Abstract
The p150 form of the RNA-specific adenosine deaminase ADAR1 is interferon-inducible and catalyzes A-to-I editing of viral and cellular RNAs. We have characterized mouse genomic clones containing the promoter regions required for Adar1 gene transcription and analyzed interferon induction of the p150 protein using mutant mouse cell lines. Transient transfection analyses using reporter constructs led to the identification of three promoters, one interferon-inducible (PA) and two constitutively active (PB and PC). The TATA-less PA promoter, characterized by the presence of a consensus ISRE element and a PKR kinase KCS-like element, directed interferon-inducible reporter expression in rodent and human cells. Interferon induction of p150 was impaired in mouse cells deficient in IFNAR receptor, JAK1 kinase or STAT2 but not STAT1. Whereas Adar1 gene organization involving multiple promoters and alternative exon 1 structures was highly preserved, sequences of the promoters and exon 1 structures were not well conserved between human and mouse.
Keywords: Interferon, Signal Transducers and Activators of Transcription (STAT), Adenosine Deaminase Acting on RNA (ADAR)
INTRODUCTION
Interferons (IFN) possess a wide range of biologic activities (Borden et al., 2007; Haller et al., 2006; Randal and Goodbourn, 2008; Samuel, 2001) that include modulation of the immune response, regulation of cell growth and death, and the induction of a potent antiviral state, the property by which interferons were discovered ~50 years ago (Isaccs and Lindenmann, 1957). Among the IFN inducible genes is ADAR1 that encodes an adenosine deaminase acting on RNA (Bass et al., 1997; Patterson and Samuel, 1995; Toth et al., 2006).
ADAR1 catalyzes the C-6 deamination of adenosine to yield inosine in RNA substrates with double-stranded (ds) character (Samuel, 2001; Valente and Nishikura, 2005; Toth et al., 2006). This A-to-I RNA modifying activity of ADAR1 is implicated in two types of processes. First, the deamination can be site-selective and occur at one or a few sites, as illustrated by the editing of viral RNAs including the hepatitis delta virus antigenome RNA (Jayan and Casey, 2002) and human herpes virus 8 kaposin K12 transcript RNA (Gandy et al., 2007) and cellular mRNA transcripts in the brain that encode receptors for L-glutamate (GluR) and serotonin (5-HT) neurotransmitters (Higuchi et al., 1993; Liu et al., 1999; Liu and Samuel, 1999). In these cases, the positional selectivity of the RNA editing events generates protein products with altered function because of the highly selective amino acid substitutions introduced when GluR, 5-HT2cR, HHV8 and HDV mRNAs are decoded by ribosomes as I is recognized as G (Bass et al., 1997; Toth et al., 2006). Second, the dsRNA-specific deamination can occur at multiple sites, as observed in the modification of viral RNA genomes during lytic and persistent infections (Bass et al., 1997). Such hypermutations of viral RNAs have been characterized during replication and subsequent persistent infection with certain single-stranded RNA viruses including measles virus, where biased A-to-I (G) hypermutations were first described (Cattaneo et al., 1988), and more recently hepatitis C virus (Taylor et al., 2007) and lymphocytic choriomeningitis (LCM) virus (Zahn et al., 2007). In the case of measles virus, persistent infection may result in subacute sclerosing panencephalitis (SSPE) and a fatal neuropathic response (Oldstone, 2008).
Although ADAR1 is IFN-inducible, a significant basal level of expression is found in cultured cells and animal tissues (Patterson et al., 1995; Shtrichman et al., 2002; George et al., 2005). Immunoblot and immunofluorescent analyses with antisera raised against recombinant human ADAR1 demonstrated the expression of two differently sized ADAR1 proteins, an IFN-inducible ~150-kDa form found in both the cytoplasm and nucleus, and a smaller ~110-kDa N-terminally truncated protein found predominantly if not exclusively in the nucleus (Patterson and Samuel, 1995). Both the inducible p150 and the constitutively expressed p110 forms of ADAR1 are active deaminases that catalyze the A-to-I editing of synthetic and naturally occurring substrates (Toth et al., 2006). Both the p150 and p110 forms of ADAR1 possess, in addition to the deaminase catalytic domain in the C-terminal region, three copies of the dsRNA-binding motif in the central region of the proteins (Patterson and Samuel, 1995; Liu and Samuel, 1996). The IFN-inducible form of ADAR1 also includes in the N-terminal region two copies of a Z-DNA binding motif (Zα, Zβ) with homology to the poxvirus E3L protein (Patterson and Samuel, 1995; Athanasiadis et al., 2005), the physiologic function of which has not yet been clearly defined. The single human ADAR1 gene spans ~40-kbp and includes 17 exons (Liu et al., 1997) on chromosome 1q21.1-21.2 (Weier et al. 1995). The IFN inducible expression of p150 and constitutive expression of p110 are achieved by a sophisticated and complex process that involves alternative promoter utilization and alternative exon 1 splicing. One promoter is IFN inducible, and at least two are constitutively active (George and Samuel, 1999 a, b; Kawakubo et al., 2000). The promoters drive the expression of human ADAR1 transcripts with alternative exon 1 structures that are spliced to a common exon 2 junction; translation initiation of the inducible p150 protein begins in alternative exon 1A, whereas neither constitutive alternative exon 1B nor 1C contain an AUG start codon and translation of the p110 constitutive form of ADAR1 begins at an in-frame AUG within exon 2 (Valente and Nishikura, 2005; Toth et al., 2006). The expression of the mouse Adar1 gene found on chromosome 3F2 (Weier et al., 2000) likewise involves the utilization of multiple promoters and alternative exon 1 splicing (George et al., 2005).
Because of the emerging role of ADAR1 as a determinant of the outcome of virus-host interaction (Toth et al., 2006) and because of the importance of the mouse model in studies of pathogenesis, it is important to define the process of transcriptional activation of the Adar1 gene. As a step toward this goal, we report herein the further characterization of the 5’-region of the mouse Adar1 gene and the identification of an IFN-inducible STAT1-independent, STAT2-dependent process of transcriptional activation of ADAR1 expression.
