding the Cryptococcus neoformans UDP-glucose dehydrogenase, is essential for growth at 37 degrees C and for capsule biosynthesis. Eukaryot Cell. 2004;3:1601–1608. doi: 10.1128/EC.3.6.1601-1608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RT, Hua J, Pryor B, Lodge JK. Identification of virulence mutants of the fungal pathogen Cryptococcus neoformans using signature-tagged mutagenesis. Genetics. 2001;157:935–947. doi: 10.1093/genetics/157.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- Nosanchuk JD, Casadevall A. The contribution of melanin to microbial pathogenesis. Cell Microbiol. 2003;5:203–223. doi: 10.1046/j.1462-5814.2003.00268.x. [DOI] [PubMed] [Google Scholar]
- Noverr MC, Williamson PR, Fajardo RS, Huffnagle GB. CNLAC1 is required for extrapulmonary dissemination of Cryptococcus neoformans but not pulmonary persistence. Infect Immun. 2004;72:1693–1699. doi: 10.1128/IAI.72.3.1693-1699.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabayashi K, Hasegawa A, Watanabe T. Microreview: capsule-associated genes of Cryptococcus neoformans. Mycopathologia. 2007;163:1–8. doi: 10.1007/s11046-006-0083-0. [DOI] [PubMed] [Google Scholar]
- Olszewski MA, Noverr MC, Chen GH, Toews GB, Cox GM, Perfect JR, Huffnagle GB. Urease expression by Cryptococcus neoformans promotes microvascular sequestration, thereby enhancing central nervous system invasion. Am J Pathol. 2004;164:1761–1771. doi: 10.1016/S0002-9440(10)63734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz HL, Dehio C. Signature-tagged mutagenesis: technical advances in a negative selection method for virulence gene identification. Curr Opin Microbiol. 2005;8:612–619. doi: 10.1016/j.mib.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Shea JM, Kechichian TB, Luberto C, Del Poeta M. The cryptococcal enzyme inositol phosphosphingolipid-phospholipase C confers resistance to the antifungal effects of macrophages and promotes fungal dissemination to the central nervous system. Infect Immun. 2006;74:5977–5988. doi: 10.1128/IAI.00768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siafakas AR, Sorrell TC, Wright LC, Wilson C, Larsen M, Boadle R, Williamson PR, Djordjevic JT. Cell wall-linked cryptococcal phospholipase B1 is a source of secreted enzyme and a determinant of cell wall integrity. J Biol Chem. 2007;282:37508–37514. doi: 10.1074/jbc.M707913200. [DOI] [PubMed] [Google Scholar]
- Vaishnav VV, Bacon BE, O'Neill M, Cherniak R. Structural characterization of the galactoxylomannan of Cryptococcus neoformans Cap67. Carbohydr Res. 1998;306:315–330. doi: 10.1016/s0008-6215(97)10058-1. [DOI] [PubMed] [Google Scholar]
- Wang P, Cutler J, King J, Palmer D. Mutation of the regulator of G protein signaling Crg1 increases virulence in Cryptococcus neoformans. Eukaryot Cell. 2004;3:1028–1035. doi: 10.1128/EC.3.4.1028-1035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock DW. Fungal diseases: an evolving public health challenge. Med Mycol. 2006;44:697–705. doi: 10.1080/13693780601009493. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.