RESULTS AND DISCUSSION
Structural organization of the mouse Adar1 gene 5’-region through exon 7 is highly conserved with its human homolog
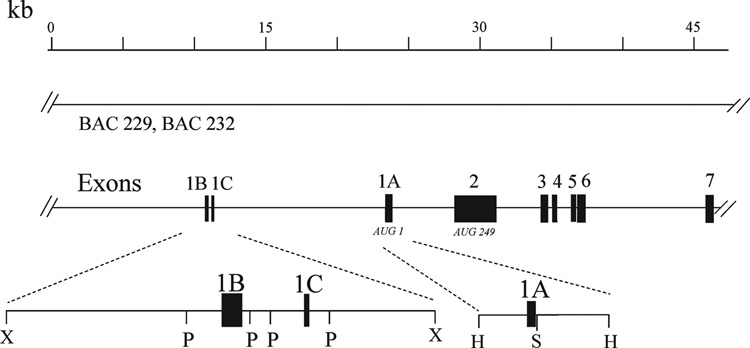
Overlapping λ-phage and bacterial artificial chromosome mouse genomic clones were isolated that contain the Adar1 gene. They were used to define the genomic organization of three alternative exon 1 structures (George et al., 2005) and to map the Adar1 gene to mouse chromosome 3F2 (Weier et al., 2000). The mouse Adar1 genomic clones were characterized by restriction enzyme analysis, by Southern blot analysis, and by direct sequence analysis. A composite map is shown in Figure 1. The exon-intron boundaries of the exons 1–7 of the mouse Adar1 gene are summarized in Table 1.
Figure 1. Physical map of the 5’-region of the mouse Adar1 gene.
The structure of the Adar1 gene is represented with regard to the organization of the exons and introns within the 5’- region of the gene as established from λ-phage and BAC genomic clones. Exons are indicated by filled boxes, numbered 1–7; introns and the 5’- and 3’-flanking regions are indicated by the solid lines. The AUG for translation initiation of the IFN inducible p150 ADAR1 protein is located in exon 1A; the AUG for initiation of the constitutively expressed p110 ADAR1 protein is located in exon 2. BAC-229 and BAC-232 genomic clones span the length of the Adar1 gene and continue into 5’-flanking region. Restriction endonuclease cleavage sites: HindIII (H), PstI (P), SacII (S), XbaI (X).
Table I.
Exon-Intron Sizes and Junction Sequences of Mouse and Human ADAR1 Genes
| Exon |
Intron |
Exon | |||||
|---|---|---|---|---|---|---|---|
| No. | Size (bp) | Junction (EXON/intron) | No. | Size (kb) | Junction (intron/EXON) | No | |
| Ms | 1A | 44 | CCACAGgtaagcccgcgg | IA | 4.2 | tgccttctacagGGGTGT | 2 |
| Hu | 1A | 201 | CGGCAGgtaagccgggct | IA | 5.4 | cttattctgcagGGGTAT | 2 |
| Ms | 1B | 106 | GCGCGGgtaagaggacct | IB | 15.8 | tgccttctacagGGGTGT | 2 |
| Hu | 1B | 107 | GACCCGgtaagagcctct | IB | 14.5 | cttattctgcagGGGTAT | 2 |
| Ms | 1C | 106 | CAGAGGgtaaggcgttct | IC | 15.2 | tgccttctacagGGGTGT | 2 |
| Hu | 1C | 107 | AACCGTgtgagtactatt | IC | 3.1 | cttattctgcagGGGTAT | 2 |
| Ms | 2 | 1432 | ACCTCGgtaagagactgc | II | 2.4 | ctttccaccaagATTTAA | 3 |
| Hu | 2 | 1586 | ACCTCGgtaagagaccac | II | 2.5 | ctttccgtcaagATTTAA | 3 |
| Ms | 3 | 184 | GAGAAGgtaggtgatctt | III | 1.0 | tcttttctccagCCAGCA | 4 |
| Hu | 3 | 184 | GAGAAGgtaggtgtcctc | III | 0.4 | cattttctctagACTGCA | 4 |
| Ms | 4 | 149 | CCCCAAgtatgtctatgt | IV | 0.6 | gtttctgctaagGTTCCA | 5 |
| Hu | 4 | 149 | ACCCAAgtatgtcctacg | IV | 0.6 | atctcctgtcagGTTCCA | 5 |
| Ms | 5 | 139 | GACCAGgtgggccgacgt | V | 0.1 | atttctccttagTCTGGA | 6 |
| Hu | 5 | 145 | AACCAGgtagggcgtttt | V | 0.1 | attctcctttagCCTGAA | 6 |
| Ms | 6 | 184 | ACCCAAgtgagtacctca | VI | 8.3 | ttcatcccaaagGTTTGT | 7 |
| Hu | 6 | 191 | GCCCAAgtgagtgtccta | VI | 6.5 | ctcatcccaaagGTTCGT | 7 |
| Ms | 7A | 226 | AAGACAgttaagacatct | VIIA | 0.2 | ctgtgtccgcagCTCCTC | 8 |
| Hu | 7A | 226 | AAGACAgttaagacgtct | VIIA | 0.3 | ttttccccacagCTCCCT | 8 |
| Ms | 7B | 148 | ACAGAGgtaaccccagta | VIIB | 0.4 | ctgtgtccgcagCTCCTC | 8 |
| Hu | 7B | 148 | ACAGAGgtaaccccagtg | VIIB | 0.4 | ttttccccacagCTCCCT | 8 |
Ms, mouse; Hu, human. Upper case font, EXON sequence at junction; lower case font, adjacent intron sequence at junction.
Comparison of the mouse Adar1 gene structure with that previously determined for the human ADAR1 gene (Liu et al. 1997) revealed that the exon-intron organization was highly conserved between the mouse and human genes for the protein coding exons 2 through 7 (Table 1). The mouse Adar1 exon 1A (see Fig. 3A), like the human ADAR1 exon 1A (George and Samuel, 1999b), includes the AUG codon that begins the long open reading frame for the IFN-inducible p150 ADAR1 protein. Neither mouse exon 1B nor 1C contained an AUG; initiation of translation of the p110 constitutive form of mouse ADAR1 would occur at AUG249 within exon 2, which conforms to AUG296 within exon 2 of human ADAR1 (George and Samuel, 1999a; George and Samuel, 2005; Kumar and Carmichael, 1997; Liu et al., 1997). The sizes of exons 2 through 7, and the adjacent introns II through VII, are very similar if not identical for the mouse and human genes. Two principal differences in size include exon 1A, which is somewhat smaller in the mouse gene, and intron IC which is significantly larger than in the human gene. Two alternative exon 7 splice variants are known in the human (Liu et al., 1997) and mouse (Shtrichman et al., 2002) Adar1 genes. The presence of alternative exon 7b occurs with exon 1A, and alternative exon 7a with exon 1B (George et al., 2005). All splice sites of mouse Adar1 (Table 1), like human ADAR1 (Liu et al., 1997), conformed to the GU-AG rule (Padgett et al., 1986) for introns I to VII.
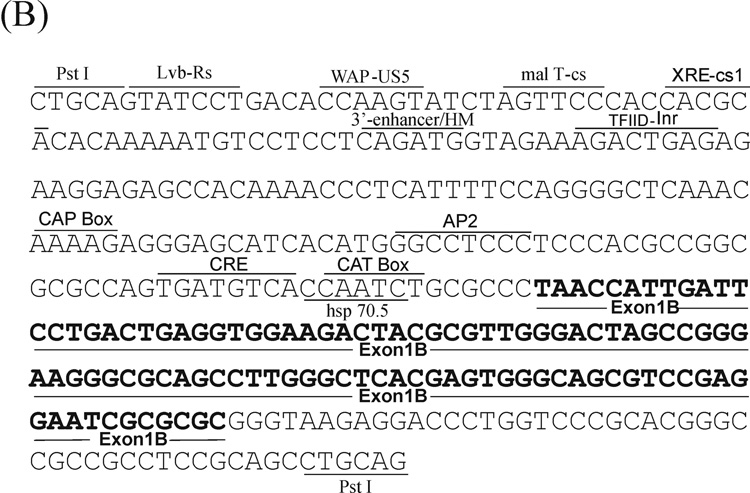
Figure 3. Nucleotide sequence of the 5’-flanking region of the mouse Adar1 gene.
The sequences of the (A) PA and (B) PB promoter regions as well as the sequences of the alternative exon 1A and exon 1B structures and adjacent introns are shown. Several potential transcription factor binding sites as described in the text are shown, along with landmark restriction endonuclease sites. The Interferon-Stimulated Response Element (ISRE) and the Kinase Consensus Sequence (KCS)-like element are boxed.
Functional identification of the interferon-inducible and constitutive mouse Adar1 promoter regions
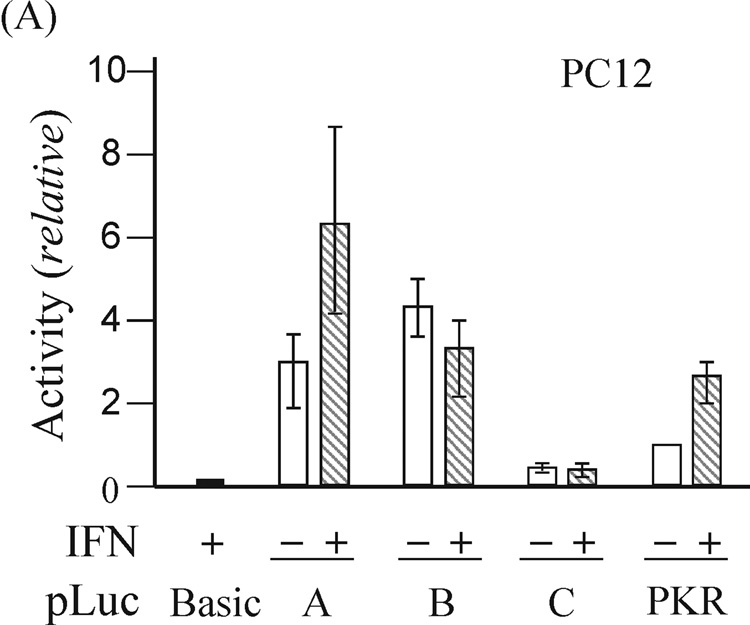
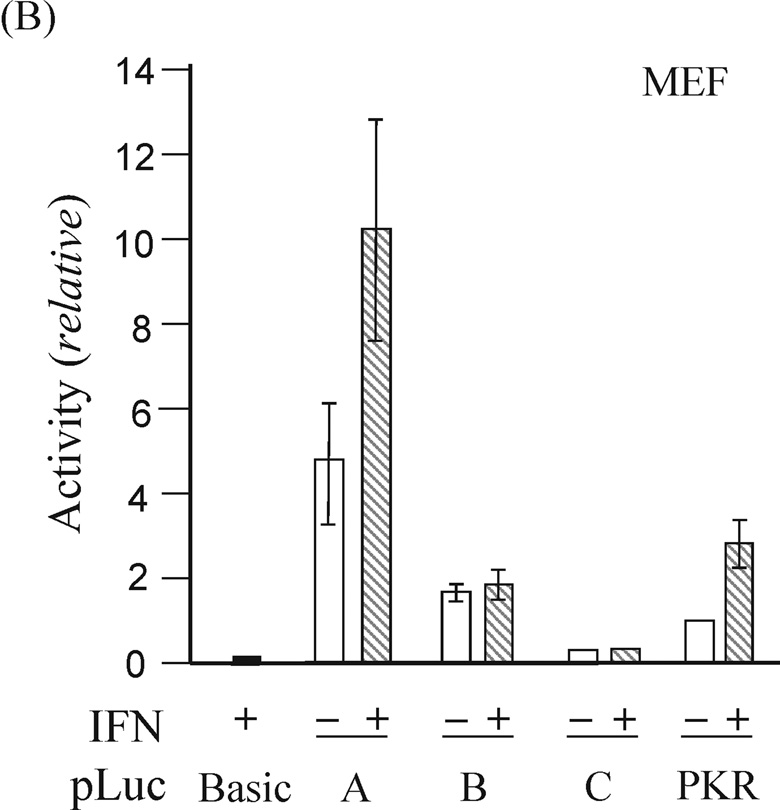
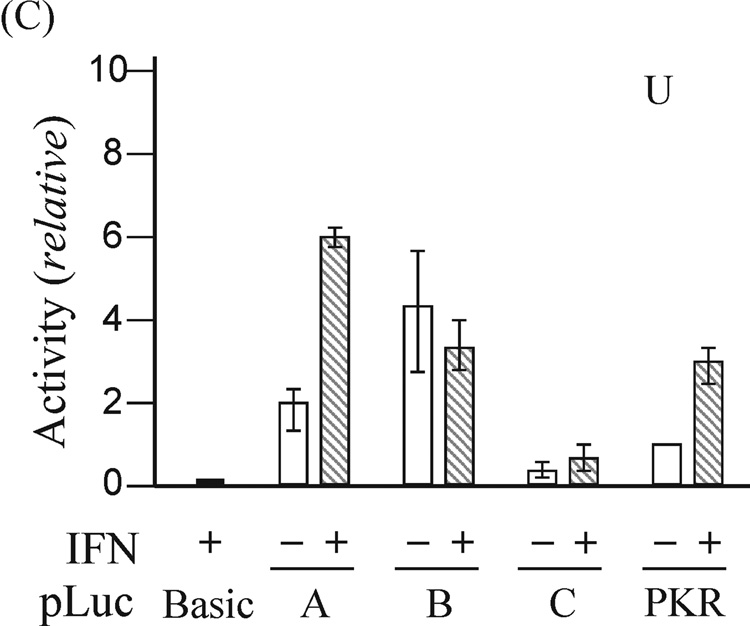
Expression of exon 1A-containing ADAR1 RNA, but not exon 1B- or 1C- containing RNA, is increased by IFN treatment of mouse fibroblasts in culture (George et al., 2005). As an approach to the functional isolation of the mouse Adar1 gene promoters, restriction fragments from the 5’-flanking region of the gene (Fig. 1) were fused to the firefly luciferase (Luc) reporter using the pGL2-basic plasmid vector. The pLuc-A construct included the 320-bp HindIII-SacII fragment flanking exon 1A, pLuc-B contained the 350-bp Pst I fragment flanking exon 1B, and pLuc-C included a 330-bp Pst I fragment flanking exon 1C. The constructs were transfected into rat PC12 cells, mouse MEF cells, and human U cells, and luciferase activity was measured in extracts prepared from untreated or IFN-treated cells. The pLuc-A construct exhibited strong and IFN-inducible promoter activity in all three cell lines, whereas the activities of pLuc-B and pLuc-C were not increased by IFN treatment in any of the cells tested (Fig. 2). The relative activities of the three reporter constructs differed among the three cell lines. Although the constitutive activity of pLuc-B was always greater than that of the relatively weak pLuc-C, neither promoter construct displayed increased activity following IFN treatment in PC12, U or MEF cells (Fig. 2). IFN treatment increased the activity of pLuc-A about 2-to-3 fold, dependent upon the type of cell transfected, similar to that seen for the human PKR promoter construct pLuc-503(WT) used as a positive control. As a negative control, the promoter-less pGL2-basic vector without inserted genomic DNA showed negligible activity even following IFN treatment.
Figure 2. Functional analysis of the mouse Adar1 gene 5’-flanking region reporter gene constructs by transient transfection.
The firefly luciferase reporter plasmids (pLuc) constructed by insertion of the indicated genomic DNA restriction fragments (see Fig. 2) from the 5’-flanking region of the Adar1 gene into the promoter-less pGL2 (pLuc-Basic) plasmid are designated pLuc-A, pLuc-B and pLuc-C. Promoter activities were measured in the following cells: (A) rat PC12; (B) mouse MEF; (C) human U cells. Open bars refer to cells left untreated, and hatched bars refer to cells treated with IFN beginning at 24 h after transfection. Transfections were repeated 3 to 5 times in independent experiments to allow for calculation of a mean value and standard deviation. pLuc-Basic, the promoter-less plasmid vector without inserted mouse genomic DNA. PKR, the pLuc-503(WT) PKR promoter consrtuct.
DNA sequence of the mouse Adar1 gene interferon-inducible promoter region includes ISRE and KCS-like elements
The genomic DNA fragment flanking exon 1A that possessed the necessary functional elements to support basal and IFN-inducible PA driven transcription in transfected cells was sequenced. The sequence is shown in Figure 3A. Among the DNA elements identified was a consensus 13-bp IFNα/β-Stimulated Response Element (ISRE) found in the promoters of most type I IFN-inducible genes that enhances transcription in response to IFN treatment (Schindler et al., 2007; Randall and Goodbourn, 2008). Immediately upstream of the ISRE was a KCS-like element similar to the 15-bp KCS element found in both the mouse and human PKR promoters that is essential for both basal and IFN-inducible PKR promoter activity (Kuhen et al., 1998; Ward and Samuel, 2002). Designated KCS for Kinase Conserved Sequence, it is exactly conserved between the mouse and human PKR promoters in sequence and position relative to the ISRE (Kuhen et al., 1998; Samuel, 2001). The KCS-like element sequence in the mouse Adar1 PA promoter was identical to the KCS-like sequence of the human IFN-inducible ADAR1 promoter (Fig 4). Eleven of the 15 bp of the KCS element present in the PKR promoter, both mouse and human, were conserved in the KCS-like element of the Adar1 promoter. However, the KCS-like element is not essential for basal or IFN-inducible activity of the human ADAR1 PI inducible promoter (Markle et al., 2003), and substitution of the human ADAR1 KCS-like element for the KCS element of the human PKR promoter results in significantly reduced bsal and IFN-inducible PKR promoter activity (Ward et al., 2002). Several candidate Sp1/Sp3 factor binding sites as well as PEA-3 and MyoD sites were present in the mouse Adar1 PA promoter, however, the inducible PA promoter sequence lacked TATA box and CATT box initiation elements (Fig. 3A).
Figure 4. Comparison of the ISRE and KCS/KCS-Like element sequences from the human and mouse Adar1 and PKR promoters.
The sequence of the mouse (Mm) Adar1 interferon inducible promoter region corresponding to the KCS-like and ISRE elements is from Figure 3; the sequence of the human (Hs) ADAR1 IFN inducible promoter is from George and Samuel (1999). The mouse Pkr promoter sequence is from Tanaka and Samuel (1994) and the human PKR promoter sequence is from Kuhen et al. (1998).
The DNA sequences were also determined for the Adar1 gene PB (Fig. 3B) and PC (data not shown) promoter regions that possessed the necessary elements to support constitutive transcription in transfected rodent and human cell lines (Fig. 2). All sequences were reported in the GenBank database. Among the binding sites found in the PB region sequence were a consensus CAAT box, initiator positioning sequence Inr and CRE-like motif (Fig. 3B), but no ISRE, GAS or KCS-like elements were present in either the PB or the PC promoter regions.
Induction of mouse ADAR1 protein by interferon is JAK1- and STAT2-dependent but STAT1-independent
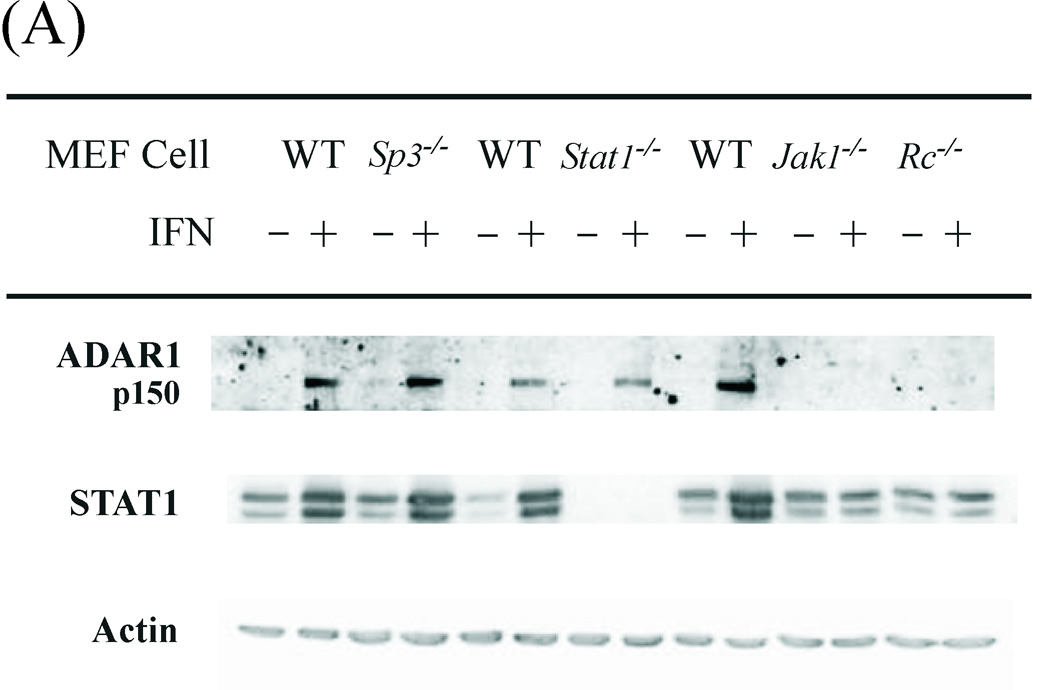
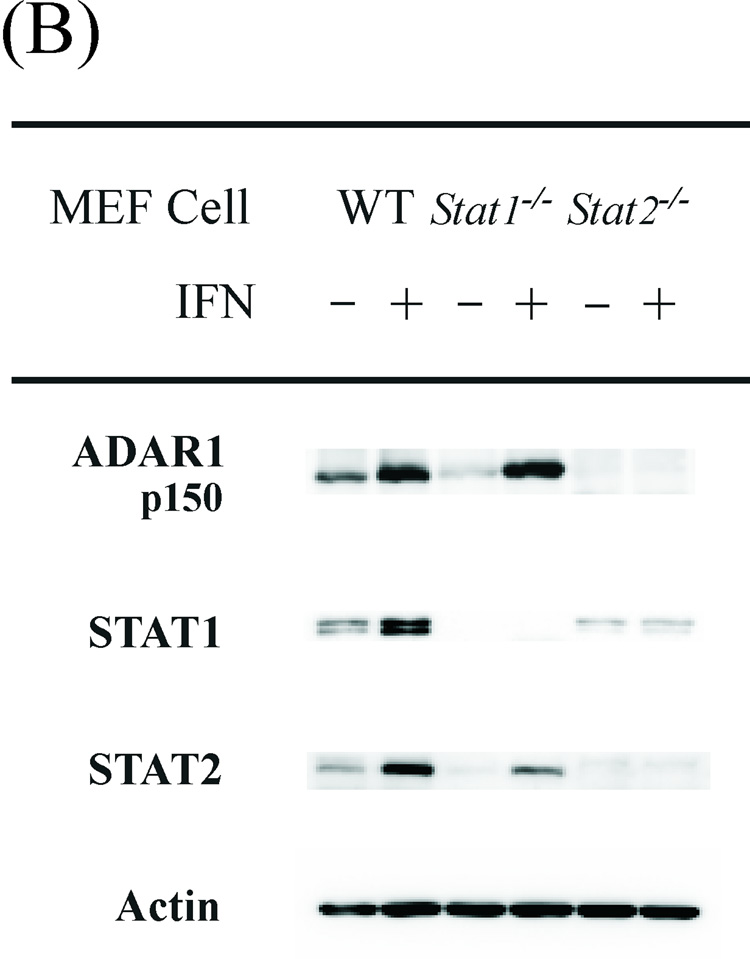
The p150 ADAR1 protein encoded by exon 1A-containing RNA is inducible by type I IFN (Patterson et al., 1995; George et al., 2005). The canonical mode of transcriptional activation by type I IFN is by JAK-STAT signaling (Borden et al., 2007; Schindler et al., 2007; Randall and Goodbourn, 2008). Following binding of IFN-α/β to the IFNAR receptor, the Tyk2 and JAK1 tyrosine kinases mediate activation of the STAT1 and STAT2 transcription factors that together with IRF9 form the heterotrimeric ISGF3 complex that binds to the ISRE element to enhance gene transcription. We used MEF cell lines with defined disruptions of genes encoding proteins involved in the canonical signaling pathway to assess their requirement for p150 ADAR1 protein induction by IFN. MEF cells were either treated with IFN or left untreated, and then the level of p150 ADAR1 protein was measured by immunoblot assay. The IFN-inducible expression of ADAR1 protein was JAK1-dependent and IFN receptor-dependent, but unexpectedly was independent of STAT1 (Fig. 5 A). Although not dependent on STAT1, induction of ADAR1 p150 by IFN was dependent on STAT2 (Fig. 5 B). Controls included blotting with antibody against actin which showed similar protein loading; antibody against STAT1 which verified the absence of STAT1 protein in the Stat1−/− MEF cells; and antibody against STAT2 which verified the absence of STAT2 protein in the Stat2−/− MEF cells (Fig. 5). Furthermore, STAT1α and β were inducible in the wild-type MEFs and Sp3−/− MEFs, but not the Jak1−/−, Stat2−/−, or Rc−/− MEFs. Finally, ADAR1 p150 induction also was independent of the transcription factor Sp3 (Fig. 5 A), a component of the KCS-binding protein complex (Das et al., 2006), as well as independent of CEBPβ and CEPBδ (data not shown).
Figure 5. JAK1 and STAT2 but not STAT1 are necessary for induction of ADAR1 expression by interferon alpha.
Western immunoblot analysis of ADAR1 p150 protein expression in WT and mutant MEF cell lines. Cells were either treated with IFN-αA/D (+) or left untreated (−). Nonidet P-40 cell free extracts were prepared and ~60 µg protein was analyzed by Western immunoblotting as described under Materials and Methods. (A) Cell-free extracts prepared from WT and mutant MEF cells genetically deficient in Sp3, Stat1, Jak1 or the IFNAR receptor (Rc) as indicated. (B) Cell-free extracts prepared from WT and mutant MEF cell lines genetically deficient in Stat1 or Stat2.
The STAT1-independent, STAT2-dependent induction of p150 ADAR1 A-to-I editing enzyme by type I IFN is intriguing. Interestingly, a second editing enzyme, APOBEC3G which catalyzes C- to -U deamination of ssDNA is likewise induced by type I IFN in a STAT1-independent, STAT2-dependent manner (Sarkis et al., 2006). The mechanism underlying STAT1- independent signaling by IFN α/β to activate ADAR1 expression remains unclear. Among the alternative signaling pathways implicated in type I IFN- mediated responses are those involving PI3K, p38, STAT3 or IRF3 (Platanias, 2005; Randall and Goodbourn, 2007; Samuel, 2001; Stark, 2007).
Viruses have evolved a range of mechanisms by which IFN signaling and the induction of an antiviral state are antagonized. Among these mechanisms is an impairment of STAT1 function, for example, by some paramyxoviruses including measles virus (Haller et al., 2006; Randall and Goodbourn, 2008). A STAT1-independent signaling response such as we find for ADAR1 would provide the host with an alternative mechanism for the induction of antiviral activities by IFN. However, viruses are known to subvert the antiviral effects of type I IFN through STAT2-dependent signaling, as has been demonstrated for both measles and LCM viruses that evade the immune system through type I IFN-mediated STAT2-dependent but STAT1-independent signaling (Hahm et al., 2005). Interestingly, both measles virus and LCM virus RNA genomes are implicated as targets of ADAR1 editing. A-to-G mutations, consistent with the action of ADAR1, are observed in LCMV genomic RNA; these mutations alter the functionality of viral glycoproteins at a high frequency (Zahn et al., 2007). A-to-G mutations of measles virus genomic RNA also are seen with virus isolated from the brains of SSPE patients (Cattaneo et al., 1988; Oldstone, 2008).
MATERIALS AND METHODS
Cell maintenance and interferon treatment
Mouse embryo fibroblast (MEF) cells, rat PC12 cells, and human amnion U cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 5% or 10% (v/v) fetal bovine serum (Hyclone), respectively, and 1% sodium pyruvate (Cambrex Bio Science), 100 µg/ml of penicillin and 100 units/ml streptomycin (GIBCO) as previously described (Samuel and Knutson 1983, Das et al., 2006). Wild-type (WT), Stat1−/−, Stat2−/−, Jak1−/−, Ifnar−/−, and Sp3−/− MEF cells were generously provided by Dr. Robert Schreiber (Washington University, St. Louis), Dr. Christian Schindler (Columbia University, New York) and Dr. Guntrum Suske (Philipps-University, Marburg) (Bouwman et al., 2000; Meraz et.al 1996, Park et al., 2000; Rodig at al, 1998). IFN treatment was with 1000 units/ml of alpha IFN (PBL) for 24 h, using recombinant IFN-αA/D (Samuel and Knutson, 1982).
Construction of reporter gene plasmids
pGL2-basic promoter-less plasmid (Promega) containing the firefly luciferase (Luc) gene as the reporter was used for construction of the Adar1 promoter plasmids. pLuc-503(WT) human PKR promoter plasmid was generously provided by Dr. P. Zhang of this laboratory. pRSV-βgal was generously provided by Dr. J. Nevins (Duke University, Durham).
Transient transfection assays
Cells were transfected with the promoter plasmid constructs containing the Luc reporter by the DEAE-dextran-chloroquine phosphate transfection method (Luthman and Magnusson, 1983) as described previously (Ward and Samuel, 2002). To normalize for variations in transfection efficiency, cells were cotransfected with the pRSV2-βgal construct as an internal reference. All DNA plasmids used in transfections were purified by cesium chloride equilibrium centrifugation; plasmid integrity was assessed by agarose gel electrophoresis. IFN treatment was initiated 24 h after transfection. Cells were harvested 48 h post-transfection, extracts prepared, and protein concentrations determined by the modified Bradford method (BioRad). Luciferase and β-galactosidase activities determined as previously described (George and Samuel, 1999; Ward and Samuel, 2002). Luciferase assays were quantified using an OPTOCOMP I luminometer. Luciferase activity values were normalized by β̃-galactosidase activity to control for variation in transfection efficiency. The data presented are the average values derived from 3 to 5 independent experiments.
Promoter cloning
Mouse genomic clones λ52, BAC229 and BAC232 of Adar1 were isolated as described (George et al., 2005) from two types of libraries, a λ-phage genomic library in the vector EMBL-3 SP6/T7 (Clontech) and the bacterial artificial chromosome (BAC) genomic library CitbCJ7 (California Institute of Technology). Restriction fragments of genomic clones were subcloned into the pBluescript plasmid (Stratagene) for detailed restriction mapping and DNA sequencing. Plasmid subclones of the genomic DNA were sequenced by the Sanger dideoxynucleotide procedure using the Sequenase protocols from United States Biochemical. Universal primer sites in the pBluescript plasmid were used as well as custom Adar1 primers obtained commercially. Sequences were analyzed using the University of Wisconsin Genetics Computer Group programs on a Silicon Graphics IRIS 4D/340VGX computer.
Western Immunoblot analysis
Cell-free extracts were prepared in the presence of 1 mM phenylmethylsulfonyl fluoride and 1% (v/v) protease inhibitor mixture (Sigma) as described previously (Das et al., 2006). Proteins were fractionated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, blocked, and then probed with an appropriate dilution of primary antibody in phosphate-buffered saline (PBS) containing 3% (w/v) skim milk and 0.05% Tween 20. Guinea pig polyclonal antibody prepared against a mouse recombinant GST-ADAR1 [aa 574–968] fusion protein expressed in E. coli was as previously described (George et al., 2005). Rabbit polyclonal antibodies from Santa Cruz Biotechnology were used to detect mouse STAT1α/β̣(M-22) and mouse STAT2 (L-22). Mouse monoclonal antibody against β-actin was from Sigma (A-5441). Western blot detection was done with horseradish peroxidase-conjugated anti-guinea pig IgG, anti-rabbit IgG or anti-mouse IgG secondary antibody using an ECL detection reagent kit (Amersham Biosciences), according to manufacturer's recommendations. Immunoreactive bands were visualized using a VersaDoc (Bio-Rad) imaging system. Alternatively, secondary IRDye-conjugated antibodies (Li-Cor) were used and immunoreactive bands were visualized using an Odyssey infrared imaging system.
Nucleotide sequence accession numbers
The sequences reported in this paper have been deposited in the GenBank database. The 5’-flanking genomic sequence of the mouse Adar1 gene including the interferon inducible promoter PA region and alternative exon 1A has been assigned the accession number AY488121. The sequences of the constitutively active promoter PB and PC regions and alternative exons 1B and 1C have been assigned the accession numbers AY488122 and AY488123, respectively.
Materials
Unless otherwise specified, materials and reagents were as described previously (George et al., 2005; Patterson and Samuel, 1995).
ACKNOWLEDGMENTS
This work was supported in part by Research Grant AI-12520 from the National Institute of Allergy and Infectious Diseases, U.S. Public Health Service. We thank Elizabeth Short for her technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations. ADAR, the RNA-specific adenosine deaminase inducible by interferon; IFN, interferon; dsRNA, double-stranded RNA; bp, base pair.
REFERENCES
- Athanasiadis A, Placido D, Maas S, Brown BA, Lowenhaupt K, Rich A. The crystal structure of the Z beta domain of the RNA-editing enzyme ADAR1 reveals distinct conserved surfaces among Z-domains. J. Mol. Biol. 2005;351:496–507. doi: 10.1016/j.jmb.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Bass BL, Nishikura K, Keller W, Seeburg PH, Emeson RB, O'Connell MA, Samuel CE, Herbert A. A standardized nomenclature for adenosine deaminases that act on RNA. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Göllner H, Elsässer, HP, Eckhoff G, Karis A, Grosveld F, Philipsen S, Suske G. Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J. 2000;19:655–661. doi: 10.1093/emboj/19.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo R. Biased (A/I) hypermutations of animal virus genomes. Curr. Op. Genetics Develop. 1994;4:895–900. doi: 10.1016/0959-437x(94)90076-0. [DOI] [PubMed] [Google Scholar]
- Cattaneo R, Schmid A, Eschle D, Baczko K, terMeulen V, Billeter MA. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell. 1988;55:255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Ward SV, Tacke RS, Suske G, Samuel CE. Activation of the RNA-dependent protein kinase PKR promoter in the absence of interferon is dependent upon Sp proteins. J. Biol. Chem. 2006;281:3244–3253. doi: 10.1074/jbc.M510612200. [DOI] [PubMed] [Google Scholar]
- Gandy SZ, Linnstaedt SD, Muralidhar S, Cashman KA, Rosenthal LJ, Casey JL. RNA editing of the HHV-8 Kaposin transcript eliminates its transforming activity and is induced during lytic replication. J. Virol. 2007;81:13544–13551. doi: 10.1128/JVI.01521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in détente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- George CX, Wagner MV, Samuel CE. Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. J. Biol. Chem. 2005;280:15020–15028. doi: 10.1074/jbc.M500476200. [DOI] [PubMed] [Google Scholar]
- George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Nat.l. Acad. Sci. U. S. A. 1999a;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, Samuel CE. Characterization of the 5'-flanking region of the human RNA-specific adenosine deaminase ADAR1 gene and identification of an interferon-inducible ADAR1 promoter. Gene. 1999b;229:203–213. doi: 10.1016/s0378-1119(99)00017-7. [DOI] [PubMed] [Google Scholar]
- Hahm B, Trifilo MJ, Zuniga EI, Oldstone MB. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity. 2005;22:247–257. doi: 10.1016/j.immuni.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Haller O, Kochs G, Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. 2006;344:119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- Jayan GC, Casey JL. Inhibition of hepatitis delta virus RNA editing by short inhibitory RNA-mediated knockdown of ADAR1 but not ADAR2 expression. J. Virol. 2002;76:12399–12404. doi: 10.1128/JVI.76.23.12399-12404.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakubo K, Samuel CE. Human RNA-specific adenosine deaminase (ADAR1) gene specifies transcripts that initiate from a constitutively active alternative promoter. Gene. 2000;27:165–172. doi: 10.1016/s0378-1119(00)00368-1. [DOI] [PubMed] [Google Scholar]
- Kuhen KL, Vessey JW, Samuel CE. Mechanism of interferon action: identification of essential positions within the novel 15-base-pair KCS element required for transcriptional activation of the RNA-dependent protein kinase pkr gene. J. Virol. 1998;72:9934–9939. doi: 10.1128/jvi.72.12.9934-9939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Carmichael GG. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl. Acad. Sci. U. S. A. 1997;15:3542–3547. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Samuel CE. Mechanism of interferon action: Functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA-specific adenosine deaminase. J. Virol. 1996;70:1961–1968. doi: 10.1128/jvi.70.3.1961-1968.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Samuel CE. Editing of glutamate receptor subunit B pre-mRNA by splice-site variants of interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J. Biol. Chem. 1999;274:5070–5077. doi: 10.1074/jbc.274.8.5070. [DOI] [PubMed] [Google Scholar]
- Liu Y, Emeson RB, Samuel CE. Serotonin-2C receptor pre-mRNA editing in rat brain and in vitro by splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J. Biol. Chem. 1999;274:18351–18358. doi: 10.1074/jbc.274.26.18351. [DOI] [PubMed] [Google Scholar]
- Liu Y, George CX, Patterson JB, Samuel CE. Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J. Biol. Chem. 1997;272:4419–4428. doi: 10.1074/jbc.272.7.4419. [DOI] [PubMed] [Google Scholar]
- Luthman H, Magnusson G. High efficiency polyoma DNA transfection of chloroquine treated cells. Nucleic Acids Res. 1983;11:1295–1308. doi: 10.1093/nar/11.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle D, Das S, Ward SV, Samuel CE. Functional analysis of the KCS-like element of the interferon-inducible RNA-specific adenosine deaminase ADAR1 promoter. Gene. 2003;304:143–149. doi: 10.1016/s0378-1119(02)01200-3. [DOI] [PubMed] [Google Scholar]
- Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- O’Connell MA, Krause S, Higuchi M, Hsuan JJ, Totty NF, Jenny A, Keller W. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol. Cell. Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara PJ, Nichol ST, Horodyski FM, Holland JJ. Vesicular stomatitis virus defective interfering particles can contain excessive genomic sequence rearrangements and base substitutions. Cell. 1984;36:915–924. doi: 10.1016/0092-8674(84)90041-2. [DOI] [PubMed] [Google Scholar]
- Oldstone MBA. Modeling subacute scherosing panencephalitis (SSPE) in a transgenic mouse system: uncoding pathogenesis of disease and illuminating components of immune control. Curr. Topics Microbiol. Immunol. 2008 doi: 10.1007/978-3-540-70617-5_2. in press. [DOI] [PubMed] [Google Scholar]
- Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA. Splicing of messenger RNA precursors. Annu. Rev. Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. Review. [DOI] [PubMed] [Google Scholar]
- Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol. Cell. Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JB, Thomis DC, Hans SL, Samuel CE. Mechanism of interferon action. Double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology. 1995;210:508–511. doi: 10.1006/viro.1995.1370. [DOI] [PubMed] [Google Scholar]
- Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signaling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signaling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM, Jr, Schreiber RD. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93:373–383. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol.Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE, Knutson GS. Mechanism of interferon action. Kinetics of induction of the antiviral state and protein phosphorylation in mouse fibroblasts treated with natural and cloned interferons. J. Biol. Chem. 1982;257:11791–11795. [PubMed] [Google Scholar]
- Samuel CE, Knutson GS. Mechanism of interferon action: human leukocyte and immune interferons regulate the expression of different genes and induce different antiviral states in human amnion U cells. Virology. 1983;130:474–484. doi: 10.1016/0042-6822(83)90101-0. [DOI] [PubMed] [Google Scholar]
- Sarkis PT, Ying S, Xu R, Yu XF. STAT1-independent cell type-specific regulation of antiviral APOBEC3G by IFN-alpha. J. Immunol. 2006;177:4530–4540. doi: 10.4049/jimmunol.177.7.4530. [DOI] [PubMed] [Google Scholar]
- Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- Shtrichman R, Heithoff DM, Mahan MJ, Samuel CE. Tissue selectivity of interferon-stimulated gene expression in mice infected with Dam(+) versus Dam(−) Salmonella enterica serovar Typhimurium strains. Infect. Immun. 2002;70:5579–5588. doi: 10.1128/IAI.70.10.5579-5588.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR. How cells respond to interferons revisited: from early history to current complexity. Cytok. Growth Factor Rev. 2007;18:418–423. doi: 10.1016/j.cytogfr.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Samuel CE. Mechanism of interferon action: structure of the mouse PKR gene encoding the interferon-inducible RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U. S. A. 1994;91:7995–7999. doi: 10.1073/pnas.91.17.7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Puig M, Darnell ME, Mihalik K, Feinstone SM. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J. Virol. 2007;79:6291–6298. doi: 10.1128/JVI.79.10.6291-6298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenoever BR, Ng SL, Chua MA, McWhirter SM, García-Sastre A, Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:369–434. doi: 10.1016/S0079-6603(06)81010-X. [DOI] [PubMed] [Google Scholar]
- Uze G, Monneron D. IL-28 and IL-29: newcomers to the interferon family. Biochimie. 2007;89:729–734. doi: 10.1016/j.biochi.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- Ward SV, Samuel CE. Regulation of the interferon-inducible PKR kinase gene: the KCS element is a constitutive promoter element that functions in concert with the interferon-stimulated response element. Virology. 2002;296:136–146. doi: 10.1006/viro.2002.1356. [DOI] [PubMed] [Google Scholar]
- Ward SV, Markle D, Das S, Samuel CE. the promoter-proximal KCS element of the PKR kinase gene enhances transcription irrespective of orientation and position relative to the ISRE element and is functionally distint from the KCS-like element of the ADAR deaminase promoter. J. Interferon & Cytokine Res. 2002;22:891–898. doi: 10.1089/107999002760274917. [DOI] [PubMed] [Google Scholar]
- Weier HU, George CX, Lersch RA, Breitweser S, Cheng JF, Samuel CE. Assignment of the RNA-specific adenosine deaminase gene (Adar) to mouse chromosome 3F2 by in situ hybridization. Cytogenet. Cell Genet. 2000;89:214–215. doi: 10.1159/000015615. [DOI] [PubMed] [Google Scholar]
- Weier HU, George CX, Greulich KM, Samuel CE. The interferon-inducible, double-stranded RNA-specific adenosine deaminase gene (DSRAD) maps to human chromosome 1q21.1-21.2. Genomics. 1995;30:372–375. doi: 10.1006/geno.1995.0034. [DOI] [PubMed] [Google Scholar]
- Zahn RC, Schelp I, Utermöhlen O, von Laer D. A-to-G hypermutation in the genome of lymphocytic choriomeningitis virus. J. Virol. 2007;81:457–464. doi: 10.1128/JVI.00067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]